Abstract
This study evaluated the influence of parameters relevant for cheese making on histamine formation by Streptococcus thermophilus. Strains possessing a histidine decarboxylase (hdcA) gene represented 6% of the dairy isolates screened. The most histaminogenic, S. thermophilus PRI60, exhibited in skim milk a high basal level of expression of hdcA, upregulation in the presence of free histidine and salt, and repression after thermization. HdcA activity persisted in cell extracts, indicating that histamine might accumulate after cell lysis in cheese.
Histamine is the biogenic amine (BA) most frequently implied in food poisoning. It interacts with the respiratory, cardiovascular, gastrointestinal, and hematological/immunological systems, causing “histamine intoxication” symptoms like hypotension, headache, diarrhea, vomiting, and skin flushing that, in the case of “scombroid poisoning,” caused by scombroid fish, are particularly severe. Moreover, “histamine intolerance,” i.e., acute toxicity even at relatively low levels of histamine (30), occurs especially in diamine oxidase (DAO)-deficient subjects (2, 17). Histamine is the only BA for which maximum specific levels in food are indicated, though only for some fresh and processed fish products with a high content of free histidine (11) that can be decarboxylated to histamine mainly by Gram-negative bacteria (17). However, some Gram-positive bacteria involved in food production, including lactic acid bacteria (LAB) strains of the species Lactobacillus buchneri, Lactobacillus curvatus, Lactobacillus helveticus, Lactobacillus hilgardii, Lactococcus lactis, Oenococcus oeni, Pediococcus damnosus, and Pediococcus parvulus (1, 4, 14, 15, 25), also possess this enzymatic activity, encoded by hdc gene clusters, and form histamine in cheeses, sausages, and fermented beverages. The presence of histamine is well documented in long-ripened and blue cheeses (19, 25) and is attributed to both nonstarter and starter LAB (4). Though Streptococcus thermophilus is the starter LAB species of major importance for the manufacture of yogurt and many cheese varieties, like Grana type, Swiss type, and pasta filata cheeses, and in spite of the wide literature concerning BA production among LAB (1, 3, 8, 28), there are few reports of histamine decarboxylase activity in this bacterial species. The presence in two S. thermophilus strains of a complete hdc gene cluster was only very recently reported (5).
The hdc gene clusters of Gram-positive bacteria usually comprise the gene hdcA, encoding a pyruvoyl-dependent decarboxylase, and hdcP, encoding a histidine/histamine antiporter, not yet identified in some histamine-producing LAB. Moreover, an hdcB gene involved in the conversion of the HdcA proenzyme to the active decarboxylase (29) and hdcRS, with functions not essential to the decarboxylation process, can also be found. In the case of S. thermophilus, the gene order is hdcAPB (5), similarly to Staphylococcus capitis and Clostridium perfringens, which, however, lack hdcB (10).
Conditions that favor histidine decarboxylation are the presence of the specific substrate or high concentrations of free amino acids and hostile environmental conditions that require the maintenance of pH homeostasis (15, 21). The first recognized role of decarboxylases was keeping a high intracellular pH as a consequence of the conversion of an amino acid to the corresponding amine. Their function as secondary energy supply processes was put in evidence by observing that some bacteria, for example O. oeni 9204, produce larger amounts of histamine under the poorest nutritional conditions (12). Indeed, a proton motive force is generated by exchange of the amine with the amino acid from the medium. In addition, histamine biosynthesis is modulated by the growth phase. Calles-Enrìquez et al. (5) observed an increase of histamine production in S. thermophilus during the stationary phase, while Landete et al. (14) found that hdcA expression in L. hilgardii was higher in the exponential phase. Moreover, histidine induced hdcA expression, whereas histamine repressed it (14).
The aims of this study were (i) establishing the occurrence of genetic determinants encoding histidine decarboxylases in S. thermophilus strains isolated from natural sources, (ii) studying hdcA expression and histamine production under conditions relevant for cheese making (presence of the substrate histidine, addition of NaCl, presence of different lactose levels, natural acidification, thermization of the inoculum), to evaluate the risk of histamine formation by S. thermophilus adventitious strains in dairy products, and (iii) evaluating the functionality of the HdcA enzyme released after cell lysis.
Screening of hdcA gene presence in S. thermophilus strains.
Eighty-three S. thermophilus strains, all (except two) dairy isolates, identified and genotypically differentiated in previous studies (16, 24), were screened by consensus PCR with oligonucleotides HIS1-F and HIS1-R (9) made more stringent by changing the annealing temperature to 58°C for 40 s. They were subcultured in M17 broth (Oxoid, Milan, Italy) added with 0.5% (wt/vol) lactose (LM17) at 37°C for 24 h, and total genomic DNA was extracted and purified from 2-ml cultures as described by Rossi et al. (26).
The strains PRI17, PRI18, and PRI21, all isolated from natural whey starter cultures for Grana-type cheese, PRI60 from mozzarella cheese, and PRI74 from traditional yogurt originated an amplification product of the expected size, close to 372 bp (data not shown). The amplicons were sequenced with primers T7 and SP6 (Promega) after purification by the Wizard SV gel and PCR clean-up system (Promega), ligation with the pGEM-T Easy vector (Promega), transformation in Escherichia coli DH5α, and plasmid extraction by the PureYield plasmid purification kit (Promega). Sequencing was done by BMR-Genomics (Padova, Italy). BLASTn (www.ncbi.nlm.nih.gov) was used for sequence similarity searches. The sequences were identical and showed the highest identity with a fragment of the hdcA gene.
The results indicated a percentage of hdcA-positive strains of 6%, somewhat higher than that reported for industrial strains by Calles-Enríquez et al. (5). Since in this case naturally occurring strains from dairy sources were screened, the frequency of hdcA presence can be considered to be more representative of the diffusion of histaminogenic strains potentially able to reach high numbers in products made from raw milk and by spontaneous fermentation.
Histaminogenic potential of hdcA-positive S. thermophilus strains.
The ability of the five S. thermophilus strains to produce histamine was confirmed and quantified by high-performance liquid chromatography (HPLC) on cultures grown in LM17 for 24 h. These were centrifuged at 8,000 × g for 10 min at 10°C. Histamine determination was done according to the method of Martuscelli et al. (18) after derivatization with dansyl chloride (Sigma-Aldrich, Gallarate, Italy) and using a PU-2089 Intelligent HPLC quaternary pump and Intelligent UV-VIS multiwavelength detector UV 2070 Plus (Jasco Corporation, Tokyo, Japan). The amount of histamine was expressed as mg liter−1 by reference to a calibration curve obtained with aqueous histamine standards of concentrations ranging between 10 and 200 mg liter−1.
The amounts of histamine produced by the S. thermophilus strains ranged between 13 and 40 mg liter−1. Strain PRI60, which produced the highest quantity of this BA, was chosen for the following experiments.
Sequencing of the S. thermophilus PRI60 hdcA gene and flanking regions.
The hdcA gene fragment of S. thermophilus PRI60 was used as a target for uneven PCR (6) to obtain the complete sequence of the gene and indications on its neighboring regions. Exact match primers STHDEC-F and STHDEC-R (Table 1) were designed on the available 372-bp hdcA gene fragment by the Primer Express 2.0 program (Applied Biosystems, Foster City, CA), also for use in reverse transcription-quantitative PCR (RT-qPCR), and were coupled with arbitrary primers. Primers RDW and Coc1 (Table 1) permitted us to determine the upstream and downstream adjacent regions, respectively. The amplification products from uneven PCR were sequenced either with specific primers or after cloning in the pGEM-T Easy vector (Promega).
Table 1.
Targets and sequences of the primer pairs used in this study
| Function | Primer | Sequence (5′→3′) | Nucleotide positions | Product size (bp) | Reference |
|---|---|---|---|---|---|
| Screening of hdcA presence in Gram-positive bacteria | HIS1-F | GGNATNGTNWSNTAYGAYMGNGCNGA | 172–197a | 372 | 9 |
| HIS1-R | ATNGCDATNGCNSWCCANACNCCRTA | 520–545a | |||
| Screening of hdcA presence in S. thermophilus strains and RT-qPCR | STDEC-F | GAATTACCGATCTATGATGC | 349–368a | 121 | This study |
| STDEC-R | ACACCTTTGTTAGCACAAAC | 451–470a | |||
| RT-qPCR on the S. thermophilusrpoA | RPOST-F | ACTGTCATTGTTGCTTGGAATG | 521–542b | 114 | 13 |
| RPOST-R | AGCTGAGGTTACTGCTGGAGAT | 613–634b | |||
| Random primer for uneven PCR | Coc-1 | AGCAGCGTGG | This study | ||
| Random primer for uneven PCR | RDW | CCGATCTTCTCGC | This study |
Positions in the S. thermophilus PRI60 hdcA sequence.
Positions in the rpoA ORF of S. thermophilus LMD-9 (accession no. CP000419; region 1755506 to 1756444).
The open reading frames (ORFs) encoding the histamine decarboxylase and the amino acid permease were identified by the ORF Finder tool at NCBI. The entire hdcA ORF, 1,313 bp preceding its ATG translation start and 466 bp following its stop codon, was determined. The complete S. thermophilus PRI60 hdcA sequence was found to share a 99% identity with those of S. thermophilus CHCC6483 and CHCC1524 (GenBank accession no. FN686790 and FN686789, respectively). In total, eight mutations, at positions 34, 78, 318, 501, 571, 645, 879, and 915, were present. The first five mutations were in common with both strains. Among these, that at position 571 resulted in a missense Asp→Asn at the amino acid level. Of the remaining mutations, two were in S. thermophilus CHCC1524 (nucleotides 879 and 915) and the other one was in S. thermophilus CHCC6483. Moreover, the hdcA gene was 71% identical to its homolog in Lactobacillus spp. 30A (GenBank accession no. DQ132891) and shared a slightly lower degree of similarity with hdcA genes from Lactobacillus reuteri (70%), Staphylococcus epidermidis, S. capitis, Lactobacillus sakei, O. oeni, Tetragenococcus halophilus, L. hilgardii (69%), Tetragenococcus muriaticus, and L. buchneri (68%).
The 458-bp portion of the hdcP ORF was 99% identical to the corresponding region in strains S. thermophilus CHCC1524 and CHCC6483. Also in this case, there were different numbers of mutations found in S. thermophilus PRI60 with respect to these two strains (three and one, respectively). One of these mutations, at position 207 of the ORF, was the same. The next most similar sequence was that of the hdcP gene of S. capitis (69% identity).
According to the prediction analysis carried out at http://www.fruitfly.org/seq_tools/promoter.html, a putative promoter (maximum score, 1.00) starting 132 nucleotides upstream of the ATG codon precedes the hdcA gene of S. thermophilus PRI60. Two and one deletions were found between the putative −35 and −10 sequences compared to those of strains S. thermophilus CHCC1524 and CHCC6483, respectively, that could account for different promoter strengths. The ribosome binding site (RBS) is probably a GGAGA motif found at −14 nucleotides from the translation start site.
The 1,313-bp sequence upstream of hdcA showed the absence of other genes belonging to the hdc cluster. BLASTn analysis revealed two regions of 614 and 220 bp, separated by a 93-bp deletion, sharing, respectively, 98% and 95% identity with portions of the S. thermophilus genomic island ΔCIME308, GenBank accession no. AJ586570.1 (20), involved in lateral gene transfer (27). The 614-bp segment comprised the first 283 bp of the hdsR gene, encoding the restriction subunit of a type I modification/restriction system. Based on these indications, the S. thermophilus hdc gene cluster is probably located on the chromosome, but the vicinity of such genetic elements suggests that hdc-positive S. thermophilus strains might confer these genes to other strains or species.
Histamine production by S. thermophilus under different conditions.
The levels of histamine produced by S. thermophilus PRI60 in different media and growth conditions were assayed. The chosen media were skim milk (Oxoid), skim milk with added 2% (wt/vol) NaCl, LM17, LM17 with 2% (wt/vol) NaCl, and LM170.1 (i.e., LM17 with a reduced amount of lactose, 0.1% [wt/vol]); when indicated, these media were supplemented after sterilization with 0.1% (wt/vol) histidine. The strain was inoculated in triplicate at 5 × 106 CFU ml−1 and incubated at 37°C for 5 days. Counts of S. thermophilus PRI60 during incubation were done by plating in duplicate serial dilutions of the cultures on LM17 agar medium and incubating at 37°C for 48 h.
Histamine was not detected when the strain was grown in skim milk, with or without 2% (wt/vol) NaCl, but it was produced when histidine was supplemented (see Fig. S1 in the supplemental material). In this case, regardless of the presence of NaCl, a maximum level of histamine, approximately 350 mg liter−1, was reached in the first 12 h. This value corresponded to the decarboxylation of about 50% of the histidine added to the medium. The growth of the strain was similar in all the tested conditions in skim milk, both with and without NaCl.
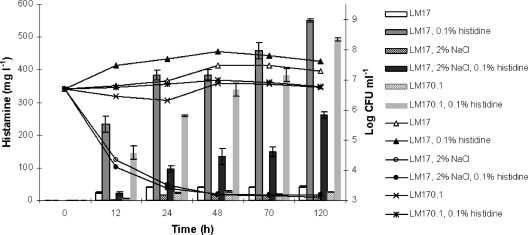
The experiment with synthetic medium LM17 was aimed at verifying the effect of low energy supply by diminishing the amount of lactose. In both LM17 and LM170.1, histamine was produced at relatively low levels (17 to 42 mg liter−1) after 24 h and remained stable for 5 days also in the samples not supplemented with histidine for the presence of this amino acid or hydrolyzable peptides containing it already in the formulation (Fig. 1). When histidine was added, the production of histamine was considerably higher. However, differences in the medium composition determined the accumulation of various amounts of the BA that are not proportional to the cell number. Particularly, in the presence of NaCl relatively large amounts of histamine were formed notwithstanding the very low count values.
Fig. 1.
Production of histamine (bars) and counts (lines) of S. thermophilus PRI60 grown in LM17 under the following conditions: LM17, LM17 with 2% (wt/vol) NaCl, LM170.1 (with 0.1% [wt/vol] lactose), all with and without the addition of 0.1% (wt/vol) histidine.
Our experiments highlighted the existence of S. thermophilus strains able to produce histamine at concentrations which can cause health concerns. In LM17 medium, after 24 h, S. thermophilus PRI60 produced approximately double the amount of histamine of S. thermophilus CHCC1524 (5). Moreover, the longer incubation time used in the present study permitted us to observe a further increase of the histamine concentration after 48 h (Fig. 1).
hdcA transcriptional analysis and histamine production under conditions relevant for cheese making.
The expression level of the hdcA gene and the histamine accumulation by S. thermophilus PRI60 were quantified by RT-qPCR and HPLC, respectively, evaluating the effect of parameters relevant for cheese making, such as thermization, presence of 0.1% (wt/vol) histidine (a level possibly found in cheese), presence of 2% (wt/vol) NaCl, and ripening temperature of 20°C. S. thermophilus PRI60 was inoculated in skim milk at an initial cell number of 5 × 106 CFU ml−1 and incubated at 37°C for 24 h. Later the samples were kept at 20°C for a further 6 days. Histidine and NaCl were added in some samples after 6 and 24 h, respectively. In other cultures an inoculum heated at 67°C for 20 s was used to study the effect of thermization.
The transcriptional level of the hdcA gene was referred to the constitutive gene rpoA, encoding the S. thermophilus RNA polymerase alpha subunit. This was chosen as a reference gene among those reported to be stably expressed in bacteria (23). The stability of expression of rpoA in S. thermophilus PRI60 was experimentally confirmed as described in a previous paper (13), with a maximum intertreatment variation of the transcript 2ΔCT = 2.0. An RT-qPCR test was assessed for each target gene using the primer pairs STHDEC-F/STHDEC-R and RPOST-F/RPOST-R (Table 1), specific for hdcA and rpoA, respectively.
The qPCRs were carried out as described previously (13) using the Platinum SYBR green Supermix UDG with Rox (6-carboxy-X-rhodamine) dye (Invitrogen, Milan, Italy). Calibration curves were constructed by using as targets duplicate 10-fold serial dilutions of the hdcA- and rpoA-specific amplicons. The linearity ranges of both tests included points between 0.3 and 6.3 log copies of target in the reaction (see Fig. S2 in the supplemental material). The efficiencies [E = 10(−1/slope) −1] of the hdcA- and rpoA-specific qPCR tests were 0.98 and 1.02, respectively.
RNA extraction from skim milk cultures and reverse transcription for the RT-qPCR tests were carried out as previously described (13). Duplicate RT-qPCR trials were carried out amplifying hdcA and rpoA in separate sets of reactions on the same cDNAs. The expression ratio of hdcA referred to rpoA was determined according to the method of Pfaffl (22), and the CT value of each gene at 4 h of incubation was used as a control.
The growth of S. thermophilus PRI60 during the trials was very similar in all the cultures (Fig. 2). A pH decrease to 4.23, or to 4.28 in the cultures with added histidine, was observed after 24 h; then the values remained stable for the whole incubation period.
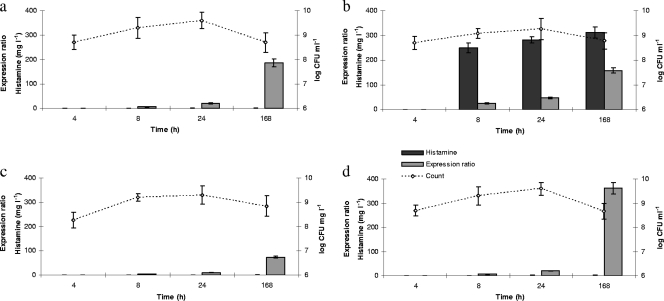
Fig. 2.
Transcription ratio of the hdcA gene referred to the reference gene rpoA (gray bars), histamine formation (black bars), and counts (lines) of S. thermophilus PRI60 during growth under the following conditions: skim milk (a), skim milk plus 0.1% (wt/vol) histidine (added after 6 h) (b), thermized (67°C for 20 s) inoculum in skim milk (c), and skim milk plus 2% (wt/vol) NaCl (added after 24 h) (d).
Based on the CT values, which depend on the transcript copy number, the hdcA gene of S. thermophilus PRI60 was highly expressed since the early stages of incubation and it further increased in the stationary phase. This is in contrast with the results of Calles-Enríquez et al. (5), who found higher expression at the onset of the stationary phase. Indeed, only at 4 h was the average copy number of the hdcA transcript (6.1 log) lower than that of the rpoA transcript (6.9 log), while after 8 h from the inoculum the copy number of the hdcA transcript was found to be equal to or higher than that of the rpoA transcript. The increase of hdcA expression during incubation suggested a positive regulation by acidification and energy source depletion. This provided evidence that the activation of HdcA reported under such conditions for O. oeni and S. capitis (7, 10) occurs at the transcriptional level in S. thermophilus.
In Fig. 2, the normalized hdcA transcription levels during the trial are reported, together with the histamine production. Histamine was detected in relevant amounts only in the samples supplemented with histidine (Fig. 2b), while hdcA was expressed in all cases. After 8 h, the expression ratio of hdcA varied from 4.9 to 24.7 compared to the control (4 h), depending on the growth conditions. However, in the samples supplemented with histidine, the expression ratio of hdcA increased faster than in the cultures without histidine, indicating that the level of transcription was enhanced by the addition of the amino acid. At 24 h the hdcA expression ratio in the presence of histidine was about 2-fold higher than in cultures without histidine (Fig. 2a and b). It must be noticed that about 80% of the total histamine produced after 7 days was accumulated in the first 2 h after the addition of histidine, when the cultures were not yet in stationary phase. The observation that a large amount of histamine is rapidly formed in the presence of histidine indicated that S. thermophilus PRI60 has a high potential of efficiently accumulating this BA in dairy substrates.
Thermization of the inoculum reduced the expression level of hdcA, and it remained lower at all time intervals compared to that in the other trials (Fig. 2c). No explanation of this finding is available from the literature. Therefore, the regulation mechanisms involved need to be clarified in order to exploit heat treatment to reduce histamine formation by S. thermophilus.
On the other hand, the addition of salt led to an upregulation of hdcA (Fig. 2d) as observed at 7 days, when the expression ratio was 1.9-fold the corresponding value in skim milk. This result suggests a potential role of hdcA in osmoprotection mechanisms, though no literature data are available on this respect. Moreover, this finding can have implications for cheese making and can promote further research on the physiological role of histidine decarboxylation in bacteria.
Effects of histidine and cell integrity on histamine production.
In order to evaluate if culturing in the presence of histidine could influence the overall decarboxylase activity of S. thermophilus PRI60 and to determine the activity of the HdcA enzyme in cell lysates, enzymatic assays were carried out with whole and disrupted cells. The strain was previously subcultured twice for 24 h in skim milk with or without the addition of 0.1% (wt/vol) histidine. Skim milk was chosen as the precultivation medium because of its inability to support histamine formation in the absence of added histidine (Fig. 2). Cultures of 100 ml were centrifuged at 4,000 × g for 10 min in the presence of two volumes of EDTA 26.8 mM, pH 12, washed twice with physiological solution (0.9% wt/vol NaCl), and resuspended in 20 ml of the same solution. Cell suspensions were subdivided into two aliquots: one was used for testing the HdcA activity of whole cells, and the other was submitted to three passages at 18,000 lb/in2 through a French pressure cell press (SLM Aminco; Spectronic Instruments, Rochester, NY) to disrupt the cells and test the activity in cell extracts. The treated cells were cooled in ice between the breaking passages. Finally, both whole-cell suspensions and cell extracts were added to phosphate buffer (pH 5.3) containing 0.1% (wt/vol) histidine in triplicate at a concentration corresponding to about 2.5 × 108 CFU ml−1, and incubated at 37°C. Samples were collected at different times (0, 0.5, 1, 1.5, 2, 3, 4, 5, 6, 7, 8, and 24 h) for the determination of histamine content. After centrifugation at 10,000 × g for 10 min at 8°C, the supernatants were used for HPLC analysis. The series of values of histamine amounts formed by whole or broken cells were compared by Student's t test.
The trial showed that histamine increased over time in both whole and broken cells, but the accumulation rates were not significantly affected by the presence of histidine in the precultivation step (see Fig. S3 in the supplemental material). In addition, the histidine decarboxylase was particularly active in the cell extracts, where histamine accumulation was faster than in the whole cells. Indeed, the histamine content after 24 h of incubation was higher for the cell extracts (approximately 220 mg liter−1 against the 160 mg liter−1 obtained with the whole cells).
Though histidine was found to enhance the expression of the hdcA gene, the increase observed at the transcriptional level did not correspond to significantly different levels of histamine produced (see Fig. S3 in the supplemental material). An explanation could be that a probable high affinity of the HdcA for its substrate and consequent high speed of the enzymatic reaction may have masked the differences in the amounts of enzyme.
It is noteworthy that the S. thermophilus HdcA was more active in cell extract. The higher activity observed can be explained by the slowing down of the decarboxylase reaction by the histamine/histidine exchange across the membrane in whole cells. The high level HdcA activity after cell lysis is a relevant concern for cheese production, since in these products HdcA could remain active for a long time during ripening and determine high histamine contents.
On the whole, results indicated that the presence of S. thermophilus strains carrying HdcA activity is a risk unknown until recently, and further research efforts should be focused on its better characterization and prevention. Remedies that can be carried out to reduce the risk of histamine formation in cheeses by hdcA gene-positive S. thermophilus strains are (i) the introduction of a screening for decarboxylases, such as a PCR-based detection of the hdcA gene, among the procedures applied for the selection of starter cultures and (ii) the possibility of using autochthonous nonhistaminogenic S. thermophilus isolates to outcompete histamine producers in artisanal dairy products.
A safer exploitation of this bacterial species can be promoted by revising its QPS (qualified presumption of safety) or GRAS (generally recognized as safe) status by taking into account the occurrence of BA-producing strains.
Nucleotide sequence accession number.
The EMBL/GenBank /DDBJ databases accession number of the sequence determined in this study is FR693807.
Supplementary Material
Acknowledgments
This research was partly supported by a grant from the Italian Ministry of University and Research within the program PRIN 2006 no. 2006072328_004.
We acknowledge Carlo Rugolotto for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Ancín-Azpilicueta C., González-Marco A., Jiménez-Moreno N. 2008. Current knowledge about the presence of amines in wine. Crit. Rev. Food Sci. Nutr. 48:257–275 [DOI] [PubMed] [Google Scholar]
- 2. Bodmer S., Imark C., Kneubühl M. 1999. Biogenic amines in foods: histamine and food processing. Inflamm. Res. 48:296–300 [DOI] [PubMed] [Google Scholar]
- 3. Bunkova L., et al. 2009. Tyramine production of technological important strains of Lactobacillus, Lactococcus and Streptococcus. Eur. Food Res. Technol. 229:533–538 [Google Scholar]
- 4. Burdychova R., Komprda T. 2007. Biogenic amine-forming microbial communities in cheese. FEMS Microbiol. Lett. 276:149–155 [DOI] [PubMed] [Google Scholar]
- 5. Calles-Enríquez M., et al. 2010. Sequencing and transcriptional analysis of the Streptococcus thermophilus histamine biosynthesis gene cluster: factors that affect differential hdcA expression. Appl. Environ. Microbiol. 76:6231–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X., Wu R. 1997. Direct amplification of unknown genes and fragments by uneven polymerase chain reaction. Gene 185:195–199 [DOI] [PubMed] [Google Scholar]
- 7. Coton E., Rollan G. C., Lonvaud-Funel A. 1998. Histidine carboxylase of Leuconostoc oenos 9204: purification, kinetic properties, cloning and nucleotide sequence of the hdc gene. J. Appl. Microbiol. 84:143–151 [DOI] [PubMed] [Google Scholar]
- 8. Coton E., Coton M. 2005. Multiplex PCR for colony direct detection of Gram-positive histamine- and tyramine-producing bacteria. J. Microbiol. Methods 63:296–304 [DOI] [PubMed] [Google Scholar]
- 9. de las Rivas B., Marcobal A., Carrascosa A. V., Muñoz R. 2006. PCR detection of foodborne bacteria producing the biogenic amines histamine, tyramine, putrescine, and cadaverine. J. Food Prot. 69:2509–2514 [DOI] [PubMed] [Google Scholar]
- 10. de las Rivas B., Rodríguez H., Carrascosa A. V., Muñoz R. 2008. Molecular cloning and functional characterization of a histidine decarboxylase from Staphylococcus capitis. J. Appl. Microbiol. 104:194–203 [DOI] [PubMed] [Google Scholar]
- 11. European Union Regulation (EC) no. 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Official Journal L 338, 22.12.2005, p. 1–26
- 12. Konings W. N. 2006. Microbial transport: adaptations to natural environments. Antonie Van Leeuwenhoek 90:325–342 [DOI] [PubMed] [Google Scholar]
- 13. La Gioia F., et al. 2011. Identification of a tyrosine decarboxylase (tdcA) gene in Streptococcus thermophilus 1TT45 and analysis of its expression and tyramine production in milk. Appl. Environ. Microbiol. 77:1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Landete J. M., Pardo I., Ferrer S. 2006. Histamine, histidine, and growth-phase mediated regulation of the histidine decarboxylase gene in lactic acid bacteria isolated from wine. FEMS Microbiol. Lett. 260:84–90 [DOI] [PubMed] [Google Scholar]
- 15. Landete J. M., de las Rivas B., Marcobal A., Muñoz R. 2008. Updated molecular knowledge about histamine biosynthesis by bacteria. Crit. Rev. Food Sci. Nutr. 48:697–714 [DOI] [PubMed] [Google Scholar]
- 16. Lazzi C., et al. 2009. Application of AFLP fingerprint analysis for studying the biodiversity of Streptococcus thermophilus. J. Microbiol. Methods 79:48–54 [DOI] [PubMed] [Google Scholar]
- 17. Lehane L., Olley J. 2000. Histamine fish poisoning revisited. Int. J. Food Microbiol. 58:1–37 [DOI] [PubMed] [Google Scholar]
- 18. Martuscelli M., Crudele M. A., Gardini F., Suzzi G. 2000. Biogenic amine formation and oxidation by Staphylococcus xylosus strains from artisanal fermented sausages. Lett. Appl. Microbiol. 31:228–232 [DOI] [PubMed] [Google Scholar]
- 19. Novella-Rodríguez S., Veciana-Nogués M. T., Roig-Sagués A. X., Trujillo-Mesa A. J., Vidal-Carou M. C. 2002. Influence of starter and nonstarter on the formation of biogenic amine in goat cheese during ripening. J. Dairy Sci. 85:2471–2478 [DOI] [PubMed] [Google Scholar]
- 20. Pavlovic G., Burrus V., Gintz B., Decaris B., Guédon G. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759–774 [DOI] [PubMed] [Google Scholar]
- 21. Pessione E., et al. 2005. A proteomic approach to studying biogenic amine producing lactic acid bacteria. Proteomics 5:687–698 [DOI] [PubMed] [Google Scholar]
- 22. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ritz M., Garenaux A., Berge M., Federighi M. 2009. Determination of rpoA as the most suitable internal control to study stress response in C. jejuni by RT-qPCR and application to oxidative stress. J. Microbiol. Methods 76:196–200 [DOI] [PubMed] [Google Scholar]
- 24. Rizzotti L., La Gioia F., Dellaglio F., Torriani S. 2009. Characterization of tetracycline-resistant Streptococcus thermophilus isolates from Italian soft cheeses. Appl. Environ. Microbiol. 75:4224–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roig-Sagués A. X., Molina A. P., Hernández-Herrero M. M. 2002. Histamine and tyramine-forming microorganisms in Spanish traditional cheeses. Eur. Food Res. Technol. 215:96–100 [Google Scholar]
- 26. Rossi F., Dellaglio F., Torriani S. 2006. Evaluation of recA gene as a phylogenetic marker in the classification of dairy propionibacteria. Syst. Appl. Microbiol. 29:463–469 [DOI] [PubMed] [Google Scholar]
- 27. Schirawski J., Hagens W., Fitzgerald G. F., Van Sinderen D. 2002. Molecular characterization of cadmium resistance in Streptococcus thermophilus strain 4134: an example of lateral gene transfer. Appl. Environ. Microbiol. 68:5508–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzzi G., Gardini F. 2003. Biogenic amines in dry fermented sausages: a review. Int. J. Food Microbiol. 88:41–54 [DOI] [PubMed] [Google Scholar]
- 29. Trip H., Mulder N. L., Rattray F. P., Lolkema J. S. 2011. HdcB, a novel enzyme catalysing maturation of pyruvoyl-dependent histidine decarboxylase. Mol. Microbiol. 79:861–871 [DOI] [PubMed] [Google Scholar]
- 30. Wohrl S., Hemmer W., Focke M., Rappersberger K., Jarisch R. 2004. Histamine intolerance-like symptoms in healthy volunteers after oral provocation with liquid histamine. Allergy Asthma Proc. 25:305–311 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.