Abstract
Macromolecular complexes containing histone deacetylase and ATPase activities regulate chromatin dynamics and are vitally responsible for transcriptional gene silencing in eukaryotes. The mechanisms that target these assemblies to specific loci are not as well understood. We show that the corepressor KAP-1, via its PHD (plant homeodomain) and bromodomain, links the superfamily of Krüppel associated box (KRAB) zinc finger proteins (ZFP) to the NuRD complex. We demonstrate that the tandem PHD finger and bromodomain of KAP-1, an arrangement often found in cofactor proteins but functionally ill-defined, form a cooperative unit that is required for transcriptional repression. Substitution of highly related PHD fingers or bromodomains failed to restore repression activity, suggesting high specificity in their cooperative function. Moreover, single amino acid substitutions in either the bromodomain or PHD finger, including ones that mimic disease-causing mutations in the hATRX PHD finger, abolish repression. A search for effectors of this repression function yielded a novel isoform of the Mi-2α protein, an integral component of the NuRD complex. Endogenous KAP-1 is associated with Mi-2α and other components of NuRD, and KAP-1-mediated silencing requires association with NuRD and HDAC activity. These data suggest the KRAB-ZFP superfamily of repressors functions to target the histone deacetylase and chromatin remodeling activities of the NuRD complex to specific gene promoters in vivo.
Keywords: KRAB-ZFPs, repression, PHD finger, bromodomain, NuRD, ATRX
The complement of transcription factors organized at a gene promoter integrates different transcriptional regulatory signals that orchestrate cellular responses such as proliferation, differentiation, and apoptosis. Regulation of RNA polymerase II involves a complex interplay between DNA–protein interactions and protein–protein interactions. Whereas the general transcription factors regulate the accurate initiation of transcription, proteins that bind gene-specific DNA elements are instrumental in either negatively or positively regulating the rate of transcription (Orphanides et al. 1996; Hampsey and Reinberg 1999). Studies aimed at understanding the mechanisms of transcriptional regulation by site-specific factors have been aided by the realization that these proteins are highly modular in architecture. In addition to sequence-specific DNA binding domains, these proteins possess an independent, functionally separable effector domain that can either activate or repress transcription. The affects of sequence-specific transcription factors in the regulation of transcription have been greatly facilitated by the identification of cofactors with which they bind and the intrinsic or associated biochemical activities of these proteins. It is clear from this strategy that these proteins possess enzymatic activities which either utilize ATP to remodel chromatin structure or covalently modify the amino-terminal tails of the core histones, creating a biochemical code that regulates gene transcription of chromatin templates (Tyler and Kadonaga 1999; Strahl and Allis 2000). Thus, sequence-specific repressor–corepressor complexes likely serve as scaffolds for the assembly of macromolecular complexes which integrate diverse transcription regulatory signals.
The Krüppel associated box (KRAB) domain is one example of an abundant amino acid sequence motif found at the N terminus of nearly one-third of all Krüppel/TFIIIA-type C2H2 zinc finger proteins (Bellefroid et al. 1991). This highly conserved domain displays potent, DNA-binding dependent repression of transcription that requires the KAP-1 corepressor (Margolin et al. 1994; Friedman et al. 1996; Kim et al. 1996; Moosmann et al. 1996, 1997). The primary amino acid sequence of KAP-1 revealed the presence of several conserved signature motifs, including a RING finger, B boxes, and a coiled-coil region (RBCC), which collectively form an integrated domain that is both necessary and sufficient to directly interact with the KRAB domain (Fig. 1A) (Peng et al. 2000a,b). The C-terminal sequence of KAP-1 revealed a conserved PHD (plant homeodomain) finger and bromodomain (Fig. 1A). This particular spatial arrangement of motifs has defined an emerging family of transcriptional regulators that include TIF1α and TIF1γ (Fig. 1A) (Le Douarin et al. 1995; Venturini et al. 1999). All members of this protein family have been shown to repress transcription when tethered to DNA, and the mechanisms by which they repress transcription are currently being defined. In this aspect, a direct interaction between KAP-1 and mammalian members of the nonhistone chromosomal protein, heterochromatin protein 1 (HP1) has been described (Le Douarin et al. 1996; Nielsen et al. 1999; Ryan et al. 1999; Lechner et al. 2000). Biochemical studies indicate that this interaction is dependent upon the chromoshadow domain of HP1 proteins and a core, hydrophobic pentapeptide sequence (PxVxL) in KAP-1 (Lechner et al. 2000). Furthermore, indirect immunofluorescence studies of interphase nuclei indicated that KAP-1 colocalizes with both heterochromatic and euchromatic HP1 proteins (Ryan et al. 1999). These particular studies suggest that the KRAB–KAP-1 complex may mediate repression in part via the maintenance or initiation of heterochromatic chromosomal environments. Moreover, it remains to be determined what role the PHD finger and bromodomain possess in KAP-1 repression, or if these domains integrate additional factors which facilitate HP1 recruitment and function.
Figure 1.
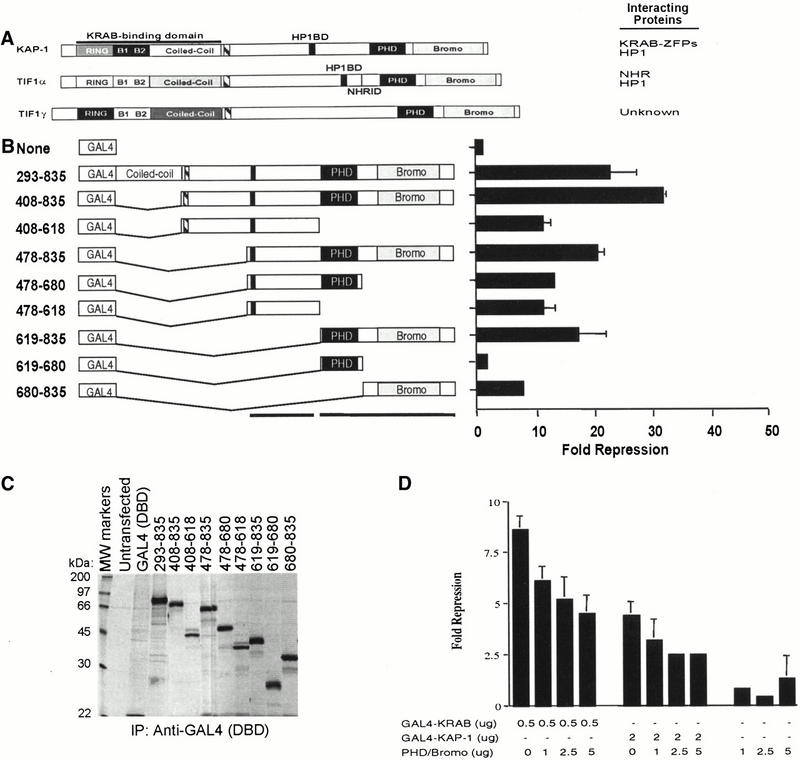
Analysis of the intrinsic repression activity of KAP-1 indicates that the PHD finger and bromodomain are required and sufficient to repress transcription. (A) Schematic illustration of the KAP-1/TIF1 family of transcriptional regulators. The KRAB binding domain in KAP-1 is indicated. (Striped box) TIF signature sequence (TSS), (black box) HP1 binding domain (HP1BD), (thin black line) nuclear hormone receptor interaction domain. (B) Schematic illustration of the heterologous GAL4–KAP-1 constructs, as defined by the amino acid numbers at the left. The intrinsic repressor activity of KAP-1 was measured in a transient assay, using a minimal TK-luciferase reporter template regulated by five consensus GAL4 UAS. All experiments were done in NIH/3T3 cells with 0.5 μg of reporter plasmid and 5 μg of the indicated heterologous GAL4–KAP-1 construct. Fold repression represents the ratio of luciferase activity measured for the reporter alone to the activity measured in the presence of the indicated effector proteins after normalization for transfection efficiency. Error bars represent the standard deviation for three independent transfections. Absence of error bars indicates a standard deviation too small to physically illustrate. (C) Each plasmid was tested for stable protein expression via transfection into COS1 cells followed by immunoprecipitation of [35S]methionine-labeled whole cell extracts with anti-GAL4 (DBD) IgG (1 μg). (D) Overexpression of the KAP-1 PHD finger and bromodomain (amino acids 619 to 835) demonstrated a dose-dependent dominant effect on GAL4–KRAB- and GAL4–KAP-1-mediated transcriptional repression. Transcriptional effects were monitored in a transient assay in which 0.5 μg of the 5x-GAL4 UAS-SV40 luciferase reporter and the indicated amounts of expression plasmids transfected into NIH/3T3 cells. Fold repression was calculated as described. Expression of the dominant negative protein was confirmed in COS1 cells (data not shown).
The PHD finger and bromodomain are two highly conserved protein motifs found in an increasing number of proteins with roles in regulating transcription via modification of chromatin structure (Aasland et al. 1995; Jeanmougin et al. 1997; Winston and Allis 1999). Furthermore, these two motifs are commonly linked architecturally in a number of proteins (Le Douarin et al. 1995, 1996; Friedman et al. 1996; Venturini et al. 1999; Bochar et al. 2000; Jones et al. 2000a,b). Although a specific function for the PHD finger remains to be fully described, naturally occurring single amino acid substitutions in the PHD finger of the hATRX and AIRE1 proteins predispose individuals to α-thalassemia, developmental defects, and autoimmune disease, respectively (Gibbons et al. 1997; Bjorses et al. 2000). These observations emphasize the biological importance of this domain in the functions of these proteins. On the other hand, structural and biochemical studies of three independent bromodomains clearly establish a role for this domain in the molecular recognition of the N termini of acetylated histone tails (Dhalluin et al. 1999; Jacobson et al. 2000; Owen et al. 2000). Moreover, mutations that specifically disrupt the bromodomain/acetyl-histone interaction have been shown to functional impair the transcriptional regulatory activities of these proteins in vivo (Cairns et al. 1999; Sterner et al. 1999; Syntichaki et al. 2000).
To understand the molecular role of the PHD finger and bromodomain in KAP-1-mediated transcriptional repression, we have used a biochemical approach to identify effector functions of this bipartite domain. Our data suggest that the spatial conservation of these motifs is essential for optimal protein function. These data are consistent with the PHD finger and bromodomain providing an interface for protein–protein interactions, and therefore may have broad implications in understanding the function of proteins that maintain this particular spatial arrangement of motifs. Comparative analyses of proteins containing these two motifs indicate that this bipartite domain is not functionally equivalent among all proteins. Thus, it is likely that the function of this integrated domain will be dependent upon those associated proteins that specifically interact with KAP-1 sequences. The interaction of KAP-1 with Mi-2α is consistent with this hypothesis and implies a role for histone deacetylases in transcriptional repression mediated by the KRAB-ZFP–KAP-1 repressor/corepressor system. Moreover, the estimated 300 to 700 KRAB zinc finger proteins in the human proteome would serve as an effective sequence-specific targeting mechanism for the histone deacetylase complex NuRD to gene promoters in vivo.
Results
Several pieces of evidence indicate that KAP-1 functions as a transcriptional corepressor for the KRAB repression module, including an intrinsic ability to repress transcription when directly tethered to DNA templates (Friedman et al. 1996; Moosmann et al. 1996; Agata et al. 1999; Nielsen et al. 1999; Ryan et al. 1999). To identify regions of KAP-1 which are required for its transcriptional repression function, we have engineered a series of KAP-1 deletion mutants with the heterologous GAL4 DNA binding domain to grossly map repression domains (Fig. 1B). Each heterologous fusion protein was tested for stable expression and nuclear localization in COS1 cells (Fig. 1C; data not shown). Nuclear extracts prepared from COS1 cells transfected with each construct demonstrated GAL4 DNA binding activity in vitro (data not shown). In general, amino acids C-terminal to the RBCC region define at least two independent repression domains within KAP-1 that appear to be additive in nature in order to obtain maximal levels of transcriptional repression. One of these domains includes the recently defined HP1 binding domain of KAP-1 (Ryan et al. 1999; Lechner et al. 2000). Optimal repression of transcription was dependent upon the PHD finger and bromodomain. When autonomously tethered to DNA, these two motifs demonstrated a significant level of repression, suggesting that they can function as an independent repression domain (Fig. 1B). This repression activity was DNA binding-dependent, dose-dependent, and universal for all promoters tested, including the SV40 immediate early enhancer, the adenovirus major late promoter, and a minimal genomic promoter fragment of the DNA polymerase β gene (data not shown). Neither the PHD finger nor the bromodomain could independently recapitulate this repression activity, indicating that these two motifs function cooperatively to repress transcription (Fig. 1B). Furthermore, expression of the segment encoding the PHD finger and bromodomain of KAP-1 relieved both GAL4–KRAB and GAL4–KAP-1 directed repression, suggesting that repression by this bipartite domain is mediated by at least one or more titratable effector molecules (Fig. 1D). The combination of these data suggests that the PHD finger and bromodomain of KAP-1 function as an integrated unit to facilitate repression of transcription by KAP-1. Moreover, these data implicate an additional repression mechanism that is independent of the recruitment of HP1 proteins (Ryan et al. 1999; Lechner et al. 2000).
Comparative analysis of the KAP-1 PHD finger and bromodomain with domains from related proteins
Because the KAP-1 orthologs, TIF1α and TIF1γ, have been reported to function as transcriptional repressors, we compared the repression properties of the PHD finger and bromodomain of all three family members. To directly test this hypothesis, we have constructed heterologous fusions between the GAL4 DNA binding domain and the PHD finger and bromodomains of TIF1α, TIF1γ, and WCRF180 (Bochar et al. 2000) (Fig. 2A). In addition, we engineered chimeric proteins in which either the PHD finger or bromodomain of TIF1α, TIF1γ, hATRX, Mi-2α, WCRF180, or hGCN5 were substituted for the KAP-1 sequences in order to address the specificity of the repression activity displayed by this bipartite domain (Fig. 2A). Stable expression and nuclear localization of each chimeric fusion protein were confirmed in COS1 cells (Fig. 2B and data not shown). Nuclear extracts prepared from COS1 cells transfected with each construct demonstrated GAL4 DNA binding activity in vitro (data not shown). When tethered to DNA, the PHD finger and bromodomain of TIF1α demonstrated a potential to activate transcription of the reporter template, whereas the PHD finger and bromodomain of TIF1γ and WCRF180 each failed to demonstrate any appreciable potentiation of transcription (Fig. 2A). Heterologous substitution of the KAP-1 PHD finger or bromodomain with similar sequences from other transcriptional regulators failed to completely complement the repression activity of the wild-type KAP-1 PHD finger and bromodomain (Fig. 2A). Interestingly, when the bromodomain of KAP-1 was tethered with the PHD finger of TIF1α, the chimeric protein functioned to repress transcription from the reporter template. In general, the KAP-1 bromodomain, when fused to any PHD finger, slightly enhanced the repression potential of the chimeric protein, suggesting that the bromodomain of KAP-1 is essential in mediating specific interactions with effectors that promote repression of transcription (Fig. 2A). The combination of these data provides additional support that the PHD finger and bromodomain of KAP-1 function as an integrated functional unit to specifically repress transcription. The data also imply that not all PHD fingers and bromodomains are functionally equivalent, despite demonstrating homology in the primary amino acid sequence. Furthermore, the spatial conservation of these two motifs architecturally in proteins is not sufficient to infer a common function for this bipartite domain.
Figure 2.
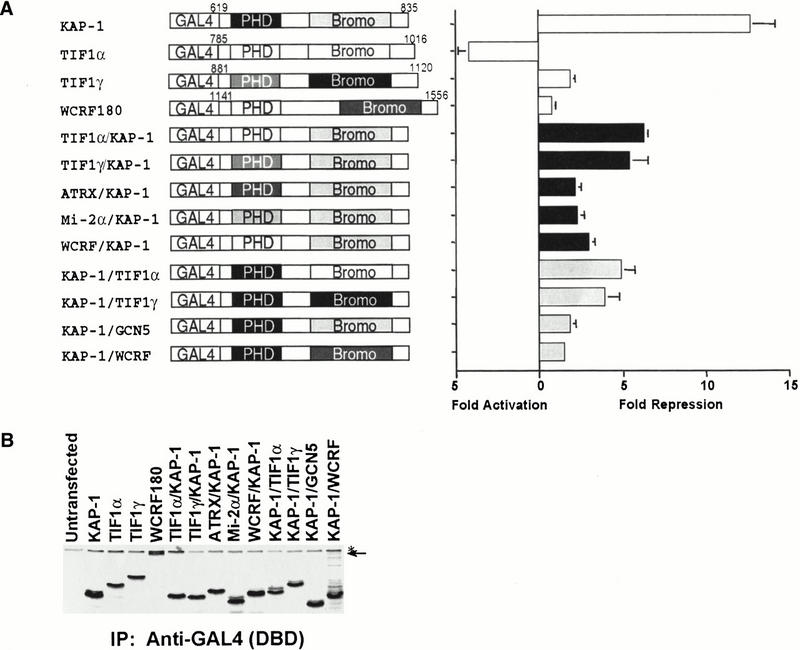
Heterologous chimeric fusions of related PHD fingers or bromodomains to KAP-1. (A) Schematic illustration of chimeric fusion proteins engineered. The chimeric fusion proteins possess the following amino acid sequences of the individual proteins, respectively: TIF1α/KAP-1 (815–876/680–835), TIF1γ/KAP-1 (880–941/680–835), ATRX/KAP-1 (210–283/680–835), Mi-2α/KAP-1 (372–423/680–835), WCRF180/KAP-1 (1141–1205/680–835), KAP-1/TIF1α (619–679/877–1016), KAP-1/TIF1γ (619–679/942–1120), KAP-1/GCN5 (619–679/714–837), and KAP-1/WCRF180 (619–679/1389–1556). All experiments were done in NIH/3T3 cells, as described in Fig. 1. (White bars) Natural PHD finger/bromodomain fusions, (black bars) chimeras with the KAP-1 bromodomain, (gray bars) chimeras with the KAP-1 PHD finger. (B) Each plasmid was tested for stable protein expression via transfection into COS1 cells followed by immunoprecipitation of [35S]methionine-labeled whole cell extracts with anti-GAL4 (DBD) IgG (1 μg). (Arrow) the migration of the heterologous GAL4–WCRF180 PHD finger/bromodomain, (asterisk) a nonspecific band retained during the immunoprecipitation.
Mutational analysis of the KAP-1 PHD finger and bromodomain in repression
In order to correlate the molecular relationship between amino acids in the PHD finger and bromodomain and transcriptional repression, we have used a site-directed mutagenesis approach. All amino acid substitutions were made in the context of the heterologous GAL4–PHD finger/bromodomain fusion protein. Stable expression and nuclear localization of all heterologous fusion proteins were confirmed in COS1 cells (Fig. 3C and data not shown). Nuclear extracts prepared from COS1 cells transfected with each construct demonstrated GAL4 DNA binding activity in vitro (data not shown). The effect of each mutation on the repression properties of this bipartite domain was determined in DNA-template based assays. As an initial strategy, we have targeted amino acid residues in the PHD finger of KAP-1 (C628R, V630S, C651F, P654C, and Q657E; Fig. 3A) which spatially parallel the position of naturally occurring mutations in hATRX that confer an inherited susceptibility to developmental defects (Gibbons et al. 1997). As illustrated in Figure 3D, the C628R and C651F mutations significantly impaired the repression activity of this domain, whereas the P654C mutation demonstrated an intermediate effect. Each of these three amino acids is highly conserved in the core consensus sequence of the PHD finger (Fig. 3A). The solution structure of the KAP-1 PHD finger indicates that these amino acids bind zinc in a cross-brace topology between anti-parallel β-strands reminiscent of RING domains (Capili et al. 2001). Both C628 and C651 are critical amino acids in the coordination of zinc that forms Site I. Chemical and physical characterization of the C651F mutation in the KAP-1 PHD finger results in an unstructured protein. The P654 residue is positioned two amino acids downstream from metal ligand-6, which completes site I zinc chelation. Introduction of an extra cysteine at this position may introduce a new metal ligand. Indeed, the chemical and physical properties of this mutant protein are consistent with an unstructured protein. The V630S and Q657E mutations had minimal effects on repression. This effect may not be entirely surprising, as these amino acids are less conserved and therefore represent residues that mediate context-specific interactions. In addition to these disease-related mutations, we have systematically analyzed the role of both conserved and non-conserved amino acid residues of the PHD finger (Fig. 3A) in KAP-1-mediated transcriptional repression (Capili et al. 2001). In general, the ability of the KAP-1 PHD domain to bind zinc and maintain the hydrophobic core are both required for its proper folding and ability to function in transcriptional repression.
Figure 3.

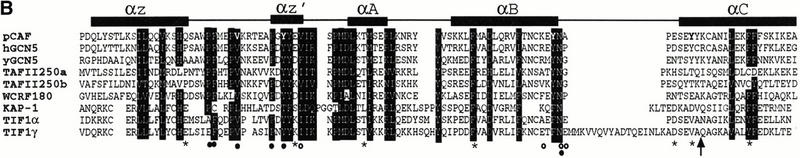

Mutations in the PHD finger and bromodomain significantly impair the intrinsic repression activity of this integrated transcriptional repression domain. (A) Amino acid sequence alignment of the KAP-1 PHD finger with related sequences from 15 independent proteins. The numbers in brackets indicate the corresponding amino acids in each protein. Strictly, conserved amino acids are shaded in black. Boxed amino acids are those in which the chemical nature of the side chain has been conserved. Asterisks indicate amino acids that were mutated to match naturally occurring mutations in the hATRX protein (Gibbons et al. 1997). Each mutation was made in the context of the GAL4–KAP-1 (619–835) expression construct. (B) Amino acid sequence alignment of the KAP-1 bromodomain with related sequences from TIF1α, TIF1γ, WCRF180, hGCN5, yGCN5, hTAFII250, and pCAF. Schematic illustration of the secondary structure and relevant position of each structural element in the bromodomain sequence are indicated above. Amino acid residues that have been conserved or where the chemical nature of the side chain has been maintained are shaded in black. Asterisks mark the amino acids that were mutated in this study. The arrow identifies the position of the 781trunc mutation. Each mutation was made in the context of the GAL4–KAP-1 (619–835) expression construct. Bolded amino acids in the pCAF sequence are those mutated in the study by Dhalliun et al. (1999). (Filled circles) Amino acids in the crystal structure that contact the acetyl moiety of acetylated histones (Owen et al. 2000). (Open circles) Amino acids in the crystal structure that contact the amide backbone of histone H4 (Owen et al. 2000). (C) Schematic diagram of the luciferase reporter and the heterologous GAL4-effector plasmid. Stable expression of each protein was determined via transfection into COS1 cells followed by immunoprecipitation of [35S]methionine-labeled whole cell extracts with anti-GAL4 (DBD) IgG (1 μg). (D) All experiments were done in NIH/3T3 cells, as described in Fig. 1. (Black bars) PHD finger mutations, (gray bars) bromodomain mutations.
We also used the information provided by the structure of the pCAF bromodomain to design six independent mutations in each of the four α-helices and one mutation in the variable loop region between helix Z and helix A of the KAP-1 bromodomain (Fig. 3B). As illustrated in Figure 3D, mutations in helix B (F761A) and helix C (DD 778,781AA, trunc 781, and F791E) significantly impaired transcriptional repression. The S730I mutation in the variable loop region between helix Z and helix A also displayed an intermediate effect on the repression potential of this domain, reaffirming the potential role for this region in macromolecular recognition (Fig. 3D). Overall, the data from this mutational analysis are consistent with the PHD finger and bromodomain of KAP-1 functioning cooperatively, as independent mutations in either domain can significantly disrupt the intrinsic repression potential of this bipartite domain. One interpretation of these data is that these two motifs provide an interface for protein–protein interactions with downstream effectors of transcriptional repression.
Identification of downstream effectors of KAP-1 PHD/bromodomain-mediated transcriptional repression
To identify potential mechanisms of transcriptional repression directed by the KAP-1 PHD finger and bromodomain, a heterologous fusion between the LEXA DNA binding domain and the PHD finger and bromodomain of KAP-1 was used as bait in a yeast two-hybrid screen. To confirm the specificity of any potential protein interactions identified in this screen, we utilized several of the amino acid substitutions engineered for the structure/function analysis of this bipartite domain. Appropriate expression of the heterologous fusion proteins was confirmed by Western blot analysis, and bait proteins displayed no ability to self-activate the selection promoters (data not shown).
From 40 million library transformants, four different gene products were recovered based on coactivation of both the integrated LEXA-responsive His3 and LacZ reporter genes. The nucleotide sequence of three independent transformants of one gene product indicated that each clone possessed an identical fusion junction to the GAL4 activation domain and that the sequence was identical to the 3′ nucleotide sequence of the dermatomyositis-specific autoantigen, Mi-2α/CHD3. Upon reintroduction into yeast, this GAL4 activation domain fusion protein displayed robust ability to activate the LacZ reporter with the wild-type LEXA–KAP-1 fusion protein (Fig. 4A), but failed to activate the reporter with other irrelevant test baits (data not shown). Two PHD finger mutants (V630S and G635A) that demonstrated nearly wild-type levels of transcriptional repression activity also displayed the ability to activate the LacZ reporter (Fig. 4A). In contrast, PHD finger mutants CC628, 631AA, C651F, and W664A, which possess impaired repression activity, either failed to activate these reporters or activated with reduced efficiency (Fig. 4A). This putative protein–protein interaction was also observed to be dependent on an intact bromodomain, as the 781 truncation mutation displayed reduced affinity for the Mi-2α in the two-hybrid assay (Fig. 4A). These observations are completely consistent with the transcriptional effects observed in transient transfection assays (Fig. 3D) in which the repression activity was dependent upon both domains. Furthermore, mutations in either the PHD finger or the bromodomain were independently sufficient to ablate the repression function of this bipartite domain. The combination of these results strengthens the argument that the PHD finger and bromodomain of KAP-1 function as an integrated functional unit which provides an interface for protein–protein interactions with molecules that facilitate repression by the KRAB–KAP-1 complex.
Figure 4.
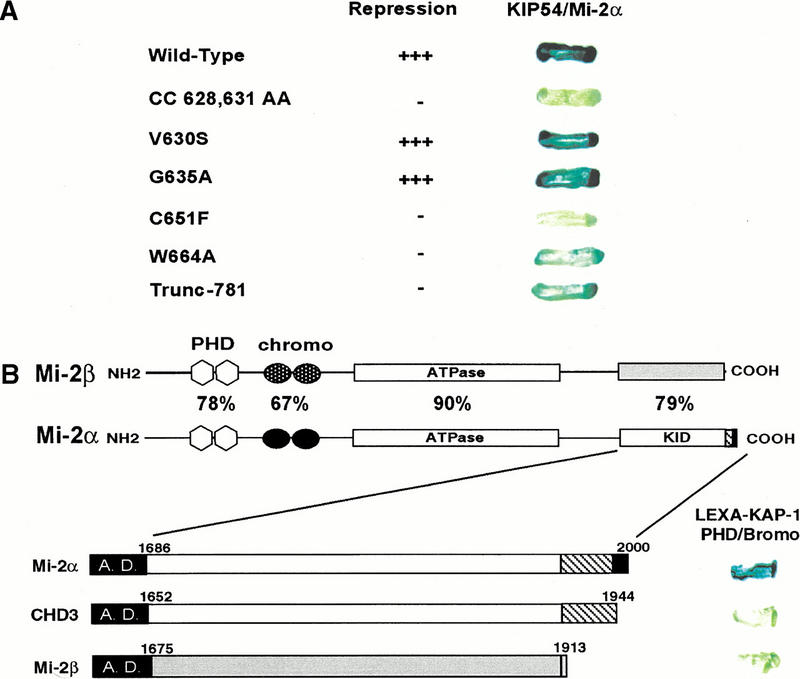

The PHD finger and bromodomain of KAP-1 specifically interact with the C-terminal amino acids of Mi-2α/CHD3. (A) The yeast strain L40 cotransformed with the indicated KAP-1 mutants as LEXA DNA binding domain fusions and the rescued Mi-2α–ct: GAL4 activation domain clone (KIP54). Positive interaction is inferred by blue yeast due to the activation of the integrated LEXA responsive LacZ reporter gene and subsequent hydrolysis of the synthetic substrate X-gal. (B) The interaction between KAP-1 and Mi-2 is specific for the C terminus of a unique isoform of Mi-2α. Schematic illustration of Mi-2α and Mi-2β. Mi-2α/CHD3 is a member of the CHD family of proteins which are characterized by the seven signature motifs of a “DEAH” box ATPase/Helicase. Other signature motifs include two PHD fingers (light gray hexagons), two chromodomains (black circles). Percentages represent percent identity between the two proteins at the indicated motifs. The putative KAP-1 interaction domain (KID) is indicated. In-frame fusion between the C-terminal sequences of hMi-2α (accession no. 3298562), hCHD3 (accession no. 2645433), hMi-2β/CHD4 (accession no. 1107696) was designed with the GAL4 activation domain. Numbers represent the corresponding amino acid in the respective sequences. L40 yeast cotransformed with the indicated GAL4 activation domain fusion proteins and the wild-type LEXA KAP-1 PHD finger and bromodomain illustrated the specificity of this interaction for Mi-2α. (C) Amino acid sequence alignment of the C-terminal residues schematically depicted in B. Identical residues indicated in black. Boxed amino acids are unique to Mi-2α.
Mi-2α/CHD3 is a member of the CHD family of proteins which were named based on the possession of several signature motifs, that is, chromodomains, ATPase/helicases, and DNA binding modules (Delmas et al. 1993; Woodage et al. 1997). Mi-2α/CHD3 is closely related to the Mi-2β/CHD4 protein. Comparison of the amino acids in the putative KAP-1 interaction domain of Mi-2α/CHD3 with Mi-2β/CHD4 revealed that these two proteins are 80% identical, yet all three KAP-1 interacting clones rescued in this screen possessed sequence specific to Mi-2α/CHD3. Based on the sequences of these clones, amino acids 1686 to 2000 harbor the minimal KAP-1 interaction domain (KID) (Fig. 4B). This sequence encodes a protein with an additional 30 amino acids at the C terminus of the protein that are not present in the putative protein described by Woodage et al. (1997) for CHD3 (accession no. AF006515), yet the remainders of the two sequences were identical (Fig. 4C). To test the functional significance of the novel 30 amino acids contained in our rescued clones versus the C-terminal sequence reported for CHD3, and the ability of Mi-2β/CHD4 to interact with KAP-1, we engineered fusions of these C-terminal sequences with the GAL4 activation domain, respectively (Fig. 4B). Upon introduction into yeast, the wild-type LEXA–PHD/bromodomain fusion of KAP-1 displayed no activation of the LacZ reporter for either protein, indicating that the C termini of these two proteins, CHD3 and Mi-2β/CHD4, are not functionally equivalent to the Mi-2α sequence we rescued in the two-hybrid screen (Fig. 4B). Closer inspection of the C-terminal amino acid residues revealed that Mi-2α/CHD3 quickly diverges from Mi-2β/CHD4 starting at amino acid 1909 and continues for an additional 91 novel amino acids (Fig. 4C). It is these amino acid residues that most likely confer the specificity of the Mi-2α/CHD3 interaction with KAP-1.
The Mi-2 family of proteins has been described as an integral component of a high molecular weight multiprotein complex containing histone deacetylase activity (Tong et al. 1998; Wade et al. 1998; Xue et al. 1998; Zhang et al. 1998a; Kim et al. 1999). We have generated polyclonal antibodies against a unique antigen in Mi-2α (amino acids 1515 to 1708, accession no. 3298562). Western blot analysis of conventionally purified NuRD from a 0.5 M phosphocellulose fractionated HeLa nuclear extract with these Mi-2α-specific antibodies demonstrated little immunoreactivity (Fig. 5A,B), consistent with previous observations that no peptides sequences specific for Mi-2α were identified in these purifications. Since all the peptide sequences reported for the NuRD/NRD complex were specific for Mi-2β, this observation suggested that Mi-2α might exist in an independent protein complex. Western blot analysis of phosphocellulose fractionated HeLa nuclear extract revealed that the majority of Mi-2α eluted in the 1.0 M fraction, while Mi-2β elutes evenly between the 0.5 M and 1.0 M fractions. KAP-1 is an abundant nuclear protein that demonstrated near equal elution in all phosphocellulose fractions (Fig. 5A).
Figure 5.
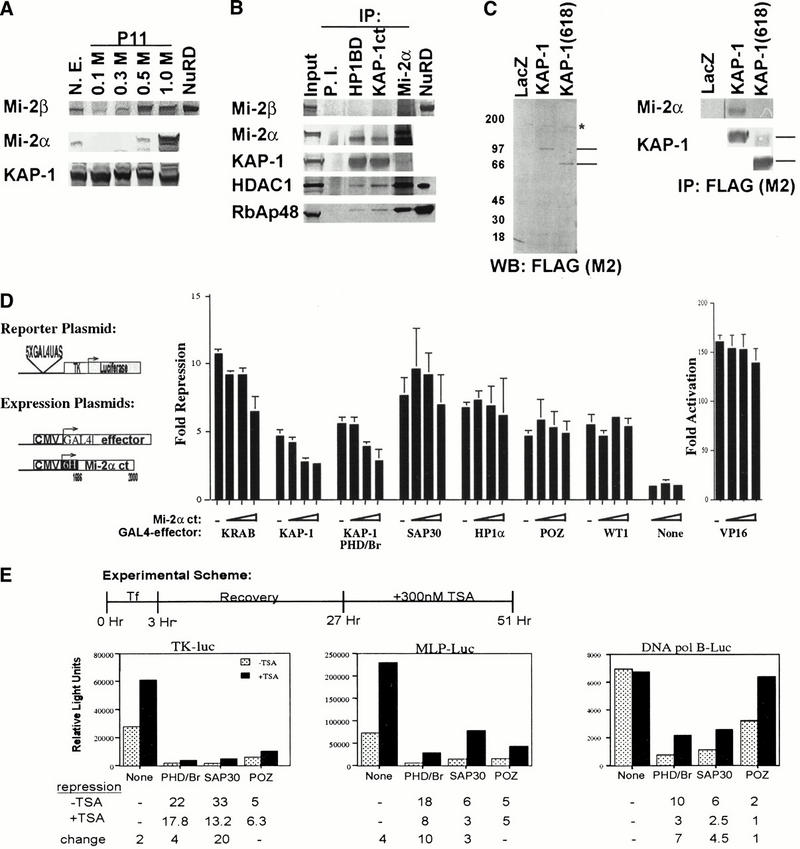
Role of Mi-2α and histone deacetylase in KAP-1 repression. (A) Western blot analysis of 10 μg of phosphocellulose (P11) fractionated HeLa nuclear extracts. NuRD, conventionally purified complex as described previously (Zhang et al. 1998a). (B) In vivo association between endogenous KAP-1 and Mi-2α. Three hundred μg of 1.0 M P11-DEAE-bound HeLa nuclear extract was immunoprecipitated by rabbit IgG (P.I.) (lane 2), anti-KAP-1 (HP1BD/423–589) antibodies (lane 3), or anti-KAP-1 (Ct/619–835) antibodies (lane 4), or anti-Mi2α antibodies (lane 5). Immunoprecipitated proteins were eluted by boiling in SDS sample buffer and then separated on a 4–12% NuPAGE gradient gel (Invitrogen), followed by immunoblot analysis with antibodies against Mi-2β, Mi-2α, KAP-1 (RBCC), HDAC1, and RbAp48. (Input) 10 μg of 1.0 M P11-DEAE bound extract, (NuRD) conventionally purified complex as described previously (Zhang et al. 1998a). (C) The in vivo association between KAP-1 and Mi-2α requires the PHD finger and bromodomain of KAP-1. Expression plasmids for LacZ, FLAG-KAP-1, or FLAG-KAP-1 (618) were transfected into 293 cells. Nuclear extracts (3–5 mg) were immunoprecipitated with anti-FLAG mAb M2 (lanes 1–3). The immune complexes were separated on a 4–12% NuPAGE gradient gel (Invitrogen) followed by immunoblot analysis with anti-Mi-2α pAb or anti-KAP-1 (RBCC/20–418) pAb. (Left panel) Anti-FLAG (M2) Western blot of transfected 293 cell nuclear extracts. Lines on right indicate the migration of full-length KAP-1 and KAP-1 (618), respectively. (Asterisk) An anti-FLAG cross-reacting species. (Right panel) The western blot analysis of anti-FLAG (M2) immunoprecipitates for Mi-2α and KAP-1 (RBCC). Lines on the right of the KAP-1 immunoblot represent the migrations of full-length KAP-1 and KAP-1 (618), respectively. (D) Schematic diagram of the luciferase reporter and the heterologous GAL4-effector plasmid. Expression of the KAP-1 interaction domain of Mi-2α (amino acids 1686 to 2000) dominantly inhibits heterologous KRAB- and KAP-1-mediated repression with minimal effects on heterologous repression observed for SAP30, HP1α, BCL6-POZ, WT1 or the activation potential of VP16. (Dashes) Repression observed in the absence of transfected Mi-2α dominant negative plasmid DNA. All experiments were done in NIH/3T3 cells with 0.1 μg of GAL4–VP16, 0.5 μg of GAL4–KRAB, 1 μg of each remaining GAL4-repressor, and 0.5 μg of the reporter plasmid. (Open triangles) Titrating amounts (1 μg, 2 μg, and 4 μg) of the dominant negative Mi-2α expression plasmid transfected. Expression of all proteins was confirmed in COS1 cells (data not shown). Fold repression was calculated as described in Fig. 1. (E) Addition of the histone deacetylase inhibitor, TSA, partially reverses the repression activity of the KAP-1 PHD finger/bromodomain. NIH/3T3 cells were transiently cotransfected with 0.5 μg of the indicated reporter plasmid and 5 μg of the heterologous GAL4 expression plasmids KAP-1 (619 to 835), SAP30, and BCL6-POZ. Twenty-four hours posttransfection, the cells were treated with 300 nM TSA (Wako) for an additional 24 h prior to harvesting. Fold repression was calculated as described in Fig. 1. Fold repression in the absence of TSA (stippled bars); fold repression in the presence of TSA (black bars).
To verify an interaction in vivo between KAP-1 and Mi-2α, we used coimmunoprecipitation experiments of endogenous proteins from the 1.0 M phosphocellulose fraction to maximize the chance to observe an interaction between these two proteins. Western blot analysis of Mi-2α immunoprecipitates revealed the retention of both HDAC1 and RbAp48, indicating that Mi-2α likely exists is some NuRD-like complex (Fig. 5B). Two independent antibodies against nonoverlapping antigens of KAP-1 specifically coprecipitated Mi-2α, HDAC1, and RbAp48 (Fig. 5B). None of these polypeptides was observed in immunoprecipitates with preimmune IgG, and no immunoreactivity for Mi-2β was observed in these immunoprecipitates, consistent with our observation in the two-hybrid screen. This result demonstrates an association between KAP-1 and Mi-2α in vivo. The significance of the association of KAP-1 exclusively with the faster migrating species in the Mi-2α immunoprecipitates is unclear at this time. Although this species is not abundant in the input, it does represent a major form of Mi-2α in the self-immunoprecipitate and may represent a degradation product. Furthermore, it is possible that this protein represents an alternatively spliced transcript of the Mi-2α gene, which has been reported previously. Whatever the nature of this species, it does not appear to disrupt the association of KAP-1 with Mi-2α, HDAC1 or RbAp48.
To determine the specificity of this interaction in vivo, we transiently transfected 293 cells with a Flag-tagged wild-type KAP-1 or a Flag-tagged delta PHD/bromodomain construct, KAP-1 (618). Anti-FLAG immunoprecipitates revealed the specific retention of endogenous Mi-2α with the wild-type, full-length KAP-1 protein, but not with the delta PHD/bromodomain KAP-1 or LacZ proteins (Fig. 5C). This result demonstrates that the association of KAP-1 with Mi-2α specifically requires sequences in the PHD finger and bromodomain.
In order to evaluate the role of Mi-2α in KAP-1-mediated transcriptional repression, we chose to disrupt endogenous KAP-1/Mi-2α interactions by expressing a dominant negative protein corresponding to amino acids 1686 to 2000 of Mi-2α (Fig. 5D). Expression of these amino acids dominantly inhibited heterologous KRAB-mediated repression of a DNA template in a dose-dependent manner (Fig. 5D). The same effect was observed for heterologous KAP-1-directed repression. Expression of the dominant negative protein had little effect on the basal activity of the minimal TK promoter, the activity obtained in combination with other non-KRAB related repression domain fusions, or compromised the activity of the VP16 activation domain (Fig. 5D). Because Mi-2α has been described as an integral component of a high-molecular-weight multiprotein complex containing histone deacetylase activity, we tested whether the addition of trichostatin A (TSA) could reverse the repression activity of the KAP-1 PHD finger and bromodomain. Figure 5E illustrates that the addition of 300 nM TSA partially reversed KAP-1 PHD/bromodomain-mediated repression of three independent promoters. These data provide evidence that Mi-2α and histone deacetylases may be one potential downstream effector of the KRAB-ZFP–KAP-1 repressor–corepressor complex in vivo.
Discussion
The recruitment of cofactors by DNA-bound transcription factors has evolved as a common, highly conserved mechanism of transcriptional regulation. Transcription factor-mediated recruitment permits gene-specific targeting of coactivator/corepressor complexes with intrinsic or associated chromatin modifying/remodeling complexes. We, and others, have previously identified a universal corepressor, KAP-1, which binds to the KRAB repression module (Friedman et al. 1996; Kim et al. 1996; Moosmann et al. 1996). Structurally, KAP-1 possesses consensus amino acid sequences for a RING finger, B-boxes, leucine zipper coiled-coil region, PHD finger, and bromodomain (Friedman et al. 1996; Kim et al. 1996; Le Douarin et al. 1996; Moosmann et al. 1996). Biochemical studies of KAP-1 indicate that the tripartite RBCC region functions as an integrated structural unit that is necessary and sufficient for KRAB recognition and KAP-1 oligomerization (Peng et al. 2000a,b). The basis of KAP-1 in transcription repression is not fully understood, but a direct interaction with members of the HP1 protein family suggests that KAP-1 may function to repress transcription via the organization of higher order chromatin structure (Le Douarin et al. 1996; Nielsen et al. 1999; Ryan et al. 1999; Lechner et al. 2000). In this report, we present data that builds upon our understanding of the function of KAP-1 in repression of transcription. We describe a novel transcriptional repression activity defined by the PHD finger and bromodomain of KAP-1. These domains are collectively required for optimal repression of DNA templates by KAP-1, and our studies suggest that these two motifs form a functionally integrated unit, as mutations or heterologous substitutions in either domain disrupt transcriptional repression. These data are consistent with this bipartite domain providing a specific interface for protein–protein interactions. In this regard, we show evidence that the PHD finger and bromodomain of KAP-1 are collectively required to mediate an interaction with the Mi-2α subunit of the histone deacetylase complex, NuRD, in vivo. This observation suggests that the KRAB–zinc finger protein superfamily functions as a sequence-specific targeting mechanism for the biochemical activities of the NuRD complex in vivo.
The recurrent theme that has emerged from studies aimed at understanding the function of transcriptional regulatory proteins is that highly conserved amino acid signature motifs often function as independent globular domains involved in macromolecular recognition. The identification of these signature amino acid sequence motifs and their spatial organization within a novel protein is often the first clue to its biochemical function. The PHD finger and bromodomain are two well-conserved signature motifs distributed widely among nuclear proteins with established roles in the formation, maintenance, or regulation of chromatin structure (Aasland et al. 1995; Jeanmougin et al. 1997; Winston and Allis 1999). Thus, the presence of these two motifs suggested that KAP-1 represses transcription via some chromatin-mediated mechanism. The conservation of the PHD finger and bromodomain architecturally in at least three independent families of proteins suggests that the two domains may biochemically function together (Le Douarin et al. 1995, 1996; Friedman et al. 1996; Venturini et al. 1999; Bochar et al. 2000; Jones et al. 2000a,b). In support of this hypothesis, we observed that optimal transcriptional repression by KAP-1 is dependent upon the integrity of this bipartite domain. This observation is similar to recent reports suggesting that the function of the bromodomain may require or be influenced by adjacent signature motifs (Cairns et al. 1999; Winston and Allis 1999). This intrinsic transcriptional repression activity could not be fully complemented by heterologous substitutions of either domain with related sequences. Thus, it appears from these results that the amino acid sequences of the KAP-1 PHD finger and bromodomain possess an inherent capacity to fold into an integrated functional unit, which provides a specific interface for protein–protein interactions with downstream effectors of transcriptional repression.
The PHD finger is a small domain of ∼60 amino acids characteristically defined by seven cysteines and a histidine that are spatially arranged in a consensus of C4HC3 of varying lengths and composition (Aasland et al. 1995). This particular arrangement of amino acids is highly homologous to the RING finger motif, an established interface for protein–protein interactions, which is created by chelating two zinc ions in a unique cross-braced fashion (Wu et al. 1996; Borden 1998, 2000; Jensen et al. 1998). Indeed, structural analysis of the KAP-1 PHD finger indicates that two atoms of zinc are bound in a cross-braced scheme between anti-parallel β-strands (Capili et al. 2001). Like the RING finger, this finding suggests that the PHD finger functions in protein–protein interactions. Our studies represent the first detailed site-directed mutagenesis approach to correlate the molecular anatomy of this domain with a biochemical function. In general, maintenance of the zinc chelation scheme and hydrophobic amino acids that stabilize the core were essential for the structural integrity of this domain and the repression activity of KAP-1 (Capili et al. 2001). This observation was expected, as mutations of analogous amino acids in the RING finger disrupt its structure and biochemical function. Although our results clearly emphasize that the PHD finger functions in conjunction with the bromodomain, we cannot preclude autonomous PHD finger functions. Recent studies have demonstrated that RING fingers act as E3 ubiquitin ligases through interactions with components of the ubiquitin conjugation machinery (Joazeiro and Weissman 2000). Because of the structural similarity to the RING finger motif, the PHD finger may also mediate similar functions with the ubiquitin or ubiquitin-like conjugation machinery. This hypothesis raises interesting possibilities that transcriptional repression by KAP-1 in part may result from ubiquitin mediated signaling (i.e., degradation). Alternatively, the potential for E3 ligase activity may in turn regulate the turnover or trafficking of KAP-1 itself. In any event, the experimental data from this panel of mutations will provide a basis for future studies aimed at defining new macromolecular interactions and functions for the PHD finger.
Similar to the PHD finger, the bromodomain is another conserved signature motif of ∼100 amino acids that is found in 30–40 independent proteins with that function in transcriptional regulation (Jeanmougin et al. 1997). Recent structural studies of three independent bromodomains indicate that these amino acids fold into a four-helical bundle with a unique left turn topology (Dhalluin et al. 1999; Jacobson et al. 2000; Owen et al. 2000). Crystals of either the TAFII250 double bromodomain or the yGCN5p with N-terminal histone H4 peptides revealed an extensive network of interactions between the two via a hydrophobic pocket formed by the ZA and BC loops of the domain (Jacobson et al. 2000; Owen et al. 2000). Moreover, synthetically derived mutations in the bromodomain of yGCN5p were observed to impair the chromatin-related functions of this protein in vitro and in vivo, without effecting the ability of GCN5 to be assembled into SAGA (Sterner et al. 1999; Syntichaki et al. 2000). These data clearly suggest that the bromodomain is involved in modulating the activity of these transcriptional regulatory complexes. Our data from the analysis of the KAP-1 bromodomain are consistent with these hypotheses. First, comparison of the primary sequence of the KAP-1 bromodomain with pCAF, yGCN5, and TAFII250 revealed conservation of amino acids involved in stabilization of the hydrophobic core (Fig. 3B). Likewise, the amino acids that comprise the recognition site for the amide backbone of the histone tail are well conserved, confirming the likelihood that histone tails are ligands of the KAP-1 bromodomain. However, the amino acids in the ZA loop that contact the carbonyl-amide moiety of acetyl-lysines are less conserved. This deviation may represent a slightly altered specificity for the type of histone modification recognized. Second, our experimental results clearly demonstrate that the bromodomain of KAP-1 has intrinsic transcriptional repression activity, which is required for optimal KAP-1 function on DNA templates. Although the F761A, F791E, and 781trunc mutations are expected to compromise the integrity of the tertiary structure of this domain, the S730I and DD778, 781AA mutations are positioned in regions that are involved in acetyl group or histone recognition, respectively. Therefore, it is reasonable to postulate that these mutations are affecting transcription without disrupting the tertiary structure of the domains. The combination of these data is consistent with the PHD finger and bromodomain functioning as a cooperative integrated unit that provides an interface for macromolecular interactions. Interestingly, the dependence of the KAP-1 interaction with Mi-2α for the PHD finger and bromodomain is similar to recent genetic evidence that implicates an essential role for the bromodomain of GCN5 in the recruitment of the SWI/SNF chromatin remodeling activities to a targeted genomic locus in yeast (Syntichaki et al. 2000).
Macromolecular complexes with histone deacetylase and ATPase chromatin remodelling activities have clearly been implicated in chromatin dynamics and transcriptional silencing. Both Mi-2α and Mi-2β have been described as integral components of a multiprotein complex containing both histone deacetylase and chromatin remodeling activities (Tong et al. 1998; Wade et al. 1998; Xue et al. 1998; Zhang et al. 1998a). The association of KAP-1 with Mi-2α builds upon our understanding of how the KRAB-ZFP–KAP-1 repressor–corepressor system mechanistically regulates gene expression. Moreover, this observation links the targeting of this multi-protein complex to specific loci via the KRAB-ZFP superfamily of repressors. We show that the KAP-1 interaction with NuRD is specifically mediated through a unique isoform of Mi-2α. Western blot analysis of conventionally purified NuRD with antibodies specific for Mi-2α demonstrated little immunoreactivity, a result consistent with no peptide sequences specific for Mi-2α identified in microsequencing from previous purifications of NuRD. However, we and others can demonstrate that Mi-2α immunoprecipitates retain HDAC1 immunoreactivity and enzymatic activity (Tong et al. 1998; Xue et al. 1998; Kim et al. 1999). These observations suggest that a Mi-2α NuRD-like complex exists in vivo that remains to be fully defined. Consistent with this hypothesis, several NuRD preparations demonstrate heterogeneity in relation to their peptide composition (Xue et al. 1998; Kim et al. 1999).
Our data suggest that the KAP-1 corepressor utilizes a Mi-2 complex and its associated biochemical activities to repress transcription. Biochemical studies indicate that a relatively small fraction of endogenous KAP-1 stably associates with Mi-2α in vivo, suggesting only a minor role for this interaction in KAP-1-mediated repression. From the data presented we cannot preclude other mechanisms that the PHD finger and bromodomain of KAP-1 use to repress transcription. In this regard we have characterized a direct interaction between the KAP-1 PHD finger and bromodomain and a novel protein containing a SET domain (D.C. Schultz and F.J. Rauscher III, unpubl.). Because significant evidence suggests that some feature of histones or nucleosomal structure serve as a ligand for the bromodomain, we speculate that the specific interaction between the PHD finger and bromodomain of KAP-1 with the Mi-2α subunit of the NuRD complex may be stabilized by some interface resulting from an interaction between the bromodomain of KAP-1 and nucleosomes. This hypothesis would be consistent with recent genetic evidence that indicates the bromodomain of GCN5 is required for the chromatin remodeling activities of the SWI/SNF complex at a genomic locus in yeast (Syntichaki et al. 2000). In this regard, both the His3 and the LacZ reporters used in the selection process for positive interacting clones in the two-hybrid assay were chromosomally integrated. Thus, it is reasonable to hypothesize that the stability of this interaction is disrupted during the extraction process. The role of Mi-2 associated histone deacetylase activity in KRAB–KAP-1 mediated repression will require the analysis of histone acetylation patterns within regulatory elements of single copy KRAB-ZFP target genes.
The interaction between KAP-1 and Mi-2α is one potential mechanism by which the biochemical activities of the NuRD complexes can be targeted in a sequence-specific manner to genes in vivo by virtue of the selectivity conferred by the C2H2 DNA binding domain of the KRAB-ZFPs. Consistent with this hypothesis, the lymphoid-specific, sequence-specific transcription factor Ikaros stably associates with the NuRD complex (Kim et al. 1999). Immuno-FISH experiments revealed that repressed genes by Ikaros were colocalized with components of NuRD and the HP1 protein in nuclear territories juxtaposed to centromeric heterochromatic during B-cell maturation (Brown et al. 1997). Similarly, we and others have reported a direct biochemical interaction between KAP-1 and mammalian homologs of the HP1 protein family, and have shown that KAP-1 and mammalian HP1 proteins can physically occupy the same spatial domains in interphase nuclei of NIH/3T3 cells (Ryan et al. 1999). The combination of these findings implies a potential link between multi-protein complexes that promote transcriptional repression by histone deacetylation and repression mediated by organization of high-order chromatin structure. In support of this hypothesis, SAP30, a novel member of the SIN3 histone deacetylase complex, has been observed to effect telomere silencing in yeast (Zhang et al. 1998b) and genetic screens for dominant suppressors of position-effect-variegation in Drosophila melanogaster have identified mutant alleles of HDAC1 (Mottus et al. 2000). Furthermore, genetic analyses in D. melanogaster demonstrate an epistatic relationship between dMi-2 and polycomb silencing of homeotic genes (Kehle et al. 1998). This apparent coupling of two independent repression mechanisms may lead to synergistic repression of their apparent target genes and promote epigenetic gene silencing.
The estimated 300 to 700 KRAB zinc finger proteins in the human proteome potentially make this superfamily of transcriptional repressors one of the master regulators of sequence-specific gene silencing in vertebrates. Transcriptional repression by a KRAB domain requires interaction with the corepressor, KAP-1. Thus, it appears that the effector molecules of KRAB-mediated transcriptional repression are likely due to a network of protein interactions with KAP-1. We suggest that the KAP-1 corepressor may function to repress transcription through several distinct mechanisms, including histone deacetylation and heterochromatinization. Unlike prokaryotic genes, eukaryotic genes exist in a restrictive state by virtue of nucleosome-based chromatin structure (Struhl 1999). In this model, the role of sequence-specific repressors like the superfamily of KRAB-ZFPs may be to create a silent gene environment completely resistant to activator and/or core transcription machinery binding. We propose that KRAB zinc finger proteins selectively bind promoter sequences and recruit the KAP-1 corepressor to the targeted locus. This complex in turn recruits the NuRD complex, which deacetylates histones in the promoter region, creating a favorable condition for HP1 proteins to nucleate a local heterochromatin environment that results in effective gene silencing. The abundance of KRAB domain proteins and the diverse capacity of the KAP-1 corepressor to repress transcription through multiple independent mechanisms emphasizes the potential of the KRAB–KAP-1 repressor–corepressor system to function as a master regulator of transcriptional gene silencing.
Materials and methods
Plasmid construction
The pM2–KAP-1 (293–835) mammalian expression vector has been described previously (Friedman et al. 1996). The GAL4–KAP-1 fusion proteins including 408–835, 408–618, 478–835, 478–680, and 478–618 were created by PCR using the primers 408for (BamHI) 5′-CCGGGATCCAGATTGTGGCAGAGC GTCCTG-3′, 478for (BamHI) 5′-CCGGGATCCAGGTGAGC GGCCTTATGCGC-3′, 618rev (HindIII) 5′-GATAAGCTTTC ACGGGCCACCACCTGGGGC-3′, 835rev (XbaI) 5′-GCTAT CTAGACTAAATGGTGGCACTGTCATCCAGG-3′, respectively. The resulting PCR products were digested with the restriction endonucleases indicated in brackets preceding the primer sequence and cloned into the corresponding restriction sites of the vector pM1. The GAL4–KAP-1 (619–835) expression plasmid was constructed by subcloning an XmaI fragment from pBL–KAP-1 into the XmaI site of pM1. The GAL4–KAP-1 (619–679) expression plasmid was created by PCR using the primers 619for (EcoRI) 5′-TAGCGAATTCGGAACCCTGGATGACAG TGC-3′ and 679rev (SalI) 5′-ATCGGTGGACATCCTCCTCC TTCAGGTCAG-3′. The resulting PCR product was digested with the restriction endonucleases indicated in brackets and cloned into the corresponding sites of the vector pM1. C-terminal to codon 679 of KAP-1 the protein reads VDASAEASR*. The GAL4–KAP-1 (674–835) expression plasmid was created by PCR, using the primers 674for (BamHI) 5′-ATCGGATCCGAC CTGAAGGAGGAGGATGGC-3′ and 835rev (HindIII) 5′-GAT CCCGGGAAGCTTTCAGGGGCCATCACCTGG-3′. The resulting PCR product was digested with the restriction endonucleases indicated in brackets and cloned into the corresponding sites of the vector pM2.
Site-directed point mutations in the KAP-1 PHD finger and bromodomain were engineered by standard overlap extension PCR-mediated mutagenesis procedures. The mutagenic primers for the described mutations contained the following codons: C628R, TGC to CGC; V630S, GTC to AGC; C651F, TGT to TTT; P654C, CCG to TGC; Q657E, CAG to GAG; E715K, GAA to AAA; S730I, TCC to ATC; L742T, CTG to ACG; F761A, TTT to GCT; D778, 781AA, GAC to GCT; F791E, TTC to GAG. The pM1-KAP-1 (619–835) trunc781 resulted from spontaneous deletion of G2444 from the nucleotide sequence during PCR of the DD 778,781 AA mutation and a reading frame shift that results in a protein with 15 novel amino acids, AKAACSPSSGCSA SSRRA* starting at codon 778 of KAP-1.
The GAL4 expression plasmids for the PHD finger and bromodomain of TIF1α (amino acids 785–1016), TIF1γ (amino acids 881–1120), and WCRF180 (amino acids 1141–1556) were created by PCR using the primers TIF1αfor (SmaI) 5′-GATCCCGGGAGGAAGGAGGATGACCCC-3′, TIF1αrev (SmaI) 5′-GTACCCGGGTTACTTAAGCAGCTGGCG-3′, TIF1γ for (SmaI)5′-GATCCCGGGAATAAAGATGATGACCC-3′, TIF1 γrev(SmaI) 5′-CTGCCCGGGTCATCTGACTTTAGGCGT-3′, WCRF180for (SmaI) 5′-GATCCCGGGATATGGTCTAAATC TATA-3′, and WCRF180rev (SmaI) 5′-CTGCCCGGGTCAGAT TCGTGACTTTTTTGC-3′, respectively. The resulting PCR products were digested with the restriction endonucleases indicated in brackets and cloned into the corresponding sites of either pM1 or pM3. The PHD finger and bromodomain chimeras were engineered by standard overlap extension PCR-mediated mutagenesis procedures. Forward primers were: ATRX 5′ (SmaI) 5′-GACCCGGGTAGCCGTGACTCAGATGG-3′, Mi-2α 5′ (SmaI) 5′-GATCCCGGGGATGGCTACGAGACGGATC-3′, and as indicated above for WCRF180 and KAP-1. Fusion primers were as follows: TIF1α/KAP-1for 5′-AAGCCAGAGGTTGAC TATGGCAGCTCAGCCTGGAT-3′, TIF1γ/KAP-1for 5′-AAG CCAGAAGTTGAATATGGCAGCCTCAGCCTGGAT-3′, AT RX/KAP-1for 5′-ACTGCATGTAACAGCGTAGGCAGCCTCA GCCTCAGC-3′, Mi-2α/KAP-1for 5′-GTCCAGTGGGAGG CCAAGGGCAGCCTCAGCCTCAGC-3′, WCRF180/KAP-1for 5′-CGTTCTAGAAGACTCTCCGGCAGCCTCAGCCTCAGC 3′, KAP-1/TIF1αfor 5′-GACCTGAAGGAGGAGGATGATTGT GATGTTCCCAGT-3′, KAP-1/TIF1γfor 5′-GACCTGAAGGA GGAGGATGATTGTGATAATTTGCAA-3′, KAP-1/WCRF180for 5′-GACCTGAAGGAGGAGGATAGATCTGTAAATATTGCT-3′, KAP-1/GCN5for 5′-GACCTGAAGGAGGAGGATACAGGC TGGAAGCCATTG-3′. Reverse primers were as follows: GCN5rev (SmaI) 5′-GATCCCGGGCTACTTGTCAATGAGGCC-3′, and as indicated above for KAP-1, TIF1α, TIF1γ, and WCRF180. Second-round PCR products were digested with restriction endonucleases as defined in the parenthesis preceding the primer sequence and cloned into the corresponding sites of either pM1 or pM3.
The LEXA–KAP-1 (amino acids 619–835) yeast expression plasmid was generated by the ligation of an XmaI fragment from pBL-KAP-1 into pBTM116. PHD finger and bromodomain mutants were subcloned from pM1 to pBTM116 as a SmaI fragment. GAL4 activation domain fusions with the C terminus of Mi-2α delta ct and Mi-2β were created by PCR using the following primers, respectively: Mi-2αfor (BamHI) 5′-GTAACGGA TCCAGGGCCTCGAGATGAGCCAC3′, Mi-2α delta ct (BamHI) 5′-GATCGGATCCTCACGTTGGTGGCGGCTGTGATGAAG-3′, Mi-2βfor (BamHI) 5′-GTAACGGATCCAAATGGAGAGAC CCCCAAGGACCTG-3′, Mi-2β rev (BamHI) 5′-GATCGGATC CTCACTGCTGCTGGGCTACCTGCTG-3′. Resultant PCR products were digested with BamHI and cloned into the corresponding restriction site of pACTII (Clontech).
The CMV–Mi-2α ct mammalian expression plasmid was created by subcloning a BamHI/BamHI DNA fragment encoding all amino acids of the rescued cDNA in-frame with an N-terminal 6His-tag possessing a consensus Kozak sequence and initiator methionine in pcDNA3.1 (Invitrogen). The GAL4–KRAB, GAL4–HP1α, and GAL4–WT1 expression plasmids have been described previously (Margolin et al. 1994; Lechner et al. 2000). The GAL4–SAP30 expression plasmid was kindly provided by Danny Reinberg (Howard Hughes Medical Institute, UMDNJ, NJ). The GAL4–BCL6–POZ expression plasmid was kindly provided by Vivian Bardwell (University of Minnesota, Minneapolis).
The FLAG–KAP-1 expression plasmid was constructed with a consensus Kozak sequence, an initiator methionine, and a FLAG-tag (DTKDDDDK) followed by amino acids 20–835 of KAP-1 in the BamHI/XbaI restriction sites of pcDNA3 (Invitrogen). Similarly, the FLAG–KAP-1 (618) expression plasmid was constructed encoding amino acids 20–618 of KAP-1 in the BamHI/XbaI restriction sites of pcDNA3 (Invitrogen).
Appropriate reading frame fusions and integrity of flanking sequences for all constructs created by PCR was confirmed by DNA sequence analysis of both strands.
Yeast two-hybrid system
The yeast two-hybrid system as modified by Stan Hollenberg was used for all yeast experiments. A human oligo-dT-primed B-cell cDNA library was screened as described previously (Jensen et al. 1998)
Transient transfection
Protein expression from all plasmids was confirmed by transient transfection of COS-1 cells followed by immunoprecipitation of [35S]methionine labeled cell extracts with anti-GAL4(DBD) IgG sc-577 (Santa Cruz), as described previously. Preparation of nuclear extracts from transfected COS1 cells was described previously. Electrophorectic mobility shift assays were done as described previously. All luciferase assays from transient transfections were done as described previously (Ryan et al. 1999).
Immunoprecipitations
Affinity-purified polyclonal antibodies raised against amino acids 20–418 (RBCC), 423–589 (HP1BD), and 618–835 (Ct) of KAP-1 and amino acids 1515 to 1708 of Mi-2α (accession no. 3298562) were a coupled to protein A-agarose (Repligen) by dimethylpimelimidate as described previously (Harlow and Lane 1988). Preparation of HeLa cell nuclear extract and phosphocellulose (P11) fractionation was done as described previously (Dignam et al. 1983). Three hundred micrograms of DEAE-bound 1.0 M P11 fractions was incubated with 50 μg of protein A-coupled antibody at 4°C for 4 h. Immune complexes were washed 3 times with BC500 (20 mM Tris-HCl at pH 8.0, 500 mM KCl, 0.2 mM EDTA, 10 % glycerol, 0.2 mM PMSF, 0.1% NP-40), once with BC100 (20 mM Tris-HCl at pH 8.0, 100 mM KCL, 0.2 mM EDTA, 10 % glycerol, 0.2 mM PMSF, 0.1% NP-40), and eluted with 0.1 M glycine (pH 2.7). Eluted proteins were resolved on a 4–12% NuPAGE gel in MOPS running buffer (Invitrogen), then transferred to PVDF, and antigens were detected as described previously (Ryan et al. 1999). Nuclear extracts from transiently transfected 293 cells were prepared as described previously (Ryan et al. 1999). Three to five milligrams of nuclear extract adjusted to 100 mM NaCl were incubated with 100 μg of anti-FLAG M2 (Sigma) at 4°C for 4 h. Bound immune complexes were washed, eluted and analyzed as described. Antibodies specific for Mi-2β (CHD4-N) were kindly provided by W. Wong (National Institute on Aging, Baltimore, MD). Antibodies against HDAC1 were kindly provided by D. Reinberg. RbAp48 was detected with mAb 13D10 (GeneTex).
Acknowledgments
D.C.S. was supported in part by the Wistar Basic Cancer Research Training Grant CA 09171 and DAMD 17-98-1-8269 grant. We acknowledge Robert F. Ryan for doing the original two-hybrid screen. We thank Trevor Woodage for Mi-2α cDNA sequences, David Picketts for the hATRX PHD finger sequence, and Shelley Berger for hGCN5 bromodomain sequences. F.J.R. is supported in part by National Institutes of Health grants CA 52009, Core grant CA 10815, DK 49210, GM 54220, DAMD 17-96-1-6141, ACS NP-954, the Irving A. Hansen Memorial Foundation, the Mary A. Rumsey Memorial Foundation, and the Pew Scholars Program in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Rauscher@wistar.upenn.edu; FAX (215) 898-3929.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.869501.
References
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Agata Y, Matsuda E, Shimizu A. Two novel Kruppel-associated box-containing zinc-finger proteins, KRAZ1 and KRAZ2, repress transcription through functional interaction with the corepressor KAP-1 (TIF1beta/KRIP-1) J Biol Chem. 1999;274:16412–16422. doi: 10.1074/jbc.274.23.16412. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Poncelet DA, Lecocq PJ, Revelant O, Martial JA. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, Perheentupa J, Ulmanen I, Peltonen L. Mutations in the AIRE gene: Effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy protein. Am J Hum Genet. 2000;66:378–392. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KLB. RING fingers and B-boxes: Zinc-binding protein–protein interaction domains. Biochem Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- Borden KLB. RING Domains: Master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Capili AD, Schultz DC, Rauscher FJ, III, Borden KLB. Solution structure of the PHD domain from the KAP-1 corepressor: Structural determinants for PHD, RING, and LIM zinc binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gibbons R J, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, Berry SA, Dahl N, Fryer A, Keppler K, et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: Mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Nezu J, Shimane M. A novel family of bromodomain genes. Genomics. 2000a;63:40–45. doi: 10.1006/geno.1999.6071. [DOI] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Shimane M. Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics. 2000b;63:35–39. doi: 10.1006/geno.1999.6070. [DOI] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kim SS, Chen YM, O'Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier JM, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ., III Molecular determinants for targeting HP1-mediated gene silencing: A direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ., III Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann P, Georgiev O, Thiesen HJ, Hagmann M, Schaffner W. Silencing of RNA polymerases II and III-dependent transcription by the KRAB protein domain of KOX1, a Kruppel-type zinc finger factor. Biol Chem. 1997;378:669–677. doi: 10.1515/bchm.1997.378.7.669. [DOI] [PubMed] [Google Scholar]
- Mottus R, Sobel RE, Grigliatti TA. Mutational analysis of a histone deacetylase in Drosophila melanogaster: Missense mutations suppress gene silencing associated with position effect variegation. Genetics. 2000;154:657–668. doi: 10.1093/genetics/154.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang J-C, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Begg GE, Harper SL, Friedman JR, Speicher DW, Rauscher FJ., III Biochemical analysis of the KRAB transcriptional repression domain: Spectral, kinetic and stoichiometric properties of the KRA–KAP-1 complex. J Biol Chem. 2000a;275:18000–18010. doi: 10.1074/jbc.M001499200. [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ., III Reconstitution of the KRAB–KAP-1 repressor complex: A model system for defining the molecular anatomy of RING-B box-coiled-coil domain- mediated protein–protein interactions. J Mol Biol. 2000b;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ., III KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: A potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Kadonaga JT. The “dark side” of chromatin remodeling: Repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken MH, Mattei MG, Ganser A, Chambon P, Losson R, de The H. TIF1γ, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Winston F, Allis CD. The bromodomain: A chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998a;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998b;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]


