Abstract
Stable high-hydrostatic-pressure (HHP)-resistant Listeria monocytogenes LO28 variants were previously isolated and characterized. These HHP variants were also more resistant to heat. In addition, nonlinear heat inactivation kinetics pointed toward the existence of heat-resistant variants, although these could not be isolated so far. In this study, we used kinetic modeling of inactivation curves of two isolated HHP variants and their wild type, and this revealed that the probability of finding resistant variants should depend on the nature of the inactivation treatment and the time of exposure. At specific heat and HHP conditions, resistant LO28 and EGDe variants were indeed isolated. Resistant LO28 variants were even isolated after a heat inactivation at 72°C in milk, and these variants showed high resistance to standard pasteurization conditions. The increased resistance of part of the isolated LO28 and EGDe variants was due to mutations in their ctsR genes. For the variants whose ctsR genes and upstream regions were not altered, the mechanisms leading to increased resistance remain to be elucidated. This research showed the strength of kinetic modeling in unraveling the causes of nonlinear inactivation and facilitating the isolation of heat-resistant L. monocytogenes variants.
INTRODUCTION
Listeria monocytogenes can cause listeriosis in animal and human populations. Human listeriosis is a rare but severe disease and is one of the leading causes of death among patients with food-borne diseases in the United States. The estimated annual rate of invasive listeriosis in the United States is 3 cases and for countries within the European Union is 2 to 10 cases per million people per year (33). Recently, several European countries experienced an apparent increase in the incidence of listeriosis (13).
A specific characteristic of L. monocytogenes that appears to be critical to its ability to cause human food-borne illness is its capacity to survive under harsh conditions. The occurrence of variants and the generation of population heterogeneity are factors that may contribute to the survival capacity of L. monocytogenes. Previous research showed that heterogeneity in L. monocytogenes populations (strains EGDe, LO28, and Scott A) affects resistance to high hydrostatic pressure (HHP). Inactivation of such heterogeneous populations resulted in survival curves with significant tailing, indicating the presence of an HHP-sensitive and an HHP-resistant fraction (32). Analysis of the cells that survived such HHP treatments revealed that the higher resistance of LO28 (32) and Scott A (17, 18) was a stable feature for part of the resistant fraction. Contrary to these results, no stable HHP-resistant isolates were obtained for EGDe (32). A significant fraction of the stably resistant variants of both Scott A and LO28 had an altered ctsR gene. This gene encodes CtsR, a DNA binding protein that regulates class III heat shock genes (7). The observed alterations in ctsR resulted not only in increased resistance to high pressure but also in increased survival to heat (17, 20, 31). Further characterization of the LO28 HHP-resistant variants without mutations in ctsR also revealed increased resistance to heat for most of these variants (31). In addition, thermal inactivation of L. monocytogenes was previously fitted with a biphasic model, indicating the presence of a heat-resistant fraction (1, 3, 5). However, so far these variants could not be isolated after heat inactivation.
For the present study, we used kinetic modeling of the inactivation of wild-type (WT) LO28 and two HHP-resistant variants as a strategy to determine the probability of detecting resistant variants. With this information, the appropriate conditions to isolate HHP- and heat-resistant variants of LO28 were established. Similar conditions were used to examine the existence of HHP- and heat-resistant variants of strain EGDe. Isolated variants of both strains were checked for mutations in the stress regulator CtsR. Finally, the significance of the occurrence of stress-resistant variants of L. monocytogenes is discussed.
MATERIALS AND METHODS
Bacterial strains and cell culturing conditions.
Listeria monocytogenes LO28 wild type and two stress-resistant variants (ctsR variant number 6 and immotile variant number 17) (32) and EGDe wild type (Laboratory of Food Microbiology, Wageningen University and Research Center, Netherlands) were used in this study. Stock cultures of all strains were kept in 15% (vol/vol) glycerol (Fluka, Buchs, Switzerland) at −80°C, and before the experiments, cells from stock were grown for 2 days at 30°C on brain heart infusion (BHI) agar (Oxoid, Hampshire, England). A single colony was used to start a preculture of 10 ml BHI broth. After 20 h of growth at 30°C in an incubator (refrigerated incubator shaker Innova 4335; New Brunswick Scientific, Edison, NJ) with shaking at 160 rpm, 0.5% (vol/vol) inoculum was added to 100 ml of BHI broth. Cells grown in BHI at 30°C from exponential growth phase (5 h) were washed twice with 50 mM N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) buffer (pH 7.0; Sigma-Aldrich, Steinheim, Germany) and resuspended in this buffer until a final concentration of approximately 109 CFU/ml was obtained. The resulting cultures were used for inactivation experiments, and each experiment was conducted at least two times on different days.
High-hydrostatic-pressure inactivation.
HHP inactivation was performed as described previously by Van Boeijen et al. (32). Cell suspensions were subjected to 350 MPa at 20°C in ACES. Before and after an HHP treatment, samples were taken and serially diluted in 0.1% peptone saline. Samples of 50 to 200 μl were plated on BHI agar using a spiral plater (Eddy Jet; LabScientific, NJ). The plates were incubated for 5 days at 30°C to allow all surviving cells to recover and form visible colonies. Survivors were enumerated, and this was considered accurate if more than 20 cells were detected. This criterion corresponds to a 2 log CFU/ml limit of quantification. The HHP inactivation data were fitted with the biphasic linear and the linear model using GInaFiT (11).
Heat inactivation.
For the heat treatment, cell suspensions of 150 μl were placed in sterile glass micropipettes (200 μl, 2-mm inner-diameter, 140-mm-length Blaubrand; Brand GmbH, Wertheim, Germany). The pipettes, with the sample in the center of the pipette, were closed by melting the tips and placed in a water bath (Thermomix ME 4P; B. Braun, Melsungen, Germany) that covered them totally. The samples were treated at 55°C and then serially diluted and plated, and colonies were enumerated. The heat inactivation data were fitted with the biphasic linear model with shoulder and the linear model with shoulder using GInaFiT (11) and the TableCurve 2D software package (version 2.03; Jandel Scientific, San Rafael, CA).
Estimation of the probability of detecting resistant variants based on kinetic modeling.
Kinetic modeling can be used to estimate the probability of detecting resistant variants. For this, the fraction of resistant variants in the population and their inactivation kinetics have to be determined. The two most prominent groups of variants found in the LO28 population comprise immotile and ctsR variants. Their specific fraction in the population (f0,var) was determined from the HHP inactivation data by dividing the number of specific variants before the HHP treatment (N0,var) (CFU/ml) by the total number of measured cells in the population before the treatment (N0,measured) (CFU/ml), as follows:
| (1) |
HHP inactivation of variants was linear. Therefore, the initial number of variants was calculated from the fraction of variants that survived HHP and by extrapolation of this fraction's survival curve, as follows:
| (2) |
where Nt,var equals the number of variants (log CFU/ml) at time t (min) and Dvar the fitted decimal reduction time or D-value (min) of the HHP-inactivated variants (31, 32).
The inactivation data were fitted with an appropriate model. For the wild type, the biphasic linear model (with shoulder) was used, and for both variants, the linear model (with shoulder) was used. The proportions of the variants in relation to the total population were calculated with equations 1 and 2 and the specific inactivation model, assuming that the fractions of these two variants for heat experiments at t = 0 were equal to the HHP-inactivated fraction. This proportion of variants is then equal to the probability of isolating variants at a specific time point.
Selection of resistant variants by stress challenge cycles.
Previously, survivors were randomly selected and individually cultured before they were subsequently assessed for stable piezotolerant phenotypes (32). Those isolates were individually subcultured five times during five consecutive days using 0.5% (vol/vol) inocula in fresh BHI medium (equivalent to approximately 40 generations) and on day 5 retested for resistance to HHP. In the present study, variants were searched by an optimized series of repetitive stress challenge cycles as described previously for the isolation of resistant variants of Escherichia coli and Staphylococcus aureus (15, 21). To increase the probability of isolating resistant variants, three challenge treatments were used, and this was shown to be effective. In the stress challenge cycle used for the experiments described herein, L. monocytogenes survivors of optimized HHP or heat experiments (ranging from 100 to 10,000 CFU) were harvested from the plates after 5 days of incubation at 30°C. Fresh BHI medium (approximately 5 ml) was added on top of the plate with the surviving colonies, and visible colonies were scraped from the plate with a spatula and resuspended in the added BHI. The cell suspension was added to 5 ml fresh BHI medium by using a pipette and grown for 20 h at 30°C. A 100-ml culture was inoculated with this preculture (0.5% [vol/vol], 5 h at 30°C) and used for a subsequent inactivation experiment. From this second inactivation, survivors were again harvested from the plate, cultured twice, and inactivated. Twenty-four isolates from the third inactivation experiment were obtained and analyzed for their stress resistance and their ctsR genes and upstream regions. This method makes it possible to examine the resistance of a large number of survivors in a relatively easy way. The previous method made it possible to examine the stable resistance of single survivors independently of their growth characteristics, whereas the current method allows the examination of the surviving fraction of the population, consisting of a large number of survivors that will compete in both their growth and resistance.
Statistical analysis.
For the wild type and both variants, the selected model for their heat or HHP inactivation was fitted to the independent reproductions individually, and the average decimal reduction times and shoulder lengths were calculated. Student's t test for two samples assuming equal variances was used (the limit of significance was set at a P value of 0.05) to determine statistical differences between decimal reduction times and shoulder lengths of the wild type and the variants and between the results for resistance of the wild type and the survivors.
Amplification and sequence analysis of the ctsR gene.
Amplification of ctsR was performed as described previously by Van Boeijen et al. (32). The sequences of the primers used were 5′-GCAGGGATAAACGCTGAAAG-3′ for the forward and 5′-ACACTCCGGACATCCAACTC-3′ for the reverse primer. The PCR products (1.2 kb) were isolated by QIAquick gel extraction (Qiagen, Venlo, Netherlands) and sent for sequence analysis (Base Clear B.V., Leiden, Netherlands).
RESULTS
HHP and heat inactivation of L. monocytogenes LO28 wild type and resistant variants.
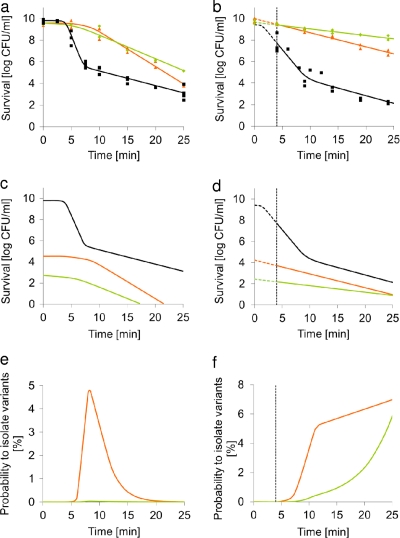
In previous research, stable HHP-resistant L. monocytogenes LO28 variants were isolated (32). Most of these variants also appeared to be heat resistant (31). Furthermore, biphasic inactivation was found not only for HHP but also for heat inactivation of WT LO28. This raised the question of whether stably resistant variants could also be isolated after a heat treatment. We selected a temperature that caused an inactivation profile similar to that found for the HHP inactivation (i.e., biphasic inactivation with a similar D-value for the resistant fraction) (Fig. 1 a and b and Table 1). One hundred two survivors were selected after 25 min of treatment at 55°C. None of these isolates was stably resistant after culturing and retesting under the same conditions (data not shown). The probability of isolating variants in the tail depends on the characteristics of the survivors. Cells can be temporarily or stably resistant as a result of physiological or genetic changes. For example, after HHP inactivation of L. monocytogenes LO28, only 25% of the survivors were stably resistant, whereas the remaining survivors were temporarily resistant (32). Consequently, some of the isolated variants will not be resistant in a subsequent inactivation treatment. Hence, we tried another approach, using the fraction of stably resistant variants in a population combined with their inactivation kinetics to estimate the probability of detecting these resistant variants during the inactivation. With this information, the best possible conditions can be established for the isolation of resistant variants. We selected representatives of the previously isolated stably resistant HHP variants that showed increased resistance to heat (31). Variant number 6 (a ctsR variant) and variant number 17 (an immotile variant) were investigated for their heat and HHP survival compared to the survival of the wild type under these conditions (Fig. 1a and b). The linear model with shoulder was used to fit their heat inactivation data, and the linear model was used to fit their HHP inactivation data (Table 1). For HHP, the application of the linear models starts at the time at which the temperature of the HHP vessel has returned to 20°C. This was the case 4 min after starting the pressurization.
Fig. 1.
(a and b) Inactivation kinetics of Listeria monocytogenes LO28 WT (black), ctsR variant number 6 (orange), and immotile variant number 17 (green) after heat treatment of exponentially growing cells at 55°C in ACES buffer (a) or HHP treatment of exponentially growing cells at 350 MPa, 20°C in ACES buffer (b). The observed values are shown as dots, and the lines represent the model-based fit. The vertical line at 4 min represents the time at which the temperature of the HHP vessel has returned to 20°C after starting the pressurization. Therefore, application of the model starts at 4 min for HHP. At t = 0 min, the measured amount of wild-type cells (N0,measured) is 9.4 log CFU/ml, whereas the extrapolated amount of wild-type cells (N0,extrapolated) is 10.3 log CFU/ml. (c and d) Predicted population inactivation of Listeria monocytogenes LO28 wild type, ctsR variant number 6, and immotile variant number 17 after heat treatment of exponentially growing cells at 55°C in ACES buffer (c) and HHP treatment of exponentially growing cells at 350 MPa, 20°C in ACES buffer (d). The number of cells at t0 of both variants is based on their estimated frequency of occurrence in the initial population. (e and f) Probability (%) of isolating resistant ctsR or immotile variants in a population of Listeria monocytogenes LO28 after heat treatment of exponentially growing cells at 55°C in ACES buffer (e) and HHP treatment of exponentially growing cells at 350 MPa, 20°C in ACES buffer (f).
Table 1.
The parameters of the fitted models for the survival curves of Listeria monocytogenes LO28 wild type and two stress-resistant variantsa
| Parameter | HHP ([biphasic] linear model) |
Heat ([biphasic] linear with shoulder model) |
||||
|---|---|---|---|---|---|---|
| Wild type | ctsR variant | Immotile variant | Wild type | ctsR variant | Immotile variant | |
| N0,measured (log CFU/ml) | 9.4 | 9.7 | ||||
| N0,extrapolated (log CFU/ml) | 10.3 | 10.0 | 9.6 | 9.7 | 9.6 | 9.6 |
| D1 (min) | 1.5 | 7.7* | 16.3** | 0.86 | 3.1** | 4.5** |
| D2 (min) | 7.1 | 7.6 | ||||
| fres | 2.1 × 10−5 | 6.5 × 10−6 | 1.0 × 10−7 | 1.6 × 10−4 | 6.5 × 10−6 | 1.0 × 10−7 |
| Sl (min) | 3.6 | 7.8*** | 5.2 | |||
Cells were treated under conditions of 350 MPa, 20°C (HHP) or at 55°C (Heat) in ACES buffer. N0,extrapolated is the model-based-fit amount of cells at t0. N0,measured is the measured amount of cells at t0. D is the decimal reduction time, where D1 and D2 are the D-values of the first and second part, respectively, of the biphasic linear inactivation curve. fres is the fraction of resistant cells in the population based on N0,measured for the wild type, where the fres given for the wild type is the total fraction of resistant cells (both stably and temporarily resistant) and the fres given for each of the two variants is the fraction of that specific variant in the wild-type population, assuming no shoulder for the two variants (see behavior in Fig. 1b) and taking into account the shoulder in the wild type by using N0,measured. Sl is the shoulder length. *, statistical analysis (P < 0.05) showed that the ctsR variant's D-value for HHP was only higher than the D-value of the sensitive fraction of the population of the wild type; **, the D-value for HHP of the immotile variant and both variants' D-values for heat were significantly different from the D-values of the sensitive and resistant fractions of the wild-type population; ***, the shoulder length of the ctsR variant was higher than that of the wild type for heat inactivation.
Estimation of the probability of detecting resistant variants based on kinetic modeling.
HHP treatment at 350 MPa of L. monocytogenes LO28 inactivated a population of 2.6 × 109 CFU/ml to 184 CFU/ml in 24 min (32) (Fig. 1a). Characterization of these cells revealed 4.9% immotile and 6.9% stably resistant ctsR variants (31, 32). Based on these surviving cell counts (9 immotile and 13 ctsR variants per ml present after 24 min at 350 MPa), together with the D-values of the inactivation of both variants (Fig. 1a and Table 1) and N0,measured, their individual fractions in the initial wild-type population can be calculated (equations 1 and 2). In a wild-type population of LO28, approximately 1 immotile and 65 ctsR variants are present per 10 million cells (Fig. 1c and d and Table 1). By combining these estimated fractions with the inactivation kinetics during a heat or HHP treatment, the probability of detecting each of these variants was calculated (Table 1 and Fig. 1e and f). The calculations implied that the probability of isolating a ctsR or immotile variant after 25 min at 350 MPa is 7 or 5%, respectively. Using the same fractions of variants at t = 0 derived from the HHP experiments, the predicted curve for heat inactivation implied that after 25 min at 55°C, the probability of isolating one of these variants was close to zero. This fits well with our observation that stable heat-resistant variants were not isolated after treating 102 isolates for 25 min at 55°C.
The data in Fig. 1c and d illustrate the differences in inactivation by the heat and HHP treatments of the variants compared to the results for the wild type. When exposed to HHP, the ctsR variant had a higher D-value than the sensitive fraction of the wild type, whereas the immotile variant had a higher D-value than both the sensitive and the resistant fractions of the wild-type population. On the other hand, for heat inactivation, only the ctsR variant had a longer shoulder length than the WT. The variants' D-values were significantly higher than that of the sensitive but lower than that of the resistant fraction of the wild-type population. The differences in heat inactivation between the wild type and the variants indicated that the probability of isolating one of the variants from the resistant fraction should be highest after approximately 8 min (5% for ctsR and 0.03% for immotile variants). At this time point, part of the wild-type population would be inactivated, in contrast to only small parts of the variants' populations. Therefore, we performed further research for the isolation of heat-resistant variants at 8 min at 55°C.
Isolation of HHP- and heat-resistant L. monocytogenes variants.
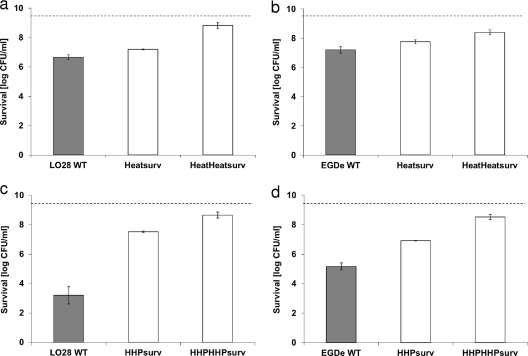
To increase the chances of isolating resistant variants, the population surviving a treatment was regrown and exposed to heat. Due to the inactivation of sensitive variants, the fraction of resistant variants in this surviving population was increased from 10−6 to 10−5 to more than 5 × 10−2. With another heat-challenge cycle, the surviving population would contain mostly heat-resistant variants. LO28 survivors of an 8-min treatment at 55°C were harvested, cultured, and retested in a heat challenge cycle. As a control, the LO28 survivors of a 24-min treatment at 350 MPa were harvested, cultured, and retested in an HHP challenge cycle as well. For both HHP and heat, these stress challenge cycles selected for resistant variants (Fig. 2 a and c). The previous isolation of HHP-resistant LO28 variants was confirmed using the stress challenge cycle method. Furthermore, this method also made it possible to isolate for the first time heat-resistant variants of LO28 and HHP- and heat-resistant EGDe variants. Because of this specific selection method, we were able to detect a small fraction of cells with resistant characteristics in a large population (Fig. 2b and d).
Fig. 2.
(a and b) Survival of exponentially growing Listeria monocytogenes cells in ACES buffer after 8 min at 55°C for LO28 (a) and EGDe (b) wild type, their recultured survivors of the heat treatment (Heatsurv), and their recultured survivors of a similar subsequent heat treatment (HeatHeatsurv). (c and d) Survival of exponentially growing L. monocytogenes cells in ACES buffer after 24 min at 350 MPa for LO28 (c) and EGDe (d) wild type, their recultured survivors of the HHP treatment (HHPsurv), and their recultured survivors of a similar subsequent HHP treatment (HHPHHPsurv). The dotted lines indicate the initial level of cells (9.5 log CFU/ml). Each inactivation experiment was conducted at least two times on different days. The error bars show one standard deviation. Results in white bars are statistically different from the results for the wild type.
Alterations in ctsR.
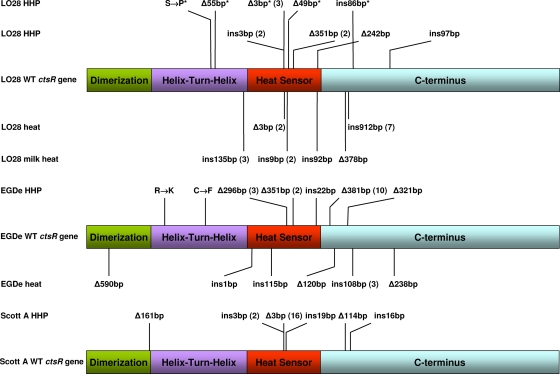
Previous research showed that a significant fraction of HHP-resistant variants had mutations in their ctsR genes. These ctsR variants comprised over 60% of Scott A HHP-resistant variants and almost 30% of LO28 HHP-resistant variants (19, 31). The newly isolated LO28 and EGDe HHP and heat variants were also tested for alterations in their ctsR genes and the related upstream regions. In all cases tested (for both strains under both inactivation conditions), ctsR mutations were found (Fig. 3). For LO28, 25% of the HHP isolates and 38% of the heat isolates were ctsR variants. The fraction of 25% HHP ctsR variants is comparable to the fraction of 29% that we found in our previous research (31). For EGDe, 79% of the HHP isolates and 33% of the heat isolates were ctsR variants. In total, 23 different mutations in ctsR genes were observed: 2 nonsynonymous single nucleotide polymorphisms, 10 inserts (ranging in size from 1 to 912 bp), and 11 deletions (ranging in size from 3 to 590 bp). All inserted DNA appeared to be sequence repeats of ctsR at the point of insertion, except for one insert that was a transposase in the C terminus of the ctsR gene. This mobile segment of DNA can insert into nonhomologous target sites, and in this case, it disrupted the ctsR gene, resulting in a truncated protein. Almost half of the mutations were found in the heat sensor domain, where a typical well-conserved glycine repeat (GGGG) is located (8, 20). One of these mutations is the CtsRΔGly found for LO28 under heat stress and previously isolated for LO28 and Scott A under HHP stress (20, 31). Karatzas et al. indicated in their research that the deletion of the glycine residue in the glycine repeat of CtsR resulted in a loss of the repressor function of this regulator (20). This was indicated by increased expression of the CtsRΔGly protein in the mutant concomitantly with increased expression of the clpP gene and the clpC operon and with increased expression of ClpC and ClpP proteins.
Fig. 3.
Observed variants of the ctsR genes of Listeria monocytogenes LO28, EGDe, and Scott A. The four putative functional domains of CtsR comprise an N-terminal dimerization domain (green), followed directly by a helix-turn-helix DNA-binding domain (purple), a central putative heat sensor domain (red), and a supposed C-terminal stabilization domain (7, 8). The glycine repeat is located in the center of the heat sensor. The position and number of base pairs inserted (ins) or deleted (Δ) are shown for all variants and inactivation conditions. ctsR variants were isolated after an HHP treatment of 24 min at 350 MPa (HHP), a heat treatment of 8 min at 55°C in ACES buffer (heat), or a heat treatment in UHT-processed whole milk of 6 s at 72°C (milk heat). Previously isolated LO28 ctsR variants are marked with an asterisk (31). Scott A ctsR variants were previously isolated by Joerger et al. (17) (Δ161bp) and Karatzas et al. (19) (ins3bp, Δ3bp, ins19bp, Δ114bp, and ins16bp). Some ctsR variants were isolated more than once under the same inactivation conditions, which is indicated in parentheses after the specific mutation.
The observed mutations for the LO28 and EGDe strains in the current study represented three major effects at the protein level: (i) in-frame, effecting a change in the glycine repeat; (ii) in-frame, effecting a change in the relative position of the C-terminal domain; and (iii) out-of-frame, effecting a truncation and loss of the C-terminal domain. It is conceivable that the observed changes will affect the function of CtsR and/or the stability of the protein, for example, by an altered interaction of the C terminus (i and iii) or by modulation and/or loss of temperature sensing (ii). Such a loss of CtsR's repression function would result in induction of its regulon, as previously observed for L. monocytogenes Scott A AK01 (20), with concomitant activation of stress defense, resulting in increased stress resistance, as observed in this study.
L. monocytogenes heat-resistant variants in milk.
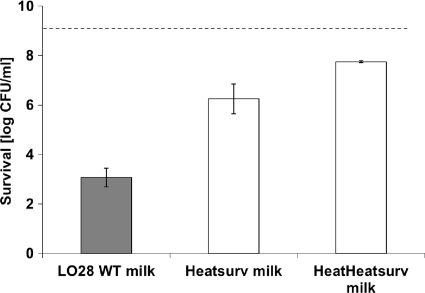
The presence of LO28 heat-resistant variants was also tested under industrially relevant conditions; i.e., by growth in ultra-high-temperature (UHT)-processed whole milk followed by the stress challenge cycle method at 72°C. Survivors of a 6-s treatment at 72°C were harvested, recultured in whole milk, and retested for their heat resistance and survival. The survivors of this second heat treatment were subsequently harvested, recultured in whole milk, and retested for their heat resistance under the same conditions (Fig. 4). From the survivors of the latter heat treatment, 10 isolates were randomly selected and tested for alterations in their ctsR genes. For 7 of the 10 variants, 4 different ctsR mutations were found (Fig. 3). For the other three variants, the ctsR genes and upstream regions were not altered. Analysis of the cultures of all three non-ctsR variants showed differences in colony sizes when plated on BHI agar. Only small colonies were found after the heat treatment, whereas both small and large colonies were found before the heat treatment. These small-colony variants appeared to be heat resistant, whereas the large colonies showed sensitivity to the heat treatment that was similar to that of the WT.
Fig. 4.
Survival (log CFU/ml) of exponentially growing cells in UHT-processed whole milk after 6 s at 72°C for Listeria monocytogenes LO28 wild-type cells, their recultured survivors of the heat treatment (Heatsurv), and their recultured survivors of a similar subsequent heat treatment (HeatHeatsurv). The dotted line indicates the initial level of cells (9 log CFU/ml). Each inactivation experiment was conducted at least two times on different days. The error bars show one standard deviation. Results in white bars are statistically different from the results for the wild type.
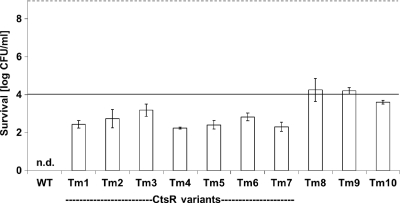
All 10 heat-resistant variants were also tested for their survival in whole milk for 15 s at 72°C, as advised by the FDA for pasteurization of whole milk (Fig. 5). The calculated D-value of WT LO28 based on linear inactivation at this temperature was 1.01 ± 0.07 s (mean ± standard deviation), whereas the mean D-value of the ctsR variants was significantly different from that of the wild type and more than 2 times higher, 2.35 ± 0.13 s. Furthermore, the heat-resistant variants without mutations in their ctsR genes had a mean D-value of 3.02 ± 0.21 s, which was three times higher even than that of the wild type. By using the heat treatment of 15 s at 72°C, the wild type and ctsR variants will have more than 5 log reductions. In contrast, the three non-ctsR variants did show reductions close to or even less than 5 log.
Fig. 5.
Survival (log CFU/ml) of exponentially growing cells in UHT-processed whole milk after 15 s at 72°C for Listeria monocytogenes LO28 wild type and 10 isolated heat-resistant variants. For LO28 WT, no cells could be detected after the heat treatment (n.d.). The first 7 heat-resistant variants are the ctsR variants. The dotted line indicates the initial level of cells (9 log CFU/ml). The solid line shows the standard 5 log reduction after 15 s at 72°C as used in the industry and advised by the FDA. Each inactivation experiment was conducted at least two times on different days. The error bars show one standard deviation. Results in white bars are statistically different from the results for the wild type.
DISCUSSION
The occurrence of stress-resistant variants in monoclonal populations of bacteria remains an important subject of debate, as some research groups have reported the isolation of such variants while others failed to demonstrate their presence and questioned their existence (1, 3, 5, 16, 17, 19, 24, 29, 31). In this study, we used a new strategy involving a kinetic modeling-based sampling scheme and an optimized stress challenge cycle method for the isolation of stress-resistant variants of L. monocytogenes. While our previous strategy led to the isolation of HHP-resistant variants for LO28 only (32), the new strategy allowed us to isolate such variants for EGDe also. In addition, the new strategy revealed the presence of heat-resistant variants within both L. monocytogenes LO28 and EGDe populations. By defining specific conditions, resistant variants could be isolated, although they represent only a small fraction (∼10−7 to 10−5) of the population. The isolation of both HHP- and heat-resistant variants of different L. monocytogenes strains illustrates that three factors play an important role in detecting these variants: first, the growth and resistance of the variants compared to their wild type; second, the frequency of revertance of variants to wild type; and finally, the chosen test conditions.
Most models of evolutionary processes imply that new variants establish in populations through mutation and selection. For each new generation, the bacterial DNA is replicated with a spontaneous mutation rate of about 0.0033 per genome. This number, however, is highly variable and depends largely on the circumstances (9). Variants with mutations that confer a competitive advantage in particular environments are selected (9). Taking into account the growth and resistance characteristics of ctsR variants, the sizes of the ctsR gene and the total genome of L. monocytogenes, and the mutation frequency as mentioned above, the chance of finding a mutation in the ctsR gene after the first inactivation step (∼13 replications) is around 7 × 10−6. This value is very close to the estimated fraction of approximately 6.5 × 10−6. By using the optimized stress challenge cycle method, the chances of isolating ctsR variants will increase with each cycle based on their growth and stress survival (31). In line with previous observations, variants with mutations in ctsR are the most frequently isolated stress-resistant variants, and in the current study, 49 of 106 tested L. monocytogenes LO28 and EGDe variants under HHP- and heat-inactivation conditions appeared to fall into this class of variants. Mutations in ctsR were previously reported for Scott A and LO28 HHP-resistant variants (17, 18, 32) (Fig. 3). The heat stress regulator CtsR plays an important role in stress survival as a repressor of class III heat shock genes. Alterations in CtsR can result in higher stress resistance, conceivably due to increased expression of the clp genes in the absence of the (active) CtsR repressor (20). Furthermore, the growth of previously characterized ctsR variants was comparable to the growth of the wild-type L. monocytogenes LO28 (31). As a result, ctsR variants are able to compete with other variants and wild-type cells, which can direct specific selection.
In previous research with L. monocytogenes Scott A, six different mutations in ctsR were found (17, 19). In this study, using L. monocytogenes LO28 and EGDe, 23 different mutations in ctsR were observed. Most of these mutations were located in or near the heat sensor domain of the gene (8, 20). The origin of these mutations could be a hot spot mutation sequence, like the Firmicutes consensus chi sequence 5′-(A/C)GCG(G/T)-3′ (14). Analysis of mutation hot spots in the ctsR gene revealed no overrepresentation of this consensus chi sequence (data not shown). Three ctsR mutations might result from strand slippage of the DNA polymerase in the GGT repeat region in the heat sensor domain. These three different mutations occurred only in the LO28 strain. None of the EGDe ctsR variants resulted from slippage in this repeat region. This contingency locus has previously been shown to be responsible for a high occurrence of mutations in L. monocytogenes Scott A (19). Another interesting possibility for the occurrence of ctsR variants is revealed by inspecting the genome context of ctsR. This context is conserved between Bacillus and Listeria. The two genes radA and yacK (lmo0234), which are located immediately downstream of the clpC operon (ctsR-mcsA-mcsB-clpC), were transcribed simultaneously with this operon under conditions of oxidative stress, indicating a strong relationship (28). Evidence for the involvement of both proteins in DNA repair and competence was obtained by phenotypic studies of mutants with changes in these two genes (23). CtsR seems to be functionally related to RadA, a recombination protein. RadA is required for the efficient repair of certain forms of DNA damage and for genetic recombination and might play a role in the stabilization and/or processing of Holliday junction intermediates (4). The role of these factors in the generation of diversity in L. monocytogenes remains to be elucidated and is currently being targeted in our laboratory.
The other half of the 106 L. monocytogenes variants were not mutated in their ctsR genes and upstream regions. The origin of resistance of these 57 variants is unknown. The underlying mechanisms of increased resistance of selected variants will be investigated in future research by comparative transcriptome analysis and genome sequencing. Three resistant variants belonging to this group were isolated after a heat treatment in milk. These variants showed different colony sizes on BHI agar before the heat treatment, and after heat inactivation, only small colonies could be recovered. Retesting the normal-sized colonies for their heat resistance revealed that these variants had reverted to the wild-type phenotype (data not shown). Future research will not only focus on the origin of mutation but will also include research into the revertance of variants to the wild-type phenotype. Revertance of resistant variants can give rise to persistence because of relatively high-frequency switches between phenotypes and genotypes (12).
The probability of detecting resistant variants depends on their frequency in the population, phenotypic characteristics (growth and resistance), and revertance. We found that the frequency of the L. monocytogenes HHP- and heat-resistant variants is below 10−5. Moreover, some previously isolated variants showed slower growth than their wild type (18, 31). Both factors make it difficult to isolate variants directly from the wild-type population. We showed that kinetic modeling can be used to find the appropriate experimental conditions with the highest probability of detecting resistant variants (Fig. 1). Furthermore, the high frequency of revertance to a wild-type phenotype of some of the isolated resistant variants might explain the difficulty of isolating resistant L. monocytogenes variants, as reported before by various research groups (5, 16).
The generation of population diversity and selection of stress-resistant variants will contribute to the survival of L. monocytogenes under constantly changing environmental conditions. In fact, resistant variants were even isolated after a heat inactivation at 72°C in milk, and a fraction of these variants showed high resistance to standard pasteurization conditions. The pasteurization of whole milk is specified by the FDA to be 72°C for 15 s and is designed to achieve at least a 5 log reduction of the most heat-resistant nonsporulating pathogen, Coxiella burnetii, in whole milk. For L. monocytogenes, a 5 log reduction in milk at 72°C in 15 s corresponds to a D-value of 3 s. This value is close to the one found for the heat-resistant variants without alterations in their ctsR genes and upstream regions. These variants are so resistant that they might become persisters in the food industry and, ultimately, cause disease. Although outbreaks of listeriosis associated with pasteurized dairy products are rare, in one of the reported outbreaks in Massachusetts, pasteurized milk was identified as the vehicle of listeriosis (10). In this case, no mistake in the pasteurization conditions could be identified, and questions were raised about the ability of the standard pasteurization process to eradicate L. monocytogenes from contaminated milk (2, 6, 10). In other cases, postpasteurization contamination of product was identified as the cause of listeriosis (30). Various studies have indicated that certain strains of L. monocytogenes survive within the food-processing environment, and the persistence of such strains in a food-processing plant is of particular concern as they have the potential to act as a source of contamination of the processed product (22, 25, 26, 27). The possible generation and establishment of stress-resistant L. monocytogenes variants in food-processing environments remains a critical challenge to the food industry.
Footnotes
Published ahead of print on 25 February 2011.
REFERENCES
- 1. Aguirre J. S., Pin C., Rodríguez M. R., García de Fernando G. D. 2009. Analysis of the variability in the number of viable bacteria after mild heat treatment of food. Appl. Environ. Microbiol. 75:6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anonymous. 1999. Grade “A” pasteurized milk ordinance 1999 revision. Public Health Service, Food and Drug Administration, U.S. Department of Health and Human Services, Washington, DC [Google Scholar]
- 3. Augustin J. C., Carlier V., Rozier J. 1998. Mathematical modelling of the heat resistance of Listeria monocytogenes. J. Appl. Microbiol. 84:185–191 [DOI] [PubMed] [Google Scholar]
- 4. Beam C. E., Saveson C. J., Lovett S. T. 2002. Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 184:6836–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchanan R. L., Golden M. H., Whiting R. C., Phillips J. G., Smith J. L. 1994. Non-thermal inactivation models for Listeria monocytogenes. J. Food Sci. 59:179–188 [Google Scholar]
- 6. Centers for Disease Control and Prevention 2008. Outbreak of Listeria monocytogenes infections associated with pasteurized milk from a local dairy—Massachusetts, 2007. MMWR Morb. Mortal. Wkly. Rep. 57:1097–1100 [PubMed] [Google Scholar]
- 7. Derré I., Rapoport G., Msadek T. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol. 31:117–131 [DOI] [PubMed] [Google Scholar]
- 8. Derré I., Rapoport G., Msadek T. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol. Microbiol. 38:335–347 [DOI] [PubMed] [Google Scholar]
- 9. Drake J. W. 2006. Chaos and order in spontaneous mutation. Genetics 173:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fleming D. W., et al. 1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 312:404–407 [DOI] [PubMed] [Google Scholar]
- 11. Geeraerd A. H., Valdramidis V. P., Van Impe J. F. 2005. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int. J. Food Microbiol. 102:95–105 (Erratum, 110:297, 2006.) [DOI] [PubMed] [Google Scholar]
- 12. Gefen O., Balaban N. Q. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 33:704–717 [DOI] [PubMed] [Google Scholar]
- 13. Goulet V., Hedberg C., Le Monnier A., de Valk H. 2008. Increasing incidence of listeriosis in France and other European countries. Emerg. Infect. Dis. 14:734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Halpern D., et al. 2007. Identification of DNA motifs implicated in maintenance of bacterial core genomes by predictive modeling. PLoS Genet. 3:1614–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hauben K. J., et al. 1997. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl. Environ. Microbiol. 63:945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hornbæk T., Brockhoff P. B., Siegumfeldt H., Budde B. B. 2006. Two subpopulations of Listeria monocytogenes occur at subinhibitory concentrations of leucocin 4010 and nisin. Appl. Env. Microbiol. 72:1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joerger R. D., Chen H., Kniel K. E. 2006. Characterization of a spontaneous, pressure-tolerant Listeria monocytogenes Scott A ctsR deletion mutant. Foodborne Pathog. Dis. 3:196–202 [DOI] [PubMed] [Google Scholar]
- 18. Karatzas K. A. G., Bennik M. H. J. 2002. Characterization of a Listeria monocytogenes Scott A isolate with high tolerance towards high hydrostatic pressure. Appl. Environ. Microbiol. 68:3183–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karatzas K. A. G., Valdramidis V. P., Wells-Bennik M. H. J. 2005. Contingency locus in ctsR of Listeria monocytogenes Scott A: a strategy for occurrence of abundant piezotolerant isolates within clonal populations. Appl. Environ. Microbiol. 71:8390–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karatzas K. A. G., et al. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility, and virulence. Mol. Microbiol. 49:1227–1238 [DOI] [PubMed] [Google Scholar]
- 21. Karatzas K. A. G., Zervos A., Tassou C. C., Mallidis C. G., Humphrey T. J. 2007. Piezotolerant small-colony variants with increased thermotolerance, antibiotic susceptibility, and low invasiveness in a clonal Staphylococcus aureus population. Appl. Environ. Microbiol. 73:1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kells J., Gilmour A. 2004. Incidence of Listeria monocytogenes in two milk processing environments, and assessment of Listeria monocytogenes blood agar for isolation. Int. J. Food Microbiol. 91:167–174 [DOI] [PubMed] [Google Scholar]
- 23. Krüger E., Msadek T., Ohlmeier S., Hecker M. 1997. The Bacillus subtilis clpC operon encodes DNA repair and competence proteins. Microbiology 143:1309–1316 [DOI] [PubMed] [Google Scholar]
- 24. Leroy F., Lievens K., De Vuyst L. 2005. Modeling bacteriocin resistance and inactivation of Listeria innocua LMG 13568 by Lactobacillus sakei CTC 494 under sausage fermentation conditions. Appl. Env. Microbiol. 71:7567–7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lianou A., Sofos J. N. 2007. A review of the incidence and transmission of Listeria monocytogenes in ready-to-eat products in retail and food service environments. J. Food Prot. 70:2172–2198 [DOI] [PubMed] [Google Scholar]
- 26. Lundén J. M., Autio T. J., Sjöberg A.-M., Korkeala H. J. 2003. Persistent and nonpersistent Listeria monocytogenes contamination in meat and poultry processing plants. J. Food Prot. 66:2062–2069 [DOI] [PubMed] [Google Scholar]
- 27. Møretrø T., Langsrud S. 2004. Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms 1:107–121 [Google Scholar]
- 28. Mostertz J., Scharf C., Hecker M., Homuth G. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497–512 [DOI] [PubMed] [Google Scholar]
- 29. Rajkovic A., et al. 2009. Resistance of Listeria monocytogenes, Escherichia coli O157:H7 and Campylobacter jejuni after exposure to repetitive cycles of mild bactericidal treatments. Food Microbiol. 26:889–895 [DOI] [PubMed] [Google Scholar]
- 30. Reij M. W., Den Aantrekker E. D., and ILSI Europe Risk Analysis in Microbiology Task Force 2004. Recontamination as a source of pathogens in processed foods. Int. J. Food Microbiol. 91:1–11 [DOI] [PubMed] [Google Scholar]
- 31. Van Boeijen I. K. H., et al. 2010. Population diversity of Listeria monocytogenes LO28: phenotypic and genotypic characterization of variants resistant to high hydrostatic pressure. Appl. Environ. Microbiol. 76:2225–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Boeijen I. K. H., Moezelaar R., Abee T., Zwietering M. H. 2008. Inactivation kinetics of three Listeria monocytogenes strains under high hydrostatic pressure. J. Food Prot. 71:2007–2013 [DOI] [PubMed] [Google Scholar]
- 33. Voetsch A. C., et al. 2007. Reduction in the incidence of invasive listeriosis in foodborne diseases active surveillance network sites, 1996–2003. Clin. Infect. Dis. 44:513–520 [DOI] [PubMed] [Google Scholar]