Abstract
Sourdough fermentation is a cereal fermentation that is characterized by the formation of stable yeast/lactic acid bacteria (LAB) associations. It is a unique process among food fermentations in that the LAB that mostly dominate these fermentations are heterofermentative. In the present study, four wheat sourdough fermentations were carried out under different conditions of temperature and backslopping time to determine their effect on the composition of the microbiota of the final sourdoughs. A substantial effect of temperature was observed. A fermentation with 10 backsloppings (once every 24 h) at 23°C resulted in a microbiota composed of Leuconostoc citreum as the dominant species, whereas fermentations at 30 and 37°C with backslopping every 24 h resulted in ecosystems dominated by Lactobacillus fermentum. Longer backslopping times (every 48 h at 30°C) resulted in a combination of Lactobacillus fermentum and Lactobacillus plantarum. Residual maltose remained present in all fermentations, except those with longer backslopping times, and ornithine was found in almost all fermentations, indicating enhanced sourdough-typical LAB activity. The sourdough-typical species Lactobacillus sanfranciscensis was not found. Finally, a nonflour origin for this species was hypothesized.
INTRODUCTION
Sourdough is a mixture of ground cereals and water that ferments spontaneously. Sourdoughs improve the properties of the bread dough, enhance bread texture and flavor, and delay bread spoilage and staling (11, 26, 40). Many studies have investigated the composition of the sourdough microbiota of traditional type I sourdoughs, which are daily backslopped, with both culture-dependent and culture-independent techniques (9–11). Lactic acid bacteria (LAB) and yeasts play a key role in the sourdough fermentation process (9, 22, 25). Unlike most other well-known food fermentation processes, sourdough is usually dominated by heterofermentative LAB, commonly belonging to the genus Lactobacillus (4, 9).
Despite changes in raw materials or the bakery environment, sourdoughs are stable ecosystems (43, 58). This stability can be ascribed to specific metabolic adaptations to the sourdough ecosystem or the production of antimicrobial compounds (10, 18). Carbohydrate fermentation targeted toward maltose catabolism (encompassing maltose phosphorylase activity), the use of alternative external electron acceptors (such as fructose), and/or the expression of the arginine deiminase (ADI) pathway are metabolic activities that favor energy production, cofactor (re)cycling, and/or tolerance toward acid stress (4, 10, 11, 18, 19, 23). Moreover, certain LAB species form a stable association with certain yeast species, due to dedicated nutritional, trophic, and metabolic interactions (4, 9, 10, 18, 23). Consequently, some LAB species that participate in sourdough fermentations are considered typical inhabitants of the sourdough ecosystem, among which Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus paralimentarius, Lactobacillus brevis, and Lactobacillus sanfranciscensis are most frequently reported (4, 11, 25).
The key sourdough LAB species is L. sanfranciscensis, which is strictly heterofermentative and maltose-positive and which, remarkably, is not yet known from any other habitat (21, 22, 26). It mainly occurs in wheat sourdoughs (3, 36, 38, 42, 43). It forms a strong association with the maltose-negative yeast Candida humilis, based on trophic and metabolic interactions (22, 36). The dominance of L. sanfranciscensis has been ascribed to its selection by the type of technology applied, i.e., backslopping practices, temperature of incubation, and/or pH of the dough (16, 17, 29, 38, 44). Although initially described in San Francisco sourdough and Panettone cake, an influence of the geographical region is not expected (11). Besides the L. sanfranciscensis-C. humilis association, associations of Saccharomyces cerevisiae and L. plantarum, Candida spp. and L. brevis, and Kazachstania barnettii and L. sanfranciscensis have been reported too (4, 27, 55). Furthermore, sourdough continues to be a source of unexplored LAB species diversity, as exemplified by the continuous reporting of new LAB species (11). This demonstrates the large microbial diversity present in sourdough. Moreover, since these species are mostly isolated in exploratory studies of a limited number of fermentations, it remains a challenge to know what the exact reasons are why one species should be dominant over another or present in one sourdough environment and not in another.
In previous studies, it has been shown that type I sourdough fermentations carried out in the laboratory with flour as the sole nonsterile ingredient harbor a different species diversity than artisan sourdoughs prepared in a bakery, with respect to both LAB and yeast species (42, 43, 49, 55, 57, 58). Laboratory sourdoughs based on wheat, rye, or spelt, daily backslopped for 10 days, whether or not initiated with a L. sanfranciscensis starter culture, reach an equilibrium through a three-step process: (i) prevalence of sourdough-atypical LAB, (ii) prevalence of sourdough-typical LAB, and (iii) prevalence of highly adapted sourdough-typical LAB, i.e., L. fermentum and L. plantarum (44, 49, 57, 58). Lactobacillus sanfranciscensis was never encountered in laboratory sourdough fermentations performed under semisterile conditions (49, 57, 58). Belgian bakery sourdoughs that are often backslopped for several years are yet characterized by stable consortia of L. paralimentarius, L. sanfranciscensis, L. plantarum, and/or Lactobacillus pontis (42). It has been shown that dominance of the final microbial species may be influenced by temperature (33, 34) and flour type (2, 45, 51, 60).
The present study aimed to unravel the differences occurring in LAB composition and dominance of sourdoughs with various temperatures and backslopping times and in particular to postulate a hypothesis for the absence of L. sanfranciscensis in laboratory sourdough fermentations performed under semisterile conditions.
MATERIALS AND METHODS
Laboratory sourdough preparation.
Liquid sourdoughs (8 kg) were prepared with sterile water and wheat flour from one local flour mill and belonging to the same production batch. The dough yield, i.e., (dough mass/flour mass) × 100, was 400. Temperature and backslopping time were varied. Sourdough fermentations were carried out in two 15-liter Biostat C fermentors (Sartorius AG; B. Braun Biotech International, Melsungen, Germany), which were presterilized with water. Fermentations were started by mixing 6 liters of sterile water with 2 kg of flour in one fermentor. This liquid dough was incubated at 23, 30, or 37°C for 24 or 48 h, depending on the experiment, during which time the mixture was kept homogeneous through stirring (300 rpm). Then, 800 g of ripe sourdough was collected in a sterile bottle to inoculate a freshwater-flour mixture (5.2 liters of sterile water and 1.8 kg of flour) as a first backslopping in a second fermentor. This dough was incubated under the same conditions as described above. This backslopping procedure was carried out during 10 days. At each refreshment step, samples were withdrawn from the ripe sourdough for culture-dependent and culture-independent microbiological analysis and metabolite target analysis. Also, the pH of each sample was measured.
Microbial community dynamics of backslopped sourdoughs analyzed through a culture-dependent approach.
Cell counts (expressed as CFU or CFU per gram of dough) were determined by mixing 10 g of fresh sourdough with 90 ml of peptone physiological solution (0.1% [wt/vol] bacteriological peptone [Oxoid, Ltd., Basingstoke, Hampshire, United Kingdom] and 0.85% [wt/vol] NaCl) and then homogenizing this mixture for 5 min (Stomacher 400; Seward, Worthington, United Kingdom). A 10-fold dilution series was made and plated on de Man-Rogosa-Sharpe-5 (MRS-5) agar containing 0.1 g of cycloheximide (Sigma, St. Louis, MO) per liter and yeast extract-glucose (YG) agar supplemented with 0.1 g of chloramphenicol (Sigma) per liter to determine LAB and yeast counts (33, 34), respectively, after incubation of the agar medium at 30°C for 48 h.
From the MRS-5 agar medium, approximately 50 colonies were picked up randomly, purified through successive transfers, and subjected to DNA extraction and (GTG)5-PCR fingerprinting for classification and identification of the isolates. DNA was isolated by using a protocol described previously (20). (GTG)5-PCR assays and analysis of the resulting DNA fingerprints were carried out as described previously (42, 47, 49).
Microbial community dynamics of backslopped sourdoughs analyzed through a culture-independent approach via denaturing gradient gel electrophoresis (DGGE) of 16S rRNA PCR amplicons.
To approximately 10 g of fresh sourdough 90 ml of peptone physiological solution was added, and 50 ml of this mixture was centrifuged (1,000 × g for 4 min at 4°C) to remove solids. The supernatant was centrifuged at 5,000 × g for 20 min at 4°C to obtain a cell pellet, which was stored at −20°C until further use. The extraction of total DNA from these cell pellets, amplification of 16S rRNA gene fragments with the universal primers F357-518R, electrophoretic separation in a denaturing gradient polyacrylamide gel, and sequencing of representative DNA bands were performed as described previously (39, 42, 49).
Culture-independent investigation of the flour microbiota.
Wheat flour (200 g) was suspended in physiological solution (800 ml) and stirred vigorously for 15 min at 4°C, after which the flour particles were removed by centrifugation at 1,000 × g for 4 min at 4°C. The supernatant was centrifuged at 5,000 × g for 20 min at 4°C to obtain a cell pellet, which was stored at −20°C until further use. DNA extraction, PCR amplification of 16S rRNA gene fragments, and DGGE were performed as described previously (42, 49). DNA from pure cultures of L. plantarum IMDO 130201, L. fermentum IMDO 130101, L. sanfranciscensis LMG 16002T, and Leuconostoc citreum LMG 6949T was included in the gel as references.
Metabolite target analysis.
Approximately 100 g of fresh sourdough was centrifuged (5,000 × g for 20 min), and the supernatant was stored at −20°C until further analysis. The concentrations of lactic acid, acetic acid, and ethanol were determined by high-performance liquid chromatography (HPLC) and quantification was performed with external standards in triplicate as described previously (32). The results are represented as the average of the three independent analyses. Errors are represented as standard deviations. The concentrations of glucose, fructose, sucrose, maltose, and mannitol were determined by high-performance anion-exchange chromatography with pulsed amperometric detection, and quantification was carried out through standard addition as described previously (52). Concentrations of arginine, citrulline, and ornithine were determined by HPLC coupled to a mass spectrometer (MS), and quantification was carried out through standard addition as described previously (53).
RESULTS
Microbial community dynamics of backslopped sourdoughs analyzed through a culture-dependent approach.
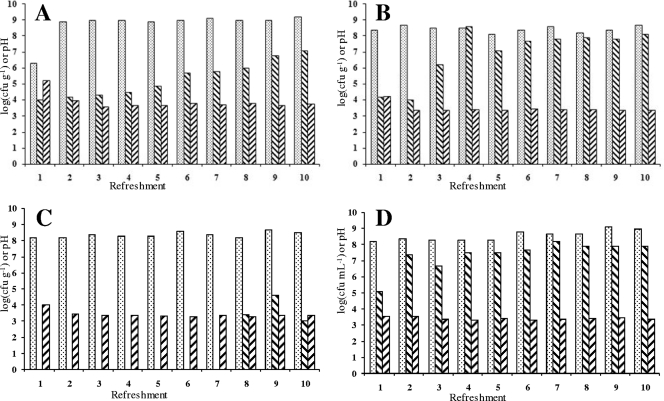
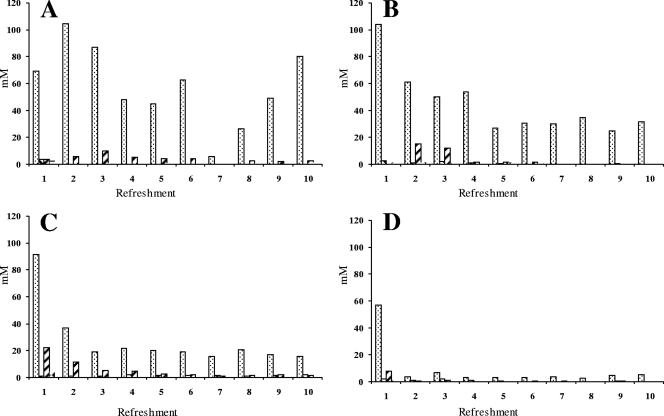
Colony counts on MRS-5 agar indicated stable LAB communities at the latest from the second backslopping on (Fig. 1). Final cell densities were generally between log 8 and log 9 CFU g−1, with the highest counts (just above log 9 CFU g−1) reached in the fermentation at 23°C with a backslopping time of 24 h and in the fermentation at 30°C with a backslopping time of 48 h. Yeast counts on YG agar generally revealed stable communities of approximately log 8 CFU g−1, albeit delayed compared to the LAB communities, except for the fermentation at 37°C with a 24-h backslopping (Fig. 1). During this fermentation, no yeasts were found, except at day 8 until day 10 with a colony count never exceeding log 5 CFU g−1. The end pH of all fermentations remained constant after about the third backslopping (Fig. 1). The end pH was usually between 3.3 and 3.4, except for the fermentation at 23°C with backslopping every 24 h that resulted in an end pH of ∼3.7.
Fig. 1.
Bacterial counts (CFU g−1) on MRS-5 agar (▩), yeast counts (CFU g−1) on YG agar ( ), and pH (▨) for fermentation A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
), and pH (▨) for fermentation A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
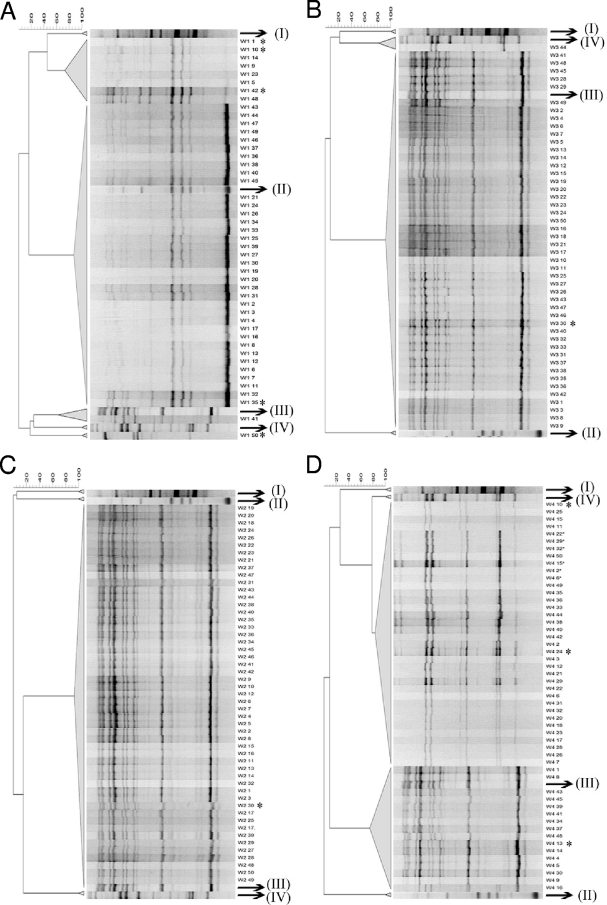
Identification of the 195 LAB isolates, picked up from MRS-5 agar, through (GTG)5-PCR fingerprinting revealed differences in the final composition of the microbiota of the four 10 times backslopped sourdoughs. Of the 47 isolates from the fermentation at 23°C with backslopping every 24 h (Fig. 2A), a single isolate was identified as L. fermentum, and 37 isolates were identified as Leuconostoc citreum. Eight isolates (W1-1, W1-5, W1-9, W1-10, W1-14, W1-23, W1-42, and W1-48) formed a separate (GTG)5-PCR cluster and were shown to be Leuconostoc citreum too, based on the 16S rRNA gene sequence of isolate W1-10 as cluster representative. A single isolate did not fit in the reference framework (W1-50) and was identified as Pediococcus pentosaceus, based on 16S rRNA gene sequencing. Of 48 isolates from the sourdough fermentation at 30°C with backslopping every 24 h (Fig. 2B), 47 were identified as L. fermentum, and a single isolate (W3-44) was identified as L. plantarum. From the sourdough fermentation at 37°C with backslopping every 24 h (Fig. 2C), all 50 isolates were identified as L. fermentum. Out of the 50 isolates from the fermentation at 30°C with backslopping every 48 h (Fig. 2D), 16 isolates were identified as L. fermentum, with the remainder ones being identified as L. plantarum.
Fig. 2.
Dendrograms derived from (GTG)5-PCR fingerprints of bacterial isolates originating from fermentation A (23°C and backslopping every 24 h), fermentation B (30°C and backslopping every 24 h), fermentation C (37°C and backslopping every 24 h), and fermentation D (30°C and backslopping every 48 h). The type strains used are Lactobacillus brevis LMG 6909T (I), Leuconostoc citreum LMG 6949T (II), Lactobacillus fermentum IMDO 130101 (III), and Lactobacillus plantarum LMG 6907T (IV). The strains marked with asterisks were subjected to sequencing of the 16S rRNA gene.
Microbial community dynamics of backslopped sourdoughs analyzed through a culture-independent approach via DGGE of 16S rRNA PCR amplicons.
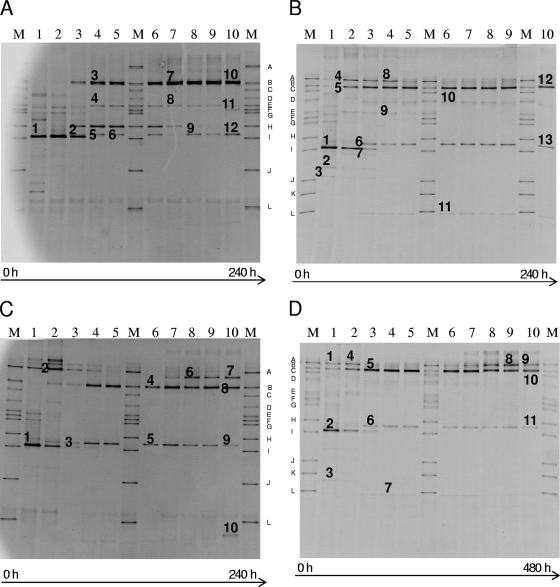
At 23°C and backslopping every 24 h (Fig. 3A and Table 1), Bacillus species were detected during the first 3 days of fermentation, after which Leuconostoc citreum appeared and remained present until the end of the fermentation. From day 3 until day 8, Lactococcus species were detected, and from day 4 until day 9, Lactobacillus sakei was found. On day 10, a faint DGGE band indicated the presence of P. pentosaceus. The sourdough fermentation at 30°C with backslopping every 24 h (Fig. 3B and Table 1) showed the presence of Bacillus, Enterobacter species, and Lactobacillus reuteri during the first day. From day 2 until day 5, Weissella cibaria was detected, as well as L. fermentum. From day 6 on, only L. fermentum was detected until the end of the fermentation. The sourdough fermentation at 37°C with backslopping every 24 h (Fig. 3C and Table 1) showed the presence of Bacillus cereus on the first day and of W. cibaria during the first and second day. From day 2 until the end of the fermentation, L. fermentum was present, and from day 7 until the end of the fermentation, Lactobacillus johnsonii was detected. At 30°C and with backslopping every 48 h (Fig. 3D and Table 1), W. cibaria and Weissella confusa were detected during the first two backsloppings, and Bacillus thuringiensis was detected during the first three backsloppings. Lactobacillus fermentum was present from the first until the last fermentation stage. From backslopping 7 until backslopping 10, L. plantarum was detected. None of the fermentations carried out revealed the presence of L. sanfranciscensis (Fig. 3).
Fig. 3.
Culture-independent 16S rRNA-PCR-DGGE analysis of fermentation samples taken at each refreshment step of a sourdough fermentation backslopped 10 times. Panels A to D correspond to fermentations A to D in Table 1. The marker ladder M consisted of Lactobacillus plantarum LMG 6907T (band A), Lactobacillus fermentum LMG 6902T (band B), Leuconostoc mesenteroides LMG 6893T (band C), Lactobacillus brevis LMG 6906T (band D), Lactobacillus curvatus LMG 69198T (band E), Pediococcus pentosaceus LMG 10478 (band F), Enterococcus faecalis LMG 7937T (band G), Lactococcus lactis LMG 19152T (band H), Lactobacillus sanfranciscensis 16002T (band I), Bifidobacterium longum BB536 (band J), Bifidobacterium adolescentis LMG 10502 (band K), and Bifidobacterium breve LMG 11084 (band L). Bands indicated by numbers have been excised and subjected to 16S rRNA gene sequence analysis. The closest relatives of the fragments sequenced are listed in Table 1.
Table 1.
Sequencing results from excised PCR-DGGE-DNA bands
| Fermentation and band no. | Identification | Identity (%) | Nearest accession no. |
|---|---|---|---|
| A (23°C and 24-h backslopping) | |||
| 1 | Bacillus thuringiensis | 100 | EF210289.1 |
| 2 | Bacillus cereus | 100 | DQ648271.1 |
| 3 | Leuconostoc citreum | 99 | AB362723.1 |
| 4 | Lactobacillus sakei/L. curvatus | 100/100 | AB362730.1/EU081014.1 |
| 5 | Chloroplast DNA | 100 | AM777385.2 |
| 6 | Chloroplast DNA | 100 | AM777385.2 |
| 7 | Leuconostoc citreum | 98 | AB362723.1 |
| 8 | Lactobacillus sakei/L. curvatus | 100/100 | AB362730.1/EU081014.1 |
| 9 | Chloroplast DNA | 100 | AM777385.2 |
| 10 | Leuconostoc citreum | 99 | AB362723.1 |
| 11 | Pediococcus pentosaceus | 100 | AB362605.1 |
| 12 | Chloroplast DNA | 100 | AM777385.2 |
| B (30°C and 24-h backslopping) | |||
| 1 | Bacillus cereus | 100 | DQ648271.1 |
| 2 | Enterobacter sakazakii | 100 | EF088360.1 |
| 3 | Enterobacter cloacae | 100 | EF185910.1 |
| 4 | Weissella cibaria | 99 | DQ885576.1 |
| 5 | Lactobacillus reuteri | 98 | AB289272.1 |
| 6 | Uncultured bacterium | 98 | DQ349083.1 |
| 7 | Bacillus cereus | 99 | EF195169.1 |
| 8 | Weissella cibaria | 100 | DQ885576.1 |
| 9 | Lactobacillus sp. | 100 | AB279927.1 |
| 10 | Uncultured bacterium | 100 | DQ235225.1 |
| 11 | Mitochondrial DNA | 99 | DQ490951.2 |
| 12 | Lactobacillus fermentum | 99 | AB289114.1 |
| 13 | Chloroplast DNA | 99 | EF115543.1 |
| C (37°C and 24-h backslopping) | |||
| 1 | Bacillus cereus | 100 | DQ648271.1 |
| 2 | Weissella cibaria/W. confusa | 100/100 | AB362617.1/EU128489.1 |
| 3 | Bacillus cereus | 100 | DQ648271.1 |
| 4 | Lactobacillus fermentum | 100 | AB362767.1 |
| 5 | Chloroplast DNA | 100 | AM777385.2 |
| 6 | Lactobacillus johnsonii | 99 | AB295648.1 |
| 7 | Lactobacillus johnsonii | 100 | AB295648.1 |
| 8 | Lactobacillus fermentum | 100 | AB362767.1 |
| 9 | Chloroplast DNA | 100 | AM777385.2 |
| 10 | Uncultured bacterium | 98 | DQ981307.1 |
| D (30°C and 48-h backslopping) | |||
| 1 | Weissella confusa | 100 | EU128489.1 |
| 2 | Bacillus thuringiensis | 100 | EF210289.1 |
| 3 | Uncultured bacterium | 98 | EU148842.1 |
| 4 | Weissella cibaria | 100 | AB362617.1 |
| 5 | Lactobacillus fermentum | 100 | AB362767.1 |
| 6 | Chloroplast DNA | 100 | AM777385.2 |
| 7 | Mitochondrial DNA | 100 | EF547250.1 |
| 8 | Lactobacillus plantarum | 97 | EF439682.1 |
| 9 | Lactobacillus plantarum | 97 | AB362982.1 |
| 10 | Lactobacillus fermentum | 97 | AB362767.1 |
| 11 | Chloroplast DNA | 99 | AM777385.2 |
Culture-independent investigation of the flour microbiota.
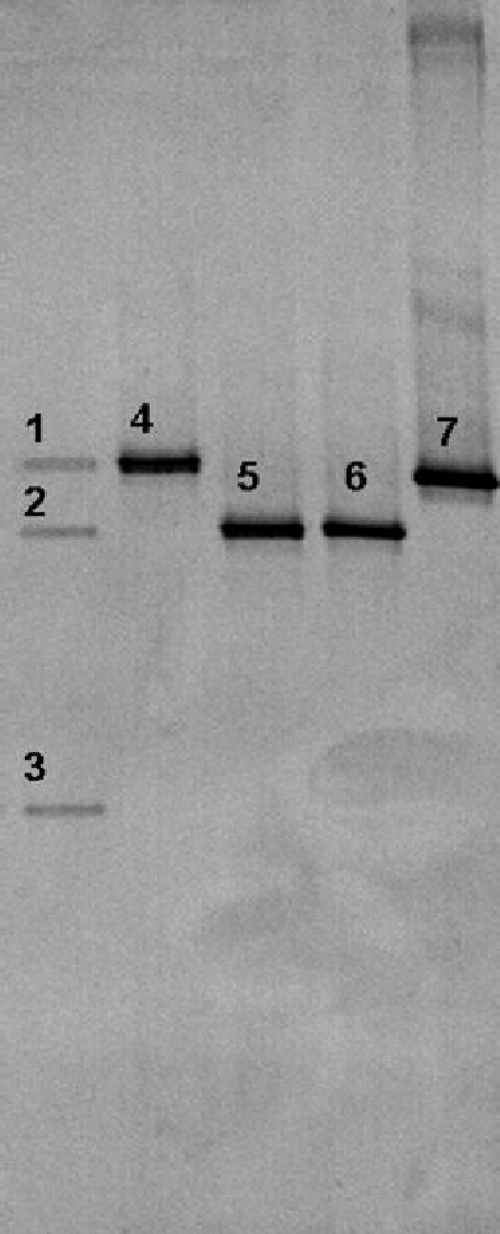
PCR-DGGE of the DNA extracted directly from the flour revealed the presence of L. plantarum and L. fermentum (Fig. 4). No L. sanfranciscensis was found. Since Leuconostoc citreum and L. fermentum presented DNA bands on the same height in the gel, the identity as L. fermentum (identity of 100%; nearest accession number HQ677597.1) was confirmed by nucleotide sequencing of the DNA fragment of the excised DGGE band.
Fig. 4.
Culture-independent 16S rRNA-PCR-DGGE analysis of a flour sample. The bands were identified as Lactobacillus plantarum (band 1), Lactobacillus fermentum (band 2), and chloroplast DNA (band 3). The reference strains were Lactobacillus plantarum IMDO 130201 (band 4), Lactobacillus fermentum IMDO 130101 (band 5), Leuconostoc citreum LMG 6949T (band 6), and Lactobacillus sanfranciscensis LMG 16002T (band 7).
Metabolite target analysis.
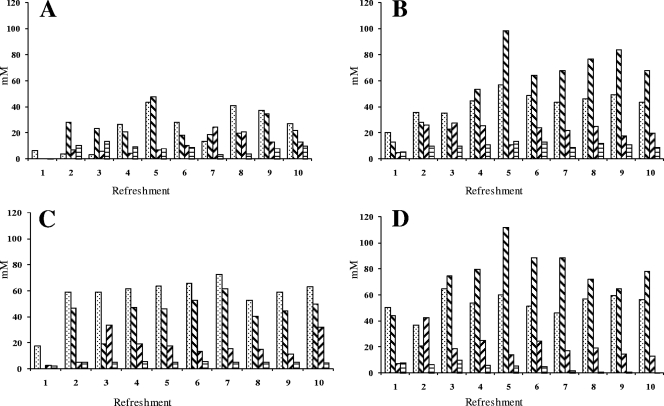
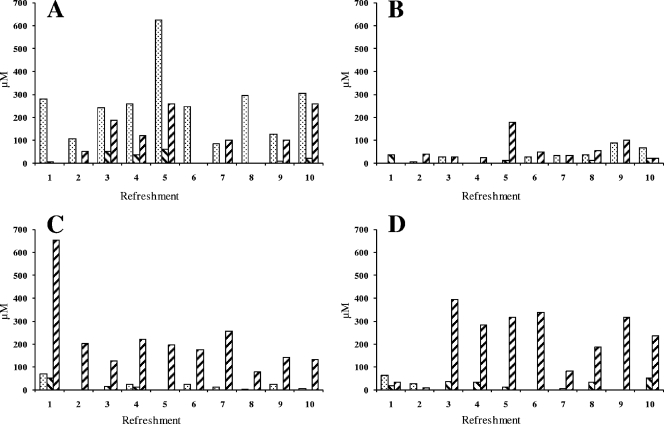
Carbohydrate analysis showed that only negligible amounts of glucose, fructose, and sucrose remained in all fermentations (Fig. 5). Maltose was found at residual concentrations varying between 15 mM (30°C and 48-h backslopping) and 80 mM (23°C and 24-h backslopping). Mannitol was found at low concentrations, except for the fermentation at 30°C and with backslopping every 48 h where hardly any mannitol was found (Fig. 6). The highest lactic acid and acetic acid concentrations were found in the fermentations at 30 and 37°C. The highest ethanol concentrations were found at 30°C with both the 24- and the 48-h backsloppings. A substantially lower ethanol concentration was found at 23°C with the 24-h backslopping. Residual arginine was present at about 300 μM at 23°C and the 24-h backslopping, while all of the other fermentations showed arginine depletion (Fig. 7). Citrulline was present in amounts beneath the detection limit, and ornithine was found in concentrations of approximately 200 μM in all fermentations, except for the fermentation at 30°C with the 48-h backslopping (300 μM).
Fig. 5.
Concentrations of maltose (▩), sucrose ( ), glucose (▨), and fructose (▤) in mM for fermentations A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
), glucose (▨), and fructose (▤) in mM for fermentations A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
Fig. 6.
Concentrations of lactic acid (▩), ethanol ( ), acetic acid (▨), and mannitol (▤) in mM for fermentations A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
), acetic acid (▨), and mannitol (▤) in mM for fermentations A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
Fig. 7.
Concentrations of arginine (▩), citrulline ( ), and ornithine (▨) in μM for fermentations A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
), and ornithine (▨) in μM for fermentations A (23°C and 24-h backslopping), B (30°C and 24-h backslopping), C (37°C and 24-h backslopping), and D (30°C and 48-h backslopping).
DISCUSSION
As reported previously with wheat flour from the same flour mill, the most common LAB dominating the stable ecosystem of laboratory sourdough fermentations carried out with wheat flour as the sole nonsterile component, are L. fermentum and L. plantarum (51). These species are typical sourdough LAB species (4, 9–11). In the present study, a modulation of the microbiota as a function of temperature could be observed going from almost exclusively L. fermentum at 30°C and 37°C with backslopping every 24 h to a codominance of L. fermentum and L. plantarum at 30°C and 48-h backslopping to the complete absence of these species at 23°C. The latter fermentations were exclusively dominated by Leuconostoc citreum. This indicates an influence of both temperature and backslopping time on the growth of LAB coming from the flour. It appears that L. fermentum needs less time to adapt to the sourdough matrix, grows faster, and is more competitive than L. plantarum in wheat sourdough fermentations, at least under the conditions tested. Also, the appearance of L. plantarum after 48 h of backslopping indicates a close adaptation to the poor nutritional and highly acidic conditions that prevail under these fermentation conditions. Alternatively, Leuconostoc citreum is more adapted to low-temperature conditions. An influence of temperature on species composition has been found before when commercially available sourdough starter mixtures were used (34).
The results of the culture-dependent and culture-independent techniques carried out during the present study were in agreement, except for the fact that in the fermentation at 37°C with 24-h backslopping, L. johnsonii was detected with the culture-independent technique, and yet no isolate of this species could be recovered through culture. Lactobacillus johnsonii has only rarely been reported from sourdough (34) and is usually associated with the vertebrate gastrointestinal tract (13).
A stable number of yeasts and LAB was obtained in all wheat sourdough fermentations, except for the fermentation at 37°C and 24-h backslopping, where yeasts were almost completely absent, possibly reflecting an increased competitiveness of the LAB compared to the yeasts or a growth limitation of the yeasts by the high temperature. Optimal growth temperatures for yeasts have been reported to be in the range of 10 to 30°C (56). For instance, for C. humilis, which is frequently encountered in sourdough fermentations, an optimal growth temperature of 27°C has been reported (17). Ornithine, the end product of the ADI pathway of certain sourdough-typical LAB species, was found in all fermentations, indicating activity of LAB species typical for sourdough, except at 30°C and a 24-h backslopping, although L. fermentum was the dominant LAB species in this fermentation. This species has been shown to convert arginine through the ADI pathway and to release either citrulline or ornithine (53, 54). However, it is possible that the yeasts present in this fermentation were responsible for either arginine depletion or ornithine conversion (37). Although ornithine was found at elevated concentrations in the fermentation at 23°C and a 24-h backslopping, the dominant species in this fermentation was Leuconostoc citreum, a species that does not possess the ADI pathway (28). Despite the fact that yeasts have only been reported to take up ornithine and not to excrete it (37), ornithine can be stored in yeast vacuoles under certain conditions, suggesting that yeast cell lysis might account for ornithine release (50). Besides the ADI pathway, factors such as inhibitory and cooperative interactions may play a role in dominance build-up (4, 9, 22).
This is the first study in which a spontaneous sourdough fermentation was dominated by Leuconostoc citreum, albeit only at a low temperature, a species thus far reported only occasionally in sourdough fermentations (3). It appears to be mostly associated with vegetable fermentations such as kimchi, where it plays an important role together with Weissella confusa, L. sakei, and Lactobacillus curvatus (1, 30). Low-temperature adaptation of Leuconostoc citreum has been described before (59). Also, higher final pH values were observed in the fermentation where this species dominated than in fermentations where L. fermentum and/or L. plantarum were dominant. It has been shown before that sourdoughs with higher pH levels are often dominated by a different microbiota, encompassing Enterococcus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, and Weissella (6, 41, 61), which are present in the cereal kernels and flour (5, 9) or during the early fermentation process before significant pH decrease (6, 49). With PCR-DGGE, Bacillus species were found during the first 3 days of fermentation. After day 4, they were no longer present because of the low pH. Bacillus species are known to be part of the microbiota of flour (9).
With the exception of a single P. pentosaceus isolate, the wheat sourdough ecosystems of the present study were composed of three prevalent species, namely, L. plantarum, L. fermentum, and Leuconostoc citreum. Under no circumstances could the well-known wheat sourdough LAB, L. sanfranciscensis, be detected. It confirms the results obtained for former laboratory wheat, rye, and spelt sourdough fermentations (49, 57, 58). This is remarkable considering the fact that it has been widely reported in Italian and Belgian artisan sourdoughs and in San Francisco sourdough fermentations, which are characterized by long fermentation times at low temperature (3, 8, 42, 43). However, its competitiveness in the sourdough ecosystem has been firmly established (6, 17, 44). Considering the semisterile conditions under which the current laboratory wheat sourdough fermentations were carried out, the only remaining possible origin of the microbiota was the flour itself, raising questions about the origin and original habitat of L. sanfranciscensis. Through PCR-DGGE, the presence of L. fermentum and L. plantarum could be shown in the flour used but not that of Leuconostoc citreum and L. sanfranciscensis. Given that the detection limit of PCR-DGGE is in the order of magnitude of 104 to 108 CFU of LAB in general g−1 (35, 46), the absence of detectable amplification can therefore mean either absence of the microorganism or presence beneath the detection limit, indicating that both species might be present in the subdominant flour microbiota, but that only for Leuconostoc citreum correct environmental circumstances that gave it a competitive edge, were encountered. Flour as a carrier of the inoculum of sourdough fermentations has been suggested before (44). If L. sanfranciscensis were absent from the flour used, an interesting report in this context is the presence of L. sanfranciscensis in the insect gastrointestinal tract (24), suggesting another possible route for this species into the flour and bakery environment and hence into the sourdoughs. Such an association of LAB dominating food fermentations and the gastrointestinal tract should come as no surprise, since L. reuteri, L. johnsonii, and L. fermentum have been shown to be inhabitants of vertebrate gastrointestinal tracts (12, 48). Some strains of these species are even used as probiotics in food and/or feed (12, 31, 48). Also, the sourdough inhabitant, Lactobacillus rossiae, has been isolated from pig feces, suggesting an intestinal habitat (7). Actually, many sourdough species have been found in pig feces before (14). The suggestion that the sourdough microbiota might be associated with contamination by mouse feces (15), and hence the gastrointestinal tract, in the grain elevators in flour mills is thus of interest. Therefore, it will be necessary to sample the whole chain from flour mill to sourdough and their environments to unravel the habitat and origin of L. sanfranciscensis ultimately.
To conclude, this is the first report of spontaneous sourdough fermentations wherein temperature and backslopping time were systematically varied. Furthermore, the present study is the first report of a sourdough fermentation dominated by a Leuconostoc species, at least at a low temperature. Also, we suggest here a nonflour origin of L. sanfranciscensis. Furthermore, these results clearly show the potential for modulation of the microbiota of a spontaneous sourdough fermentation through manipulation of the environmental conditions such as temperature and backslopping time.
ACKNOWLEDGMENTS
We acknowledge the financial support of the Research Council of the Vrije Universiteit Brussel (RC-VUB) and the Fund for Scientific Research-Flanders (FWO-Vlaanderen).
T.R. is supported by a predoctoral fellowship of the FWO-Vlaanderen. G.V. is supported by a postdoctoral fellowship of the RC-VUB.
Footnotes
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Cho J., et al. 2006. Microbial population dynamics of kimchi, a fermented cabbage product. FEMS Microbiol. Lett. 257:262–267 [DOI] [PubMed] [Google Scholar]
- 2. Coda R., et al. 2010. Spelt and emmer flours: characterization of the lactic acid bacteria microbiota and selection of mixed starters for bread making. J. Appl. Microbiol. 108:925–935 [DOI] [PubMed] [Google Scholar]
- 3. Corsetti A., et al. 2001. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 64:95–104 [DOI] [PubMed] [Google Scholar]
- 4. Corsetti A., Settanni L. 2007. Lactobacilli in sourdough fermentation. Food Res. Int. 40:539–558 [Google Scholar]
- 5. Corsetti A., et al. 2007. A taxonomic survey of lactic acid bacteria isolated from wheat (Triticum durum) kernels and non-conventional flours. Syst. Appl. Microbiol. 30:561–571 [DOI] [PubMed] [Google Scholar]
- 6. Corsetti A., Settanni L., Valmorri S., Mastrangelo M., Suzzi G. 2007. Identification of subdominant sourdough lactic acid bacteria and their evolution during laboratory-scale fermentations. Food Microbiol. 24:592–600 [DOI] [PubMed] [Google Scholar]
- 7. De Angelis M., et al. 2006. Selection of potential probiotic lactobacilli isolated from pig feces to be used as additives in pelleted feeding. Res. Microbiol. 157:792–801 [DOI] [PubMed] [Google Scholar]
- 8. Decock P., Cappelle S. 2005. Bread technology and sourdough technology. Trends Food Sci. Technol. 16:113–120 [Google Scholar]
- 9. De Vuyst L., Neysens P. 2005. The sourdough microflora: biodiversity and metabolic interactions. Trends Food Sci. Technol. 16:43–56 [Google Scholar]
- 10. De Vuyst L., Vancanneyt M. 2007. Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 24:120–127 [DOI] [PubMed] [Google Scholar]
- 11. De Vuyst L., Vrancken G., Ravyts F., Rimaux T., Weckx S. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26:666–675 [DOI] [PubMed] [Google Scholar]
- 12. Dommels Y. E. M., et al. 2009. Survival of Lactobacillus reuteri DSM 17938 and Lactobacillus rhamnosus GG in the human gastrointestinal tract with daily consumption of a low-fat probiotic spread. Appl. Environ. Microbiol. 75:6198–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du Plessis E. M., Dicks L. M. T., Vescovo M., Torriani S., Dellaglio F. 1996. Lactobacillus acidophilus and related species: a review. Ann. Microbiol. Enzim. 46:319–340 [Google Scholar]
- 14. Du Toit M., Dicks L. M. T., Holzapfel W. H. 2003. Identification of heterofermentative lactobacilli isolated from pig faeces by numerical analysis of total soluble cell protein and RAPD patterns. Lett. Appl. Microbiol. 37:12–16 [DOI] [PubMed] [Google Scholar]
- 15. Ehrmann M. A., Vogel R. F. 2005. Molecular taxonomy and genetics of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:31–42 [Google Scholar]
- 16. Ferchichi M., Valcheva R., Prévost H., Onno B., Dousset X. 2007. Molecular identification of the microbiota of French sourdough using temporal temperature gradient gel electrophoresis. Food Microbiol. 24:678–686 [DOI] [PubMed] [Google Scholar]
- 17. Gänzle M., Ehmann M., Hammes W. P. 1998. Modeling of the growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of sourdough fermentation. Appl. Environ. Microbiol. 64:2616–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gänzle M. G., Vermeulen N., Vogel R. F. 2007. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 24:128–138 [DOI] [PubMed] [Google Scholar]
- 19. Gänzle M. G., Zhang C., Monang B. S., Lee V., Schwab C. 2009. Novel metabolites from cereal-associated lactobacilli: novel functionalities for cereal products? Food Microbiol. 26:712–719 [DOI] [PubMed] [Google Scholar]
- 20. Gevers D., Huys G., Swings J. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31–36 [DOI] [PubMed] [Google Scholar]
- 21. Gobbetti M., Corsetti A. 1997. Lactobacillus sanfrancisco, a key sourdough lactic acid bacterium: a review. Food Microbiol. 14:175–187 [Google Scholar]
- 22. Gobbetti M. 1998. The sourdough microflora: interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 9:267–274 [Google Scholar]
- 23. Gobbetti M., De Angelis M., Corsetti A., Di Cagno R. 2005. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:57–69 [Google Scholar]
- 24. Groenewald W. H., et al. 2006. Identification of lactic acid bacteria from vinegar flies based on phenotypic and genotypic characteristics. Am. J. Enol. Vit. 57:519–525 [Google Scholar]
- 25. Hammes W. P., et al. 2005. Microbial ecology of cereal fermentations. Trends Food Sci. Technol. 16:4–11 [Google Scholar]
- 26. Hansen A. S. 2004. Sourdough bread, p. 729–755In Hui Y. H., et al. (ed.), Handbook of food and beverage fermentation technology. Marcel Dekker, New York, NY [Google Scholar]
- 27. Iacumin L., et al. 2009. Description of the microflora of sourdoughs by culture-dependent and culture-independent methods. Food Microbiol. 26:128–135 [DOI] [PubMed] [Google Scholar]
- 28. Kim J. F., et al. 2008. Complete genome sequence of Leuconostoc citreum KM20. J. Bacteriol. 190:3093–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kitahara M., Sakata S., Benno Y. 2005. Biodiversity of Lactobacillus sanfranciscensis strains isolated from five sourdoughs. Lett. Appl. Microbiol. 40:353–357 [DOI] [PubMed] [Google Scholar]
- 30. Lee J. S., et al. 2005. Analysis of kimchi microflora using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 102:143–150 [DOI] [PubMed] [Google Scholar]
- 31. Lin W. H., Yu B., Jang S. H., Tsen H. Y. 2007. Different probiotic properties for Lactobacillus fermentum strains isolated from swine and poultry. Anaerobe 13:107–113 [DOI] [PubMed] [Google Scholar]
- 32. Makras L., Van Acker G., De Vuyst L. 2005. Lactobacillus paracasei subsp. paracasei 8700:2 degrades inulin-type fructans exhibiting different degrees of polymerization. Appl. Environ. Microbiol. 71:6531–6537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meroth C. B., Hammes W. P., Hertel C. 2003. Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:7453–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meroth C. B., Walter J., Hertel C., Brandt M., Hammes W. P. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogier J.-C., Son O., Gruss A., Tailliez P., Delacroix-Buchet A. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ottogalli G., Galli A., Foschino R. 1996. Italian bakery products obtained with sour dough: characterization of the typical microflora. Adv. Food Sci. 18:131–144 [Google Scholar]
- 37. Ough C. S., Huang Z., Stevens D. 1991. Amino acid uptake by 4 commercial yeasts at 2 different temperatures of growth and fermentation: effects on urea excretion and reabsorption. Am. J. Enol. Vit. 42:26–40 [Google Scholar]
- 38. Picozzi C., D'Anchise F., Foschino R. 2006. PCR detection of Lactobacillus sanfranciscensis in sourdough and Panettone baked product. Eur. Food Res. Technol. 222:330–335 [Google Scholar]
- 39. Pitcher D. G., Saunders N. A., Owen R. J. 1989. Rapid extraction of bacterial genomic DNA with guanidinium thiocyanate. Lett. Appl. Microbiol. 8:151–156 [Google Scholar]
- 40. Rehman S. U., Paterson A., Piggott J. R. 2006. Flavour in sourdough breads: a review. Trends Food Sci. Technol. 17:557–566 [Google Scholar]
- 41. Rocha J. M., Malcata X. F. 1999. On the microbial profile of traditional Portuguese sourdough. J. Food Prot. 62:1416–1429 [DOI] [PubMed] [Google Scholar]
- 42. Scheirlinck I., et al. 2007. Influence of geographical origin and flour type on diversity of lactic acid bacteria in traditional Belgian sourdoughs. Appl. Environ. Microbiol. 73:6262–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scheirlinck I., et al. 2008. Taxonomic structure and stability of the bacterial community in Belgian sourdough ecosystems as assessed by culture and population fingerprinting. Appl. Environ. Microbiol. 74:2414–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siragusa S., et al. 2009. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl. Environ. Microbiol. 75:1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sterr Y., Weiss A., Schmidt H. 2009. Evaluation of lactic acid bacteria for sourdough fermentation of amaranth. Int. J. Food Microbiol. 136:75–82 [DOI] [PubMed] [Google Scholar]
- 46. Temmerman R., Scheirlinck I., Huys G., Swings J. 2003. Culture-dependent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vancanneyt M., et al. 2006. Reclassification of Lactobacillus brevis strains LMG 11494 and LMG 11984 as Lactobacillus parabrevis sp. nov. Int. J. Syst. Evol. Microbiol. 56:1553–1557 [DOI] [PubMed] [Google Scholar]
- 48. Van Coillie E., et al. 2007. Identification of lactobacilli isolated from the cloaca and vagina of laying hens and characterization for potential use as probiotics to control Salmonella enteritidis. J. Appl. Microbiol. 102:1095–1106 [DOI] [PubMed] [Google Scholar]
- 49. Van der Meulen R., et al. 2007. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl. Environ. Microbiol. 73:4741–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Velasco I., Tenreiro S., Calderon I. L., André B. 2004. Saccharomyces cerevisiae Aqr1 is an internal-membrane transporter involved in excretion of amino acids. Eukaryot. Cell 3:1492–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vogelmann S. A., Seitter M., Singer U., Brandt M. J., Hertel C. 2009. Adaptability of lactic acid bacteria and yeasts to sourdoughs prepared from cereals, pseudocereals and cassava and use of competitive strains as starters. Int. J. Food Microbiol. 130:205–212 [DOI] [PubMed] [Google Scholar]
- 52. Vrancken G., Rimaux T., De Vuyst L., Leroy F. 2008. Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128:58–66 [DOI] [PubMed] [Google Scholar]
- 53. Vrancken G., Rimaux T., Weckx S., De Vuyst L., Leroy F. 2009. Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int. J. Food Microbiol. 135:216–222 [DOI] [PubMed] [Google Scholar]
- 54. Vrancken G., Rimaux T., Wouters D., Leroy F., De Vuyst L. 2009. The arginine deiminase pathway of Lactobacillus fermentum IMDO 130101 responds to growth under stress conditions of both temperature and salt. Food Microbiol. 26:720–727 [DOI] [PubMed] [Google Scholar]
- 55. Vrancken G., et al. 2010. Yeast species compostion differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 10:471–481 [DOI] [PubMed] [Google Scholar]
- 56. Walker G. M. 1998. Yeast physiology and biotechnology. John Wiley & Sons, Inc., San Francisco, CA [Google Scholar]
- 57. Weckx S., et al. 2010. Community dynamics of bacteria in sourdough fermentations as revealed by their metatranscriptome. Appl. Environ. Microbiol. 76:5402–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weckx S., et al. 2010. Lactic acid bacteria community dynamics and metabolite production of rye sourdough fermentations share characteristics of wheat and spelt sourdough fermentations. Food Microbiol. 27:1000–1008 [DOI] [PubMed] [Google Scholar]
- 59. Yim C. Y., Eom H. J., Jin Q., Kim S. Y., Han N. S. 2008. Characterization of low temperature-adapted Leuconostoc citreum HJ-P4 and its dextransucrase for the use of kimchi starter. Food Sci. Biotechnol. 17:1391–1395 [Google Scholar]
- 60. Zannini E., et al. 2009. Microbiological and technological characterization of sourdoughs destined for bread-making with barley flour. Food Microbiol. 26:744–753 [DOI] [PubMed] [Google Scholar]
- 61. Zotta T., Piraino P., Parente E. 2008. Characterization of lactic acid bacteria isolated from sourdoughs for Cornetto, a traditional bread produced in Basilicata (Southern Italy). World J. Microbiol. Biotechnol. 24:1785–1795 [Google Scholar]