Abstract
We studied how soil pH (pHs 4 to 8) influenced the mineralization of low-molecular-weight (LMW)-dissolved organic carbon (DOC) compounds, and how this compared with differences in microbial community structure. The mineralization of LMW-DOC compounds was not systematically connected to differences in soil pH, consistent with soil respiration. In contrast, the microbial community compositions differed dramatically. This suggests that microbial community composition data will be of limited use in improving the predictive power of soil C models.
The primary connection between soil microbial activity and carbon (C) cycling is well established (29, 46, 51); however, there has recently been a call for a more detailed understanding and integration of the microbial community into models for soil organic C (SOC) cycling to improve their predictive power (5, 6, 10, 11, 20, 29, 35, 44, 47). To accomplish this, we need to study the drivers of respiration and composition of microbial communities together (36).
Low-molecular-weight (LMW)-C compounds dominate the C used in soil respiration (13, 18, 22, 31, 48, 49, 50). Although observations regarding factors that affect LMW-DOC cycling have started to emerge (13, 23, 31), the canonical environmental factors influencing their mineralization remain unclear. This study's aims were to test how one of the most influential factors for the composition of the soil microbial community, soil pH (4, 8, 26, 32, 39, 40, 41, 42), influenced the mineralization of LMW-C compounds across a wide pH gradient and to compare our findings with microbial community composition.
Soil was obtained from the Hoosfield pH gradient at Rothamsted Research, United Kingdom (1, 2, 3, 7, 39). Thirty topsoil samples (0- to 23-cm depth) were tested along the gradient in March 2010, sieved (<2 mm), and characterized (39). The 30 individual soil samples were used for microbial and chemical analyses (e.g., organic C, total N, pH, phospholipid fatty acid [PLFA] composition, and bacterial growth), while the gradient was split into four pH levels for the soil solution analysis and C substrate mineralization assays. For these, independent replicates (n = 3) were used for each of the four pH levels (pH 4.1 ± 0.04, pH 5.0 ± 0.07, pH 6.0 ± 0.05, and pH 7.1 ± 0.08). Soil solution was extracted by centrifugal drainage (19, 43; see also Supplement S1 in the supplemental material), and the free amino acid and sugar concentrations of the solutions were determined (21, 30).
Soil (5 g) from each pH level was weighed into polypropylene tubes. Soil solution from each replicate (450 μl) was then individually spiked with one of eight different 14C-labeled substrates (50 μl) at a trace level and added to the soil and the mineralization monitored using 1 M NaOH CO2 traps at 22°C for 7 days (see Supplement S1 in the supplemental material). The 14CO2 level in the NaOH traps was determined by liquid scintillation. Sorption of the added 14C LMW-DOC compounds across the pH gradient was determined in sterilized soil samples (see Supplement S1). Bacterial growth was estimated using leucine incorporation (9, 25), and microbial community structure was determined from PLFA patterns (16, 17, 32).
14C substrate mineralization and half-lives (t1/2) were modeled by fitting a double-first-order decay equation to the experimental results (13, 23, 33, 48, 49; see also Supplement S1 in the supplemental material). The PLFA composition (mol% of the 26 most abundant PLFAs) was analyzed by principal-component analysis (PCA) after values were standardized to unit variance. Analysis of variance (ANOVA) with Tukey's honestly significant difference test post hoc comparisons were used to determine differences in variables with soil pH, while regression analyses were used to describe relationships with pH across all 30 samples. Results were compared to those for an identical sampling of the pH gradient from 2008 (40). In addition, a 454-pyrosequencing-based analysis of the bacterial community composition was also previously performed on the same 2008 samples (42; based on analyses from references 14, 27, 28, and 34). A type II major-axis regression analysis was used to investigate the connection between the sequence composition (the principal coordinate of variation of the sequence composition [see Supplement S1]) of the bacterial community with microbial PLFA composition.
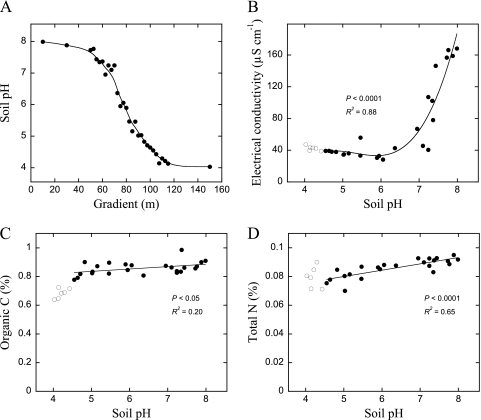
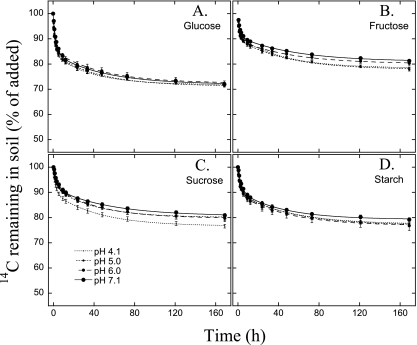
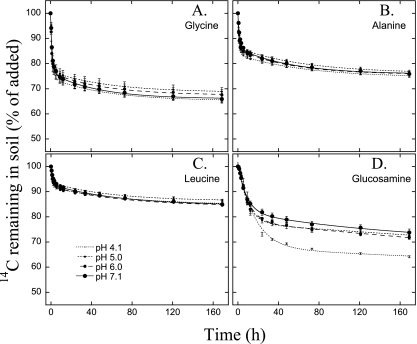
Soil measurements showed a smooth pH gradient between pH 4 and 8 with only small differences in most other chemical variables (39) (Fig. 1). DOC and dissolved organic nitrogen (DON) concentrations did not systematically change over the pH gradient (see Table S1 in the supplemental material). The DOC and DON concentrations derived from both sugars and amino acids were similar over the gradient (P > 0.05) (Table S1), averaging 53.5 ± 7.1 nmol of C derived from sugar (sugar C) g−1 soil and 11.4 ± 2.1 nmol amino acid C g−1 soil. The mineralization patterns conformed well to a double-first-order kinetic model (R2 > 0.99 for all substrates) (Fig. 2 and 3). The individual amino acids and sugars degraded at different rates (Fig. 2); however, this did not systematically change with pH (see Table S2 in the supplemental material).
Fig. 1.
Soil pH along the Hoosfield acid strip (A), the effect of soil pH on electrical conductivity (B), the organic C (C), and total N (D). Data points below pH 4.5 (open symbols) were not used in the regression analyses (see Materials and Methods).
Fig. 2.
Mineralization of sugars. Amounts of 14C-labeled glucose (A), fructose (B), sucrose (C), and starch (D) remaining in soils of the four different pH levels after the injection of the isotopically labeled soil solution into the soil. Values represent means ± 1 SE (n = 3). The lines represent the best fits of double-first-order exponential-decay functions (R2 > 0.99 for all curve fits).
Fig. 3.
Mineralization of amino acids and amino sugars. Amounts of 14C-labeled glycine (A), alanine (B), leucine (C), and glucosamine (D) remaining in soils of the four different pH levels after the injection of the isotopically labeled soil solution into the soil. Values represent means ± 1 SE (n = 3). The lines represent the best fits of double-first-order exponential-decay functions (R2 > 0.99 for all curve fits).
Combining our estimates of free sugar and amino acid concentrations with estimates of their turnover rate enabled estimation of the contribution of LMW-C mineralization to soil respiration. We estimate that approximately 0.32 and 0.09 μg of C derived from CO2 (CO2 C) g−1 h−1 of soil respiration were derived from sugar and amino acid degradation, respectively. Taken together, this is within the range of previous assessments of the basal respiration rate at the site (0.20 to 0.50 μg CO2 C h−1 g−1) (3, 39, 41).
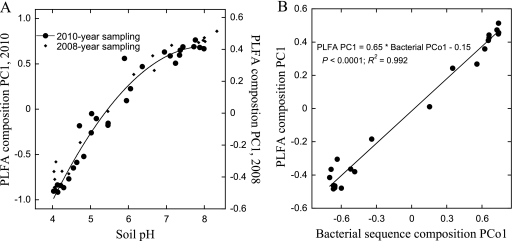
Bacterial growth increased about 6-fold between pH 4.5 and 8.0 (P < 0.0001) (see Fig. S1 in the supplemental material). The first component from a PCA of the microbial PLFA composition explained 40% of the variation in the data and was closely related to soil pH (P < 0.0001, R2 = 0.96) (see Fig. S2A in the supplemental material). The variation of microbial PLFA composition across the Hoosfield site was highly reproducible, with the major component of variation from a PCA aligning identically with soil pH (Fig. 4A). There was also a very close relationship between the variation in the sequence composition of the bacterial community (42) (Fig. 4B).
Fig. 4.
(A) Comparison of the levels of pH dependence of the microbial PLFA compositions across the Hoosfield acid strips in the soil samples of the present study compared to those sampled in 2008 (data are from reference 40); (B) first principal component (PC1) of the variation in the PLFA compositions (explaining 39.6% of the variation in PLFA composition) across the Hoosfield acid strip from the 2008 sampling (data are from reference 40) compared with the first component of a principal-coordinate (PCo1) analysis (explaining 29.1% of the total sequence variation) of the compositions of bacterial sequences obtained with a 454-pyrosequencing assessment (data are from reference 42). A type II major-axis regression was used to fit the line.
The Hoosfield site effectively isolated soil pH from most other variables associated with microbial community differences. In addition, it was previously shown that excess Al (or other unidentified inhibitors) or a lack of P or N did not limit bacterial and fungal growth across the pH gradient (41). Soluble LMW-C compound concentrations were also not systematically affected by pH. Similarly, rates of turnover of individual LMW-C compounds did not change consistently with pH. In contrast, the microbial community composition radically differed along the pH gradient. Direct comparison between the microbial PLFA composition and its sequence composition (obtained by 454 pyrosequencing [42]) (Fig. 4B) indicated that the difference in microbial PLFA composition across the gradient was a consequence of a difference in bacterial species composition. The enormous pH-related differences in microbial community composition did not affect the mineralization of LMW C, the dominant source of soil respiration, across the pH gradient. Therefore, we could not find support for a connection between microbial community structure and function (the turnover of the LMW-DOC compounds that govern soil respiration). This is consistent with conjectures derived from theoretical models of soil organic matter (SOM) turnover that have suggested high redundancy in the microbial processing of LMW C (12, 24, 45). We cannot discount the possibility that processes transforming SOM to LMW DOC are closely related to microbial community structure. However, there was only a minor discrepancy between basal respiration (30% difference across the gradient) and LMW-DOC mineralization across the gradient (no difference across the gradient), setting the upper limit for the functional relevance of the microbial community difference. This does not lessen the need to identify and incorporate the rate-limiting mechanisms in models for SOC turnover (6, 29), but it does suggest that adding information about the microbial community composition may be of limited use.
Supplementary Material
Acknowledgments
J.R. was supported by a Swedish Research Council (Vetenskapsrådet) Postdoctoral Fellowship (project no. 623-2009-7343), and D.L.J. was supported by funds from the Natural Environment Research Council (NERC), United Kingdom. Rothamsted Research is an institute of the Biotechnology and Biological Sciences Research Council (BBSRC), United Kingdom.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Aciego-Pietri J. C. 2001. Effects of soil pH on microbial biomass, microbial activity and microbial community structure. Ph.D. thesis. University of Nottingham, Nottingham, United Kingdom [Google Scholar]
- 2. Aciego Pietri J. C., Brookes P. C. 2008. Nitrogen mineralisation along a pH gradient of a silty loam UK soil. Soil Biol. Biochem. 40:797–802 [Google Scholar]
- 3. Aciego Pietri J. C., Brookes P. C. 2008. Relationships between soil pH and microbial properties in a UK arable soil. Soil Biol. Biochem. 40:1856–1861 [Google Scholar]
- 4. Aciego Pietri J. C., Brookes P. C. 2009. Substrate inputs and pH as factors controlling microbial biomass, activity and community structure in an arable soil. Soil Biol. Biochem. 41:1396–1405 [Google Scholar]
- 5. Allison S. D., Treseder K. K. 2008. Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Global Change Biol. 14:2898–2909 [Google Scholar]
- 6. Allison S. D., Wallenstein M. D., Bradford M. A. 2010. Soil-carbon response to warming dependent on microbial physiology. Nat. Geosci. 3:336–340 [Google Scholar]
- 7. Avery B. W., Catt J. A. 1995. The soil at Rothamsted, Lawes Agricultural Trust. IACR-Rothamsted, Harpenden, Hertfordshire, United Kingdom [Google Scholar]
- 8. Bååth E., Anderson T. H. 2003. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35:955–963 [Google Scholar]
- 9. Bååth E., Pettersson M., Söderberg K. H. 2001. Adaptation of a rapid and economical microcentrifugation method to measure thymidine and leucine incorporation of soil bacteria. Soil Biol. Biochem. 33:1571–1574 [Google Scholar]
- 10. Barcenas-Moreno G., Gomez-Brandon M., Rousk J., Bååth E. 2009. Adaptation of soil microbial communities to temperature: comparison of fungi and bacteria in a laboratory experiment. Global Change Biol. 15:2950–2957 [Google Scholar]
- 11. Bardgett R. D., Freeman C., Ostle N. J. 2008. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2:805–814 [DOI] [PubMed] [Google Scholar]
- 12. Bengtson P., Bengtsson G. 2007. Rapid turnover of DOC in temperature forests accounts for increased CO2 production at elevated temperatures. Ecol. Lett. 10:783–790 [DOI] [PubMed] [Google Scholar]
- 13. Boddy E., Hill P. W., Farrar J., Jones D. L. 2007. Fast turnover of low molecular weight components of the dissolved organic carbon pool of temperate grassland field soils. Soil Biol. Biochem. 39:827–835 [Google Scholar]
- 14. Fierer N., Hamady M., Lauber C. L., Knight R. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:17994–17999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reference deleted.
- 16. Frostegård Å., Tunlid A., Bååth E. 1993. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 59:3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frostegård Å., Bååth E. 1996. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fert. Soils 22:59–65 [Google Scholar]
- 18. Fujii K., Hayakawa C., van Hees P. A. W., Funakawa S., Kosaki T. 2010. Biodegradation of low molecular weight organic compounds and their contribution to heterotrophic soil respiration in three Japanese forest soils. Plant Soil 334:475–489 [Google Scholar]
- 19. Giesler R., Lundström U. 1993. Soil solution chemistry: effects of bulking soil samples. Soil Sci. Soc. Am. 57:1283–1288 [Google Scholar]
- 20. Heimann H., Reichstein M. 2008. Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 451:289–292 [DOI] [PubMed] [Google Scholar]
- 21. Jones D. L., Owen A. G., Farrar J. F. 2002. Simple method to enable the high resolution determination of total free amino acids in soil solutions and soil extracts. Soil Biol. Biochem. 34:1893–1902 [Google Scholar]
- 22. Jones D. L., Hodge A., Kuzyakov Y. 2004. Plant and mycorrhizal regulation of rhizodeposition. New Phytol. 163:459–480 [DOI] [PubMed] [Google Scholar]
- 23. Jones D. L., et al. 2005. Rapid intrinsic rates of amino acid biodegradation in soils are unaffected by agricultural managements strategy. Soil Biol. Biochem. 37:1267–1275 [Google Scholar]
- 24. Kemmitt S. J., et al. 2008. Mineralization of native soil organic matter is not regulated by the size, activity or composition of the soil microbial biomass—a new perspective. Soil Biol. Biochem. 40:61–73 [Google Scholar]
- 25. Kirchman D., K'nees E., Hodson R. 1985. Leucine incorporation and its potential as a measure of protein-synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lauber C. L., Hamady M., Knight R., Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lozupone C., Hamady M., Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lozupone C., Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McGuire K. L., Treseder K. K. 2010. Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol. Biochem. 42:529–535 [Google Scholar]
- 30. Myklestad S. M., Skanoy E., Hestmann S. 1997. A sensitive and rapid method for analysis of dissolved mono- and polysaccharides in seawater. Mar. Chem. 56:279–286 [Google Scholar]
- 31. Nambu K., van Hees P. A. W., Jones D. L., Vinogradoff S., Lundström U. S. 2008. Composition of organic solutes and respiration in soils derived from alkaline and nonalkaline parent materials. Geoderma 144:468–477 [Google Scholar]
- 32. Nilsson L.-O., Bååth E., Falkengren-Grerup U., Wallander H. 2007. Growth of ectomycorrhizal mycelia and composition of soil microbial communities in oak forest soils along a nitrogen deposition gradient. Oecologia 153:375–385 [DOI] [PubMed] [Google Scholar]
- 33. Paul E. A., Clarke F. E. 1996. Soil microbiology and biochemistry, 2nd ed Academic Press, New York, NY [Google Scholar]
- 34. Price M. N., Dehal P. S., Arkin A. P. 2009. FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rinnan R., Michelsen A., Bååth E., Jonasson S. 2007. Fifteen years of climate change manipulations alter soil microbial communities in a subarctic heath ecosystem. Global Change Biol. 13:28–39 [Google Scholar]
- 36. Rinnan R., Rousk J., Yergeau E., Kowalchuk G. A., Bååth E. 2009. Temperature adaptation of soil bacterial communities along an Antarctic climate gradient: predicting responses to climate warming. Global Change Biol. 15:2615–2625 [Google Scholar]
- 37. Reference deleted.
- 38. Reference deleted.
- 39. Rousk J., Brookes P. C., Bååth E. 2009. Contrasting soil pH effects on fungal and bacterial growth suggests functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 75:1589–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rousk J., Brookes P. C., Bååth E. 2010. The microbial PLFA composition as affected by pH in an arable soil. Soil Biol. Biochem. 42:516–520 [Google Scholar]
- 41. Rousk J., Brookes P. C., Bååth E. 2010. Investigating the opposing pH relationships of fungal and bacterial growth in soil. Soil Biol. Biochem. 42:926–934 [Google Scholar]
- 42. Rousk J., et al. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351 [DOI] [PubMed] [Google Scholar]
- 43. Rousk J., Jones D. L. 2010. Loss of low molecular weight dissolved organic carbon (DOC) and nitrogen (DON) in H2O and 0.5 M K2SO4 soil extracts. Soil Biol. Biochem. 42:2331–2335 [Google Scholar]
- 44. Schimel J. P., Balser T. C., Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88:1386–1394 [DOI] [PubMed] [Google Scholar]
- 45. Schimel J. P., Bennett J. 2004. Nitrogen mineralization: challenges of a changing paradigm. Ecology 85:591–602 [Google Scholar]
- 46. Schlesinger W. H. 1997. Biogeochemistry: an analysis of global change. Academic Press, San Diego, CA [Google Scholar]
- 47. Strickland M. S., Lauber C., Fierer N., Bradford M. A. 2009. Testing the significance of microbial community composition. Ecology 90:441–451 [DOI] [PubMed] [Google Scholar]
- 48. van Hees P. A. W., Jones D. L., Finlay R., Godbold D. L., Lundström U. S. 2005. The carbon we do not see—the impact of low molecular weight compounds on carbon dynamics and respiration in forest soils: a review. Soil Biol. Biochem. 37:1–13 [Google Scholar]
- 49. van Hees P. A. W., et al. 2005. Modelling low molecular weight organic acid dynamics in forest soils. Soil Biol. Biochem. 37:517–531 [Google Scholar]
- 50. van Hees P. A. W., Johansson E., Jones D. L. 2008. Dynamics of simple carbon compounds in two forest soils as revealed by soil solution concentrations and biodegradation kinetics. Plant Soil 310:11–23 [Google Scholar]
- 51. Waksman S. A., Tenney F. G., Stevens K. R. 1928. The role of microorganisms in the transformation of organic matter in forest soils. Ecology 9:126–144 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.