Abstract
The objective of this study was to systematically evaluate and compare the effects of select antimethanogen compounds on methane production, feed digestion and fermentation, and populations of ruminal bacteria and methanogens using in vitro cultures. Seven compounds, including 2-bromoethanesulphonate (BES), propynoic acid (PA), nitroethane (NE), ethyl trans-2-butenoate (ETB), 2-nitroethanol (2NEOH), sodium nitrate (SN), and ethyl-2-butynote (EB), were tested at a final concentration of 12 mM. Ground alfalfa hay was included as the only substrate to simulate daily forage intake. Compared to no-inhibitor controls, PA, 2NEOH, and SN greatly reduced the production of methane (70 to 99%), volatile fatty acids (VFAs; 46 to 66%), acetate (30 to 60%), and propionate (79 to 82%), with 2NEOH reducing the most. EB reduced methane production by 23% without a significant effect on total VFAs, acetate, or propionate. BES significantly reduced the propionate concentration but not the production of methane, total VFAs, or acetate. ETB or NE had no significant effect on any of the above-mentioned measurements. Specific quantitative-PCR (qPCR) assays showed that none of the inhibitors significantly affected total bacterial populations but that they did reduce the Fibrobacter succinogenes population. SN reduced the Ruminococcus albus population, while PA and 2NEOH increased the populations of both R. albus and Ruminococcus flavefaciens. Archaeon-specific PCR-denaturing gradient gel electrophoresis (DGGE) showed that all the inhibitors affected the methanogen population structure, while archaeon-specific qPCR revealed a significant decrease in methanogen population in all treatments. These results showed that EB, ETB, NE, and BES can effectively reduce the total population of methanogens but that they reduce methane production to a lesser extent. The results may guide future in vivo studies to develop effective mitigation of methane emission from ruminants.
INTRODUCTION
Methane (CH4) emissions from ruminants can result in a significant loss of feed efficiency: up to a 12% loss of gross energy intake for forage-fed cattle and 4% for concentrate-fed cattle (14). Because methane is 25 times more potent than carbon dioxide as a greenhouse gas (11), methane emitted from ruminants amounted to 141 teragrams of CO2 equivalents (Tg CO2 eq), accounting for 25% of total methane emissions from anthropogenic activities in the United States in 2008 (26). To mitigate the negative impact on climate change and to improve feed efficiency, numerous strategies for reducing methane emission from ruminant livestock have been tested. Plant extracts (7, 9), vaccines (28), ionophores (27), and dietary strategies (21) have been evaluated for their efficacy in reducing ruminal methane emission. However, only monensin has been used in animal-feeding operations, and it typically achieves only transient reductions in methane production (12). More importantly, the monensin-driven reduction in methane reduction is largely attributable to decreased feed digestibility (4, 19).
Recent studies showed that some nitrocompounds (2, 3, 5), lauric acid and monolaurin (Lauricidin) (5), and 2-bromochloromethane (8) can be more potent than the aforementioned substances in reducing methane production in in vitro cultures. These studies also documented changes in fermentation and profiles of volatile fatty acids (VFAs). Conceivably, these antimethanogen compounds can affect both ruminal bacteria and archaea, but such potential effects have not been reported. In this study, we systematically evaluated and compared the potencies of seven inhibitors in reducing methane production, which include 2-bromoethanesulphonate (BES), propynoic acid (PA), nitroethane (NE), ethyl trans-2-butenoate (ETB), 2-nitroethanol (2NEOH), sodium nitrate (SN), and ethyl-2-butynote (EB), using in vitro ruminal cultures. Their impact on the major cultured cellulolytic bacteria (i.e., Fibrobacter succinogenes, Ruminococcus albus, and Ruminococcus flavefaciens) and methanogens was also examined.
MATERIALS AND METHODS
In vitro ruminal cultures.
Ruminal fluid was collected from a cannulated Jersey bull fed rye grass before its morning feeding. The fluid was filtered through four layers of sterilized cheesecloth and clarified by centrifugation at 4°C and 10,000 × g for 20 min. The medium consisted of the clarified rumen fluid and artificial saliva in a 1:2 ratio (17). Each culture tube received 9 ml medium and 1 ml fresh ruminal fluid (obtained from the same bull) as an inoculum. Finely ground alfalfa hay, which is one of the most common forages for dairy cattle, was added (0.2 g dry mass [DM] per culture tube) as the only forage substrate (2, 3, 5). Each of the following inhibitors was added to a final concentration of 12 mM: 2-bromoethanesulfonate (BES), propynoic acid (PA; as free acid), sodium nitroethane (NE), ethyl trans-2-butenoate (ETB), 2-nitroethanol (2NEOH), sodium nitrate (SN), and ethyl-2-butynoate (EB). The control cultures were set up in the same way as the treatments but received no inhibitor. Each treatment and control culture was prepared in triplicate. All the cultures were prepared in an anaerobic chamber containing N2 (85%), H2 (10%), and CO2 (15%). Each culture tube was fitted with a butyl rubber stopper that was fastened with an aluminum crimp. All the cultures were incubated at 39°C for 48 h without agitation.
Biogas and VFA analyses.
After 48 h of incubation, the gas volume produced in each tube was determined by volume displacement using a system consisting of a needle, tubing, and another 20-ml culture tube. Composition of the biogas collected from the headspace and concentrations of VFAs in the cultures were determined using gas chromatography as described previously (20).
PCR, DGGE, and quantitative PCR (qPCR).
Metagenomic DNA was extracted using the repeated bead beating and column purification method (33), which results in efficient recovery of PCR-quality DNA from microbiome samples. The quality of the DNA was assessed using agarose gel (1.0%) electrophoresis, and DNA concentrations were determined using a Quant-iT kit (Invitrogen, Carlsbad, CA). The primers and their corresponding annealing temperatures used in this study are listed in Table 1. PCR-denaturing gradient gel electrophoresis (DGGE) for bacteria and archaea was done using a 40 to 60% denaturant gradient essentially as described previously (30, 32). To eliminate the potential impact of different amounts of DNA template on DGGE profiles, 100 ng metagenomic DNA was used in each PCR mixture, and the same volumes of PCR products were resolved on DGGE gels for all the cultures. A 30-min final elongation step was added to the PCR to eliminate artifactual double DGGE bands (13). After normalization of the gels against ladders, only those bands with a peak intensity exceeding 2.0% of the strongest band in each lane were included in further analyses.
Table 1.
PCR primers, targeted hypervariable regions, annealing temperatures, and amplicon length
| Primer | Target taxon | Sequence (5′→3′) | Target region(s) | Annealing temp (°C) | Amplicon length (bp) | Reference |
|---|---|---|---|---|---|---|
| GC-A357fa | Bacteria | CCC TAC GGG GCG CAG CAG | V3 | 56 | 194 | 32 |
| 519r | GWA TTA CCG CGG CKG CTG | |||||
| GC-RC344fa | Archaea | ACG GGG YGC AGC AGG CGC GA | V3 | 56 | 201 | 30 |
| 519r | GWA TTA CCG CGG CKG CTG | |||||
| Eub358f | TCC TAC GGG AGG CAG CAG T | V3-V4 | 60 | 467 | ||
| Eub806r | GGA CTA CCA GGG TAT CTA ATC CTG TT | 22 | ||||
| TaqMan probe | Bacteria | 6-FAM-5′-CGT ATT ACC GCG GCT GCT GGC AC-3′-TAMRAb | 70 | |||
| ARC787F | Archaea | ATT AGA TAC CCS BGT AGT CC | V3-V4 | 60 | 272 | 29 |
| ARC1059R | GCC ATG CAC CWC CTC T | |||||
| Rf154f-K | Ruminococcus flavefaciens | TCT GGA AAC GGA TGG TA | V3-V4 | 55 | 295 | 16 |
| Rf425r-K | CCT TTA AGA CAG GAG TTT ACA A | |||||
| Fs-f | Fibrobacter succinogenes | GGT ATG GGA TGA GCT TGC | V3-V4 | 63 | 446 | 16 |
| Fs-r | GCC TGC CCC TGA ACT ATC | |||||
| Ra1281f | Ruminococcus albus | CCC TAA AAG CAG TCT TAG TTC G | V3-V4 | 55 | 175 | 16 |
| Ra1439r | CCT CCT TGC GGT TAG AAC A |
A primer with a 40-bp GC clamp (CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G) attached to the 5′ end was used in PCR-DGGE.
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
The population sizes of total bacteria, total archaea, and Fibrobacter succinogenes, Ruminococcus albus, and R. flavefaciens cultures were quantified using qPCR assays with respective specific primers and probes (Table 1). The qPCR standard for F. succinogenes was prepared by PCR using its specific primers and genomic DNA from F. succinogenes S85. One sample-derived qPCR standard each was prepared for the other species, total bacteria, and total archaea using respective specific PCR primers and a composite DNA sample that was pooled from equal amounts of metagenomic DNAs extracted from all the cultures as described previously (6, 31). For each of the standards, copy number concentration was calculated based on the length of the PCR product and the mass concentration. Tenfold serial dilutions were made in Tris-EDTA (TE) buffer prior to qPCR assays.
The conditions of the qPCR assays were the same as reported previously (6, 31) except for the primer annealing temperature (Table 1). All the qPCR assays were performed using an Mx3000p qPCR system (Stratagene, La Jolla, CA). Fluorescence resulting from possible primer dimers was excluded by using the fluorescence signal that was acquired at 86°C, at which temperature primer dimers were completely denatured, as verified by melting curve analysis (31). Following qPCR, the amplicon products were confirmed by agarose gel (1.2%) electrophoresis. To minimize variations, the qPCR assay for each species or group was done in triplicate for both the standards and the metagenomic DNA samples using the same master mix and the same PCR plate.
Statistical analysis.
The data were analyzed using the General Line Model Procedure of SAS 8.1 (SAS Institute, Cary, NC). Means separation was conducted using the Student-Newman-Keuls test of SAS, with significance declared at a P of ≤0.05. The population size of each microbial species or group was expressed as the number of 16S rRNA gene copies per ml of culture.
RESULTS
Effects of inhibitors on total biogas and methane production.
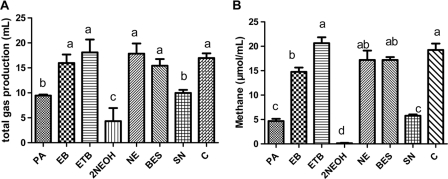
After 48 h of incubation of the in vitro ruminal cultures, biogas production was significantly inhibited in the PA, 2NEOH, and SN treatments, with 2NEOH inhibiting the most; other inhibitors did not significantly reduce biogas production (Fig. 1A). The inhibition of methane production followed the same pattern as the inhibition of biogas, except with EB, which also reduced methane production, though the magnitude was much smaller than with PA, 2NEOH, and SN (Fig. 1B). Specifically, relative to the methane produced in the no-inhibitor control, methane production was reduced by treatment with PA by 75.7%, with EB by 23.3%, with 2NEOH by 99.3%, and with SN by 70.1%. The remaining inhibitors tested did not significantly affect methane production in the in vitro cultures.
Fig. 1.
Amounts of biogas (A) and methane (B) produced per ml of in vitro culture after 48 h of incubation. PA, propynoic acid; BES, 2-bromoethanesulphonate; NE, nitroethane; ETB, ethyl trans-2-butenoate; 2NEOH, 2-nitroethanol; SN, sodium nitrate; EB, ethyl 2-butynote; C, control containing no inhibitor. Error bars indicate standard deviations (n = 3), with different letters designating significant differences (P < 0.05).
Effects of inhibitors on VFA production.
Analysis of VFAs at the end of the 48 h of incubation revealed significantly reduced production of total VFAs in the PA, 2NEOH, and SN cultures but not in the EB, ETB, BES, or NE cultures, compared to that of the no-inhibitor control (Table 2). The production of major individual VFAs was affected as well; PA, 2NEOH, and SN reduced the production of all the individual VFAs analyzed; EB reduced only valerate, isobutyrate, and isovalerate; NE reduced valerate only; BES reduced propionate and valerate; and ETB did not reduce any of the VFAs analyzed. Compared to the no-inhibitor control, the PA, 2NEOH, BES, and SN treatments significantly elevated the acetate/propionate ratio but to different magnitudes, with SN increasing it the most and BES the least. No significant difference in acetate/propionate ratios was seen among the no-inhibitor control and the EB, ETB, and NE treatments.
Table 2.
Effects of antimethanogenic compounds on VFA concentrations in the ruminal cultures
| VFA(s) | Mean concn (μmol ml−1) witha: |
SEM | P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PA | EB | ETB | 2NEOH | NE | BES | SN | C | |||
| Acetate | 41.58 c | 72.85 a,b | 68.41 a,b | 26.50 d | 65.44 b | 78.18 a | 46.74 c | 66.75 a,b | 2.87 | <0.0001 |
| Propionate | 5.67 c | 27.44 a | 27.20 a | 4.93 c | 26.18 a | 17.69 b | 5.49 c | 26.94 a | 0.94 | <0.0001 |
| Butyrate | 3.20 b | 7.26 a | 8.84 a | 2.69 b | 7.39 a | 8.44 a | 2.94 b | 7.33 a | 0.40 | <0.0001 |
| Valerate | 0.24 b,c | 0.33 b,c | 0.38 a,b | 0.20 c | 0.28 b,c | 0.24 b,c | 0.20 c | 0.47 a | 0.03 | 0.0003 |
| Isobutyrate | 0.33 c | 0.45 b,c | 0.64 a,b,c | 0.27 c | 0.80 a,b | 0.65 a,b,c | 0.27 c | 0.87 a | 0.11 | 0.0008 |
| Isovalerate | 0.26 b | 0.33 b | 0.45 a,b | 0.21 b | 0.80 a | 0.68 a,b | 0.19 b | 0.81 a | 0.11 | 0.0017 |
| Total VFAs | 51.28 b | 108.66 a | 105.92 a | 34.80 c | 100.89 a | 105.88 a | 55.84 b | 103.17 a | 3.96 | <0.0001 |
| Acetate/propionate | 7.32 b | 2.66 e | 2.52 e | 5.37 c | 2.51 e | 4.43 d | 8.57 a | 2.48 e | 0.14 | <0.0001 |
PA, propynoic acid; EB, ethyl-2-butynote; ETB, ethyl trans-2-butenoate; 2NEOH, 2-nitroethanol; NE, nitroethane; BES, 2-bromoethanesulphonate; SN, sodium nitrate (SN); C, control containing no inhibitor. Means within rows with different letters differ significantly (P < 0.05).
Effects of inhibitors on bacteria and methanogens.
In agreement with the total biogas production, DGGE profiling revealed similar microbial communities among the NE, EB, BES, and no-inhibitor control cultures. However, the PA, SN, 2NEOH, and ETB treatments exhibited profiles that differed from those of the no-inhibitor control and the NE, EB, and BES treatments (Fig. 2), suggesting significant effects on the bacterial population structure. In the ETB, 2NEOH, and PA treatments, several DGGE bands were absent in the low-gradient part of the gel. Intense DGGE bands were observed in the SN and PA treatments in the high-gradient part of the DGGE gel.
Fig. 2.
DGGE profiles of total bacteria in the in vitro cultures with a denaturant gradient from 40 to 60% (left to right of the gel). See the legend of Fig. 1 for lane labeling.
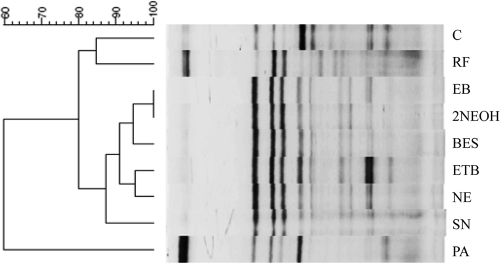
Overall, all the inhibitors except PA produced similar archaeal DGGE profiles, which were different from that of the control culture (Fig. 3). The addition of PA resulted in a DGGE profile that differed from those of other inhibitors and the no-inhibitor control, especially in the presence of intense doublet bands near the left margin of the DGGE gel. These doublet bands were also present in the rumen fluid inoculum. The no-inhibitor control culture also exhibited a DGGE profile different than that of the rumen fluid inoculum, suggesting temporal successions in methanogens during the in vitro cultivation.
Fig. 3.
DGGE profiles of methanogens in the in vitro cultures with a denaturant gradient from 40 to 60% (left to right of the gel). RF, rumen fluid inoculum. See the legend of Fig. 1 for lane labeling.
The abundances of total bacteria and of the three major cellulolytic species of bacteria in all the in vitro cultures were determined. Except for PA, which reduced the total bacterial population by nearly 24%, none of the inhibitors at the tested dose significantly affected the abundances of total bacteria in the in vitro cultures (Table 3). All the inhibitors significantly reduced the population of F. succinogenes but to different magnitudes, with SN and ETB reducing this species the greatest, by more than 2 orders of magnitude, and NE the least (by less than 1 log). With respect to R. flavefaciens, 2NEOH and PA significantly increased its population, by 217% and 725%, respectively, whereas the other inhibitors significantly reduced the population of this species by various magnitudes. The population of R. albus was significantly increased in the 2NEOH treatment (by 6-fold) but substantially decreased in the SN treatment. The other inhibitors did not appear to affect the population of R. albus significantly. All the tested compounds considerably reduced the abundance of methanogens, with PA reducing total methanogens by 77.2%, SN by nearly 2 logs, and the other compounds by 93% to 97%.
Table 3.
Comparison of population changes of total methanogens and select bacteria
| Microbe(s) or methanogen | Mean population (16S rRNA gene copies/ml culture)a with: |
SEM | P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PA | EB | ETB | 2NEOH | NE | BES | SN | C | |||
| Total bacteria | 1.3 × 1010 c | 4.6 × 1010 a | 2.5 × 1010 a,b | 2.1 × 1010 a,b | 1.8 × 1010 a,b | 4.1 × 1010 a,b | 1.9 × 1010 a,b | 1.7 × 1010 a,b | 6.2 × 109 | 0.0137 |
| F. succinogenes | 1.5 × 105 c | 3.8 × 104 c | 3.2 × 104 c | 3.3 × 105 c | 1.6 × 106 b | 1.4 × 105 c | 1.4 × 104 c | 8.2 × 106 a | 9.6 × 104 | <0.0001 |
| R. flavefaciens | 2.0 × 106 b | 7.5 × 105 c | 2.7 × 105 c | 5.2 × 106 a | 4.5 × 105 c | 1.4 × 105 c | 2.2 × 104 c | 6.3 × 105 c | 1.7 × 105 | <0.0001 |
| R. albus | 3.1 × 105 b | 3.1 × 105 b | 1.8 × 105 b | 1.4 × 106 a | 3.4 × 105 b | 1.3 × 105 b | NDb | 2.0 × 105 b | 7.5 × 104 | <0.0001 |
| Methanogen | 1.6 × 107 b | 4.0 × 106 b,c | 3.6 × 106 b,c | 4.0 × 106 b,c | 4.8 × 106 b,c | 2.2 × 106 b,c | 9.4 × 104 c | 6.9 × 107 a | 2.9 × 106 | <0.0001 |
Means within rows with different letters differ significantly (P < 0.05).
ND, not detected.
DISCUSSION
Methane production can be affected by the availability of methanogenesis substrates in the rumen (primarily hydrogen and carbon dioxide), inhibition of the methanogenesis pathway, or toxicity to methanogens. In this study, we compared several categories of compounds with respect to their effects on methane production, fermentation, and several key species/groups of rumen microbes: terminal electron acceptors that can compete with carbon dioxide for hydrogen (i.e., nitroethane, 2-nitroethanol, nitrate, ethyl trans-2-butenoate), a coenzyme M analogue that directly inhibits the methanogenesis pathway (2-bromoethanesulfonate), and compounds toxic to methanogens (i.e., propynoic acid and ethyl-2-butynote [24]). Of the seven inhibitors tested, only propynoic acid, ethyl-2-butynote, 2-nitroethanol, and sodium nitrate reduced methane production by a large margin, and 2-nitroethanol appeared to be the most potent. Except in the ethyl-2-butynote treatment, reduced methane production was accompanied by a reduced production of total VFAs, acetate, and propionate. These results suggest that reduced fermentation activities are among the possible reasons for the reduced methane production in the rumen cultures, a finding that corroborates previous studies using different inhibitors (4, 10, 25). It should be noted that the effect of antimethanogenic compounds on total and individual VFAs may be affected by other factors, such as the rumen fluid used and substrates added. For example, nitrocompounds were shown not to significantly reduce VFA concentrations in ruminal in vitro cultures when the medium consisted entirely of rumen fluid and contained formate or hydrogen only as added substrates (1, 3). In the latter studies, there might be little fermentation or VFA production due to the lack of fermentable sugars, and the VFAs detected might be those present in the original rumen fluid. Therefore, effects of antimethanogenic compounds on digestion and fermentation by ruminal cultures should be interpreted by taking into account the substrates available in the cultures.
Sodium nitroethane and nitroethanol were shown to reduce methane production to similar magnitudes (>97%) in in vitro ruminal cultures after a 24-h incubation (3, 5). In this study, however, sodium nitroethane did not significantly reduce methane production. It is not certain if the longer incubation period (48 h versus 24 h) is a factor. Ethyl-2-butynoate [CH3C CC(O)OCH2CH3] and ethyl trans-2-butenoate (CH3CH=CHCOOC2H5) can serve as terminal electron acceptors and thus can potentially inhibit methanogenesis. Although shown to reduce methane production significantly by methanogen species in pure cultures (24), ethyl-2-butynoate reduced methane production only by less than 25% in the in vitro ruminal cultures, while ethyl trans-2-butenoate had no inhibition. The lack of inhibitor or limited inhibition by these two compounds are consistent with those observed for fumarate (HO2CCH=CHCO2H) (23).
CC(O)OCH2CH3] and ethyl trans-2-butenoate (CH3CH=CHCOOC2H5) can serve as terminal electron acceptors and thus can potentially inhibit methanogenesis. Although shown to reduce methane production significantly by methanogen species in pure cultures (24), ethyl-2-butynoate reduced methane production only by less than 25% in the in vitro ruminal cultures, while ethyl trans-2-butenoate had no inhibition. The lack of inhibitor or limited inhibition by these two compounds are consistent with those observed for fumarate (HO2CCH=CHCO2H) (23).
2-Bromoethanesulfonate decreased propionate production, but it did not significantly reduce total VFA or methane production. Although single cultures of ruminal methanogens are sensitive to low concentrations of BES (24), methanogens present in complex microbiomes are much more tolerant to BES (34). Indeed, BES did not decrease methane production in continuous ruminal cultures at 250 μM (15) and decreased methane production only by about 70% in batch in vitro ruminal cultures at a 30 mM concentration (18). As seen in Table 3, however, BES reduced total methanogens by nearly 1 log. The discord between the dynamics of methanogen population and the production of methane might be partially attributable to the insensitivity of some ruminal methanogens to BES and the high rate of the methanogenesis pathway.
The bacterial DGGE profiles in the SN, ETB, 2NEOH, and PA treatments were altered, while the DGGE profiles in the NE, EB, and BES treatments remained similar to that of the no-inhibitor control. It is interesting to note that the SN, 2NEOH, and PA treatments, but not the ETB treatment, also significantly reduced VFAs and methane production. These results suggest that these three inhibitors can affect certain bacterial populations in the ruminal cultures. This conclusion was confirmed by the qPCR analysis of R. albus, R. flavefaciens, and F. succinogenes: all of the tested inhibitors dramatically reduced the population of F. succinogenes but not of R. albus (except nitrate) or R. flavefaciens. Interestingly, propynoic acid and 2-nitroethanol increased the populations of both ruminococci. These three cellulolytic species have been regarded as major cultured cellulolytic bacteria in the rumen, but because of the presence of other cryptic cellulolytic species and our lack of knowledge of their in situ cellulolytic activities, the depressed fermentation cannot be attributed solely to the decreased F. succinogenes population. This premise is also supported by the fact that the F. succinogenes population was reduced in all treatments, while the concentrations of total VFAs, acetate, and propionate were not affected in the ETB, NE, or BES treatments. It is also interesting to note that F. succinogenes, a Gram-negative bacterial species, was reduced by all the inhibitors tested, while either the two Gram-positive cellulolytic ruminococcal species were unaffected or their abundance increased. Although it is attempting to conclude that Gram-negative bacteria might be more susceptible to these inhibitors than Gram-positive species, future studies using multiple bacterial species are required to verify this hypothesis.
The DGGE profiles of archaea in all treatments differed from that of the no-inhibitor control. Comparisons between the no-inhibitor control and the original rumen fluid inoculum also revealed temporal changes in archaeal population structure during the in vitro incubation. However, it is evident that all the inhibitors resulted in different DGGE profiles that are different from that of the no-inhibitor control, suggesting that these inhibitors can affect different methanogen species differently. Sequencing of 16S rRNA genes is required to identify the species affected by each of the inhibitors. All the inhibitors decreased the population of total archaea. As mentioned above, however, only methane production in the PA, 2NEOH, and SN treatments was substantially reduced. Apparently, decreases in methanogen populations may not necessarily lead to a reduction in methane production, and vise versa, at least within a short period of time.
Collectively, the seven antimethanogen inhibitors evaluated in this study can be grouped into two groups: one group containing propynoic acid, 2-nitroethanol, and nitrate, which inhibited VFAs, methanogens, and methane production, and another group containing ethyl-2-butynote, ethyl trans-2-butenoate, nitroethane, and BES, which reduced methanogens but not VFAs or methane production. The two groups of inhibitors might differently affect bacteria, feed digestion and fermentation, methanogens, and methane production. Future studies are needed to determine the concentrations of each inhibitor and combinations thereof that inhibit methane production but not feed digestion or fermentation.
ACKNOWLEDGMENT
A scholarship awarded to Z.Z. from the China Scholarship Council partially supported his tenure at The Ohio State University and the research reported in this paper.
Footnotes
Published ahead of print on 25 February 2011.
REFERENCES
- 1. Anderson R. C., et al. 2003. Effect of select nitrocompounds on ruminal fermentation; an initial look at their potential to reduce economic and environmental costs associated with ruminal methanogenesis. Bioresour. Technol. 90:59–63 [DOI] [PubMed] [Google Scholar]
- 2. Anderson R. C., et al. 2010. Effect of nitroethane, dimethyl-2-nitroglutarate and 2-nitro-methyl-propionate on ruminal methane production and hydrogen balance in vitro. Bioresour. Technol. 101:5345–5349 [DOI] [PubMed] [Google Scholar]
- 3. Anderson R. C., et al. 2008. Effects of select nitrocompounds on in vitro ruminal fermentation during conditions of limiting or excess added reductant. Bioresour. Technol. 99:8655–8661 [DOI] [PubMed] [Google Scholar]
- 4. Beauchemin K. A., McGinn S. M. 2006. Methane emissions from beef cattle: effects of fumaric acid, essential oil, and canola oil. J. Anim. Sci. 84:1489–1496 [DOI] [PubMed] [Google Scholar]
- 5. Bozic A. K., et al. 2009. Effects of the methane-inhibitors nitrate, nitroethane, lauric acid, Lauricidin and the Hawaiian marine algae Chaetoceros on ruminal fermentation in vitro. Bioresour. Technol. 100:4017–4025 [DOI] [PubMed] [Google Scholar]
- 6. Chen J., Yu Z., Michel F. C., Jr., Wittum T., Morrison M. 2007. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 73:4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goel G., Makkar H. P., Becker K. 2008. Changes in microbial community structure, methanogenesis and rumen fermentation in response to saponin-rich fractions from different plant materials. J. Appl. Microbiol. 105:770–777 [DOI] [PubMed] [Google Scholar]
- 8. Goel G., Makkar H. P., Becker K. 2009. Inhibition of methanogens by bromochloromethane: effects on microbial communities and rumen fermentation using batch and continuous fermentations. Br. J. Nutr. 101:1484–1492 [DOI] [PubMed] [Google Scholar]
- 9. Guo Y. Q., et al. 2008. Effect of tea saponin on methanogenesis, microbial community structure and expression of mcrA gene, in cultures of rumen micro-organisms. Lett. Appl. Microbiol. 47:421–426 [DOI] [PubMed] [Google Scholar]
- 10. Holtshausen L., et al. 2009. Feeding saponin-containing Yucca schidigera and Quillaja saponaria to decrease enteric methane production in dairy cows. J. Dairy Sci. 92:2809–2821 [DOI] [PubMed] [Google Scholar]
- 11. Intergovernmental Panel on Climate Change 2007. Climate change 2007. Fourth assessment report. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 12. Ipharraguerre I. R., Clark J. H. 2003. Soyhulls as an alternative feed for lactating dairy cows: a review. J. Dairy Sci. 86:1052–1073 [DOI] [PubMed] [Google Scholar]
- 13. Janse I., Bok J., Zwart G. 2004. A simple remedy against artifactual double bands in denaturing gradient gel electrophoresis. J. Microbiol. Methods 57:279–281 [DOI] [PubMed] [Google Scholar]
- 14. Johnson K. A., Johnson D. E. 1995. Methane emissions from cattle. J. Anim. Sci. 73:2483–2492 [DOI] [PubMed] [Google Scholar]
- 15. Karnati S. K., Sylvester J. T., Ribeiro C. V., Gilligan L. E., Firkins J. L. 2009. Investigating unsaturated fat, monensin, or bromoethanesulfonate in continuous cultures retaining ruminal protozoa. I. Fermentation, biohydrogenation, and microbial protein synthesis. J. Dairy Sci. 92:3849–3860 [DOI] [PubMed] [Google Scholar]
- 16. Koike S., Kobayashi Y. 2001. Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. FEMS Microbiol. Lett. 204:361–366 [DOI] [PubMed] [Google Scholar]
- 17. Krishnamoorthy U., Steingass H., Menke K. H. 1991. Preliminary observations on the relationship between gas production and microbial protein synthesis in vitro. Arch. Tierernahr. 41:521–526 [DOI] [PubMed] [Google Scholar]
- 18. Martin S. A., Macy J. M. 1985. Effects of monensin, pyromellitic diimide and 2-bromoethanesulfonic acid on rumen fermentation in vitro. J. Anim. Sci. 60:544–550 [DOI] [PubMed] [Google Scholar]
- 19. McGinn S. M., Beauchemin K. A., Coates T., Colombatto D. 2004. Methane emissions from beef cattle: effects of monensin, sunflower oil, enzymes, yeast, and fumaric acid. J. Anim. Sci. 82:3346–3356 [DOI] [PubMed] [Google Scholar]
- 20. Meiser H., Hagedorn H. W., Schulz R. 2000. Development of a method for determination of cyanide concentrations in serum and rumen fluid of cattle. Am. J. Vet. Res. 61:658–664 [DOI] [PubMed] [Google Scholar]
- 21. Min B. R., Pinchak W. E., Anderson R. C., Fulford J. D., Puchala R. 2006. Effects of condensed tannins supplementation level on weight gain and in vitro and in vivo bloat precursors in steers grazing winter wheat. J. Anim. Sci. 84:2546–2554 [DOI] [PubMed] [Google Scholar]
- 22. Nadkarni M. A., Martin F. E., Jacques N. A., Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 23. Ungerfeld E. M., Kohn R. A., Wallace R. J., Newbold C. J. 2007. A meta-analysis of fumarate effects on methane production in ruminal batch cultures. J. Anim. Sci. 85:2556–2563 [DOI] [PubMed] [Google Scholar]
- 24. Ungerfeld E. M., Rust S. R., Boone D. R., Liu Y. 2004. Effects of several inhibitors on pure cultures of ruminal methanogens. J. Appl. Microbiol. 97:520–526 [DOI] [PubMed] [Google Scholar]
- 25. Ungerfeld E. M., Rust S. R., Burnett R. 2007. Increases in microbial nitrogen production and efficiency in vitro with three inhibitors of ruminal methanogenesis. Can. J. Microbiol. 53:496–503 [DOI] [PubMed] [Google Scholar]
- 26. U.S. Environmental Protection Agency 2010. Inventory of U.S. greenhouse gas emissions and sinks: 1990-2008. U.S. Environmental Protection Agency, Washington, DC: http://www.epa.gov/climatechange/emissions/usinventoryreport.html [Google Scholar]
- 27. Weimer P. J., Stevenson D. M., Mertens D. R., Thomas E. E. 2008. Effect of monensin feeding and withdrawal on populations of individual bacterial species in the rumen of lactating dairy cows fed high-starch rations. Appl. Microbiol. Biotechnol. 80:135–145 [DOI] [PubMed] [Google Scholar]
- 28. Williams Y. J., et al. 2009. A vaccine against rumen methanogens can alter the composition of archaeal populations. Appl. Environ. Microbiol. 75:1860–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu Y., Lee C., Kim J., Hwang S. 2005. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89:670–679 [DOI] [PubMed] [Google Scholar]
- 30. Yu Z., Garcia-Gonzalez R., Schanbacher F. L., Morrison M. 2008. Evaluations of different hypervariable regions of archaeal 16S rRNA genes in profiling of methanogens by archaea-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 74:889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu Z., Michel F. C., Jr., Hansen G., Wittum T., Morrison M. 2005. Development and application of real-time PCR assays for quantification of genes encoding tetracycline resistance. Appl. Environ. Microbiol. 71:6926–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu Z., Morrison M. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Z., Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812 [DOI] [PubMed] [Google Scholar]
- 34. Zinder S. H., Anguish T., Cardwell S. C. 1984. Selective inhibition by 2-bromoethanesulfonate of methanogenesis from acetate in a thermophilic anaerobic digestor. Appl. Environ. Microbiol. 47:1343–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]