Abstract
The synthesis of bacterial polyhydroxyalkanoates (PHA) is very much dependent on the expression and activity of a key enzyme, PHA synthase (PhaC). Many efforts are being pursued to enhance the activity and broaden the substrate specificity of PhaC. Here, we report the identification of a highly active wild-type PhaC belonging to the recently isolated Chromobacterium sp. USM2 (PhaCCs). PhaCCs showed the ability to utilize 3-hydroxybutyrate (3HB), 3-hydroxyvalerate (3HV), and 3-hydroxyhexanoate (3HHx) monomers in PHA biosynthesis. An in vitro assay of recombinant PhaCCs expressed in Escherichia coli showed that its polymerization of 3-hydroxybutyryl-coenzyme A activity was nearly 8-fold higher (2,462 ± 80 U/g) than that of the synthase from the model strain C. necator (307 ± 24 U/g). Specific activity using a Strep2-tagged, purified PhaCCs was 238 ± 98 U/mg, almost 5-fold higher than findings of previous studies using purified PhaC from C. necator. Efficient poly(3-hydroxybutyrate) [P(3HB)] accumulation in Escherichia coli expressing PhaCCs of up to 76 ± 2 weight percent was observed within 24 h of cultivation. To date, this is the highest activity reported for a purified PHA synthase. PhaCCs is a naturally occurring, highly active PHA synthase with superior polymerizing ability.
INTRODUCTION
Unlike petrochemical polymers, the synthesis of biologically based polymers is very much dependent on the catalytic activities of the various enzymes involved, as well as on the carbon feedstock from which the monomers are produced. Polymerization rates and yields vary based on the biosynthetic pathway of the organism and the available monomer supply. One such biopolymer that has attracted widespread interest is polyhydroxyalkanoate (PHA). Owing to its thermoplastic and biodegradable properties, PHA makes an excellent candidate for the biodegradable replacement of conventional plastics (7). PHA has garnered a great deal of interest for applications in various industries, including medicine, pharmacology, agriculture, packaging, and cosmetics (2, 27, 45). PHA has been produced using wild-type as well as recombinant microorganisms (17–19). The biosynthesis of this bacterial polymer is controlled by both the enzymes that supply usable monomers and PHA synthase (commonly known as PhaC), the key enzyme involved in polymerization (29, 36, 38).
The dominant role played by PhaC in determining polymer composition and properties provided an impetus to extensive investigations of PHA synthases. Four major classes of PHA synthases have been classified with respect to their primary structures, substrate specificities, and subunit composition (28, 29). So far, the PhaC of Cupriavidus necator (PhaCCn) (class I) has been studied in some mechanistic detail and is the benchmark commonly used to evaluate the performance of other synthases (12, 14, 34). Some studies also have been carried out on the synthase of Allochromatium vinosum (class III) (13, 23). Nevertheless, PhaC is a complex enzyme, and its complete structure and properties are not yet fully understood. It is known that the affinity and polymerization activity toward different hydroxyalkanoate-coenzyme A (CoA) substrates vary based on the different classes of PHA synthases. Efforts have been taken to alter and improve the properties of natural synthase enzymes via enzymatic evolution, with the goal of engineering a more active enzyme with broader substrate specificity (38). Several successful studies have reported the engineering of mutant synthases with up to 4-fold increased activity (1, 26, 42). Nevertheless, the search for a natural synthase with comparable properties still is widespread.
The property of PHA is dependent on its monomeric composition, which is determined in part by PhaC (29, 36). Recently, Bhubalan et al. cloned the PHA synthase gene (phaCCs) from an organism termed Chromobacterium sp. USM2, isolated from Malaysian sources, and heterologously expressed the synthase gene in a PHA-negative mutant of C. necator, PHB−4 (5). In this study, poly(3-hydroxybutyrate-co-3-hydroxyvalerate) [P(3HB-co-3HV)] copolymer with the high-3-hydroxyvalerate (3HV) fraction was synthesized from mixtures of fructose and sodium valerate. Furthermore, 3-hydroxyhexanoate (3HHx) monomer was successfully incorporated when crude palm kernel oil (CPKO) was fed as the sole carbon source, resulting in the production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) [P(3HB-co-3HHx)] copolymer. P(3HB-co-3HV) and P(3HB-co-3HHx) copolymers are known to possess improved mechanical and thermal properties compared to those of the P(3HB) homopolymer (9, 10). When a combination of sodium valerate, or sodium propionate, with CPKO was fed to the C. necator PHB−4 strain containing heterologously expressed phaCCs, high intracellular contents of polymer comprising 3HB, 3HV, and 3HHx monomers were produced (4). This P(3HB-co-3HV-co-3HHx) terpolymer produced was found to possess elastomeric properties. However, not many microorganisms express a native PHA synthase with the ability to incorporate both short-chain-length (scl) and medium-chain-length (mcl) monomers. The ability of PhaCCs to produce PHA-containing monomers of mixed chain lengths highlighted the potential of this synthase.
In this study, the PHA synthase of Chromobacterium sp. USM2 was further characterized by in vitro and in vivo assays using Escherichia coli JM109 to fully understand its PHA-synthesizing ability. We also purified a heterologously expressed, Strep2-tagged version of PhaCCs to examine the unique abilities of this enzyme. The results obtained in this work showed that PhaCCs is a highly active enzyme in its natural form, and it is expressed at high levels in E. coli. The ability to produce high concentrations of active synthase in vivo might facilitate overcoming one of the bottlenecks in the crystallization of the PhaC enzyme, which is producing and isolating an abundant amount of pure protein. Once this is possible, attempts can be made to determine the three-dimensional structure of this complex enzyme that, to date, still remains an impenetrable barrier.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli JM109 was used for all standard genetic engineering, and its transformants were used for PHA biosynthesis. The plasmids used in this study are listed in Table 1. E. coli JM109 was grown at 37°C in LB broth consisting of the following components (per liter): 10 g casein enzymatic hydrolysate, 5 g yeast extract, and 10 g NaCl at pH 7.0. To determine the functional expression of the cloned gene in vivo, PHA biosynthesis was carried out by transferring 1.5 ml (3% [vol/vol]) of inoculum from a preculture grown for 12 h in LB into 50 ml of fresh LB in 250-ml Erlenmeyer flasks supplemented with 2% (wt/vol) glucose. The cultures were incubated at 30 and 37°C for 72 h on a reciprocal shaker at 180 rpm. Ampicillin was added at a final concentration of 100 μg/ml to maintain plasmid stability. For maintenance purposes, bacterial cultures from the exponential growth phase were stored at −20°C in 20% (vol/vol) glycerol.
Table 1.
Strains and plasmids used in this study
| Bacterial strains and plasmids | Relevant phenotype | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli JM109 | E14-(mcrA) recA1 gryA96 thi-1 hsdR17(rk−, mk+) supE44 relA1 Δ(lac-proAB) [F′ traD36, proAB, lacIqZΔM15] | Stratagene |
| BL21(DE3) | F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Novagen |
| Plasmids | ||
| pBBR1MCS-C2 | pBBR1MCS-2 derivative harboring an ∼2.0-kb fragment of phaCCs from Chromobacterium sp. USM2 with putative promoter | 5 |
| pGEM′CAB | pGEM-T derivative with C. necator promoter and terminator harboring phaCCn, phaACn, phaBCn | 21 |
| pGEM″C1AB | pGEM-T derivative with C. necator promoter and terminator harboring phaC1Ps, phaACn, phaBCn | 20 |
| pGEM″AB | pGEM-T derivative with C. necator promoter and terminator harboring phaACn, phaBCn | This study |
| pGEM″AB(L) | pGEM-T derivative with C. necator promoter and terminator harboring phaACn, phaBCn, synthetic XbaI-EcoRI-EcoRV-Asp718-HindIII-PstI linker | This study |
| pGEM″AB(phaCCs) | pGEM-T derivative with C. necator promoter and terminator harboring phaCCs, phaACn, phaBCn | This study |
| pET51b | Protein expression vector for N-terminally Strep2-tagged proteins | Novagen |
| pET-phaCCs | pET51b derivative with phaCCs open reading frame inserted into BamHI/HindIII restriction site | This study |
For the extraction of crude protein, E. coli JM109 transformants were grown at 30°C in 2 ml of LB broth for 14 h. An aliquot of 17.5 μl (1% [vol/vol]) was inoculated to 1.75 ml of fresh LB broth and was incubated at 30°C for 9 h. Ampicillin was added at a final concentration of 100 μg/ml for plasmid maintenance. For Strep2-PhaCCs expression and purification, E. coli BL21(DE3) was used as a host strain. The expression of Strep2-PhaCCs was performed as follows. Cells with Strep2-PhaCCs expression plasmid were grown in 1 liter of LB broth supplemented with 100 μg/ml ampicillin until an optical density at 600 nm (OD600) of 0.6. Enzyme synthesis was induced by the addition of 0.1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) and allowed to incubate for 2 h at 30°C on a reciprocal shaker at 180 rpm. Cells then were pelleted, and protein was purified as described below.
DNA manipulation and plasmid construction.
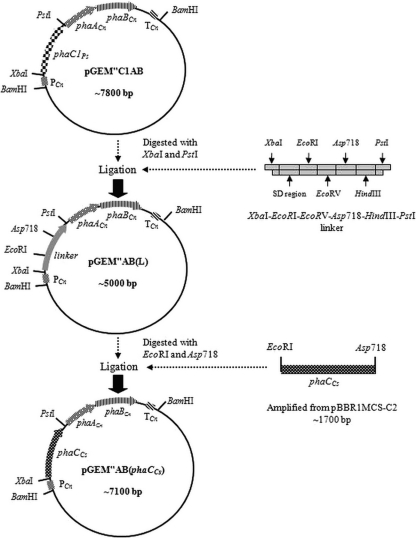
Plasmid isolation and DNA manipulation was carried out according to standard procedures (30). All of the restriction enzymes (TaKaRa, Toyobo, and Roche) were used according to the manufacturers' protocols. All other chemicals used were of analytical grade. The plasmid pGEM″AB(phaCCs) in Fig. 1 was constructed to determine the expression and activity of PhaCCs in E. coli JM109 via in vitro and in vivo experiments. First, the plasmid vector pGEM″C1AB was digested with XbaI and PstI to remove the Pseudomonas sp. 61-3 phaC1 gene. The vector then was ligated with a synthetic linker, XbaI-EcoRI-EcoRV-Asp718-HindIII-PstI, which was derived by annealing a set of complementary primers (FXbaIPstILK and RXbaIPstIL) (nucleotide sequences are shown in Table 2). The resultant vector was named pGEM″AB(L). The phaCCs gene then was cloned using the forward primer FEcoRICs and the reverse primer RAsp718Cs (Table 2) from the plasmid vector pBBR1MCS-C2. The resulting 1.7-kb gene fragment, flanked with EcoRI and Asp718 restriction sites, was purified and then digested with the corresponding enzymes and ligated into the pGEM″AB(L) vector, which was digested with the same enzymes. The resultant vector was named pGEM″AB(phaCCs). DNA sequencing for the confirmation of new plasmid constructs was carried out by the dideoxy chain termination method with a Prism 310 genetic analyzer DNA sequencer (Applied Biosystems) and the CEQ2000XL DNA analysis system (Beckman Coulter) using a BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems) and Dye terminator cycle sequencing with quick start kit (Beckman Coulter).
Fig. 1.
Construction of the pGEM″AB(phaCCs) expression plasmid harboring the PHA synthase gene of Chromobacterium sp. USM2 with promoter (PCn)-, terminator (TCn)-, and monomer-supplying genes phaACn and phaBCn of C. necator. pGEM″C1AB harbors the PHA synthase gene of Pseudomonas sp. 61-3 with promoter (PCn)-, terminator (TCn)-, and monomer-supplying genes phaACn and phaBCn of C. necator. pGEM″AB(L) harbors the synthetic XbaI-EcoRI-EcoRV-Asp718-HindIII-PstI linker with promoter (PCn)-, terminator (TCn)-, and monomer-supplying genes phaACn and phaBCn of C. necator.
Table 2.
Primer sequences used in this studya
| Primer name | Sequence (5′–3′) |
|---|---|
| FEcoRICs | GCGGCCAACCAGGAATTCATGC |
| RAsp718Cs | GGGACGGTACCTTCGGTTCAG |
| FXbaIPstILK | CTAGATAAGAAGGAGATGAATTCGATATCGGTACCAAGCTTCTGCA |
| RXbaIPstIL | GAAGCTTGGTACCGATATCGAATTCATCTCCTTCTTAT |
| Strep2phaCCsFW | CAAGGATCCGATGCAGCAGTTTGTCAATTCCCT |
| Strep2phaCCsRV | CTTAAGCTTTCAGTTCAAGGCGGCGA |
Restriction sites are underlined.
Plasmid pET-phaCCs was constructed using pET51b (Novagen) as the parent plasmid. For the construction of strep2-phaCCs, phaCCs on the plasmid pBBR1MCS-C2 was amplified by PCR using the forward primer Strep2phaCCsFW and the reverse primer Strep2phaCCsRV (Table 2) to introduce the unique restriction site BamHI 5′ to the phaCCs open reading frame and the unique restriction site HindIII 3′ to the phaCCs open reading frame. The amplified gene was digested with BamHI and HindIII, followed by ligation with BamHI- and HindIII-cut pET51b to produce the plasmid pET-phaCCs. The portion of pET-phaCCs containing the tagged phaCCs gene was sequenced by MIT Biopolymers Laboratory.
Preparation of crude protein samples.
E. coli JM109 harboring either pGEM″AB(phaCCs), pGEM′CAB, or pGEM″AB(L) was cultured as discussed above. Cells were harvested by centrifugation, and whole-cell extracts of each transformant were prepared by resuspending the cells in 2 ml of ice-cold 40 mM potassium phosphate buffer (pH 7.5) and subsequent disruption by sonication (three cycles, 5 s each) on ice using a Tomy UD-200 sonicator. A soluble fraction was obtained from the resulting supernatant when the disrupted cells were centrifuged at 13,700 × g for 10 min at 4°C, and the insoluble fraction was obtained from the subsequent precipitate. Protein was measured using the Bradford assay (6).
Expression and purification of Strep2-tagged PhaCCs.
E. coli BL21(DE3)/pET-phaCCs was cultured as discussed above. Cells (6.5 to 7.8 g wet weight) were pelleted by centrifugation at 2,988 × g at 4°C. The cell pellet was resuspended in 25 ml buffer A (100 mM Tris-HCl, pH 8.0) and lysed using a French pressure cell (two passes at 12,000 lb/in2). The resulting cell lysate was centrifuged at 100,000 × g to remove cell debris. The clarified lysate was loaded onto a Strep-tactin column (IBA, GmbH, Göttingen, Germany; 10-ml column volume) preequilibrated with 80 ml buffer B (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA). The lysate and column were incubated at 4°C for 15 to 20 min. The column was eluted and washed with 5 × 10 ml buffer B. Strep2-PhaCCs was eluted from the column with six 5-ml fractions of buffer C (buffer B plus 2.5 mM desthiobiotin). The Strep-tactin column was regenerated according to the manufacturer's instructions. The protein concentration of each fraction was determined by Bradford assay. The pooled fractions then were concentrated using a Vivaspin 15R concentrator (Sartorius AG, Göttingen, Germany) to 5.5 to 25.5 mg protein/ml and dialyzed twice against 100 mM Tris-HCl (pH 8.0), containing 0.5 mM EDTA and 0.5 mM dithiothreitol, for 12 to 16 h using a Slide-a-Lyzer dialysis cassette (Thermo Scientific). Aliquots of 100 μl of the protein preparation were stored at −80°C. Protein concentrations of pooled, concentrated fractions were determined by Bradford assay and confirmed spectrometrically at A280 using the molar absorption coefficient 110,810 M−1 cm−1. Strep2-PhaCCs purification was performed three separate times.
In vitro enzymatic assay of crude PhaCCs.
The activity of PHA synthase from crude extract was determined by measuring the amount of CoA released from 3HB-CoA during polymerization (41). The assay mixture contained 2 mM 3HB-CoA, 40 mM potassium phosphate buffer (pH 7.5), 10 mM 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB), and 1 mg/ml bovine serum albumin (BSA). The reaction was initiated by adding 35 to 40 μg of protein obtained from the soluble fraction of disrupted cells into the reaction mixture described above, and the absorbance at 412 nm was measured at 30°C. The concentration of CoA was determined spectrometrically (12) using a molar absorption coefficient of 13,600 M−1 cm−1 at 412 nm with a Hitachi U-3900H spectrophotometer. One unit of enzyme activity is defined as the amount of enzyme that catalyzed the release of 1.0 μmol CoA/min. Enzyme assays were performed in triplicate.
In vitro enzymatic assay of Strep2-PhaCCs.
Assays were carried out as previously described (43). Final enzyme concentrations of 7.5 to 30 nM Strep2-PhaCCs and 600 μM 3HB-CoA were used. The concentration of CoA was determined spectrometrically (12) using a molar absorption coefficient of 13,600 M−1 cm−1 at 412 nm with an Agilent 8453 spectrophotometer. Preparations of Strep2-PhaCCn were used as a control and purified as described elsewhere (M. Cho, C. Brigham, A. Sinskey, and J. Stubbe, unpublished data). One unit of enzyme activity is defined as described above. Enzyme assays were performed in triplicate.
Western blot analysis.
A total of 10 μg of proteins prepared from both soluble and insoluble fractions of the disrupted E. coli JM109 transformants were separated using 12.5% SDS-PAGE. Separated proteins from the soluble fraction then were transferred onto a polyvinylidene fluoride (PVDF) membrane (Immun-blot PVDF membrane [Bio-Rad]) using a Criterion blotter (Bio-Rad). An immunoblot analysis of PHA synthase was carried out using specific rabbit antiserum raised against the C-terminal region of PhaCCn as described by Murata et al. (24). PhaC protein was detected using goat anti-rabbit IgG conjugated with alkaline phosphatase as a secondary antibody.
GC and polymer isolation.
The methanolysis of the lyophilized cells in the presence of 15% (vol/vol) sulfuric acid and 85% (vol/vol) methanol was carried out prior to determining P(3HB) content through gas chromatography (GC) analysis (8). P(3HB) was extracted by refluxing lyophilized cells with chloroform for 4 h at 60°C. The polymer solution then was purified by precipitation with chilled methanol. The purified polymer then was air dried in a fume cupboard.
GPC.
The average molecular weight of P(3HB) produced was estimated using a Shimadzu 10A gel permeation chromatography (GPC) system and a 10A refractive index detector with Shodex K-806 M and K-802 columns. Chloroform was used as the eluent at a flow rate of 0.8 ml/min, and analysis was carried out at 40°C. A sample concentration of 1.0 mg/ml was used. The calibration curve was generated using low-polydispersity polystyrene standards.
TEM.
E. coli JM109 harboring pGEM″AB(phaCCs) was cultured for 24 h in LB supplemented with 2% (wt/vol) glucose as mentioned above. Transmission electron microscopy (TEM) analysis was carried out to observe the accumulation of PHA granules and the changes in cell morphology under the electron microscope (Philip CM 12/STEM and JLM-2000FX11). Cells were harvested and fixed in McDowell-Trump fixative at 4°C for 24 h (22). The cell pellets then were postfixed with 1% osmium tetroxide (OsO4) at room temperature. Cells were dehydrated in an increasing ethanol series (50, 75, 95, and 100%) and then transferred to 100% acetone. Cells were embedded at 60°C for 24 to 48 h in Spurr's low-viscosity resin (35). Ultrathin sections were prepared, mounted on copper grids, and stained with uranyl acetate and lead citrate for electron microscope examination at an acceleration voltage of 80 kV (Philip CM 12/STEM and JLM-2000FX11).
RESULTS
In vitro assay of crude PhaCCs in E. coli.
The ability of the C. necator PHB−4 transformant harboring phaCCs (GenBank accession no. HM989943) to utilize CPKO and 3HV precursors for the biosynthesis of PHA polymers containing 3HB, 3HV, and 3HHx monomers (4, 5) served as groundwork to investigate the interesting properties of this synthase further. Hence, in this study, PhaCCs was characterized using in vivo and in vitro assays to understand better its PHA-synthesizing ability. The phaCCs gene was cloned into plasmid pGEM″AB harboring the monomer-supplying genes of C. necator H16 (phaACn and phaBCn) and subsequently expressed in E. coli JM109. E. coli harboring pGEM′CAB plasmid, which contains the PHA biosynthetic genes (phaCAB) of C. necator H16, was used as the positive control. The construction of pGEM″AB(phaCCs) is shown in Fig. 1.
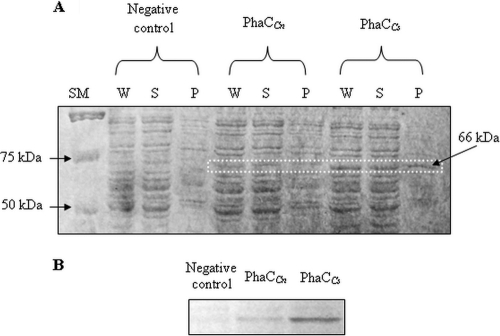
The in vivo level of PhaCCs was evaluated through a series of in vitro assays. From the SDS-PAGE analysis of the crude cell lysates (Fig. 2A), it can be seen that the in vivo level of PhaCCs protein appeared to be higher than that of PhaCCn using the same background strain. In Fig. 2A, distinct bands of approximately 66 kDa in size corresponded to the sizes of the synthases. The detection of similar-size protein bands in the precipitate (cell pellets) confirmed the presence of an insoluble population of synthase protein. A more distinctly observed elevation in the concentration of PhaCCs was seen through Western blot analysis. As shown in Fig. 2B, the intensity of the PhaCCs band was much greater than that of PhaCCn. This suggested the presence of a higher concentration of PhaCCs in the bacterial cells. As expected, no protein band was detected in the negative-control sample, whereby plasmid pGEM″AB(L) harboring only monomer-supplying genes without the presence of PHA synthase gene was used.
Fig. 2.
(A) SDS-PAGE analysis of crude extracts of PhaCCn and PhaCCs. E. coli cells harboring plasmid pGEM″AB(L) were used as a negative control. For each sample, a total of 10 μg of protein was loaded into each well. SM, size marker; W, whole-cell extract; S, supernatant (soluble fraction); P, precipitate (cell pellets). (B) Comparison of the expression levels of PhaCCn and PhaCCs in E. coli transformants using Western blot analysis. E. coli cells harboring plasmid pGEM″AB(L) were used as a negative control. A total of 10 μg of protein from the supernatant was used for the analysis.
To investigate the activity of PhaCCs, 3HB-CoA was used as the substrate, and the release of CoA during polymerization was measured to determine the total enzyme activity. The total activity of PhaCCs was measured using the soluble fraction of the crude extract. PhaCCs demonstrated a superior ability in polymerizing 3HB-CoA compared to that of PhaCCn. The total synthase activity of cell extracts containing PhaCCs (2,462 ± 80 U/g) was nearly 8-fold higher than that of cells expressing PhaCCn (307 ± 24 U/g). The high activity of PhaCCs could be associated with its elevated level of expression in E. coli. Given these results, it was assumed that the availability of higher concentrations of PhaCCs in the cells would ensure more efficient and faster accumulation of polymer upon the addition of a carbon substrate. Results obtained from the in vivo evaluation of PhaCCs confirmed this hypothesis, as shown below.
In vivo evaluation of PhaCCs.
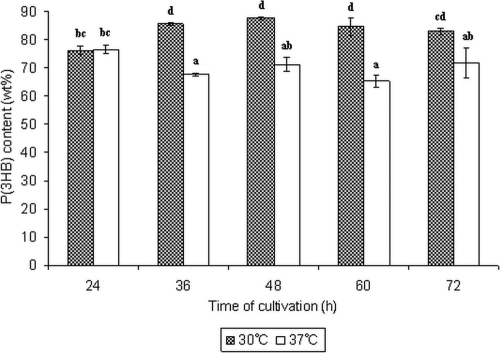
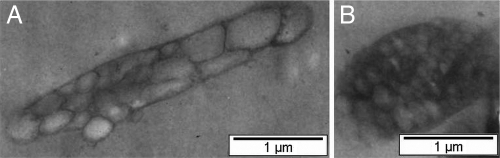
The E. coli transformant harboring phaCCs was found to accumulate large amounts of intracellular P(3HB) (76 ± 2 weight percent [wt%]) within 24 h of cultivation using glucose as the carbon source. The total P(3HB) concentration reached a maximum value of 7.2 ± 0.2 g/liter after 48 h of cultivation. It is well known that the optimal growth temperature for E. coli is 37°C. However, the optimal temperature for growth and PHA accumulation by Chromobacterium sp. USM2 previously had been identified as 30°C (5). Therefore, an E. coli transformant harboring phaCCs was cultivated at 30°C to determine the effect of lowered temperature on the overall growth and productivity. A significant difference in the P(3HB) accumulation was noticed at 30°C compared to that at 37°C (Fig. 3). P(3HB) content of up to 88 ± 1 wt% (48 h) was accumulated by this transformant at 30°C, whereas there was a maximum of 76 ± 2 wt% (24 h) at 37°C. The polymerization of P(3HB) by PhaCCs expressed in E. coli appeared to be better at 30°C. As shown in Fig. 4A, the E. coli transformant was packed with granules of various sizes. Some cells contained mainly smaller granules, as shown in Fig. 4B. The molecular mass of P(3HB) produced averaged 5 × 105 Da, with a high polydispersity of 6.0.
Fig. 3.
Comparison of P(3HB) production at 30 and 37°C by an E. coli JM109 transformant harboring phaCCs. Cells were incubated for 72 h in LB medium supplemented with 2% (wt/vol) glucose and 100 μg/ml of ampicillin. Data shown are means from triplicates. Means with different letters are significantly different (Tukey's honestly significant difference test; P < 0.05).
Fig. 4.
TEM showing P(3HB) granules in an E. coli JM109 transformant harboring pGEM″AB(phaCCs). Cells were cultured for 24 h at 30°C in LB supplemented with 2% (wt/vol) glucose and 100 μg/ml ampicillin. It could be observed that some cells contained granules of various sizes (A), while others contained a large number of smaller granules (B).
Enzymatic activity of purified Strep2-PhaCCs.
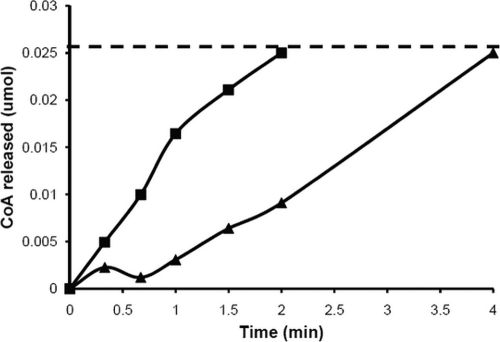
To further investigate the polymerization ability of PhaCCs, we constructed a Strep2-tagged PhaCCs for expression in and purification from E. coli. Strep2-PhaCCs was cloned into E. coli BL21(DE3) and purified as described in Materials and Methods. The specific activity of this highly purified Strep2-PhaCCs was 238 ± 98 U/mg, which is significantly greater than that of purified synthase from C. necator from previously published results (specific activity, 40 U/mg) (43). It could be argued that the difference in N-terminal epitope tags (Strep2 tag for the enzyme purified in this work and an oligonucleotide-HIS tag in reference 43) could result in differences of activity. A Strep2-PhaCCn enzyme was expressed previously and purified from E. coli BL21(DE3), and the activity was determined to be 38.5 ± 8.1 U/mg (M. Cho et al., unpublished), which is similar to that of the previously published His6-PhaCCn activity (43). The purified Strep2-PhaCCs enzyme also exhibited a lag phase in activity (Fig. 5), which is consistent with previous results using purified class I PhaC proteins (such as PhaC from C. necator) isolated from E. coli (12, 43, 44). While the lag phase of Strep2-PhaCCn is more prevalent in Fig. 5, the lag phase in activity of Strep2-PhaCCs is more prevalent when lower concentrations of enzyme are used (data not shown).
Fig. 5.
Time course of CoA release from 3HB-CoA catalyzed by purified Strep2-PhaCCs (black boxes) and purified Strep2-PhaCCn (black triangles). In the experiment represented by this figure, 30 nM Strep2-PhaCCs and 30 nM Strep2-PhaCCn were used. The dashed line indicates when the 3HB-CoA substrate was completely used up.
DISCUSSION
In vitro and in vivo characterization of PhaCCs in E. coli was carried out by the heterologous expression of phaCCs along with phaACn and phaBCn under the control of the C. necator promoter in a pGEM″AB(phaCCs) expression plasmid. The heterologously expressed synthase showed an increased level of expression and enzyme activity. As seen in Fig. 2A in the crude cell lysates, a distinct band of approximately 66 kDa in size was observed that corresponded to the class I PHA synthases (28, 29). In SDS-PAGE gel analysis, a more distinct band exhibited by PhaCCs compared to that of PhaCCn suggests that PhaCCs was expressed at a higher level in this system. This finding was further confirmed by Western blot analysis (Fig. 2B). The total activity of cell extracts containing PhaCCs toward the polymerization of 3HB-CoA was nearly 8-fold higher than those containing PhaCCn. This suggested that the total enzymatic activity of PhaCCs can be partially correlated with its elevated level of expression in vivo.
The ability to polymerize 3HB-CoA varies among the different classes of PHA synthases, with class I, III, and IV synthases showing higher preference toward the polymerization of 3HB-CoA (36, 38). The expression and activity of these genes in E. coli are commonly used as benchmarks to compare the performance of other heterologous PHA biosynthesis genes. In this study, the activity of the heterologous PhaCCn from cell extracts of E. coli was comparable to that observed in extracts of wild-type C. necator, whereby its activity is known to range from 180 to 330 U/g during PHA accumulation stages (11, 15, 31).
The activities of several PHA synthases recombinantly expressed in E. coli have been documented previously. Alterations in the expression level and specific activity of some of these enzymes were achieved through enzyme evolution studies. The PhaCCn enzyme harboring an F420S mutation has a 2.4-fold higher specific activity for the polymerization of 3HB-CoA than wild-type PhaCCn (39). Meanwhile, PhaCCn harboring a double mutation (G4D and F420S) exhibited an increased synthase concentration in vivo and enhanced polymer accumulation (26). In a similar study, the synthase activities in cell extracts of Aeromonas punctata wild-type and mutant strains were found to be in the range of 118 to 768 U/g (1). The mutant synthases exhibited up to 5-fold-increased activity compared to that of the wild-type synthase. On the other hand, wild-type and mutant synthases of Pseudomonas sp. 61-3, which belongs to the class II PHA synthase, exhibited activity of less than 50 U/g toward the polymerization of 3HB-CoA (42). In a recent study, the enzymatic activities of the PHA synthase of Aeromonas caviae (PhaCAc) and some of its mutants, when expressed in C. necator PHB−4 grown on fructose, were reported to be in the range of 18 to 249 U/g (41). Compared with the activity levels of these wild-type and mutant PHA synthases, PhaCCs clearly exhibited a much higher 3HB-CoA polymerizing activity (2,462 ± 80 U/g).
It is interesting that PhaCCs revealed a homology of 46% with PhaCCn (GenBank accession no. P23608) but only 34% with PhaCAc (GenBank accession no. BAA21815), even though PhaCCs also is known to incorporate the 3HHx monomer into PHA (4, 5). As mentioned earlier, engineered PhaCCn synthases are known to exhibit improved levels of synthase activity and polymer accumulation. The mutant synthases that harbor mutations at F420S, G4D, or G4D/F420S showed improved activities and higher in vivo concentrations of enzyme (25, 26, 39). It was found that the amino acid sequences at positions 4 and 420 in PhaCCs did not contain the altered residues mentioned above. The amino acids at these positions both were identified as phenylalanine. This indicated that the PHA synthase of Chromobacterium sp. USM2 was highly active in its natural form. The characterization of PhaCCs in vitro showed that this enzyme was produced at high concentrations in E. coli cells. Such a high level of expression exhibited by PhaCCs might be correlated with the efficient translation capability of E. coli due to optimal codon usage (33).
Besides the high level of expression, PhaCCs also exhibited a very high level of activity, approximately 8-fold higher than that of PhaCCn. Furthermore, the activity of purified Strep2-PhaCCs was shown to be at least three to five times greater than the activity of pure Strep2-tagged PhaCCn. Preliminary enzymatic assay experiments using 3HV-CoA also suggested that the specific activity of Strep2-PhaCCs is roughly twice as great as that of Strep2-PhaCCn using this substrate (data not shown). It was reported previously that a mutant synthase of A. caviae had an increased specific activity toward 3HB-CoA of approximately 1.6-fold compared to that of the wild-type (0.016 U/mg) (16). PhaCCs exhibits a much higher preference toward 3HB-CoA than to other class I PHA synthases, such as PhaCCn and PhaCAc. The characteristics of PhaCCs in its native strain, Chromobacterium sp. USM2, or in the C. necator PHB−4 transformant have yet to be investigated to determine if these elevated levels of gene expression and enzymatic activity are strain dependent. Nevertheless, the findings from this study have given us invaluable insights on the interesting properties of this synthase.
Results of in vivo evaluation on PhaCCs correlated with results obtained from the in vitro experiments. The synthase efficiently polymerized P(3HB) when glucose was the carbon source. Cells were able to rapidly accumulate P(3HB) to 76 ± 2 wt% within 24 h of cultivation. The highest P(3HB) content of 88 ± 1 wt% was accumulated at 48 h. This resulted in a P(3HB) concentration of 7.2 ± 0.2 g/liter. Previously, E. coli transformants harboring phaCCn were shown to accumulate P(3HB) in the range of 60 to 70 wt% (37). The residual cell biomass was in the range of 1 ± 0.1 to 1.4 ± 0.1 g/liter throughout 72 h of cultivation. An increase in total cell biomass of up to 8.2 ± 0.2 g/liter at 60 h was caused by increasing amounts of intracellular P(3HB) accumulation. The average molecular mass of P(3HB) synthesized by PhaCCs (5 × 105 Da) was found to be lower than that produced by some E. coli transformants harboring different PHA synthases, such as PhaCCn (9.7 × 105 Da) and mutant PHA synthase of Pseudomonas sp. 61-3 (7.2 × 105 Da) (3, 40). The smaller molecular size of the resulting polymer could be attributed to the high in vivo concentration of the synthase (32). As observed in Fig. 4B, many small granules were present in some of the transformants. It is possible that higher concentrations of synthase in vivo could have resulted in the formation of many small granules, which leads to the formation of shorter P(3HB) chains, thus increasing the polydispersity.
The Chromobacterium sp. USM2 synthase produced larger amounts of P(3HB) at 30°C than at 37°C. A significant difference was noticed in the polymer accumulation compared to that at 30°C, whereby a reduction in P(3HB) content of approximately 18% was noticed at the end of cultivation at 37°C (Fig. 3). Nevertheless, residual biomass values (data not shown) indicated that cell growth was not affected by the different cultivation temperatures. The higher accumulation of P(3HB) at a lower temperature could be correlated with the temperature optimum of PhaCCs. Since wild-type Chromobacterium sp. USM2 is known to grow and accumulate PHA at an optimum temperature of 30°C, the synthase potentially is more active at this temperature. The performance of PHA synthases is known to be affected by various temperatures (26).
The PHA synthase of Chromobacterium sp. USM2 has been successfully characterized via in vitro and in vivo assays in E. coli. The synthase exhibited high levels of expression and specific activity toward the polymerization of 3HB-CoA compared to that of the PHA synthase of model strain C. necator. The activity of this natural synthase was found to be higher than that of some of the engineered mutant synthases. This finding raises the possibility that organisms with other such PHA synthases are present in nature and have yet to be discovered. The naturally active PhaCCs can be developed as a model synthase to compare the activity of other synthases.
ACKNOWLEDGMENTS
This study was supported by the Techno Fund, provided by The Ministry of Science, Technology and Innovation, Malaysia (MOSTI). K.B. and J.C. acknowledge a National Science Fellowship awarded by MOSTI and USM's Fellowship Scheme, respectively, for financial support.
We are grateful for the guidance and suggestions provided by Ken'ichiro Matsumoto and Miwa Yamada. We also thank JoAnne Stubbe and Ping Li for the generous gift of some of the 3HB-CoA used in this study and Jingnan Lu for helpful assistance with protein purification.
Footnotes
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Amara A. A., Steinbüchel A., Rehm B. H. A. 2002. In vivo evolution of the Aeromonas punctata polyhydroxyalkanoate (PHA) synthase: isolation and characterization of modified PHA synthases with enhanced activity. Appl. Microbiol. Biotechnol. 59:477–482 [DOI] [PubMed] [Google Scholar]
- 2. Anderson A. J., Haywood G. W., Dawes E. A. 1990. Biosynthesis and composition of bacterial poly(hydroxyalkanoates). Int. J. Biol. Macromol. 12:102–105 [DOI] [PubMed] [Google Scholar]
- 3. Antonio R. V., Steinbüchel A., Rehm B. H. A. 2000. Analysis of in vivo substrate specificity of the PHA synthase from Ralstonia eutropha: formation of novel copolyesters in recombinant Escherichia coli. FEMS Microbiol. Lett. 182:111–117 [DOI] [PubMed] [Google Scholar]
- 4. Bhubalan K., Rathi D. N., Abe H., Iwata T., Sudesh K. 2010. Improved synthesis of P(3HB-co-3HV-co-3HHx) terpolymers by mutant Cupriavidus necator using the PHA synthase gene of Chromobacterium sp. USM2 with high affinity towards 3HV. Polym. Degrad. Stab. 95:1436–1442 [Google Scholar]
- 5. Bhubalan K., Yong K. H., Kam Y. C., Sudesh K. 2010. Cloning and expression of the PHA synthase gene from a locally isolated Chromobacterium sp. USM2. Malays. J. Microbiol. 6:81–90 [Google Scholar]
- 6. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 7. Braunegg G., Lefebvre G., Genser K. F. 1998. Polyhydroxyalkanoates, biopolyesters from renewable resources: physiological and engineering aspects. J. Biotechnol. 65:127–161 [DOI] [PubMed] [Google Scholar]
- 8. Braunegg G., Sonnleitner B., Lafferty R. M. 1978. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur. J. Appl. Microbiol. 6:29–37 [Google Scholar]
- 9. Doi Y., Kitamura S., Abe H. 1995. Microbial synthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Macromolecules 28:4822–4828 [Google Scholar]
- 10. Du G. C., Chen J., Yu J., Lun S. 2001. Feeding strategy of propionic acid for production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) with Ralstonia eutropha. Biochem. Eng. J. 8:103–110 [Google Scholar]
- 11. Fukui T., Doi Y. 1997. Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J. Bacteriol. 179:4821–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerngross T. U., et al. 1994. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry 33:9311–9320 [DOI] [PubMed] [Google Scholar]
- 13. Jia Y., Kappock T. J., Frick T., Sinskey A. J., Stubbe J. 2000. Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry 39:3927–3936 [DOI] [PubMed] [Google Scholar]
- 14. Jia Y., et al. 2001. Mechanistic studies on class I polyhydroxybutyrate (PHB) synthase from Ralstonia eutropha: class I and III synthases share a similar catalytic mechanism. Biochemistry 40:1011–1019 [DOI] [PubMed] [Google Scholar]
- 15. Kichise T., Fukui T., Yoshida Y., Doi Y. 1999. Biosynthesis of polyhydroxyalkanoates (PHA) by recombinant Ralstonia eutropha and effects of PHA synthase activity on in vivo PHA biosynthesis. Int. J. Biol. Macromol. 25:69–77 [DOI] [PubMed] [Google Scholar]
- 16. Kichise T., Taguchi S., Doi Y. 2002. Enhanced accumulation and changed monomer composition in polyhydroxyalkanoate (PHA) copolyester by in vitro evolution of Aeromonas caviae PHA synthase. Appl. Environ. Microbiol. 68:2411–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S. Y. 1996. Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 49:1–14 [DOI] [PubMed] [Google Scholar]
- 18. Lee S. Y. 1996. Plastic bacteria? Progress and prospects for polyhydroxyalkanoate production in bacteria. Trends Biotechnol. 14:431–438 [Google Scholar]
- 19. Li R., Zhang H., Qi Q. 2007. The production of polyhydroxyalkanoates in recombinant Escherichia coli. Bioresour. Technol. 98:2313–2320 [DOI] [PubMed] [Google Scholar]
- 20. Matsumoto K., Takase K., Aoki E., Doi Y., Taguchi S. 2005. Synergistic effects of Glu130Asp substitution in the type II polyhydroxyalkanoate (PHA) synthase: enhancement of PHA production and alteration of polymer molecular weight. Biomacromolecules 6:99–104 [DOI] [PubMed] [Google Scholar]
- 21. Matsusaki H., Abe H., Taguchi K., Fukui T., Doi Y. 2000. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant bacteria expressing the PHA synthase gene phaC1 from Pseudomonas sp. 61-3. Appl. Microbiol. Biotechnol. 53:401–409 [DOI] [PubMed] [Google Scholar]
- 22. McDowell E. M., Trump B. F. 1976. Histologic fixatives suitable for diagnostic light and electron-microscopy. Arch. Pathol. Lab. Med. 100:405–414 [PubMed] [Google Scholar]
- 23. Müh U., Sinskey A. J., Kirby D. P., Lane W. S., Stubbe J. 1999. PHA synthase from Chromatium vinosum: cysteine 149 is involved in covalent catalysis. Biochemistry 38:826–837 [DOI] [PubMed] [Google Scholar]
- 24. Murata T., Takase K., Yamato I., Igarashi K., Kakinuma Y. 1997. Purification and reconstitution of Na+-translocating vacuolar ATPase from Enterococcus hirae. J. Biol. Chem. 272:24885–24890 [DOI] [PubMed] [Google Scholar]
- 25. Normi Y. M., et al. 2005. Characterization and properties of G4X mutants of Ralstonia eutropha PHA synthase for poly(3-hydroxybutyrate) biosynthesis in Escherichia coli. Macromol. Biosci. 5:197–206 [DOI] [PubMed] [Google Scholar]
- 26. Normi Y. M., et al. 2005. Site-directed saturation mutagenesis at residue F420 and recombination with another beneficial mutation of Ralstonia eutropha polyhydroxyalkanoate synthase. Biotechnol. Lett. 27:705–712 [DOI] [PubMed] [Google Scholar]
- 27. Park S. J., Choi J. I., Lee S. P. 2005. Short-chain-length polyhydroxyalkanoates: synthesis in metabolically engineered Escherichia coli and medical applications. J. Microbiol. Biotechnol. 15:206–215 [Google Scholar]
- 28. Pötter M., Steinbüchel A. 2005. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules 6:552–560 [DOI] [PubMed] [Google Scholar]
- 29. Rehm B. H. A. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Schubert P., Steinbüchel A., Schlegel H. G. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J. Bacteriol. 170:5837–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sim S. J., et al. 1997. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat. Biotechnol. 15:63–67 [DOI] [PubMed] [Google Scholar]
- 33. Slater S. C., Voige W. H., Dennis D. E. 1988. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthetic pathway. J. Bacteriol. 170:4431–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song J. J., Zhang S., Lenz R. W., Goodwin S. 2000. In vitro polymerization and copolymerization of 3-hydroxypropionyl-CoA with the PHB synthase from Ralstonia eutropha. Biomacromolecules 1:433–439 [DOI] [PubMed] [Google Scholar]
- 35. Spurr A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31–43 [DOI] [PubMed] [Google Scholar]
- 36. Steinbüchel A., Lütke-Eversloh T. 2003. Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates in microorganisms. Biochem. Eng. J. 16:81–96 [Google Scholar]
- 37. Taguchi K., et al. 2002. Metabolic pathways and engineering of PHA biosynthesis, p. 217–247 In Doi Y., Steinbüchel A. (ed.), Biopolymer handbook. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 38. Taguchi S., Doi Y. 2004. Evolution of polyhydroxyalkanoate (PHA) production system by “enzyme evolution”: successful case studies of directed evolution. Macromol. Biosci. 4:145–156 [DOI] [PubMed] [Google Scholar]
- 39. Taguchi S., Nakamura H., Hiraishi T., Yamato I., Doi Y. 2002. In vitro evolution of a polyhydroxybutyrate synthase by intragenic suppression-type mutagenesis. J. Biochem. 131:801–806 [DOI] [PubMed] [Google Scholar]
- 40. Taguchi S., et al. 2008. A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme. Proc. Natl. Acad. Sci. U. S. A. 105:17323–17327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takase K., Matsumoto K., Taguchi S., Doi Y. 2004. Alteration of substrate chain-length specificity of type II synthase for polyhydroxyalkanoate biosynthesis by in vitro evolution: in vivo and in vitro enzyme assays. Biomacromolecules 5:480–485 [DOI] [PubMed] [Google Scholar]
- 42. Tsuge T., et al. 2007. Variation in copolymer composition and molecular weight of polyhydroxyalkanoate generated by saturation mutagenesis of Aeromonas caviae PHA synthase. Macromol. Biosci. 7:846–854 [DOI] [PubMed] [Google Scholar]
- 43. Yuan W., et al. 2001. Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch. Biochem. Biophys. 394:87–98 [DOI] [PubMed] [Google Scholar]
- 44. Zhang S., Yasuo T., Lenz R. W., Goodwin S. 2000. Kinetic and mechanistic characterization of the polyhydroxybutyrate synthase from Ralstonia eutropha. Biomacromolecules 1:244–251 [DOI] [PubMed] [Google Scholar]
- 45. Zinn M., Witholt B., Egli T. 2001. Occurrence, synthesis and medical application of bacterial polyhydroxyalkanoate. Adv. Drug Deliv. Rev. 53:5–21 [DOI] [PubMed] [Google Scholar]