Abstract
Microbial mats in marine cold seeps are known to be associated with ascending sulfide- and methane-rich fluids. Hence, they could be visible indicators of anaerobic oxidation of methane (AOM) and methane cycling processes in underlying sediments. The Napoli mud volcano is situated in the Olimpi Area that lies on saline deposits; from there, brine fluids migrate upward to the seafloor. Sediments associated with a brine pool and microbial orange mats of the Napoli mud volcano were recovered during the Medeco cruise. Based on analysis of RNA-derived sequences, the “active” archaeal community was composed of many uncultured lineages, such as rice cluster V or marine benthic group D. Function methyl coenzyme M reductase (mcrA) genes were affiliated with the anaerobic methanotrophic Archaea (ANME) of the ANME-1, ANME-2a, and ANME-2c groups, suggesting that AOM occurred in these sediment layers. Enrichment cultures showed the presence of viable marine methylotrophic Methanococcoides in shallow sediment layers. Thus, the archaeal community diversity seems to show that active methane cycling took place in the hypersaline microbial mat-associated sediments of the Napoli mud volcano.
INTRODUCTION
More than 200 mud volcanoes have been found along the northern flank of the Mediterranean Ridge in the eastern Mediterranean Sea (12). The formation of the Mediterranean Ridge is linked to the collisional tectonics between the African and Eurasian plates, resulting in intensive faulting (18). Within the Mediterranean Ridge, the Olimpi area, situated south of Crete, is a dynamic environment containing active seepage of mud, fluid, and brines. During the Messinian salinity crisis, evaporites were deposited in the Mediterranean Basins (69), resulting in continuous evaporite dissolution and brines migrating upwards in the Olimpi area (13). Mud volcanism is often associated with brine seeps in this area (69). The Napoli mud volcano is a submarine circular dome situated in the Olimpi area (Fig. 1). Subsurface brines reaching the seafloor of the mud volcano create brine pools and lakes with diameters ranging from centimeters to meters (12). The highest fluid flows are located near the physical center of the mud volcano, where mud mixed with brine enriched in biogenic methane is mostly expelled (12).
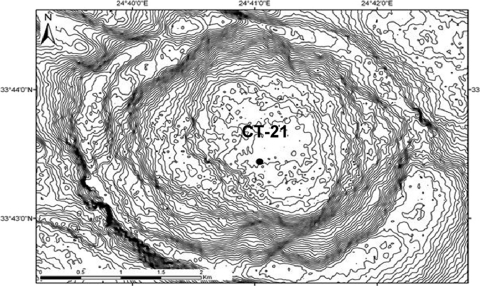
Fig. 1.
Closer view of the Napoli mud volcano and the position of the sampled push core (Bénédicte Ritt, unpublished data, Medeco cruise 2007 [reprinted with permission from Ifremer]).
Most of the methane rising up does not reach the seafloor because it is mainly consumed by an efficient microbially mediated process known as anaerobic oxidation of methane (AOM) (36). AOM has been documented in various anoxic marine sediments, such as sediments of mud volcanoes (44), hydrothermal vents (61), and hypersaline environments (35). AOM is driven by anaerobic methanotrophs (ANME) of the Archaea and is mainly coupled to sulfate reduction driven by sulfate-reducing bacteria. ANME are divided into three phylogenetic groups: ANME-1, ANME-2, and ANME-3. The ANME-1 Archaea are distantly affiliated with the methanogenic orders Methanosarcinales and Methanomicrobiales, the ANME-2 are affiliated with the methanogenic order Methanosarcinales, and the ANME-3 are affiliated with the methanogenic genera Methanococcoides and Methanolobus. Alternative electron acceptors such as NO2− (53), Fe3+, and Mn4+ (4) have been recently reported to be coupled to AOM with higher energy yields, based on thermodynamic estimations. Thus far, no pure culture or defined consortium of ANME has been isolated, and the biochemical pathways of AOM remain unknown. In the current reverse methanogenesis hypothesis, i.e., CO2 reduction to CH4, methane oxidation is catalyzed by a modified methyl coenzyme M reductase (MCR) (21, 22, 31), which in methanogens catalyzes the final step of methanogenesis (62). The mcrA gene, encoding the MCR, is unique and found in all methanogens and anaerobic methanotrophic Archaea (62). Phylogenetic mcrA-based trees mirror the phylogeny of the 16S rRNA genes for all known methanogens (19, 37). Moreover, the mcrA genes are conserved, making them specific and useful functional gene markers, targeting methanogens and methanotrophs in the environment.
Dense filamentous microbial mats on the seafloor of cold seep sediments are visible to the naked eye. These mats are mainly composed of multicellular filaments (diameters of 12 to 160 μm [40]) and are either pigmented (e.g., orange or white) or unpigmented. Microbial communities in sediments underlying microbial mats have been shown to support high rates of sulfate reduction (7, 26), sulfur oxidation (49), nitrate reduction (7, 26), and anaerobic methane oxidation (7, 26). Members of these communities have been previously identified as filamentous sulfur-oxidizing bacteria of the Beggiatoa, Thioplaca, Leucothrix, Thirotrix, and Desmanthos genera (23), as well as diverse Proteobacteria (23, 42) and Archaea (41, 42). Interestingly, the archaeal communities in sediments underlying seep-associated microbial mats are dominated by methanogens and methane oxidizers (34). Thus, sediments underlying mats provide alternative niches for diverse active archaeal communities adapted to dynamic changes of fluid flow regimens.
We analyze here archaeal community structure and diversity with depth in hypersaline sediments associated with orange-pigmented mats of the Napoli mud volcano. The main objectives were to characterize the archaeal communities in hypersaline sediments underlying dense microbial mats. Vertical distribution patterns of archaeal communities were assessed by using PCR-DGGE. Total RNA was extracted from 0 to 4 cm below seafloor (cmbsf) and 6- to 10-cmbsf sediment layers and subjected to reverse transcription-PCR with primers specific to the archaeal 16S rRNA genes. The archaeal methanotroph and methanogen diversity was then determined based on mcrA genes from two different sediment depths (2 to 4 cmbsf and 8 to 10 cmbsf). Finally, since methane was previously shown to mainly have a biogenic origin in the Napoli mud volcano, enrichment cultures for methanogens were carried out at all depths.
MATERIALS AND METHODS
Sediment sampling and porewater analysis.
Sediment samples were collected in the Napoli Mud Volcano in the Eastern Mediterranean Sea during the Ifremer Medeco cruise with the research vessel Pourquoi Pas? in October and November 2007. Sediment push-core CT-21 (Fig. 1) was recovered during dive PL 331-10 by the remotely operated vehicle (ROV) VICTOR 6000 (Ifremer) at 1,938 meters of water depth (N33°43.4397, E24°41.0385). In the sampled area, sediments were recovered with dense orange microbial mats. Brine pools and rivers were observed in close proximity to the microbial orange mats. The sediment push-core sample contained bacterial orange filaments that penetrated the first 2- to 3-cm layers. Immediately after retrieval, the sediment core (10 cm long) was sectioned aseptically in 2-cm-thick layers in the cooling room (4°C) and then frozen at −80°C for nucleic acid extractions.
Depth profiles of dissolved pore water sulfate and chloride were quantified from diluted pore waters. Pore water was obtained by centrifuging ∼10 g of crude sediment (15 min, 3,000 × g) at 4°C. The pore water was then stored at −20°C. Depth profiles of dissolved pore water sulfate and chloride were quantified from diluted pore waters. Sulfate and chloride concentrations were measured by using ion-exchange chromatography, with an isocratic DX120 ion chromatography system (Dionex Corp., Sunnyvale, CA) fitted with Ionpas AS9-SC columns and a suppressor (ASRS-ultra II) unit in combination with a DS4-1 heated conductivity cell. Components were separated by using a sodium carbonate gradient, with a flow of 1.5 ml/min.
Culture media for enrichment of methanogens.
One volume of sediment subsample (10 cm3) was transferred into an anaerobic cabinet and then into 50-ml vials containing 1 volume (10 ml) of sterile and reduced artificial seawater (ASW) medium. ASW corresponded to medium 141 from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) devoid of organic carbon substrates. Enrichments were performed anaerobically in 50-ml vials according to the method of Balch and Wolfe (2). Medium 141 from the DSMZ was used with slight modifications: organic substrates were omitted, except for yeast extract with a concentration adjusted to 0.2 g/liter. The medium was prepared and sterilized under 80% N2 and 20% CO2 gas atmosphere. In order to enrich CO2-reducing, acetoclastic, and methylotrophic methanogens, three enrichment media supplemented with H2 (200 kPa), acetate (10 mM), and trimethylamine (TMA; 20 mM) were used. One gram of sediment from the different sections of CT-21 was inoculated into 9 ml of medium (pH 7), the suspension was mixed and serially diluted to 10−3. The cultures were incubated at 15°C to mimic in situ conditions. Cultures were periodically checked for methane production for 1 year. Methane was detected directly in the headspace of vial cultures by a micro MTI M200 gas chromatograph equipped with an MS-5A capillary column and a Poraplot U capillary column. Positive enrichment dilutions of methanogens were monitored by PCR-denaturing gradient gel electrophoresis (DGGE). For dilutions showing one DGGE band on the fingerprint, 16S rRNA genes were amplified, cloned, and sequenced using the A8F and A1492R primers.
Nucleic acid extraction and purification.
Total genomic DNA was directly extracted and purified from 5 g of wet sediment for all sections in duplicates, using the Zhou et al. (68) method with modifications. Sediment samples were mixed with DNA extraction buffer as described previously (68), followed by three cycles of freezing in liquid N2 and thawing at 65°C. The pellet of crude nucleic acids obtained after centrifugation was washed with cold 80% ethanol and resuspended in sterile deionized water to give a final volume of 100 μl. Crude DNA extracts were then purified by using a Wizard DNA cleanup kit (Promega, Madison, WI). DNA extracts were separated into aliquots and stored at −20°C until required for PCR amplification. Total RNA was directly extracted and purified from 2 g of wet sediment from pooled sediment sections from 0 to 4 cmbsf and 6 to 10 cmbsf, using an RNA PowerSoil total RNA isolation kit (MO-Bio Labs., Inc., Carlsbad, CA) according to the manufacturer's recommendations. Aliquots of RNA extracts were treated by Turbo DNase (Applied Biosystems, Foster City, CA) and purified by using an RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The quality of the RNA samples was examined by agarose gel electrophoresis, and concentrations were determined by using spectrophotometry (Nanodrop ND-100; NanoDrop Technologies, Wilmington, DE).
Archaeal 16S rRNA PCR-DGGE amplification.
Archaeal 16S rRNA genes were amplified by PCR from purified DNA extracts using the primer pair 8F (5′-CGGTTGATCCTGCCGGA-3′) and 1492R (5′-GGCTACCTTGTTACGACTT-3′) (9). All PCRs (total volume of reaction mixture, 25 μl) contained 1 μl of purified DNA template, 1× PCR buffer (Promega), 2 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphates, 0.4 mM concentrations of each primer (Eurogentec), and 0.6 U of GoTaq DNA polymerase (Promega). Amplification was carried out by using a GeneAmp PCR 9700 System (Applied Biosystems). The PCR conditions were as follows: denaturation at 94°C for 1 min, annealing at 49°C for 1 min 30 s, and extension at 72°C for 2 min for 30 cycles. All of the archaeal 16S rRNA gene PCR products were then reamplified in a nested PCR with primer 340F (5′-CCCTACGGGGYGCASCAG-3′) (64) containing a GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-3′) at the 5′ end and primer 519R (5′-TTACCGCGGCKGCTG-3′) (50). The PCR conditions were as follows: denaturation at 94°C for 30 s, annealing at 72 to 62°C (touchdown, −0.5°C per cycle) for 30 s, and extension at 72°C for 1 min for 20 cycles, followed by denaturation at 94°C for 30 s, annealing at 62°C for 30 s, and extension at 72°C for 1 min for 10 cycles, with a final extension at 72°C for 30 min (24).
To restrict contamination to a minimum, PCR experiments was carried out under aseptic conditions (Captair Bio, Erlab; Fisher Bioblock Scientific) using autoclaved and UV-treated plasticware and pipettes, and only sterile nuclease-free molecular-grade water (MP Biomedicals, Solon, OH). Positive (DNA extracted from pure cultures) and negative (molecular-grade water) controls were used in all PCR amplifications.
DGGE fingerprinting analysis.
DGGE was carried out as described by Toffin et al. (63) with some modifications. PCR products were separated by DGGE using a D-Gene System (Bio-Rad Laboratories, Hercules, CA) on 8% (wt/vol) polyacrylamide gels (40% acrylamide/bis solution, 37.5:1 [Bio-Rad]) with a gradient of denaturant between 20 and 60% (100% denaturant consists of 7 M urea and 40% [vol/vol] formamide). Gels were poured with the aid of a 30-ml volume gradient mixer (Hoefer SG30; GE Healthcare, Buckinghamshire, United Kingdom) and prepared with 1× TAE buffer (MP Biomedicals, Solon, OH). Electrophoresis was carried out at 60°C and 200 V for 5 h (with an initial electrophoresis for 10 min at 80 V) in 1× TAE buffer. Polyacrylamide gels were stained with SYBR Gold nucleic acid gel stain (Invitrogen, San Diego, CA) for 30 min and viewed using a Typhoon 9400 variable mode imager (GE Healthcare).
Construction of RNA-derived 16S rRNA gene libraries.
RNA-derived cDNA was synthesized by reverse transcription using the 16S rRNA archaeal primer 915R (5′-GTGCTCCCCCGCCAATTCCT-3′) (8) and Moloney murine leukemia virus reverse transcriptase (M-MuLV; MP Biomedicals, Irvine, CA) according to the manufacturer's protocol. Purified RNA (100 to 150 ng) was initially denatured at 65°C for 10 min, and 7.7 μM primer 915R was added to the denatured RNA. The mixture was incubated at 70°C for 10 min. The reverse transcription reaction mixture (total volume, 22 μl) consisted of denatured RNA, 1× M-MuLV buffer, 200 μM deoxynucleoside triphosphate mix, and 10 mM dithiothreitol. The reverse transcription reaction mix was incubated at 42°C for 2 min. A 200-U aliquot of M-MuLV reverse transcriptase was added prior to an 80-min incubation at 42°C for the reverse transcription of the RNA into cDNA. The reaction was then stopped by heating at 70°C for 15 min. The cDNA end product was used as a template for archaeal 16S rRNA gene-based PCR using the primer set 340F/915R. The PCR amplification involved 20 cycles of 94°C for 1 min, 71 to 61°C (touchdown, −1°C per cycle) for 1 min, and 72°C for 2 min. PCR products were purified with the QIAquick gel extraction kit (Qiagen), analyzed on 1% (wt/vol) agarose gels in 1× TAE buffer, stained with ethidium bromide, and UV illuminated. Purified PCR products were cloned into a TOPO XL PCR cloning kit and transformed into Escherichia coli TOP10 One Shot cells (Invitrogen) according to the manufacturer's recommendations. Control PCR using the purified RNA and the same primers was performed to monitor possible DNA contamination of the RNA templates. No contaminating DNA was detected in any of these reaction mixtures.
Construction of mcrA environmental gene libraries.
The mcrA genes were amplified using the primers ME1 (5′-GCMATGCARATHGGWATGTC-3′) and ME2 (5′-TCATKGCRTAGTTDGGRTAGT-3′) (19). The PCR conditions were as follows: denaturation at 94°C for 40 s, annealing at 50°C for 1 h 30 min, and extension at 72°C for 3 min for 30 cycles. PCR products were purified on a 1% agarose gel by using a QIAquick gel extraction kit (Qiagen) and cloned using a TOPO XL PCR cloning kit (Invitrogen) according to the manufacturer's protocols.
Phylogenetic analysis of DNA.
The gene sequencing was performed by Taq cycle sequencing and determined on a ABI Prism 3100 genetic analyzer (Applied Biosystems) using the M13R universal primer (5′-CAGGAAACAGCTATGAC-3′). RNA-derived cDNA, DNA-derived mcrA, and enrichment culture-derived 16S rRNA gene sequences were analyzed by using the NCBI BLASTN search program within GenBank (http://blast.ncbi.nlm.nih.gov/Blast) (1). Potential chimeric sequences in the clone libraries were identified with the CHIMERA CHECK program of the Ribosomal Database Project II (Center for Microbial Ecology, Michigan State University [http://wdcm.nig.ac.jp/RDP/html/analyses.html]). Potential chimeras were eliminated before phylogenetic trees were constructed. The RNA-derived 16S rRNA sequences and the enrichment culture-derived 16S rRNA gene sequences were then edited in the BioEdit v7.0.5 program (20) and aligned using the SINA webaligner (http://www.arb-silva.de/) (52). The mcrA sequences were translated into amino acid sequences by using BioEdit and aligned by using ClustalX (32). Sequence data were analyzed with the MEGA4.0.2 program (60). The phylogenetic trees were calculated by using the neighbor-joining method. The robustness of the inferred topology was tested by bootstrap resampling (1,000 replicates). Rarefaction curves were calculated for the RNA-derived 16S rRNA and mcrA gene libraries by using the RarFac program (http://www.icbm.de/pmbio/) and a 97% similarity cutoff value for sequence-based operational taxonomic units (OTUs). Gene library coverage (C) was calculated using the following formula: C = [1 − (n1/N)] × 100, where n1 is the number of unique OTUs and N is the number of clones in the library (57).
Statistical analyses of DGGE banding patterns.
The DGGE profile was analyzed as described by Fry et al. (14), using a presence or absence scoring of the DGGE bands. After a grid was made to determine whether bands were present (score = 1) or absent (score = 0) for each lane on a same line of the DGGE profile, a presence/absence matrix was obtained. This matrix was then used to build a similarity matrix based on the Jaccard coefficient, using the vegan package within the R software (54). Finally, a dendrogram was obtained using the Ward agglomeration method within the hierarchical clustering package of the R software.
Nucleotide sequence accession numbers.
The sequence data reported here will appear in the GenBank nucleotide sequence database under the accession numbers HM004785 to HM004825 and HQ443429 to HQ443514 for RNA-derived 16S rRNA gene sequences, HM004828 to HM004903 and HQ454430 to HQ454493 for mcrA gene sequences, and HM004826 to HM004827 for enrichment culture-derived 16S rRNA gene sequences.
RESULTS AND DISCUSSION
Geochemical and biological characteristics.
Observation of large orange-pigmented mats on the surface of the sampled sediment core and direct microscopic examination of filamentous morphology indicated that the bacteria were possibly members of the genus Beggiatoa or the genus Thioploca, as reported elsewhere (45, 55).
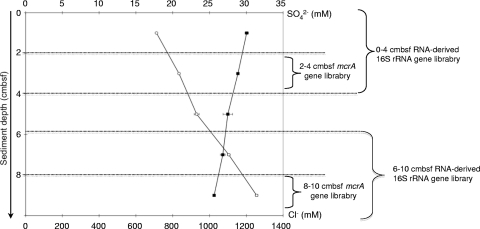
Profiles of sulfate and chloride concentrations in pore water sediments underlying the orange microbial mats were anticorrelated. The chloride pore water profile showed an increase in concentration with depth (Fig. 2), reaching 1,200 mM at 10 cmbsf, which is more than two times higher than normal seawater concentrations (600 mM) (12). The sodium concentrations showed a similar increase with depth (see Fig. S1 in the supplemental material), reaching 1,224 mM at 10 cmbsf, which is also more than two times higher than normal seawater concentrations (492 mM) (12). Furthermore, Charlou et al. characterized the brines in Napoli as being enriched in Cl and Na. Hence, increases in chloride and sodium were presumably linked to the upflowing brines from deep sources in the Napoli mud volcano. The surface sediment layers colonized by the orange-pigmented mat bacteria showed chloride concentrations of 700 mM and could be influenced by the brine pools contiguous to the filamentous bacteria on the seafloor. Moreover, bacterial mats are a common feature found in habitats influenced by hypersaline brine fluid intrusions (42, 47).
Fig. 2.
Sulfate (■) and chloride (○) pore water concentrations (mean ± the standard deviation) in mM with depth for core CT-21 of the Napoli mud volcano sediments given in cmbsf. The scale represents the sediment depth below the seafloor. Sediment sections dedicated to mcrA- and RNA-derived 16S rRNA are indicated on the right.
The sulfate pore water concentrations showed a slight decrease with depth (Fig. 2), which could suggest sulfate reduction. Sediments associated with orange- and white-pigmented Beggiatoa mats have been shown to host high rates of sulfate reduction, probably a combination of increased substrate availability in the seep fluids, and of rapid sulfate recycling within the sulfur-oxidizing bacterial mat (42, 47). However, the Napoli sediments did not seem to have a clear sulfate-reducing zone. Profiles of the Mg2+ and Ca2+ pore water concentrations (see Fig. S1 in the supplemental material) showed a decrease with depth. Concentrations reached 34 and 3 mM for Mg2+ and Ca2+, respectively, at 10 cmbsf (whereas seawater concentrations are typically 56 and 11 mM) (12), suggesting authigenic carbonate precipitation in these sediment layers. Indeed, anaerobic oxidation of methane increases alkalinity in pore water fluids by producing HCO3−, which in turn reacts with and precipitates Mg2+ and Ca2+ cations (10, 29).
Vertical distribution of the archaeal communities.
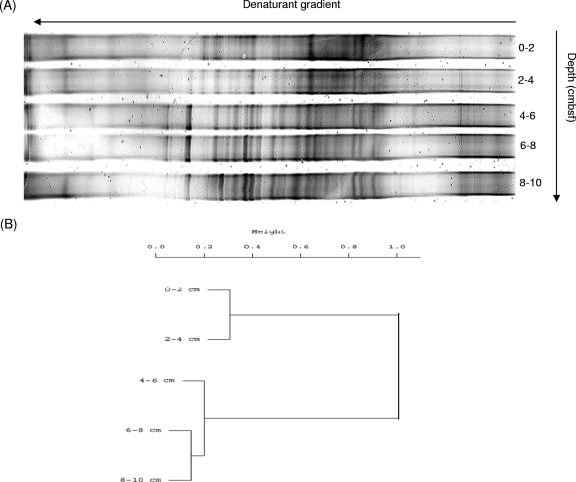
The DGGE fingerprints (Fig. 3A) generated from DNA samples extracted from sediment layers associated with orange-pigmented microbial mats of the Napoli mud volcano displayed a complex and diverse distribution of the archaeal communities. The resulting dendrogram (Fig. 3B) of the DGGE pattern highlighted clear changes in populations with depth in two separate clusters. The first cluster grouped depths of 0 to 4 cmbsf, and the second grouped the 4- to 10-cmbsf sediment layers, suggesting a change in archaeal populations at 4 cmbsf, with increasing depth and salinity. This shift could be linked to the presence of the bacterial filaments in these sediments. Indeed, these filaments belonging to what are commonly called “big bacteria” could have locally modified the geochemical conditions in the surrounding sediments (55), therefore affecting the microbial community diversity in the upper 4 cm. Salt concentrations might also have affected the depth distribution of the microbial communities, as reported elsewhere (27).
Fig. 3.
(A) DGGE analysis of archaeal 16S rRNA genes obtained by nested PCR in the Napoli mud volcano. (B) Dendrogram obtained from clustering analysis of DGGE banding profiles, with bands scored as present or absent using the R software. The bar indicates the dissimilarity values.
According to the statistical analysis of the DGGE pattern indicating a shift in the archaeal community at 4 cmbsf, we constructed two RNA-derived 16S rRNA gene libraries for depths of 0 to 4 cmbsf and 6 to 10 cmbsf. A total of 55 archaeal RNA-derived 16S rRNA gene sequences were analyzed for the 0- to 4-cmbsf section and 72 for the 6- to 10-cmbsf section. Rarefaction curves generated for the RNA-derived 16S rRNA genes indicated saturation (see Fig. S2 in the supplemental material), while the percent coverage was determined to be 40 and 72.2% for the 0- to 4-cmbsf and 6- to 10-cmbsf sections, respectively. Hence, coverage analysis suggests that the full diversity of archaeal 16S rRNA sequences was not exhausted and that a greater diversity remains to be detected within these sediments. Simpson's diversity indices (56) were calculated for each section, and these indicated a decrease in archaeal diversity with depth (D = 0.9554 for the 0- to 4-cmbsf section and 0.9109 for the 6- to 10-cmbsf section).
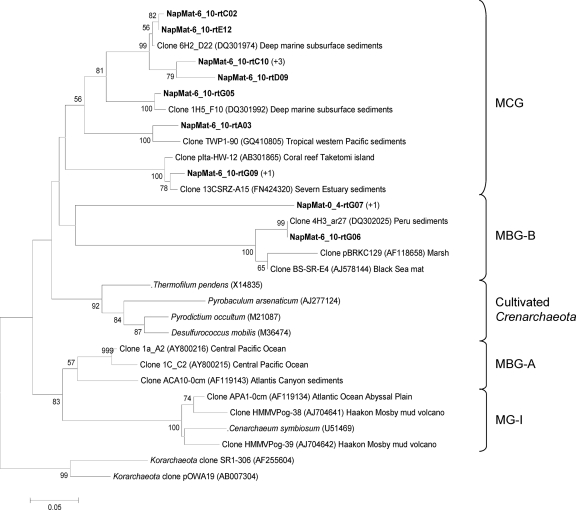
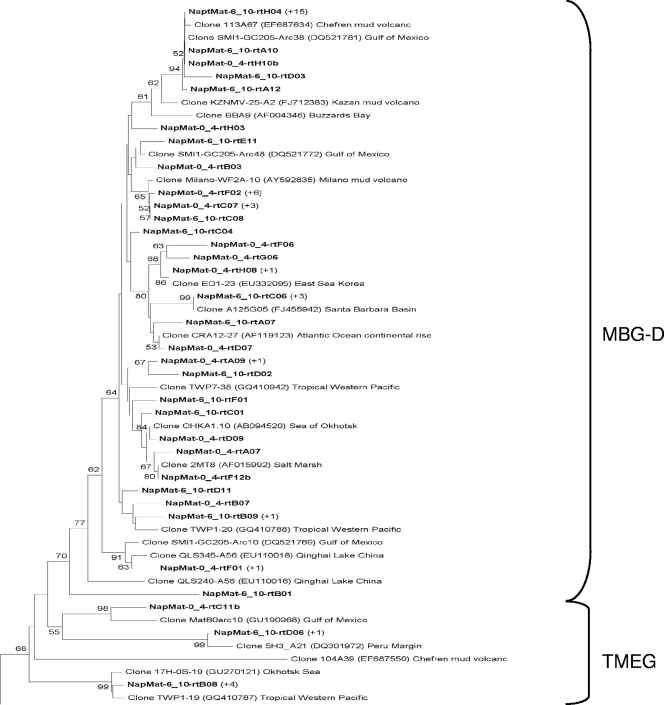
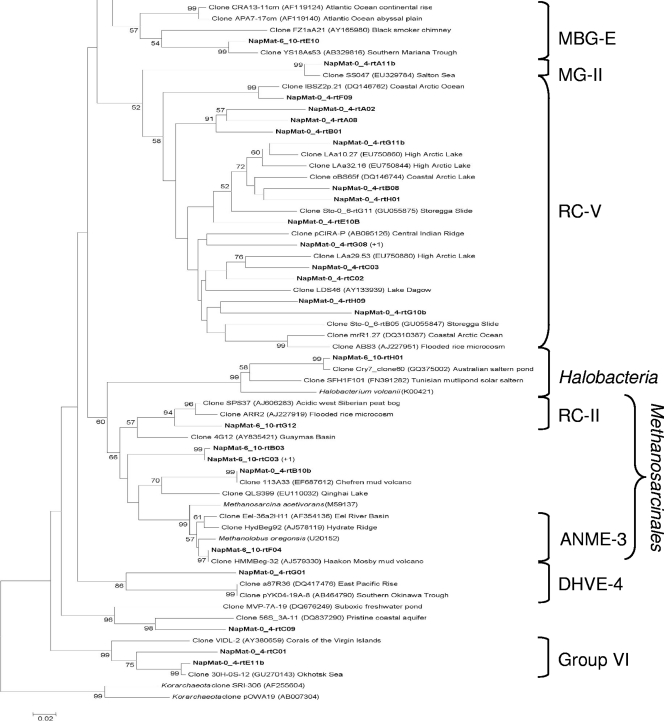
The phylogenetic trees of the RNA-derived 16S rRNA gene libraries showed high archaeal diversity, and a majority of sequences were most closely related to environmental clones from mud volcano sediments of the Mediterranean Sea (i.e., Milano, Kazan, Chefren), marine sediments, and subseafloor sediments (i.e., Peru margin). Thirty-eight OTUs belonging to archaeal uncultured groups were detected in the 0- to 4-cmbsf sediment layer 16S gene library and belonged to two major groups (Fig. 4 and 5): marine benthic group D (MBG-D; 40.5%) and rice cluster V (RC-V; 40.5%). The other minor groups that were detected also belong to uncultured archaeal lineages (Fig. 4 and 5), i.e., deep-sea hydrothermal vent euryarchaeotic group 4 (DHVE-4), group VI, the terrestrial miscellaneous euryarchaeotal group (TMEG), marine group II (MG-II), the miscellaneous crenarchaeotic group (MCG), and marine benthic group B (MBG-B). One clone (NapMat-0_4-rtC09) was not related to any known group. The only sequence closely related to cultured prokaryotes (NapMat-0_4-rtB11b) was related to the methanogenic order of the Methanosarcinales (see Table S1 in the supplemental material). Thirty-one OTUs also belonging to archaeal uncultured groups were detected in the 6- to 10-cmbsf sediment layer 16S gene library and belonged to two major groups (Fig. 4 and 5): MBG-D (55%) and MCG (17.5%). Other minor groups related to uncultured archaeal groups were detected as well (Fig. 4 and 5), i.e., TMEG, marine benthic group E (MBG-E), and MBG-B. Two clones were affiliated with the methanogenic order Methanosarcinales, one with the anaerobic methanotrophic group ANME-3 and one with the rice cluster II, which are surmised to be involved in methane production (17), and finally one clone (NapMat-6_10-rtH01) was affiliated with an extreme halophilic Archaea of the Halobacteria (46). Most of the sequences only detected in the 6- to 10-cmbsf section (i.e., MBG-B, MCG, and TMEG) were closely related with sequences retrieved from subsurface sediments of the Peru margin (6, 59).
Fig. 4.
Phylogenetic analysis of the crenarchaeal RNA-derived 16S rRNA genes of the Napoli mud volcano sediments based on the neighbor-joining method with 575 homologous positions. The percent bootstrap values are based on 1,000 replicates and are indicated at the nodes for branch values with ≥50% bootstrap support. Gene sequences recovered in the present study from Napoli mud volcano sediments are in boldface type. Clones with designation beginning NapMat-0_4 and NapMat-6_10 are from the sediment sections from 0 to 4 cmbsf and from 6 to 10 cmbsf, respectively. Numbers in parentheses indicate the number of analyzed clones that have >97% sequence identity. The scale bar indicates five substitutions per 100 nucleotides. MCG, miscellaneous crenarchaeotic group; MBG-B, marine benthic group B; MBGA, marine benthic group A; MG-I, marine group I.
Fig. 5.
Phylogenetic analysis of the euryarchaeal RNA-derived 16S rRNA genes of the Napoli mud volcano sediments based on the neighbor-joining method with 575 homologous positions. The percent bootstrap values are based on 1,000 replicates and are indicated at the nodes for branch values with ≥50% bootstrap support. Gene sequences recovered in the present study from Napoli mud volcano sediments are in boldface type. Clones with designations beginning with NapMat-0_4 and NapMat-6_10 are from the sediment sections from 0 to 4 cmbsf and from 6 to 10 cmbsf, respectively. Numbers in parentheses indicate the number of analyzed clones that have >97% sequence identity. The scale bar indicates two substitutions per 100 nucleotides. RCV, rice cluster V; MBG-D, marine benthic group D; TMEG, terrestrial miscellaneous euryarchaeotal group; MBG-E, marine benthic group E; MG-II, marine group II; RC-II, rice cluster II; DHVE-4, deep sea hydrothermal vent euryarchaeotal group 4.
Intriguingly, RC-V members were presumably active in the 0- to 4-cmbsf sediments underlying orange-pigmented mats. The RC-V organisms were first discovered in an anoxic flooded rice paddy soil (17), detected in many freshwater sediments (11, 16, 66, 67), and were recently shown to be active in tubeworm populated sediments of the Storegga Slide (33). The intralineage levels of rRNA gene similarity of the RC-V sequences were low, highlighting that this group seems to be phylogenetically very diverse. This could suggest diverse metabolic activities and physiologies. RC-V group organisms were also previously observed in cold coastal waters of the Mackenzie River in northwestern Canada, which are rich in suspended particles (15). The authors of that study suggested that RC-V organisms were linked to detrital decomposition. The Napoli sediments in which the RC-V organisms were detected had a high organic matter content (data not shown), which could support the hypothesis of an organotrophic metabolism. The present study is the first to report the occurrence of probable active members of the RC-V group in hypersaline sediments. Also, sequences affiliated with the RC-V group were not detected in the 6- to 10-cmbsf sediment layers, where the salinity reached 1,300 mM, suggesting that members of the RC-V group probably do not tolerate high salt concentrations.
Sequences affiliated with the archaeal uncultured MBG-D were detected in many saline or hypersaline environments (5, 25, 35, 47, 58). Jiang et al. proposed that salinity could play a role in controlling the distribution of marine benthic groups, such as the MBG-D (25). Furthermore, the MBG-D group was the main archaeal group presumably “active” in both sections (0 to 4 cmbsf and 6 to 10 cmbsf) where Cl− and Na+ pore water concentrations were high (834 and 792 mM, respectively).
Diversity and distribution of the ANME.
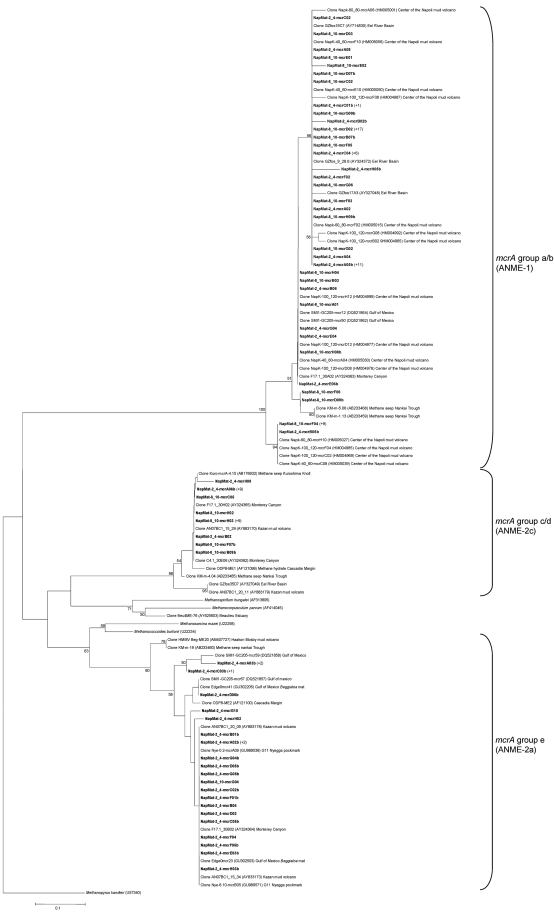
In order to demonstrate whether ANME- and/or methanogen-affiliated sequences in hypersaline sediments are phylogenetically distinct from sequences in nonhypersaline conditions, mcrA gene libraries were constructed for representative sediment sections characterized by increasing Cl− and Na+ pore water concentrations, i.e., 2 to 4 cmbsf and 8 to 10 cmbsf. A total of 75 mcrA sequences were analyzed for the 2- to 4-cmbsf sediment section, and 65 mcrA sequences were analyzed for the 8- to 10-cmbsf layers (Fig. 6). Rarefaction curves generated for the mcrA clones of the two libraries indicated saturation (see Fig. S2 in the supplemental material), whereas the percent coverages were determined to be 81.3 and 87.7% for the 2- to 4-cmbsf and 8- to 10-cmbsf gene libraries, respectively. Simpson's diversity indices (56) were calculated for each section, and these indicated a decrease in the methanotrophic/methanogenic diversity with depth (D = 0.7952 for the 2- to 4-cmbsf section and 0.7096 for the 8- to 10-cmbsf section).
Fig. 6.
Phylogenetic analysis of MCR amino acid sequences from the Napoli mud volcano sediments based on the neighbor-joining method with approximately 258 amino acid positions. The percent bootstrap values are based on 1,000 replicates and are indicated at the nodes for branch values with ≥50% bootstrap support. Gene sequences from the Napoli mud volcano sediments obtained in the present study are in boldface type. Clones with designations beginning with NapMat-2_4 are from the sediment section from 2 to 4 cmbsf, and clones with designations beginning with NapMat-8_10 are from the sediment section from 8 to 10 cmbsf. Numbers in parentheses indicate the number of analyzed clones that have >97% nucleotide sequence identity and >99% amino acid sequence identity. The scale bar indicates the 10% estimated difference. ANME, anaerobic methanotroph.
Three dominant mcrA phylotypes were present (see Table S2 in the supplemental material), i.e., mcrA group a/b (ANME-1 as defined by Hallam et al. [21]), mcrA group c/d (ANME-2c), and mcrA group e (ANME-2a). The majority of the mcrA clones was related to the ANME-2a at 2 to 4 cmbsf (58.3%), followed by the ANME-1. The ANME-2c sequences represented only a small portion of the ANME in the gene library. At the 8- to 10-cmbsf level, the ANME-1 became the dominant group (65.6%), followed by the ANME-2c, and the ANME-2a represented only a small proportion of the ANME in the gene library.
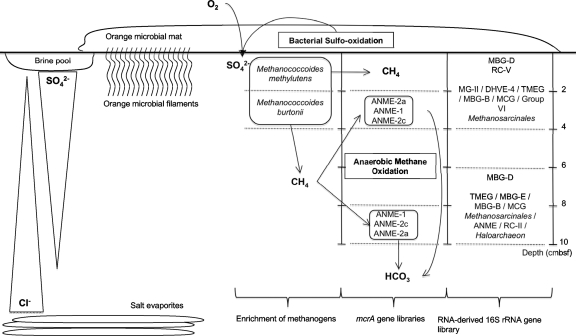
Sediments underlying bacterial mats seem to constitute hot spots for AOM (42), as a consequence of high methane flux in the upwardly moving subsurface fluids, combined with sulfate availability in the surficial sediments aided by the mats (34). ANME-2a Archaea have been found as the unique methanotrophic representative in sediments of the active center of the Napoli mud volcano (C. S. Lazar et al., unpublished data) and dominated marine Skagerrak sediments (51) and sediments covered with white-pigmented mats in the Gulf of Mexico (42). It has been suggested that ANME-2 may be more active at low temperatures compared to ANME-1 (43). Temperature gradient measurements of the active center of the Napoli mud volcano showed little increase in temperature with depth down to 160 cmbsf, with an average temperature of 14°C (Lazar et al., unpublished). It has also been suggested that ANME-2 dominates sediment layers with high sulfate concentrations, whereas ANME-1 seems to be found in sediment layers with low sulfate concentrations (30). Therefore, the distribution of the ANME-2 in the Napoli mud volcano sediments could support these assessments (Fig. 7).
Fig. 7.
Schematic illustrating the potential interactions between anaerobic methanotrophic Archaea (ANME) probably mediating anaerobic oxidation of methane, methanogens, and uncultured Archaea in different hypersaline sediment sections of the Napoli mud volcano. The sediment depth below the seafloor is indicated at the right of the illustration.
ANME-1 Archaea have been detected in various environments. In hypersaline sediments of the Gulf of Mexico, the community of ANME was found to be limited to the ANME-1b, which were probably active (35). The authors of that study suggested that the ANME-1b preponderance could be explained by the high salinity of the site (the chloride concentration was 2,200 mM) and that the ANME-1b could be a high-salinity-adapted subpopulation. The increase in ANME-1 sequences with depth and with chloride concentrations (at 8 to 10 cmbsf, the chloride concentration was 1,256 mM) in the Napoli sediments could support this assumption.
Culturable methanogenic diversity.
Methane production was detected in media designed to enrich methylotrophic methanogens on trimethylamines (TMA) in the shallow sulfate-rich 0- to 2-cmbsf and 2- to 4-cmbsf sediment sections. Microscopic observations of positive enrichments from the medium designed to enrich hydrogenotrophs suggested that methanogens were coccus-shaped cells. Under UV light, autofluorescent cells were detected as free cells. The total genomic DNA was extracted from the TMA-enriched medium inoculated with the 0- to 2-cmbsf and 2- to 4-cmbsf sediment sections. Phylogenetic affiliation analysis of the clone NapMat-0_2-enr30 showed a 99% sequence similarity to clone Tommo05_1274_3_Arch90 of the Euryarchaeota (FM179838) recovered from the Tommeliten methane seep in the North Sea (65) and a 98% sequence similarity to the closest cultured methanogen Methanococcoides methylutens (M59127). Phylogenetic affiliation analysis of clone NapMat-2_4-enr31 showed 98% sequence similarity to the cultured methanogen Methanococcoides burtonii (CP000300).
These results are in agreement with previous studies detecting methylotrophic Methanoccoides-type methanogens in saline or hypersaline habitats such as a brackish lake (3), marine sediments in Skan Bay (28), anaerobic sediments of mangroves (38, 39), brine seeps of the Gulf of Mexico (27), and recently sediments of the center of the Napoli mud volano (Lazar et al., unpublished). Representative species of methylotrophic methanogens in culture collections take up methylated compounds as substrates that are not used by other competitive microorganisms, such as sulfate-reducing bacteria. Methylated compounds could derive from organic detritus from the microbial mat (Fig. 7). Moreover, known cultured methylotrophic methanogens, belonging to the Methanohalophilus and Methanohalobium genera, have been shown to efficiently tolerate high-salinity environments (up to 25% NaCl) (48).
Surprisingly, only one 16S rRNA gene sequence of a known halophilic Archaea of the Halobacteria was detected in the present study, despite the high measured chloride concentrations. The same observation was also reported for brines from the Gulf of Mexico (27). However, since most of the sequences detected (i.e., DHVE4, MBG-D, MCG, RC-V, and ANME) belong to as-yet-uncultured archaeal lineages, we can assume that some of the Napoli sequences represent unknown halophilic or halotolerant microorganisms. Moreover, Simpson's diversity indices indicated a decrease in archaeal and methanotrophic/methanogenic diversity with depth and hence with increasing salinity. This could suggest that salt-adapted Archaea dominated the deeper layers of the Napoli mud volcano.
Conclusion.
In the present study, culture-dependent and -independent techniques were used in order to assess the distribution of the “active” RNA-derived 16S rRNA archaeal sequences in sediments associated with orange-pigmented mats of the brine impacted Napoli mud volcano. In the shallow sulfate-rich sediment layers of the Napoli mud volcano, the active fraction of the archaeal community was mainly represented by sequences belonging to as-yet-uncultured lineages similar to those present in cold seeps and deep subseafloor sediments but also, unexpectedly, in rice paddies. mcrA gene libraries suggested that AOM might have occurred in the Napoli mud volcano sediments. Enrichment cultures indicated that viable methanogens were present in the shallow sulfate-rich sediment layers. Therefore, a complex archaeal community was observed in this hypersaline habitat, possibly intertwining sulfur and methane cycles.
Supplementary Material
ACKNOWLEDGMENTS
We thank Josée Sarrazin and Catherine Pierre, the chief scientists of the Medeco cruise, the ROV team, and the officers and crew of the RV Pourquoi Pas?, as well as the shipboard scientific community for their help at sea. We also thank Andreas Teske (University of North Carolina) and Christian Jeanthon (Station Biologique of Roscoff) for their helpful scientific comments.
This study was funded by the HERMES project contract GOCE-CT-2005-511234-1 and the ANR Deep Oases.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Balch W. E., Wolfe R. S. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banning N., et al. 2005. Investigation of the methanogen population structure and activity in a brackish lake sediment. Environ. Microbiol. 7:947–960 [DOI] [PubMed] [Google Scholar]
- 4. Beal E. J., House C. H., Orphan V. J. 2009. Manganese- and iron-dependent marine methane oxidation. Science 325:184–187 [DOI] [PubMed] [Google Scholar]
- 5. Benlloch S., et al. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349–360 [DOI] [PubMed] [Google Scholar]
- 6. Biddle J. F., et al. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U. S. A. 103:3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boetius A., et al. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626 [DOI] [PubMed] [Google Scholar]
- 8. Casamayor E. O., et al. 2002. Changes in archaeal, bacterial, and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338–348 [DOI] [PubMed] [Google Scholar]
- 9. Casamayor E. O., Schäfer H., Baneras L., Salio C. P., Muyzer G. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaduteau C. 2008. Origin and movement of fluids in sediments of the margins: contribution of helium and methane in understanding the process, a study of two active areas. Ph.D. thesis Université de Bretagne Occidentale, Brest, France: (In French.) http://archimer.ifremer.fr/doc/00000/3724 [Google Scholar]
- 11. Chan O. C., et al. 2005. Vertical distribution of structure and function of the methanogenic archaeal community in Lake Dagow sediment. Environ. Microbiol. 7:1139–1149 [DOI] [PubMed] [Google Scholar]
- 12. Charlou J. L., et al. 2003. Evidence of methane venting and geochemistry of brines on mud volcanoes of the eastern Mediterranean Sea. Deep-Sea Res. Part I 50:941–958 [Google Scholar]
- 13. Dählmann A., de Lange G. J. 2003. Fluid-sediment interactions at Eastern Mediterranean mud volcanoes: a stable isotope study from ODP Leg 160. Earth Planet. Sci. Lett. 212:377–391 [Google Scholar]
- 14. Fry J. C., Webster G., Cragg B. A., Weightman A. J., Parkes R. J. 2006. Analysis of DGGE profiles to explore the relationship between prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol. Ecol. 58:86–98 [DOI] [PubMed] [Google Scholar]
- 15. Galand P. E., Lovejoy C., Vincent W. F. 2006. Remarkably diverse and contrasting archaeal communities in a large arctic river and the coastal Arctic Ocean. Aquat. Microb. Ecol. 44:115–126 [Google Scholar]
- 16. Glissmann K., Chin K.-J., Casper P., Conrad R. 2004. Methanogenic pathway and archaeal community structure in the sediment of eutrophic Lake Dagow: effect of temperature. Microb. Ecol. 48:389–399 [DOI] [PubMed] [Google Scholar]
- 17. Grosskopf R., Stubner S., Liesack W. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haese R. R., Hensen C., de Lange G. J. 2006. Pore water geochemistry of eastern Mediterranean mud volcanoes: implications for fluid transport and fluid origin. Mar. Geol. 225:191–208 [Google Scholar]
- 19. Hales B. A., Titchie E. C. D. A., Hall G. P. R. W., R S. J. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 21. Hallam S. J., Girguis P. R., Preston C. M., Richardson P. M., DeLong E. F. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing Archaea. Appl. Environ. Microbiol. 69:5483–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hallam S. J., et al. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462 [DOI] [PubMed] [Google Scholar]
- 23. Heijs S. K., Sinninghe Damsté J. S., Forney L. J. 2005. Characterization of a deep-sea microbial mat from an active cold seep at the Milano mud volcano in the Eastern Mediterranean Sea. FEMS Microbiol. Ecol. 54:47–56 [DOI] [PubMed] [Google Scholar]
- 24. Janse I., Bok J., Zwart G. 2004. A simple remedy against artifactual double bands in denaturing gradient gel electrophoresis. J. Microbiol. Methods 57:279–281 [DOI] [PubMed] [Google Scholar]
- 25. Jiang H., et al. 2008. Dominance of putative marine benthic Archaea in Qinghai Lake, north-western China. Environ. Microbiol. 10:2355–2367 [DOI] [PubMed] [Google Scholar]
- 26. Joye S. B., et al. 2004. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem. Geol. 205:219–238 [Google Scholar]
- 27. Joye S. B., et al. 2009. Metabolic variability in seafloor brines revealed by carbon and sulfur dynamics. Nat. Geosci. 2:349–354 [Google Scholar]
- 28. Kendall M. M., et al. 2007. Diversity of Archaea in marine sediments from Skan Bay, Alaska, including cultivated methanogens, and description of Methanogenium boonei sp. nov. Appl. Environ. Microbiol. 73:407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knittel K., Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63:311–334 [DOI] [PubMed] [Google Scholar]
- 30. Krüger M., et al. 2008. A novel, multi-layered methanotrophic microbial mat system growing on the sediment of the Black Sea. Environ. Microbiol. 10:1934–1947 [DOI] [PubMed] [Google Scholar]
- 31. Krüger M., et al. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878–881 [DOI] [PubMed] [Google Scholar]
- 32. Larkin M. A., et al. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 33. Lazar C. S., Dinasquet J., Pignet P., Prieur D., Toffin L. 2010. Active archaeal communities at cold seep sediments populated by siboglinidae tubeworms from the Storegga Slide. Microb. Ecol. doi:10.1007/s00248-010-9654-1 [DOI] [PubMed] [Google Scholar]
- 34. Lloyd K. G., et al. 2010. Spatial structure and activity of sedimentary microbial communities underlying a Beggiatoa spp. mat in a Gulf of Mexico hydrocarbon seep. PLoS One 5:e8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lloyd K. G., Lapham L., Teske A. 2006. An anaerobic methane-oxidizing community of ANME-1b Archaea in hypersaline Gulf of Mexico sediments. Appl. Environ. Microbiol. 72:7218–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lösekann T., et al. 2007. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73:3348–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luton P. E., Wayne J. M., Sharp R. J., Riley P. W. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521–3530 [DOI] [PubMed] [Google Scholar]
- 38. Lyimo T. J., Pol A., Jetten M. S. M., Op den Camp H. J. M. 2009. Diversity of methanogenic archaea in a mangrove sediment and isolation of a new Methanococcoides strain. FEMS Microbiol. Ecol. 291:247–253 [DOI] [PubMed] [Google Scholar]
- 39. Lyimo T. J., Pol A., Op den Camp H. J. M., Harhangi H. R., Vogels G. D. 2000. Methanosarcina semesiae sp. nov., a dimethylsulfide-utilizing methanogen from mangrove sediment. Int. J. Syst. Evol. Microbiol. 50:171–178 [DOI] [PubMed] [Google Scholar]
- 40. McHatton S. C., Barry J. P., Jannasch H. W., Nelson D. C. 1996. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62:954–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Michaelis W., et al. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013–1015 [DOI] [PubMed] [Google Scholar]
- 42. Mills H. J., Martinez R. J., Story S., Sobecky P. A. 2004. Identification of members of the metabolically active microbial populations associated with Beggiatoa species mat communities form Gulf of Mexico cold-seep sediments. Appl. Environ. Microbiol. 70:5447–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nauhaus K., Treude T., Boetius A., Krüger M. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7:98–106 [DOI] [PubMed] [Google Scholar]
- 44. Niemann H., et al. 2006. Microbial methane turnover at mud volcanoes of the Gulf of Cadiz. Geochim. Cosmochim. Acta 70:5336–5355 [Google Scholar]
- 45. Nikolaus R., Ammerman J. W., MacDonald I. R. 2003. Distinct pigmentation and trophic modes in Beggiatoa from hydrocarbon seeps in the Gulf of Mexico. Aquat. Microb. Ecol. 32:85–93 [Google Scholar]
- 46. Oh D., Porter K., Russ B., Burns D., Dyall-Smith M. 2010. Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 14:161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Omoregie E. O., et al. 2008. Biogeochemistry and community composition of iron- and sulfur-precipitating microbial mats at the Chefren Mud Volcano (Nile Deep Sea Fan, Eastern Mediterranean). Appl. Environ. Microbiol. 74:3198–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oren A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otte S., et al. 1996. Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl. Environ. Microbiol. 65:3148–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ovreas L., Forney L., Daae F. L., Torsvik V. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parkes R. J., et al. 2007. Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark). Environ. Microbiol. 9:1146–1161 [DOI] [PubMed] [Google Scholar]
- 52. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned rRNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Raghoebarsing A. A., et al. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921 [DOI] [PubMed] [Google Scholar]
- 54. R Development Core Team 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0 http://www.R-project.org [Google Scholar]
- 55. Schulz H. N., Jørgensen B. B. 2001. Big bacteria. Annu. Rev. Microbiol. 55:105–137 [DOI] [PubMed] [Google Scholar]
- 56. Simpson E. H. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 57. Singleton D. R., Furlong M. A., Rathbun S. L., Whitman W. B. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sørensen K. B., Canfield D. E., Teske A. P., Oren A. 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 70:7352–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sorensen K. B., Teske A. 2006. Stratified communities of active Archaea in deep marine subsurface sediments. Appl. Environ. Microbiol. 72:4596–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 61. Teske A., et al. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thauer R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377–2406 [DOI] [PubMed] [Google Scholar]
- 63. Toffin L., Webster G., Weightman A. J., Fry J. C., Prieur D. 2004. Molecular monitoring of culturable bacteria from deep-sea sediment of the Nankai Trough, Leg 190 Ocean Drilling Program. FEMS Microbiol. Ecol. 48:357–367 [DOI] [PubMed] [Google Scholar]
- 64. Vetriani C., Jannasch H. W. M. B. J., Stahl D. A., Reysenbach A.-L. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wegener G., et al. 2008. Biogeochemical processes and microbial diversity of the Gullfaks and Tommeliten methane seeps (Northern North Sea). Biogeosciences 5:1127–1144 [Google Scholar]
- 66. Wu L., Ma K., Li Q., Ke X., Lu Y. 2009. Composition of archaeal community in a paddy field as affected by rice cultivar and N fertilizer. Microb. Ecol. 58:819–826 [DOI] [PubMed] [Google Scholar]
- 67. Zepp Falz K., et al. 1999. Vertical distribution of methanogens in the anoxic sediment of Rotsee (Switzerland). Appl. Environ. Microbiol. 65:2402–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhou J., Bruns M. A., Tiedje J. M. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zitter T. A. C., Huguen C., Woodside J. M. 2005. Geology of mud volcanoes in the eastern Mediterranean from combined sidescan sonar and submersible surveys. Deep-Sea Res. I. 52:457–475 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.