Abstract
Mupirocin is an antibiotic commonly used in selective media for the isolation of bifidobacteria. However, little is known about the genetic traits responsible for bifidobacterial resistance to mupirocin. Our investigation demonstrates that all of the bifidobacteria tested exhibit a phenotype of generally high resistance to this antibiotic. The genotypic reason for bifidobacterial mupirocin resistance was further characterized by sequencing of the isoleucyl-tRNA synthetase gene (ileS) coupled with three-dimensional modeling of the encoded protein and cloning of the ileS gene of Bifidobacterium bifidum PRL2010 in a mupirocin-sensitive Escherichia coli strain. These analyses revealed key amino acid residues of the IleS protein that apparently are crucial for conferring a mupirocin resistance phenotype to bifidobacteria.
INTRODUCTION
Mupirocin is a narrow-spectrum antibiotic produced by Pseudomonas fluorescens (4, 17) and is active against certain Gram-negative and Gram-positive bacteria, including microorganisms that are used in fermented dairy products and functional foods, such as Streptococcus spp., Lactococcus spp., and Lactobacillus spp. (16). The antibacterial activity of mupirocin is due to competitive inhibition, as it competes with isoleucine as a substrate for isoleucyl-tRNA synthetase (6). The chemical structure of mupirocin resembles that of the isoleucyl-adenylate complex, and thus, the biochemical reason for its antagonistic activity corresponds to inhibition of the aminoacylation process in which isoleucyl-adenylate is synthesized.
Mupirocin is widely used as a selective agent for the isolation of bifidobacteria from complex ecosystems such as the human gut (16, 19). Bifidobacteria are Gram-positive microorganisms belonging to the Actinobacteria phylum that are natural inhabitants of the mammalian gastrointestinal tract (for a review, see reference 20). Bifidobacteria have recently generated growing scientific interest due to their presumed activity in maintaining gastrointestinal health and other beneficial or probiotic properties (13). Thus, significant efforts have been made to understand the genetic and ecological properties of this group of bacteria, including their susceptibility to different antibiotics (2, 9–11). So far, very little is known about the spectrum of bifidobacterial susceptibility to mupirocin, which will be of great interest in terms of developing a novel selective mupirocin-based medium for this group of microorganisms. Moreover, nothing is known about the molecular mechanisms responsible for the susceptibility/resistance of bifidobacteria to this antibiotic. Such knowledge may be useful for the development of genetic tools for bifidobacteria (e.g., gene knockout systems), while bifidobacterial mutants displaying higher/lower resistance to this antibiotic may also be used to monitor bifidobacterial colonization in clinical settings or in vivo trials.
Here, we provide an extensive analysis of mupirocin resistance in the majority of currently described bifidobacterial species, taking into consideration the genetic location of the mupirocin resistance determinant and linking the different levels of resistance identified in bifidobacteria to sequence variability of the isoleucyl-tRNA synthetase gene. Furthermore, we propose a structural mechanism to explain mupirocin resistance in bifidobacteria.
Identification of MICs.
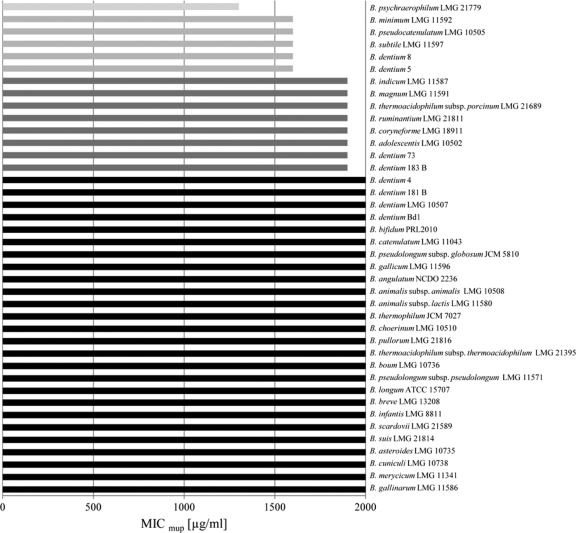
Bifidobacterial susceptibility to mupirocin was analyzed by MIC assays, which consisted of cultivating different bifidobacterial strains in the presence of various concentrations of this antibiotic ranging from 200 to 2,000 μg/ml. The bifidobacterial strains utilized were selected in order to represent the majority of the bifidobacterial species described so far (Fig. 1). Bifidobacteria were cultivated anaerobically using an anaerobic cabinet (Concept 400; Ruskin, West Yorkshire, United Kingdom) in de Man-Rogosa-Sharpe medium (Scharlau Chemie, Barcelona, Spain) supplemented with 0.05% (wt/vol) l-cysteine hydrochloride and incubated at 37°C for 16 h. The MIC for each strain employed in this study was determined (Fig. 1) according to a previously described method (11). All of the bifidobacterial species tested exhibited high mupirocin resistance (MIC values in excess of 2,000 μg/ml), with the exception of a small number of species, which were shown to be susceptible to this antibiotic at concentrations higher than 1,800 μg/ml (Fig. 1) and which were thus considered to exhibit reduced mupirocin resistance relative to the majority of the bifidobacterial strains tested. Such findings indicate that most, if not all, bifidobacteria exhibit stable resistance to mupirocin at moderate-to-high levels. Interestingly, for a small number of bifidobacterial species, the observed level of mupirocin susceptibility was shown to vary at the intraspecies level. This occurred within the species Bifidobacterium dentium, which includes strains that are moderately resistant to mupirocin (e.g., strains 5 and 8), as well as strains that are highly resistant to this antibiotic (e.g., strains 181 and 4) (Fig. 1). This suggests that moderate or high resistance to mupirocin is a strain-dependent feature rather than a species-dependent characteristic.
Fig. 1.
MIC values identified in the different bifidobacterial species.
Such different levels of mupirocin susceptibility shown by bifidobacteria resemble those described for Staphylococcus aureus (3). In the latter case, the resistance of S. aureus to high levels of mupirocin (above 2,000 μg/ml) was due to the presence of an additional isoleucyl-tRNA synthetase-encoding gene, called mupA or ileS2, specifying a mupirocin-resistant isoleucyl-tRNA synthetase, which is similar to eukaryotic IleS (5). In contrast, bioinformatic screening for the ileS genes in currently available complete bifidobacterial genome sequences (B. dentium Bd1 [21], B. longum subsp. longum NCC2705 [15], B. longum subsp. longum DJO10A [8], and B. animalis subsp. lactis DSM10140 [1]), as well as incomplete bifidobacterial genome sequences (B. dentium ATCC 27678, B. longum subsp. infantis CCUG 52486, B. bifidum PRL2010, B. bifidum NCIMB 41171, B. adolescentis L2-32, and B. catenulatum DSM 16992 [NCBI sources]), showed that such bifidobacteria harbor a single chromosomal copy of ileS in their genomes.
Introduction of the ileS gene of B. bifidum PRL2010 into E. coli.
In order to verify if the ileS gene product would be responsible for conveying resistance to mupirocin in bifidobacteria as it does in other bacteria (7), the coding region of the ileS gene of B. bifidum PRL2010 (here ileSbifidum) was amplified by PCR using primers ileS1 (5′-GGCCAAGCTTGGTGAGCGAAACCACCAATTCC-3′) and ileS2 (5′-GGCGCTGCAGTCACGCCTTGGCGACCTCCAC-3′), including the HindIII and PstI restriction sites (underlined), respectively, cloned into the pUC19 vector under the control of the lac promoter, and subsequently electroporated into E. coli strain DH10B. The presence of recombinant plasmid pUC19-ileSbifidum was confirmed for transformants by PCR and sequencing of the PCR amplicons obtained. E. coli DH10B is susceptible to a low level of mupirocin (MIC of 15 μg/ml), but it was converted to mupirocin resistance (MIC of 120 μg/ml) by transformation with pUC19-ileSbifidum. Such results were also confirmed by the overlay disc diffusion test (data not shown). Although pUC19 is a high-copy-number plasmid, it has been previously demonstrated that overexpression of the E. coli mupirocin-sensitive ileS gene through cloning in pUC19 did not increase mupirocin resistance in E. coli (22), suggesting that high levels of a sensitive IleS protein do not provoke resistance to mupirocin. These results therefore clearly indicate that the ileSbifidum gene represents the genetic determinant of the mupirocin resistance displayed by bifidobacteria.
Structural analysis of the IleS proteins encoded by mupirocin-resistant bifidobacteria.
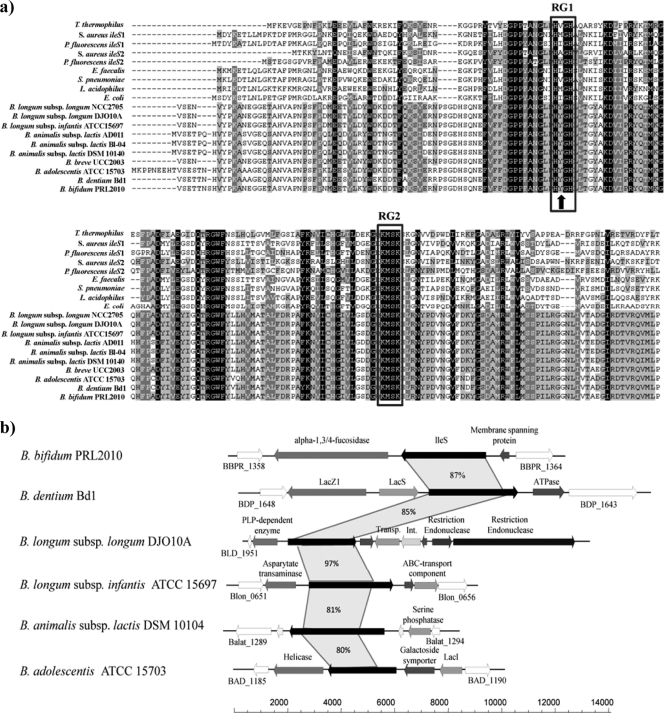
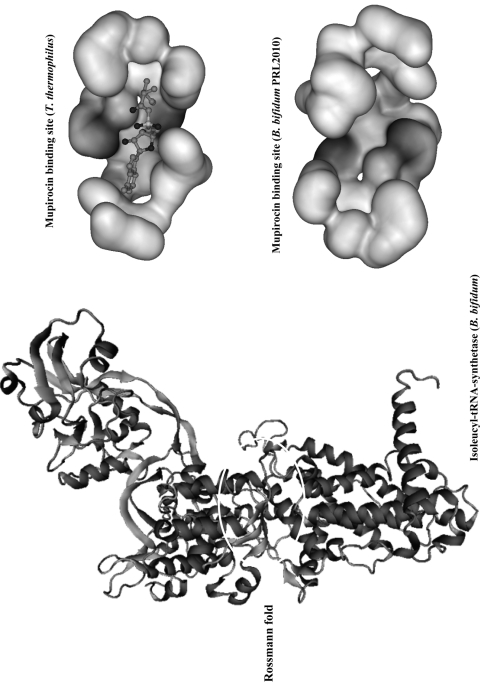
As previously mentioned, it is well known that the protein encoded by ileS represents the target site of mupirocin (for reviews, see references 14 and 23). Multiple-sequence alignment of the ileS protein products from susceptible microorganisms (e.g., Lactobacillus acidophilus, Enterococcus spp., Streptococcus spp., E. coli, Staphylococcus aureus, and Thermus thermophilus) and mupirocin-resistant microorganisms such as Bifidobacterium spp. and Pseudomonas spp. highlights the presence of amino acid residues that may be responsible for structural differences in the IleS protein (Fig. 2) and consequently for the mupirocin resistance/susceptibility phenotype. BLASTP analysis of the IleS protein sequence encoded by B. bifidum PRL2010 against the Protein Data Bank (PDB) database allowed the identification of a reliable structural template (IleS from Thermus thermophilus) (12). No other IleS structural model phylogenetically close to bifidobacteria was found in the PDB database. Although the phylogenetic distance between B. bifidum and T. thermophilus is considerable, the level of sequence identity between the IleS protein of B. bifidum PRL2010 and the template is still significantly high (E value of 2e−170). Based on the structural analysis of T. thermophilus IleS (12), the predicted three-dimensional (3D) structure of IleS of B. bifidum PRL2010 (SWISS-MODEL at http://swissmodel.expasy.org/) contains a reactive site which is presumed to act as the binding site of Ile-AMP during the aminoacylation process. Employing the online tool First Glance in Jmol version 1.45, we further analyzed the IleS Ile-AMP binding site sequence from the mupirocin-sensitive microorganism T. thermophilus, which highlighted 18 residues that are believed to be involved in specific interactions with the Ile-AMP ligand. A further comparative analysis including a larger data set of IleS sequences from different microorganisms exhibiting various levels of susceptibility to mupirocin highlighted a number of amino acid residues that may be responsible for the mupirocin resistance displayed by bifidobacterial strains. In order to probe the molecular effects of changes in amino acid residues (mutations) in the IleS sequences of both mupirocin-resistant and mupirocin-susceptible bacteria upon the interaction of IleS with mupirocin, we used the publicly available 3D structure of IleS from T. thermophilus (PDB code 1GAX [12]) as a template for the prediction of the structural model of IleS of B. bifidum PRL2010 (Fig. 3). Notably, the susceptibility of these bacteria to mupirocin is totally different (sensitive versus resistant) and thus the in-depth analysis of the 3D structures of IleS from T. thermophilus and B. bifidum PRL2010 was used to study the molecular effects of amino acid substitutions at various positions within different IleS sequences (RG1 and RG2). Notably, the RG1 and RG2 sequences partially overlap those regions believed to be involved in specific interaction with the Ile-AMP ligand.
Fig. 2.
Comparative analysis of isoleucyl-tRNA synthetase sequences. Multiple-sequence alignment of bacterial isoleucyl-tRNA synthetases displaying various levels of susceptibility to mupirocin (resistant versus susceptible) (a). When there are multiple ileS gene copies in the same organism (e.g., ileS2 of S. aureus and ileS2 of P. fluorescens), both of the protein sequences encoded are displayed. The amino acid residues constituting the RG1 and RG2 regions are highlighted. (b) Comparative schematic representation of the ileS locus of B. bifidum PRL2010 and those of various other bifidobacterial strains. Each arrow indicates an open reading frame, the size of which is proportional to the length of the arrow. The predicted function of the protein is indicated above each arrow.
Fig. 3.
Predicted 3D structure of the isoleucyl-tRNA synthetase from B. bifidum PRL2010 obtained by homology modeling using T. thermophilus as the template. This image is based on a docking model. The magnified sections represent the active site characterized by a Rossmann-type folding of the isoleucyl-tRNA synthetase from T. thermophilus as retrieved from the PDB and interacting with the mupirocin molecule, as well as the homologous region of the isoleucyl-tRNA synthetase from B. bifidum PRL2010. These structures are depicted as ball-and-stick structures shaded according to atom type. The different levels of interaction with the IleS binding cavity are indicated by various levels of gray shading.
The binding site investigation showed that the lack of a hydrophobic interaction at the apolar mupirocin long tail can cause a decrease in binding energy and the release of this molecule from the active site in RG1. In fact, the hydrophobic valine residue of T. thermophilus is replaced in bifidobacteria with a tyrosine's hydrocarbon benzene ring, which is predicted to cause reduced stability of the ligand in the active site. Interestingly, in the mupirocin producer P. fluorescens, a tyrosine residue is present in one of the products of the two ileS genes carried on its genome, which confers self-immunity (18, 22). Notably, all organisms possessing high resistance to mupirocin contain a tyrosine residue or an alanine residue in the RG1 region of their IleS sequence, e.g., IleS2 of S. aureus.
Sequence analyses of ileS genes in bifidobacteria.
A set of 34 bifidobacterial strains was subjected to PCR amplification of their ileS genes using primers ileS1 (5′-GAGTTCGTGTTCTTCGAC-3′) and ileS2 (5′-GACACGGTGGTGTCCTG-3′) and primers ileS7 (5′-CAGTTCGGTAAGTGGCT-3′) and ileS4 (5′-GTAGTAGGAGCTCCACAC-3′), followed by DNA sequencing. We analyzed two ileS gene sequence regions consisting of 54 and 234 bp, respectively, corresponding to codons 215 to 264 and 1737 to 1971 of ileS of B. bifidum PRL2010. These regions, named RG1 and RG2, are directly involved in forming the ligand binding site, and for this reason the amino acid residues encompassing these regions are considered to be crucial for resistance to the ligand mupirocin (Fig. 3). Notably, the alignments of RG1 and RG2 display high conservation at the amino acid level among all of the species tested (RG1 consensus sequence, HYGH; RG2 consensus sequence, KMSK). All of the IleS sequences analyzed display a tyrosine residue in RG1 that, based on structural analyses of the IleS-mupirocin structures, is believed to be responsible for providing resistance to mupirocin (see above).
Conclusion.
This study investigated the genetic basis of the intrinsic mupirocin resistance displayed by bifidobacteria. For this reason, mupirocin is widely used as a selective agent for the isolation of bifidobacteria from environmental samples (e.g., fecal samples, intestinal biopsy specimens, and food products) by the addition of mupirocin to different synthetic media used for cultivation of bifidobacteria. However, so far, little is known about the mupirocin susceptibility exhibited by the currently recognized bifidobacterial species. In this report, we show that bifidobacteria display variable levels of susceptibility to mupirocin, and this knowledge will be important for the development of novel and effective selection protocols for bifidobacteria based on antibiotic inclusion in selective media. Furthermore, we investigated the genetic basis of resistance to mupirocin in bifidobacteria and provided molecular evidence as to how the intrinsic mupirocin resistance of bifidobacteria is supported by the product encoded by the ileS gene and how a particular amino acid, tyrosine, in the targeted enzyme (IleS) may be responsible for the high level of mupirocin resistance observed in bifidobacteria. Bifidobacteria are generally not or only poorly genetically accessible, which at this time prevents us from performing particular confirmatory experiments. Future functional genomic investigations directed to the silencing of the ileS gene, as well as whole-genome transcription profiling experiments involving bifidobacteria grown in the presence of mupirocin, will allow us to identify the precise genetic requirements responsible for the high resistance to this antibiotic displayed by bifidobacteria. Furthermore, sequence analyses of the DNA regions surrounding the ileS gene in the genomes of bifidobacteria have not revealed any genetic features (e.g., atypical codon usage or atypical GC content or dinucleotide frequencies) or, except for the genome of B. longum subsp. longum NCC2705, the presence of nearby mobile elements (e.g., in the genome of B. bifidum PRL2010, the closest mobile element is represented by a transposases placed 203 kb downstream of the ileS gene), which indicates that mupirocin resistance in bifidobacteria is not mediated by a transferable genetic factor, minimizing the risk for horizontal transmission of resistance to other microorganisms in the digestive tract.
Acknowledgments
This work was financially supported by the Italian Award for Outstanding Young Researcher scheme Incentivazione alla mobilità di studiosi stranieri e italiani residenti all'estero 2005-2009, a Marie Curie reintegration grant (MERG-CT-2005-03080), Spinner 2013, Regione Emilia Romagna, and EFSE. This work was also financially supported by an IRCSET Embark postgraduate fellowship to F.B. D.V.S. is a member of The Alimentary Pharmabiotic Centre, which is a Centre for Science and Technology funded by Science Foundation Ireland (grants 02/CE/B124 and 07/CE/B1368) through the Irish Government's National Development Plan.
We thank all students and coworkers for contributing data and for their enthusiasm.
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Barrangou R., et al. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191:4144–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charteris W. P., Kelly P. M., Morelli L., Collins J. K. 1998. Antibiotic susceptibility of potentially probiotic Bifidobacterium isolates from the human gastrointestinal tract. Lett. Appl. Microbiol. 26:333–337 [DOI] [PubMed] [Google Scholar]
- 3. Eltringham I. 1997. Mupirocin resistance and methicillin-resistant Staphylococcus aureus (MRSA). J. Hosp. Infect. 35:1–8 [DOI] [PubMed] [Google Scholar]
- 4. Hill R. L., Duckworth G. J., Casewell M. W. 1988. Elimination of nasal carriage of methicillin-resistant Staphylococcus aureus with mupirocin during a hospital outbreak. J. Antimicrob. Chemother. 22:377–384 [DOI] [PubMed] [Google Scholar]
- 5. Hodgson J. E., et al. 1994. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother. 38:1205–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hughes J., Mellows G. 1980. Interaction of pseudomonic acid A with Escherichia coli B isoleucyl-tRNA synthetase. Biochem. J. 191:209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hurdle J. G., O'Neill A. J., Ingham E., Fishwick C., Chopra I. 2004. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob. Agents Chemother. 48:4366–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J. H., et al. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Margolles A., Florez A. B., Moreno J. A., van Sinderen D., de los Reyes-Gavilan C. G. 2006. Two membrane proteins from Bifidobacterium breve UCC2003 constitute an ABC-type multidrug transporter. Microbiology 152:3497–3505 [DOI] [PubMed] [Google Scholar]
- 10. Margolles A., Moreno J. A., van Sinderen D., de Los Reyes-Gavilan C. G. 2005. Macrolide resistance mediated by a Bifidobacterium breve membrane protein. Antimicrob. Agents Chemother. 49:4379–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Masco L., Van Hoorde K., De Brandt E., Swings J., Huys G. 2006. Antimicrobial susceptibility of Bifidobacterium strains from humans, animals and probiotic products. J. Antimicrob. Chemother. 58:85–94 [DOI] [PubMed] [Google Scholar]
- 12. Nakama T., Nureki O., Yokoyama S. 2001. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J. Biol. Chem. 276:47387–47393 [DOI] [PubMed] [Google Scholar]
- 13. Ouwehand A. C., Salminen S., Isolauri E. 2002. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82:279–289 [PubMed] [Google Scholar]
- 14. Patel J. B., Gorwitz R. J., Jernigan J. A. 2009. Mupirocin resistance. Clin. Infect. Dis. 49:935–941 [DOI] [PubMed] [Google Scholar]
- 15. Schell M. A., et al. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simpson P. J., Fitzgerald G. F., Stanton C., Ross R. P. 2004. The evaluation of a mupirocin-based selective medium for the enumeration of bifidobacteria from probiotic animal feed. J. Microbiol. Methods 57:9–16 [DOI] [PubMed] [Google Scholar]
- 17. Sutherland R., et al. 1985. Antibacterial activity of mupirocin (pseudomonic acid), a new antibiotic for topical use. Antimicrob. Agents Chemother. 27:495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomas C. M., Hothersall J., Willis C. L., Simpson T. J. 2010. Resistance to and synthesis of the antibiotic mupirocin. Nat. Rev. Microbiol. 8:281–289 [DOI] [PubMed] [Google Scholar]
- 19. Turroni F., et al. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75:1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ventura M., et al. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ventura M., et al. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yanagisawa T., Kawakami M. 2003. How does Pseudomonas fluorescens avoid suicide from its antibiotic pseudomonic acid? Evidence for two evolutionarily distinct isoleucyl-tRNA synthetases conferring self-defense. J. Biol. Chem. 278:25887–25894 [DOI] [PubMed] [Google Scholar]
- 23. Yanagisawa T., Lee J. T., Wu H. C., Kawakami M. 1994. Relationship of protein structure of isoleucyl-transfer RNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as inhibition of isoleucyl-tRNA synthetase. J. Biol. Chem. 269:24304–24309 [PubMed] [Google Scholar]