Abstract
A series of 100 Staphylococcus aureus isolates ascribed to sequence type 398 (ST398) and recovered from different sources (healthy carrier and diseased pigs, dust from pig farms, milk, and meat) in Germany were investigated for their virulence and antimicrobial resistance genetic background. Antimicrobial resistance was determined by the disk diffusion method. Virulence and resistance determinants (37 and 31 genes, respectively) were tested by PCR. Only two virulence profiles, including the accessory gene regulator agrI and three or four hemolysin-encoding genes, were detected. In contrast, 33 resistance profiles were distinguished (only 11 were shown by more than one isolate). Fifty-nine isolates were multiresistant (four or more antimicrobial classes), and 98 were methicillin resistant (mecA positive). All of the ST398 isolates showed resistance to tetracycline [encoded by tet(M) alone or together with tet(K) and/or tet(L)]. In addition, 98% were resistant to other antimicrobials, including macrolide-lincosamine-streptogramin B (70%, encoded by ermA, ermB, and ermC, alone or in combination), trimethoprim (65%, mostly due to dfrK and dfrG), kanamycin and gentamicin [29% and 14%, respectively, mainly related to aac(6′)-Ie-aph(2″)-Ia and/or ant(4′)-Ia but also to aph(3′)-IIIa], chloramphenicol (9%, fexA or cfr), quinupristin-dalfopristin (9%), ciprofloxacin (8%), and trimethoprim-sulfamethoxazole (4%). The heterogeneity of the resistance profiles underlines the ability of the ST398 clone to acquire multiple antimicrobial resistance genes. However, the virulence gene content of the tested isolates was low. Continuous surveillance is needed to clarify whether its pathogenicity potential for animals and humans will increase over time.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) of sequence type 398 (ST398) has gained particular attention during recent years because of its association with pigs and its ability to colonize pig farmers and other people in close contact with pigs (7, 12, 47). The MRSA isolates of ST398 usually lack important virulence determinants that are typical in other community and hospital MRSA isolates. The majority of the ST398 isolates analyzed so far carry only hemolysin-encoding genes (13, 21, 31, 32), although a small number of cases in which the isolates carried the bicomponent leukotoxin Panton-Valentine (lukPV genes) (43, 49) or staphylococcal enterotoxins (SEs, se genes) (21, 26) have also been reported. Genes for other toxins, like exfoliatins (ET, et genes), leukotoxins, and toxic shock syndrome toxin (TSST-1, tst gene) have not been found yet in ST398 isolates (13, 21, 31, 32, 44). The regulation of the expression of most extracellular virulence factors in S. aureus is under the control of a two-component signaling system called the accessory gene regulator (agr), which is polymorphic and divided into four distinct genetic groups (I to IV). A correlation exists between some agr groups and certain pathotypes and clonal complexes (CCs) (48), and CC398 seem to be associated with agr group I (agrI) (31, 32).
Typically, ST398 strains display a multiresistant phenotype. The majority of the isolates have been reported as MRSA, but in most of the epidemiological studies, the results were influenced by the use of selective isolation methods (8, 17). Methicillin-susceptible S. aureus (MSSA) strains have also been described (3, 17, 28, 43), and in the latest S. aureus surveillance studies of humans in Europe, all ST398 isolates detected were MSSA (16). Resistance to methicillin and other β-lactam antibiotics is mediated by mecA carrier elements called staphylococcal cassette chromosome mec (SCCmec). Together with methicillin resistance in ST398 isolates from swine, resistance to tetracycline, erythromycin, clindamycin, quinupristin-dalfopristin, ciprofloxacin, sulfonamides, trimethoprim, sulfamethoxazole-trimethoprim, and aminoglycosides has been reported (5, 9, 21, 38, 39).
The aim of the present work was to study the virulence and antimicrobial resistance genetic repertoire of a collection of 100 S. aureus strains previously ascribed to ST398 which were recovered from different sources (mainly livestock and food of animal origin) in Germany.
MATERIALS AND METHODS
Bacterial strains.
A series of 100 ST398 S. aureus isolates, previously characterized by spa typing, SCCmec typing, pulsed-field gel electrophoresis (PFGE) analysis with SmaI and its neoschizomer Cfr9I, and multilocus sequence typing (MLST) (3), were included in this study. The isolates were selected from the strain collection of the National Reference Laboratory for Coagulase-Positive Staphylococci (NRL-Staph) at the Federal Institute for Risk Assessment (BfR). They had been isolated between 2004 and 2008 at different German regional laboratories or the NRL-Staph as part of various monitoring surveys and research projects. The selection of the isolates was done to cover a variety of animal species and tissues and different stages of the food chain. When several isolates with the same source were available, isolates from different farms, slaughter batches, and/or spa/SCCmec types were selected in order to cover a broad range of ST398 subtypes. The ST398 isolates had been collected from healthy asymptomatic carrier pigs at slaughter (42 isolates), clinical samples from pigs (29 isolates; information is available for 19 that were collected at postmortem from different altered tissues where no other S. aureus isolate was present), dust from pig farms (10 isolates), cow's milk (5 isolates from individual milk samples from quarters with subclinical mastitis), and meat from food-producing animals (14 isolates, including 8 from pigs, 4 from turkeys, 1 from a broiler chicken, and 1 from a cow).
Five previously characterized ST398 isolates collected from fecal samples of pigs in the Netherlands (3) were included as ST398 controls. Six non-ST398 S. aureus isolates from meat (turkey and chicken), pig, and human samples, as well as S. aureus NCTC 8325 and S. aureus ATCC 29213, were used as outgroup controls. The eight outgroup isolates had been previously characterized by spa typing, SCCmec typing, and SmaI PFGE (3, 4). MLST (10) of these isolates was performed for the present study.
Positive controls for PCR amplification analyses were taken from previously analyzed collections (2) or were provided by the European Community Reference Laboratory for Antimicrobial Resistance (EURL-AR, Food-DTU, Copenhagen, Denmark) and the German National Reference Centre for Staphylococcus (NRZ-Staph, Robert Koch Institute [RKI], Wernigerode, Germany).
Antimicrobial susceptibility testing.
All isolates were tested for antimicrobial susceptibility by the disk diffusion method, using Mueller-Hinton agar and commercially available discs (Oxoid, Wesel, Germany). The antimicrobial agents tested were methicillin, oxacillin, tetracycline, gentamicin, kanamycin, rifampin, erythromycin, clindamycin, fusidic acid, quinupristin-dalfopristin, teicoplanin, chloramphenicol, ciprofloxacin, linezolid, trimethoprim, and trimethoprim-sulfamethoxazole. The methicillin-oxacillin-susceptible isolates were also tested for ampicillin and penicillin. The results were interpreted by following the CLSI breakpoints (6). For vancomycin susceptibility, the isolates were tested by agar broth dilution using a concentration range of 4 to 1 μl/ml (susceptible MICs are ≤2 μg/ml [6]). For selected isolates, the MICs for kanamycin, gentamicin and quinupristin-dalfopristin were also determined by broth microdilution (6). The MICs for these antimicrobials were interpreted using both CLSI (6) and EUCAST epidemiological cutoff values (www.eucast.org) (for kanamycin, susceptible MICs are ≤8 mg/ml; for gentamicin, ≤2 mg/ml; and for quinupristin-dalfopristin, ≤1 mg/ml). In all experiments, the strain S. aureus ATCC 29213 was used for quality assurance.
Virulence and resistance gene typing.
All isolates were screened for virulence and resistance determinants by PCR amplification using primers previously described or designed for this study (see Table S1 in the supplemental material). The resistance determinants tested conferred resistance to ampicillin-penicillin (blaZ), methicillin-oxacillin (mecA and SCCmec type), macrolides (msrA and msrB), lincosamides (linA/linA′), streptogramins A (vatA, vatB, vatC, vgaA, vgaB, and vgaC), streptogramins B (vgbA and vgbB), macrolides-lincosamides-streptogramins B (MLSB [ermA, ermB, and ermC]), tetracyclines [tet(K), tet(L), tet(M), and tet(O)], aminoglycosides (aac(6′)-Ie-aph(2″)-Ia, ant(4′)-Ia, and aph(3′)-IIIa), phenicols (cat::pC194, cat::pC221, cat::pC223, and fexA), trimethoprim (dfrD, dfrK, dfrG, and dfrS1), and phenicols-lincosamides-oxazolidinones-pleuromutilin-streptogramins A (cfr). The virulence determinants tested encoded hemolysins (hla, hlb, hld, hlg, and hlg variant), leukotoxins (lukED, lukM, and lukPV), exfoliatins (eta, etb, and etd), toxic shock syndrome toxin (tst), and SEs (sea, seb, sec, sed, see, seg, seh, sei, sej, sek, sel, sem, sen, seo, sep, seq, ser, and seu) or were markers of pathogenicity islands (ear, splF, and bsaB) or identified the type of the regulatory system agr (agrI, agrII, agrIII, and agrIV).
Dendrograms of similarity showing the clustering of the isolates according to virulence or the resistance gene profiles were generated with Bionumerics (BioNumerics version 5.1; Applied Maths, St.-Martens-Latem, Belgium) using the Jaccard's coefficient of similarity.
RESULTS
Virulence genes.
The 100 isolates assigned to ST398 were negative for leukotoxins, exfoliatins, and superantigen toxins. They displayed two different virulence profiles: hla hlb hld hlg agrI (98 isolates) and hla hlb hld agrI (2 isolates) (Table 1). The ST398 isolates from the Netherlands displayed the hla hlb hld hlg agrI (3 isolates) and hla hlb hld hlg seb agrI (2 isolates) profiles, while the outgroup isolates showed profiles with a higher number of virulence genes (Table 1). A dendrogram of similarity built on the basis of the presence or absence of the screened virulence genes clearly separated the outgroup controls from the ST398 isolates, which clustered together in subcluster A1 (Fig. 1).
Table 1.
Virulence profiles of S. aureus ST398 isolates and non-ST398 outgroup isolates and their relationships with spa types and PFGE profilesa
| Virulence profile | Virulence genotypeb | Strain group | Source(s)c | spa type(s) | PFGE profile(s)d |
|---|---|---|---|---|---|
| V1 (101) | hla hlb hld hlg agrI | ST398 | AP (42), CP (29), D (10), FPe (3), M (5), MC, MB, MP (7), MT (3) | t011 (37), t034 (31), t108 (10), t571 (2), t779, t1250, t1255, t1451 (4), t1457, t1580, t1793, t1928, t1985, t2011, t2346 (2), t2510, t2576 (4), t2970 | C1 (11), C2 (2), C3 (3), C4, C5 (8), C6 (16), C7, C8 (2), C9 (6), C10 (2), C11 (3), C12 (8), C13, C14 (6), C15 (3), C16 (4), C17, C18 (6), C19, C20 (2), C21–C29, C30 (2), C31–C33 |
| V2 (2) | hla hlb hld agrI | ST398 | MT, MP | t108, t5210 | C12, C24 |
| V3 (2) | hla hlb hld hlg seb agrI | ST398 | FPe (2) | t011 (2) | C34, C35 |
| V4 | hla hlb hld hlg lukM egc-like splF agrIII | ST433 | CP | t318 | S1 |
| V5 (2) | hla hlb hld hlg-v egc agrII | ST9 | CP, MB | t337, t1430 | S2, S3 |
| V6 | hla hld hlg-v lukED egc splF agrII | ST5 | MT | t002 | S4 |
| V7 | hla hlb hld hlg sec egc agrI | ST217 | CH | t032 | S5 |
| V8 | hla hlb hld hlg-v lukED egc sed sej ser splF agrII | ST225 | CH | t003 | S6 |
| V9 | hla hlb hld hlg-v lukED seb ear agrIV | ST8 | NCTC 8325 | t211 | Control PFGE 1 |
| V10 | hla hlb hld hlg-v lukED sea egc ear splF agrII | ST5 | ATCC 29213 | t002 | Control PFGE 2 |
The number of isolates is shown in parentheses when n > 1. In total, 113 isolates were analyzed.
The egc and egc-like clusters include the genes seg, sei, sem, sen, and seo. The egc-like cluster includes also the gene seu.
ATCC, American Type Culture Collection; CH, human clinical sample; AP, asymptomatic carrier pig; CP, pig clinical sample; D, dust from pig farms; FP, Dutch pig fecal sample (isolates included as ST398 controls); M, milk; MC, meat from cattle; MB, meat from broiler; MP, meat from pig; MT, meat from turkey; NCTC, National Collection of Type Cultures.
C, Cfr9I PFGE profile; S, SmaI PFGE profile.
ST398 isolates kindly provided by D. Mevius (Central Veterinary Institute of Waningen, Lelystad, Netherlands), used as control strains.
Fig. 1.
Dendrogram showing the relatedness between virulence gene profiles (V-profile) for 30 exotoxin genes, 3 island markers, and the agr group of the ST398 and outgroup S. aureus isolates tested. The number of isolates is shown in parentheses when n > 1. Clusters are described in the text. The scale shows genetic similarity using Jaccard's coefficient (J). Dashed lines separate clusters/subclusters grouping ST398 isolates.
Phenotypic antimicrobial resistance.
The ST398 isolates showed resistance to oxacillin (95%), tetracycline (100%), erythromycin-clindamycin (70%), trimethoprim (65%), kanamycin (29%), gentamicin (14%), quinupristin-dalfopristin, chloramphenicol (9% each), ciprofloxacin (8%), and trimethoprim-sulfamethoxazole (4%), arranged in 33 phenotypic resistance patterns (denoted by “R” and a number or alphanumeric) (Table 2; also see Table S2 in the supplemental material). Most of these included resistance to β-lactams and tetracyclines, together with trimethoprim and/or macrolide/lincosamide resistance. Only 13 of the resistance patterns were represented by more than one isolate (Table 2; also see Table S2 in the supplemental material), the most frequent being R21 (21 isolates), R8 (13 isolates), R11 (11 isolates), R28 (10 isolates), R25 and R29 (6 isolates each), and R15 (5 isolates). Only 14% of the series showed resistance to less than three classes of antimicrobials, whereas 21% were resistant to three classes and 30%, 33%, and 4% to four, five, and more than five classes, respectively. Within the outgroup isolates, eight additional resistance patterns (R7, R16, R17, R19, R22, R36, R37, and R38a) were identified (Table 2; also see Table S2 in the supplemental material).
Table 2.
Resistance profiles of ST398 and outgroup S. aureus isolatesa
| Genotypic resistance patternb | Phenotypic resistance patternc | Strain group(s) | Source(s)d |
|---|---|---|---|
| R0 (2) | Susceptible | ST8, ST433 | NCTC, CP |
| R1 | Amp-Pen | ST5 | ATCC |
| R2e | Tet | ST398 | M |
| R3e | Amp-Pen Tet | ST398 | CP |
| R4 | Amp-Pen Tet Ery-Cli | ST398 | CP |
| R5 | Amp-Pen Tet Ery-Cli-Syn | ST398 | CP |
| R6e | Amp-Pen Tet Ery-Cli Tmp | ST398 | CP |
| R7 | Amp-Pen Gen Tet Ery-Cli | ST9 | CP |
| R8 (13) | Met-Oxa Tet | ST398 | AP (6), CP (2), FPf, M (4) |
| R9 | Met-Oxa Kan Tet | ST398 | D |
| R10 | Met-Oxa Gen-Kan Tet | ST398 | D |
| R11 (11) | Met-Oxa Tet Ery-Cli | ST398 | AP (4), CP (2), MC, MP (3), FPf |
| R12 | Met-Oxa Kan Tet | ST398 | CP |
| R13 | Met-Oxa Tet Chl | ST398 | AP |
| R14 | Met-Oxa Tet Cip | ST398 | AP |
| R15 (5) | Met-Oxa Tet Tmp | ST398 | AP (2), D (2), MP |
| R16 | Met-Oxa Syn Cip | ST217 | CH |
| R17 | Met-Oxa Gen Tet Ery-Cli | ST398 | FPf |
| R18 (2) | Met-Oxa Kan Tet Ery-Cli | ST398 | D, MB |
| R19 | Met-Oxa Kan Tet Tmp | ST398 | FPf |
| R20 (2) | Met-Oxa Gen-Kan Tet Tmp | ST398 | AP (2) |
| R21 (21) | Met-Oxa Tet Ery-Cli Tmp | ST398 | AP (9), CP (7), D (2), MP (3) |
| R22 | Met-Oxa Tet Cip Tmp | ST398 | FPf |
| R23 | Met-Oxa Tet Sxt Tmp | ST398 | CP |
| R24 | Met-Oxa Gen Tet Ery-Cli Tmp | ST398 | AP |
| R25 (6) | Met-Oxa Kan Tet Ery-Cli Tmp | ST398 | AP (4), D, CP |
| R26 | Met-Oxa Kan Tet Cip Tmp | ST398 | AP |
| R27 | Met-Oxa Kan Tet Ery-Cli Chl | ST398 | CP |
| R28 (10) | Met-Oxa Gen-Kan Tet Ery-Cli Tmp | ST398 | AP (5), D, CP (2), MT (2) |
| R29 (6) | Met-Oxa Tet Ery-Cli Syn Tmp | ST398 | AP, D, CP, MP, MT (2) |
| R30 | Met-Oxa Tet Ery-Cli Sxt-Tmp | ST398 | AP |
| R31 (2) | Met-Oxa Tet Ery-Cli Cip Tmp | ST398 | AP, CP |
| R32 | Met-Oxa Tet Ery-Cli Chl Cip | ST398 | AP |
| R33 | Met-Oxa Tet Ery-Cli Chl Tmp | ST398 | CP |
| R34 | Met-Oxa Tet Ery-Cli-Syn Sxt-Tmp | ST398 | AP |
| R35 (2) | Met-Oxa Tet Chl Cip Tmp | ST398 | CP (2) |
| R36 | Met-Oxa Kan Tet Ery-Cli Cip | ST5 | MT |
| R37 | Met-Oxa Kan Ery-Cli Cip Mup | ST225 | CH |
| R38 (2) | Met-Oxa Kan Tet Ery-Cli Cip Tmp | ST9, ST398 | CP, MB |
| R39 | Met-Oxa Kan Tet Ery-Cli Chl Tmp | ST398 | CP |
| R40 | Met-Oxa Kan Tet Ery-Cli-Syn Chl Tmp | ST398 | AP |
| R41 | Met-Oxa Kan Tet Ery-Cli Chl Sxt-Tmp | ST398 | CP |
The number of isolates is shown in parentheses when n > 1. In total, 113 isolates were analyzed.
The subtyping of the resistance pattern according to the presence of a genetic background is shown in Fig. 2.
Met, methicillin; Oxa, oxacillin; Tet, tetracycline; Gen, gentamicin; Kan, kanamycin; Ery, erythromycin; Cli, clindamycin; Mup, mupirocin; Syn, quinupristin-dalfopristin; Chl, chloramphenicol; Cip, ciprofloxacin; Tmp, trimethoprim; Sxt, trimethoprim-sulfamethoxazole. Intermediate susceptibility was considered resistance in this table (see Table S2 in the supplemental material for details). Isolates susceptible to methicillin and oxacillin were also tested for resistance to ampicillin and penicillin (Amp-Pen).
ATCC, American Type Culture Collection; CH, human clinical sample; AP, asymptomatic carrier pig; CP, pig clinical sample; D, dust (from pig farms); FP, Dutch pig fecal sample; M, milk; MC, meat from cattle; MB, meat from broiler; MP, meat from pig; MT, meat from turkey; NCTC, National Collection of Type Cultures.
mecA-positive isolates (MRSA).
ST398 isolates kindly provided by D. Mevius (Central Veterinary Institute of Waningen, Lelystad, Netherlands), used as control strains.
Genotypic basis of resistance.
The results for the genotypic bases of resistance of ST398 and outgroup isolates are summarized in Table 3. Among the German ST398 isolates, the following results were found. The ampicillin-penicillin resistance gene blaZ was detected in 94% of the isolates. For aminoglycoside resistance determinants, the gentamicin-kanamycin resistance gene aac(6′)-Ie-aph(2″)-Ia was found in 12 gentamicin-kanamycin-resistant isolates (12% of the total and 92% of those resistant to gentamicin-kanamycin) and in one gentamicin-resistant isolate (1% of the total and 7% of the gentamicin-resistant isolates), while the kanamycin resistance genes ant(4′)-Ia and aph(3′)-IIIa were detected in 37 (37% of the total and 76% of kanamycin-resistant isolates) and 2 (2% of the total and 7% of kanamycin-resistant isolates) isolates, respectively. Some of the isolates (with resistance profiles R4, R8a, R15c, R21h-R21n, R29b, R31a, and R33) were positive for the presence of genes conferring aminoglycoside resistance but did not express this resistance (MICs between 32 and 8 μg/ml) in assays using the CLSI clinical breakpoints. In one kanamycin-resistant isolate, none of the tested genes was present. For tetracycline resistance, the tet(M) gene was present in all ST398 isolates, alone (9%) or together with tet(K) (51%), tet(L) (22%), or both genes (18%). The genes encoding resistance to MLSB, ermA, ermB, and ermC, were present in 35 (35%), 32 (32%), and 41 (41%) isolates, respectively, each gene being found alone or in combination in the following percentages of isolates: ermA (7%), ermB (9%), ermC (26%), ermA ermB (14%), ermA ermC (6%), ermB ermC (1%), and ermA ermB ermC (8%). No isolate was positive for the erythromycin resistance genes msrA and msrB. Three isolates (3%) carried the clindamycin resistance linA/linA′ gene, but two of them were phenotypically susceptible (using the CLSI breakpoints). The remaining isolate was erythromycin and clindamycin resistant and also carried ermB. All quinupristin-dalfopristin-resistant isolates were negative for the tested genes conferring resistance to streptogramins of type A (vatA, vatB, vatC, vgaA, vgaB, and vgaC) and B (vgbA and vgbB). No isolate carried the tested chloramphenicol acetyltransferase genes (cat::pC194, cat::pC221, and cat::pC223), whereas two (2%) isolates were positive for fexA. One of these was also positive for the multidrug resistance gene cfr. The following four genes conferring trimethoprim resistance were detected, alone or in combination: dfrS1 dfrD (1%), dfrK dfrG (2%), dfrG (21%), and dfrK (39%).
Table 3.
Genetic determinants conferring resistance to different classes of antimicrobials in ST398 and outgroup isolates of S. aureusa
| Class of antimicrobial agent | Phenotypic resistance vs susceptibilityb | Resistance genotypec | Strain group(s) | Source(s)d |
|---|---|---|---|---|
| β-Lactams | Amp-Pen-Met-Oxar (104) | blaZ SCCmec II | ST225 | CH |
| blaZ SCCmec IVa (5) | ST9, ST398 (4) | AP (3), MB, MT | ||
| blaZ SCCmec V (73) | ST398 (73) | AP (25), CP (21), D (9), FPe (5), M (3), MB, MC, MP (6), MT (2) | ||
| blaZ SCCmec V* (10) | ST398 (10) | AP (4), CP (4), D, MP | ||
| blaZ SCCmec nt (9) | ST5, ST217, ST398 (7) | AP (6), CH, MP, MT | ||
| SCCmec IVa | ST398 | MT | ||
| SCCmec V (4) | ST398 (4) | AP (3), M | ||
| SCCmec V* | ST398 | AP | ||
| Amp-Penr Met-Oxas (3) | blaZ SCCmec V (2) | ST398 (2) | CP (2) | |
| Amp-Penr (2) | blaZ (4) | ST5, ST9, ST398 (2) | ATCC, CP (3) | |
| Amp-Pen-Met-Oxas (3) | blaZ SCCmec V | ST398 | M | |
| None (2) | ST8, ST433 | CP, NCTC | ||
| Tetracycline | Tetr (108) | tet(M) (10) | ST9, ST398 (9) | AP (5), CP (3), MP (2) |
| tet(L) | ST9 | MB | ||
| tet(M) tet(K) (56) | ST5, ST398 (55) | AP (16), CP (15), D (6), FPe (4), M (5), MB, MC, MP (5), MT (3) | ||
| tet(M) tet(L) (22) | ST398 (22) | AP (13), CP (5), D, MP, MT (2) | ||
| tet(M) tet(K) tet(L) (19) | ST398 (19) | AP (8), CP (7), D (3), FPe | ||
| Tets (5) | tet(M) tet(K) | ST225 | CH | |
| None (4) | ST5, ST8, ST217, ST433 | ATCC, CH, CP, NCTC | ||
| Aminoglycosides | Genr (3) | aac(6′)-Ie-aph(2″)-Ia ant(4′)-Ia | ST398 | AP |
| ND (2) | ST9, ST398 | CP, FPe | ||
| Kanr (20) | ant(4′)-Ia (15) | ST9, ST398 (14) | AP (4), CP (5), D (3), FPe, MB (2) | |
| aph(3′)-IIIa (3) | ST5, ST398 (2) | AP (2), MT | ||
| ND (2) | ST225, ST398 | CH, CP | ||
| Gen-Kanr (13) | aac(6′)-Ie-aph(2″)-Ia (5) | ST398 (5) | AP (3), CP, D | |
| aac(6′)-Ie-aph(2″)-Ia ant(4′)-Ia (7) | ST398 (7) | AP (3), CP, D, MT (2) | ||
| ant(4′)-Ia | ST398 | AP | ||
| Gen-Kans (77) | ant(4′)-Ia (15) | ST398 (15) | AP (6), CP (6), D, M, MP | |
| None (62) | ST 5, ST8, ST217, ST398 (58), ST433 | AP (22), ATCC, CH, CP (16), D (4), FPe (3), M (4), MC, MP (7), MT (2), NTCT | ||
| MLSB groupf | Ery-Clir (74) | erm(A) (9) | ST5, ST398 (8) | AP (2), CP (3), FPe, MP (2), MT |
| erm(B) (8) | ST9, ST398 (7) | AP (5), D, MB, MT | ||
| erm(C) (28) | ST9, ST225, ST398 (26) | AP (12), CH, CP (9), D (2), FPe, MB, MP (2) | ||
| erm(A) erm(B) (14) | ST398 (14) | AP (6), CP (3), D, MP (2), MT (2) | ||
| erm(A) erm(C) (6) | ST398 (6) | AP, CP (3), D, MC | ||
| erm(B) erm(C) | ST398 | CP | ||
| erm(B) linA/linA′ | ST398 | CP | ||
| erm(A) erm(B) erm(C) (6) | ST398 (6) | AP (3), CP, D, MP | ||
| ND (3) | ST398 (3) | CP (2), MT | ||
| Ery-Clis (37) | erm(A) | ST217 | CH | |
| erm(B) | ST398 | D | ||
| erm(C) | ST398 | D | ||
| linA/linA′ (3) | ST398 (3) | CP (2), FPe | ||
| erm(A) erm(B) erm(C) (2) | ST398 (2) | AP (2) | ||
| None (29) | ST5, ST8, ST398 (26), ST433 | AP (11), ATCC, CP (6), D (2), NCTC, M (5), MP, FPe (2) | ||
| Chloramphenicol | Chlr (9) | fexA | ST398 | CP |
| fexA cfr | ST398 | CP | ||
| ND (7) | ST398 (7) | AP (3), CP (4) | ||
| Chls (104) | None (104) | ST5 (2), ST8, ST9 (2), ST217, ST225, ST398 (97), ST433 | AP (39), ATCC, CH (2), CP (25), D (10), FPe (5), M (5), MB (2), MC, MP (8), MT (5), NTCT | |
| Trimethoprim | Tmpr (65) | dfrK (40) | ST9, ST398 (39) | AP (22), CP (11), D (2), FPe, MB, MP, MT (2) |
| dfrG (22) | ST398 (22) | AP (5), CP (9), D (2), FPe, MP (3), MT (2) | ||
| dfrK dfrG (2) | ST398 (2) | D, MP | ||
| dfrD dfrS1 | ST398 | AP | ||
| Tmps (48) | dfrK (2) | ST398 (2) | D (2) | |
| ND (3) | ST398 (3) | AP, D (2) | ||
| None (43) | ST5 (2), ST8, ST9, ST217, ST225, ST398 (33), ST433, | AP (13), ATCC, CH (2), CP (11), D, FPe (3), M (5), MB, MC, MP (3), MT, NCTC |
The number of isolates is shown in parentheses when n > 1. In total, 113 isolates were analyzed.
Amp, ampicillin; Pen, penicillin; Met, methicillin; Oxa, oxacillin; Tet, tetracycline; Gen, gentamicin; Kan, kanamycin; Ery, erythromycin; Cli, clindamycin; Chl, chloramphenicol; Tmp, trimethoprim. Superscript “r” and “s” denote resistance and susceptibility, respectively. Intermediate susceptibility was considered resistance in this table. Met-Oxa-resistant isolates were considered Amp-Pen resistant.
Gene or genes related to a determined resistance pattern. SCCmec V* corresponds to isolates that were SCCmec III by the Zhang et al. (50) typing protocol but amplified the ccrC of SCCmec type V (3). ND, not determined; nt, not typeable.
ATCC, American Type Culture Collection; CH, human clinical sample; AP, asymptomatic carrier pig; CP, pig clinical sample; D, dust (from pig farms); FP, Dutch pig fecal sample; M, milk; MC, meat from cattle; MB, meat from broiler; MP, meat from pig; MT, meat from turkey; NCTC, National Collection of Type Cultures.
ST398 isolates kindly provided by D. Mevius (Central Veterinary Institute of Waningen, Lelystad, Netherlands), used as control strains.
MLS, macrolide, lincosamide, and streptogramin B group.
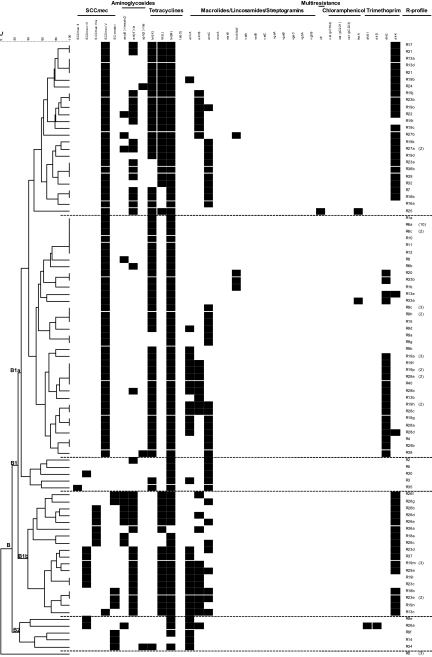
A dendrogram of similarity was established on the basis of the presence or absence of the screened resistance genes (Fig. 2). The dendrogram separated susceptible isolates from the resistant isolates, which grouped in a large cluster (B) comprising two subclusters (B1 and B2). The resistant Dutch ST398 isolates and the outgroup isolates clustered together with the resistant ST398 isolates from Germany.
Fig. 2.
Dendrogram showing the relatedness between resistance gene profiles (R-profile) for 31 resistance genes or cassettes of the ST398 and outgroup S. aureus isolates tested. The scale shows genetic similarity using Jaccard's coefficient (J). Dashed lines separate clusters/subclusters.
DISCUSSION
The emerging MRSA ST398 clonal group is widespread in the domestic pig population and can also be identified in other livestock species, such as veal calves, dairy cattle, and poultry (1, 12). Farmers and other livestock professionals handling carrier animals are at high risk of being colonized (7). Although infrequently, clinical infections with MRSA ST398 have been described, being in most cases related to these high-risk groups (12, 27, 45, 47). Human infections with MSSA ST398 have also been recorded, and they seem to be more frequent than those caused by MRSA ST398 (16). While the prevalence of the pathogen in farm animals and food has been studied extensively during the last few years, there is not much information on the molecular epidemiology of virulence and resistance determinants in this clonal group. The present work has investigated the virulence and resistance gene contents in a collection of 100 ST398 strains of nonhuman origin, representative of the German NRL-Staph collection (years 2004 to 2008), which was well characterized by several molecular methods previously (3).
The tested isolates of spa types assigned to ST398 lack several clinically important S. aureus-associated virulence factors regardless of their source. This is in line with the results of other studies (13, 21, 31, 32) and with the fact that clinical disease in swine and humans colonized by ST398 MRSA is rarely observed (16, 45, 47). However, cases of clinical and subclinical mastitis associated with this type of MRSA in dairy herds have been reported (13, 40), as have pathological lesions of pigs (28). From the 37 genes tested in this study that are related to virulence, the presence of pathogenicity islands, and regulators, we could only detect hemolysin-encoding genes (hla, hlb, hld, and hlg) and the agr group. None of the German isolates carried enterotoxins, but seb was present in two of the Dutch ST398 isolates. Low levels of occurrence of either seb sek and seq, or sed and seg were also reported for MRSA ST398 isolates from Germany (21) and France (26). It has been speculated that the environment contributes to the low presence of virulence determinants in this clone. However, some studies focused on the pig population have shown how in other S. aureus isolates (i.e., MSSA isolates from undetermined CCs), the presence of enterotoxins can be high (33). The Panton-Valentine leukocidin (PVL) was absent in our isolates. Genes encoding this toxin, which is frequently present in community-acquired S. aureus isolates, have also been detected in ST398 isolates from hospitalized patients (49) and, at a low frequency, in pig isolates (43). The majority of PVL-positive ST398 isolates described so far has been obtained from humans without exposure to animal husbandry (46, 49).
The low number of virulence factors in ST398 isolates is in clear contrast to the high number of resistance determinants. In this respect, type I and II restriction-modification systems detected in the genome of ST398 (36) may have interfered so far with the acquisition of virulence factors, which are mainly encoded by pathogenicity islands or phages (34). However, this does not explain the high number of resistance genes carried by the clone. In S. aureus, resistance genes are mainly located in plasmids and, depending on size and GC content, their chances to escape restriction might be high. Moreover, selection pressure on ST398 isolates induced by the use of antimicrobials in livestock production could have potentiated positive selection for resistance determinants, while infrequent contact with pathogenic bacteria, more common in hospitals, could explain why the clone still lacks many virulence determinants.
In different countries, different antimicrobial usage habits can influence the antimicrobial resistance patterns (regarding phenotypes and mobile genetic determinants) observed in the bacterial populations. The nonhuman German ST398 isolates analyzed in this study were highly heterogeneous with regard to antimicrobial resistance properties. This may be due to the livestock being raised with different types of antimicrobials, but data on the antimicrobial usage on the farms from which the isolates came are not available. A high percentage (59%) of multiresistance was present in the series. Almost 37% of the isolates, 43% of them recovered from healthy carrier pigs, showed resistance to five or more classes of antimicrobials, whereas only 28% did in a study of 55 German ST398 isolates from diseased swine (21). All 100 isolates were resistant to tetracycline, a property of the ST398 clone also found by other authors (9, 41). This resistance is mostly related to the presence of the tet(M) gene located in transposons Tn5801 and Tn916. Only a few ST398 isolates lacking tet(M) have been described so far (21, 22). Nearly all isolates (91%) in this study also carried additional tetracycline resistance genes, like tet(K) and/or tet(L), that are normally harbored on plasmids. The latter gene, which has rarely been described in non-ST398 MRSA isolates (14, 42), was relatively common in our series (40%), as well as in other isolates from the ST398 clone (24%) (13, 21). The prevalence of trimethoprim resistance was also high (65%), and the dfrK and dfrG genes, which have rarely been detected among other MRSA isolates, seem to be frequent in German ST398 isolates (13, 21). Colocalization of the tet(L) and dfrK genes on the pKKS2187 plasmid was described in a German MRSA ST398 isolate from a diseased pig (22). In 36% of our isolates, both genes appeared together, with 4% and 6% being positive for only tet(L) or only dfrK, respectively. Chloramphenicol resistance was expressed in nine isolates (9%), but only two of them were positive for fexA, a gene that has rarely been detected in staphylococci from animal sources (25). Among these fexA-positive isolates, one isolate also carried the multidrug resistance determinant cfr, which was previously found, associated with fexA, in individual ST398 and ST9 isolates (24). This indicates that, currently, both genes are infrequent in the German ST398 population.
In the present study, the percentage of resistance to gentamicin (14%) was similar to that reported for diseased swine by Kadlec et al. (21) but, in comparison to that report, the percentage of resistance to kanamycin was significantly higher (29% versus 7%; χ2 test, P = 0.003) in our study. High percentages of gentamicin and/or kanamycin resistance (22% and 41%, respectively) were reported in ST398 isolates from cases of bovine mastitis (13). The genes implicated in the aminoglycoside resistance of the isolates from our series were aac(6′)-Ie-aph(2″)-Ia and/or ant(4′)-Ia, very frequently present in S. aureus isolates (14) and ST398 isolates from other series (13, 21). The aph(3′)-IIIa gene, infrequent in S. aureus isolates, was found in only 2% of our series. In 16 isolates (16%) that had repeatedly been shown to be susceptible to kanamycin in disc diffusion assays (corresponding to MICs of ≤16 μg/ml), ant(4′)-Ia could be detected. With their MICs having been tested by broth microdilution and considering the EUCAST cutoff values (susceptible range, ≤8 μg/ml), the same isolates showed either susceptibility or resistance (various MICs between 32 and 8 μg/ml). This phenomenon has also been noticed in other studies (11, 19, 30) and could be due to a requirement for the induction of transcription or the presence of an inactive ant(4′)-Ia. It has been demonstrated that a rearrangement between the blaZ gene and a Tn4001-IS257 hybrid structure has developed an aac(6′)-Ie-aph(2″)-Ia gene inducible by β-lactams (20). In our series, all except one of the aac(6′)-Ie-aph(2″)-Ia-positive strains were also positive for blaZ, but in the present work, no further investigations into the presence of this hybrid structure were conducted. Regarding the MLSB resistance, several erm genes were found (prevalences of 30 to 40%), appearing either alone or in all possible combinations. This is interesting because, in S. aureus, the ermA gene is the most frequent determinant that confers constitutive resistance to MLSB, while ermC is more common in strains with an inducible phenotype (14, 15, 18). The ermB gene has been identified in multiple bacterial genera (35). It has been found more frequently in staphylococci from animal sources than from human sources (14, 15, 18, 37). None of the nine (9%) quinupristin-dalfopristin-resistant isolates carried the genes coding for resistance to type A and B streptogramins tested in the study. The novel gene vgaC found in the ST398 clone (23) was also absent. This suggests that the clone possesses other mechanisms responsible for this resistance.
When looking at the clustering analyses (Fig. 2), there was a correlation between the resistance profiles of the isolates, their Cfr9I PFGE profiles, and their SCCmec types previously described by Argudín et al. (3). Isolates with SCCmec type V usually carried tet(K) and clustered together in subcluster B1a. These isolates also clustered together at a similarity of 0.63 when analyzed by Cfr9I PFGE (3). Conversely, subcluster B1b grouped mainly isolates with SCCmec IVa or V* without tet(K), and their PFGE profiles were also grouped within a different cluster (similarity of 0.64) (3). Some of the SCCmec V carriers also have dfrK, like most of the SCCmec IVa or V* carriers. Despite the differences, all of the ST398 Cfr9I PFGE profiles clustered together and were separate from the outgroup strains of different CCs.
So far, the virulence gene content of the ST398 clone appears to be low. However, continuous surveillance is needed in order to clarify whether the pathogenicity potential of the clone will evolve in the coming years. Moreover, MRSA ST398 is a reservoir for multiple determinants (13, 21, 26, 29) conferring resistance to several antimicrobials of clinical relevance. For the same phenotypic resistance, one or more genes, frequently found on plasmids [blaZ, ant(4′)-Ia, tet(K), tet(L), ermC, dfrD, and dfrK] or transposons [aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa tet(M), ermA, ermB, and dfrS1], are present in these isolates. Many of them may be colocalized on the same genetic element (22, 34). The high prevalence of this type of S. aureus in livestock and food, together with the carriage of multiple resistance determinants with a high risk of spread, underlines the importance of controlling this emergent bacterium in the animal population and its spread to exposed humans.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Kowall, U. Kämpe, W. Barownick, and J. Beutlich (Federal Institute for Risk Assessment, BfR, Berlin) for their support. We also are grateful to the different German laboratories, as well as to D. Mevius and K. Veldman (Central Veterinary Institute of Wageningen, Lelystad, Netherlands) for providing the isolates or samples that were included in this study, to W. Witte and G. Werner (Robert Koch Institute, Wernigerode, Germany) and F. Aarestrup and H. Hasman (Food-DTU, Lyngby, Denmark) for the control strains, and to the Department of Biological Safety of the BfR for their hospitality toward M. A. Argudín during a short-term visit.
The project was supported by the BFR (grants BfR-45-004, BfR-46-001, and BfR-41-001), the German Ministry for Food, Agriculture, and Consumer Protection (grant 2808HS032), and the Fondo de Investigaciones Sanitarias (grant FIS-PI080656) of the Spanish Ministry of Science and Innovation. M. A. Argudín, Ph.D. student, was the recipient of grant FPU AP-2004-3641 from the Ministry of Science and Innovation, Spain, cofunded by the European Social Fund. She performed a short stay at the BfR supported by the same grant.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Anonymous 2009. Joint scientific report of ECDC, EFSA and EMEA on methicillin-resistant Staphylococcus aureus (MRSA) in livestock, companion animals and food. EFSA-Q-2009-00612 [EFSA Scientific Report (2009) 301, 1-10] and EMEA/CVMP/SAGAM/62464/2009. European Food Safety Agency, Parma, Italy: www.health.gov.mt/fsc/fschome_files/efsa_biohaz_report_301_joint_mrsa_en.pdf [Google Scholar]
- 2. Argudín M. A., et al. 2009. Clonal complexes and diversity of exotoxin gene profiles in methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from patients in a Spanish hospital. J. Clin. Microbiol. 47:2097–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Argudín M. A., et al. 2010. High heterogeneity within methicillin-resistant Staphylococcus aureus ST398 isolates, defined by Cfr9I macrorestriction-pulsed-field gel electrophoresis profiles and spa and SCCmec types. Appl. Environ. Microbiol. 76:652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Argudín M. A., Rodicio M. R., Guerra B. 2010. The emerging methicillin-resistant Staphylococcus aureus ST398 clone can easily be typed using the Cfr9I SmaI-neoschizomer. Lett. Appl. Microbiol. 50:127–130 [DOI] [PubMed] [Google Scholar]
- 5. Battisti A., et al. 2010. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet. Microbiol. 142:361–366 [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing (M100-S19), 19th informational supplement, vol. 29, no. 3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Cuny C., et al. 2009. Nasal colonization of humans with methicillin-resistant Staphylococcus aureus (MRSA) CC398 with and without exposure to pigs. PLoS One 4:e6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Boer E., et al. 2009. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 134:52–56 [DOI] [PubMed] [Google Scholar]
- 9. de Neeling A. J., et al. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366–372 [DOI] [PubMed] [Google Scholar]
- 10. Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fatholahzadeh B., et al. 2009. Characterisation of genes encoding aminoglycoside-modifying enzymes among methicillin-resistant Staphylococcus aureus isolated from two hospitals in Tehran, Iran. Int. J. Antimicrob. Agents 33:264–265 [DOI] [PubMed] [Google Scholar]
- 12. Ferber D. 2010. From pigs to people: the emergence of a new superbug. Science 329:1010–1011 [DOI] [PubMed] [Google Scholar]
- 13. Feßler A., et al. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619–625 [DOI] [PubMed] [Google Scholar]
- 14. Fluit A. C., Visser M. R., Schmitz F. J. 2001. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev. 14:836–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gherardi G., De Florio L., Lorino G., Fico L., Dicuonzo G. 2009. Macrolide resistance genotypes and phenotypes among erythromycin-resistant clinical isolates of Staphylococcus aureus and coagulase-negative staphylococci, Italy. FEMS Immunol. Med. Microbiol. 55:62–67 [DOI] [PubMed] [Google Scholar]
- 16. Grundmann H., et al. 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7:e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guardabassi L., Stegger M., Skov R. 2007. Retrospective detection of methicillin resistant and susceptible Staphylococcus aureus ST398 in Danish slaughter pigs. Vet. Microbiol. 122:384–386 [DOI] [PubMed] [Google Scholar]
- 18. Gul H. C., et al. 2008. Macrolide-lincosamide-streptogramin B resistant phenotypes and genotypes for methicillin-resistant Staphylococcus aureus in Turkey, from 2003 to 2006. Pol. J. Microbiol. 57:307–312 [PubMed] [Google Scholar]
- 19. Hauschild T., et al. 2008. Aminoglycosides resistance in clinical isolates of Staphylococcus aureus from a university hospital in Bialystok, Poland. Folia Histochem. Cytobiol. 46:225–228 [DOI] [PubMed] [Google Scholar]
- 20. Ida T., et al. 2002. Antagonism between aminoglycosides and β-lactams in a methicillin-resistant Staphylococcus aureus isolate involves induction of an aminoglycoside-modifying enzyme. Antimicrob. Agents Chemother. 46:1516–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadlec K., et al. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156–1164 [DOI] [PubMed] [Google Scholar]
- 22. Kadlec K., Schwarz S. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob. Agents Chemother. 53:776–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kadlec K., Schwarz S. 2009. Novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 53:3589–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kehrenberg C., Cuny C., Strommenger B., Schwarz S., Witte W. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53:779–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kehrenberg C., Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laurent F., Jouy E., Granier S., et al. 2009. Molecular characterization of antimicrobial resistance genes and virulence genes by using microarrays in representative ST398 MRSA isolates from pigs in France, abstr. S5:2. ASM-ESCMID Conference on Methicillin-Resistant Staphylococci in Animals, London, England, 22 to 25 September 2009 [Google Scholar]
- 27. Lewis H. C., et al. 2008. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14:1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meemken D., et al. 2010. Livestock associated methicillin-resistant Staphylococcus aureus (LaMRSA) isolated from lesions of pigs at necropsy in northwest Germany between 2004 and 2007. Zoonoses Public Health 57:e143–e148 [DOI] [PubMed] [Google Scholar]
- 29. Mevius D. J., Wit B., van Pelt W. (ed.). 2007. MARAN 2007: monitoring of antimicrobial resistance and antibiotic usage in animals in the Netherlands in 2006/2007. Veterinary Antibiotic Usage and Resistance Surveillance Working Group. Central Veterinary Institute of Wageningen UR, Lelystad, Netherlands [Google Scholar]
- 30. Monecke S., Ehricht R. 2005. Rapid genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolates using miniaturised oligonucleotide arrays. Clin. Microbiol. Infect. 11:825–833 [DOI] [PubMed] [Google Scholar]
- 31. Monecke S., Kuhnert P., Hotzel H., Slickers P., Ehricht R. 2007. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet. Microbiol. 125:128–140 [DOI] [PubMed] [Google Scholar]
- 32. Monecke S., Jatzwauk L., Weber S., Slickers P., Ehricht R. 2008. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from eastern Saxony. Clin. Microbiol. Infect. 14:534–545 [DOI] [PubMed] [Google Scholar]
- 33. Nitzsche S., Zweifel C., Stephan R. 2007. Phenotypic and genotypic traits of Staphylococcus aureus strains isolated from pig carcasses. Vet. Microbiol. 120:292–299 [DOI] [PubMed] [Google Scholar]
- 34. Novick R. P., Subedi A. 2007. The SaPIs: mobile pathogenicity islands of Staphylococcus. Chem. Immunol. Allergy 93:42–57 [DOI] [PubMed] [Google Scholar]
- 35. Roberts M. C. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282:147–159 [DOI] [PubMed] [Google Scholar]
- 36. Schijffelen M. J., Boel C. H., van Strijp J. A., Fluit A. C. 2010. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitz F. J., et al. 2000. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45:891–894 [DOI] [PubMed] [Google Scholar]
- 38. Schwarz S., Kadlec K., Strommenger B. 2008. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius detected in the BfT-GermVet monitoring programme 2004–2006 in Germany. J. Antimicrob. Chemother. 61:282–285 [DOI] [PubMed] [Google Scholar]
- 39. Smith T. C., et al. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in Midwestern U.S. swine and swine workers. PLoS One 4:e4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spohr M., et al. 11 July 2010. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in Southwest Germany. Zoonoses Public Health. doi:10.1111/j.1863-2378.2010.01344.x [DOI] [PubMed] [Google Scholar]
- 41. Tenhagen B.-A., et al. 2009. Prevalence of MRSA types in slaughter pigs in different German abattoirs. Vet. Rec. 165:589–593 [DOI] [PubMed] [Google Scholar]
- 42. Trzcinski K., Cooper B. S., Hryniewicz W., Dowson C. G. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45:763–770 [DOI] [PubMed] [Google Scholar]
- 43. van Belkum A., et al. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Duijkeren E., et al. 2008. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 126:383–389 [DOI] [PubMed] [Google Scholar]
- 45. van Loo I., et al. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg. Infect. Dis. 13:1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Welinder-Olsson C., et al. 2008. Infection with Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus t034. Emerg. Infect. Dis. 14:1271–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Witte W., Strommenger B., Stanek C., Cuny C. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, central Europe. Emerg. Infect. Dis. 13:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright J. S., et al. 2005. The agr radiation: an early event in the evolution of staphylococci. J. Bacteriol. 187:5585–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu F., et al. 2008. Prevalence of Staphylococcus aureus carrying Panton-Valentine leukocidin genes among isolates from hospitalised patients in China. Clin. Microbiol. Infect. 14:381–384 [DOI] [PubMed] [Google Scholar]
- 50. Zhang K., McClure J. A., Sameer E., Louie T., Conly J. M. 2005. Novel multiple PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to IV in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.