Abstract
1-Butanol, an important chemical feedstock and advanced biofuel, is produced by Clostridium species. Various efforts have been made to transfer the clostridial 1-butanol pathway into other microorganisms. However, in contrast to similar compounds, only limited titers of 1-butanol were attained. In this work, we constructed a modified clostridial 1-butanol pathway in Escherichia coli to provide an irreversible reaction catalyzed by trans-enoyl-coenzyme A (CoA) reductase (Ter) and created NADH and acetyl-CoA driving forces to direct the flux. We achieved high-titer (30 g/liter) and high-yield (70 to 88% of the theoretical) production of 1-butanol anaerobically, comparable to or exceeding the levels demonstrated by native producers. Without the NADH and acetyl-CoA driving forces, the Ter reaction alone only achieved about 1/10 the level of production. The engineered host platform also enables the selection of essential enzymes with better catalytic efficiency or expression by anaerobic growth rescue. These results demonstrate the importance of driving forces in the efficient production of nonnative products.
INTRODUCTION
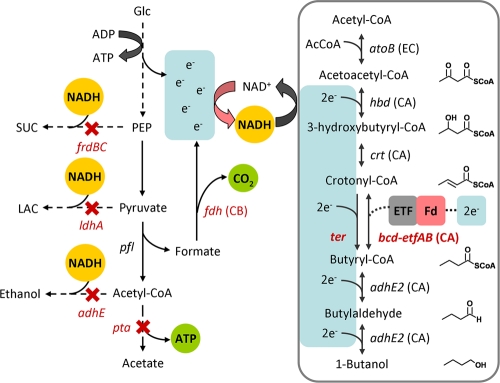
1-Butanol, a potential fuel substitute and an important C4 chemical feedstock (30), is naturally synthesized by Clostridium species using a pathway that involves multiple coenzyme A (CoA)-activated intermediates (hereinafter called the CoA-dependent pathway). Various attempts have been made to transfer the CoA-dependent 1-butanol pathway to more tractable organisms, such as Escherichia coli (0.55 to 1.2 g/liter) (2, 25), Saccharomyces cerevisiae (2.5 mg/liter) (38), Lactobacillus brevis (300 mg/liter) (7), Pseudomonas putida (580 mg/liter), and Bacillus subtilis (120 mg/liter) (32). These low titers of heterologous 1-butanol production demonstrate the difficulty in transferring this pathway to nonnative hosts and are in sharp contrast to the high titers of related compounds, such as ethanol (50 g/liter) (27, 45), isobutanol (20 to 50 g/liter) (3, 5), and isopropanol (40 to 140 g/liter) (24), produced by recombinant E. coli strains using related pathways. Examination of these high-titer production processes revealed that all of the pathways involve a decarboxylation reaction near the end products, in which the irreversible release of CO2 serves as a driving force to pull fluxes to the desired compounds. Such a strategy is also present in fatty acid synthesis, which involves decarboxylation of malonyl-CoA in the chain elongation step. In contrast, when the clostridial CoA-dependent 1-butanol pathway is transferred to E. coli, no significant driving force exists to direct the carbon flux through the five reversible steps starting from acetyl-CoA (Fig. 1).
Fig. 1.
Synthetic build-up of NADH and acetyl-CoA (shown on the left) in the 1-butanol production system. Shown on the right is the 1-butanol production pathway engineered in E. coli from C. acetobutylicum (boxed). Acetyl-CoA initiates the NADH-consuming reactions. A total of four NADH molecules is needed to make one 1-butanol molecule. Enzymes that utilize NADH directly as the electron donor (Ter) are coupled more tightly to the NADH driving force than the enzymes that require additional electron transfer mediators (Bcd). Elimination of Pta, involved in acetate production, not only increased the pools of available acetyl-CoA but also reduced ATP synthesis, both of which can be used as driving forces to increase 1-butanol production. SUC, succinate; PEP, phosphoenolpyruvate; LAC, lactate; ETF, electron transfer flavoprotein; Fd, ferrodoxin. CB, C. boidinii; CA, C. acetobutylicum; EC, E. coli.
We reason that a significant driving force is necessary to achieve high-titer production, whereas driving forces are not as important in proof-of-concept production approaches. For 1-butanol, we define high titers as those greater than the toxicity level, which is about 10 g/liter (4). When examined in this light, driving forces can be found in almost all successful metabolic engineering strategies and may exist in several different forms, including (i) release of a gaseous molecule (e.g., CO2) which escapes out of the cell through diffusion and becomes diluted in the gas phase, (ii) irreversible reactions or polymerization (e.g., glycogen and polyhydroxyalkanoate), (iii) ATP draining by futile cycling (13, 14, 33) or disruption of ATP synthase (12), (iv) transfer of electrons to an external sink (e.g., respiration and hydrogen production), or (v) product removal by phase separation. Therefore, metabolic engineering or synthetic biology strategies for high-titer production can be summarized and recast in two steps: (i) the creation of a driving force and (ii) coupling of the driving force to the desired pathway.
Since the clostridial CoA-dependent pathway for 1-butanol synthesis is reversible on both thermodynamic (47) and enzymatic (43) grounds, the presence of an artificial driving force would be essential to achieve high-titer production. The clostridial 1-butanol synthesis pathway utilizes both NADH and reduced ferredoxin (Bcd-EtfAB complex) (31) as sources of reducing power (Fig. 1). If it is possible to reconstruct the 1-butanol pathway to utilize only NADH as the reducing cofactor, then the accumulation of NADH can be used as a driving force for 1-butanol production. The NADH driving force (8, 9, 26, 46, 48) could be established by deleting the mixed-acid fermentation reactions (ethanol, lactate, and succinate) in E. coli. The resulting strain, JCL166 (ΔadhE ΔldhA Δfrd), lost its ability to grow anaerobically due to the lack of NADH-consuming pathways as an electron sink (Fig. 1). Such a strain is unable to recycle NADH, thereby creating a driving force for reactions that consume NADH. Similar strategies have been successfully applied to the production of (d/l)-lactate (49), succinate (26), ethanol (29, 44), and l-alanine (46) in E. coli.
To couple the NADH driving force effectively to the target 1-butanol pathway, the crotonyl-CoA reduction step catalyzed by Clostridium acetobutylicum butyryl-CoA dehydrogenase complex (Bcd-EtfAB) must be modified to utilize NADH as the direct reducing equivalent (Fig. 1). Challenges in the detection of Bcd-EtfAB in vitro activity (2, 10, 22) and measurement of intracellular pathway intermediates (19) also suggest that this step is deficient in the production of 1-butanol in E. coli. Another class of enzymes available to catalyze the reduction of crotonyl-CoA is the trans-enoyl-CoA reductase (Ter) (23, 41). The Treponema denticola Ter (41) has been shown to possess significant specific activity toward crotonyl-CoA reduction using NADH as a direct reducing equivalent without flavoproteins or ferredoxin. This NADH-dependent enzyme could potentially facilitate tighter coupling of 1-butanol production with the NADH driving force (Fig. 1). In addition, our in vitro enzyme assays suggest that the Ter-catalyzed reduction is irreversible, which may serve as an irreversible driving force to channel the carbon flux toward 1-butanol. However, when the T. denticola Ter was utilized in E. coli (11) for 1-butanol production, only 0.25 g/liter of 1-butanol was accumulated. This result suggests that additional factors need to be considered to drive high flux into the 1-butanol pathway.
The condensation reaction of acetyl-CoA to make acetoacetyl-CoA in the CoA-dependent 1-butanol pathway may be another limiting step because of its unfavorably high Gibbs energy change (47). To facilitate this reaction, in this work we chose E. coli's acetyl-CoA acetyltransferase (AtoB), involved in acetoacetate degradation, instead of the Clostridium acetoacetyl-CoA thiolase (Thl), because of its higher specific activity (AtoB, 1,078 U/mg, versus Thl, 216 U/mg) (17, 43). These enzymes all exhibit reversible kinetics that strongly favor the acetoacetyl-CoA cleavage reaction (17, 43), therefore highlighting the need for a strong driving force to channel carbon flux into the recombinant 1-butanol pathway. In addition to the synthetic NADH driving force, the major acetyl-CoA-consuming enzyme phosphate acetyltransferase, encoded by pta, was also deleted to build an acetyl-CoA driving force (42) coupled with the use of E. coli AtoB to provide a better link to the 1-butanol pathway.
Using the NADH and acetyl-CoA driving forces coupled with the irreversible Ter reaction, we attained high-titer production of 1-butanol (15 g/liter or 30 g/liter with in situ product removal). In contrast, our previous work (2) used the Bcd-EtfAB complex instead of Ter. Without effective coupling to the driving forces, only 550 mg/liter of 1-butanol was achieved in semianaerobic conditions and about 40 mg/liter was attained under anaerobic conditions. Another study (11) that used the T. denticola Ter without a significant acetyl-CoA driving force resulted in about 250 mg/liter of 1-butanol anaerobically. These results suggest the importance of driving forces in achieving high-titer production.
MATERIALS AND METHODS
Reagents.
Restriction enzymes and Antarctic phosphatase were purchased from New England BioLabs (Ipswich, MA). The Rapid DNA ligation kit was from Roche (Mannheim, Germany). KOD DNA polymerase was from EMD Chemicals (San Diego, CA). Oligonucleotides were purchased from IDT (San Diego, CA). All chemicals used were obtained from Sigma-Aldrich (St. Louis, MO) or Fisher Scientifics (Pittsburgh, PA) unless specified otherwise. Reagents for cell lysis, Bugbuster and Lysonase, were purchased from Novagen (San Diego, CA).
Bacterial strains.
Escherichia coli BW25113 (rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) was designated as the wild type (16). XL-1 Blue (Stratagene, La Jolla, CA) was used to propagate all plasmids. The construction of strains JCL16 (BW25113 with lacIq provided on F′), JCL166, and JCL299 was described previously (2).
Plasmid construction.
All plasmid constructs were sequenced for verification with Genewiz. A list of plasmids and primers used is shown in Table 1. Fdh from Candida boidinii was amplified with primers fdhfacc65 and fdhrsal by PCR from genomic DNA, digested, and cloned into an empty medium-copy-number plasmid cut with the same restriction enzymes, resulting in pCS102. To make it a low-copy-number plasmid, pCS102 was digested with AvrII and AatII and the piece containing the p15A origin and Kanr was removed by gel purification and replaced with the pSC101 origin and Cmr cut with the same enzymes. The resulting plasmid is pCS138.
Table 1.
Strains, plasmids, and oligonucleotides used
| Strain, plasmid, or primer | Genotype or sequencea | Reference or source | Plasmid constructed |
|---|---|---|---|
| E. coli strains | |||
| BW25113 | rrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 16 | |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene | |
| JCL16 | BW25113/F′ [traD36 proAB+lacIqZΔM15 (Tetr)] | 2 | |
| JCL166 | JCL16 ΔldhA ΔadhE ΔfrdBC | 2 | |
| JCL299 | JCL16 ΔldhA ΔadhE ΔfrdBC Δpta | 2 | |
| Plasmids | |||
| pCS102 | PLlacO1::fdhCB p15A ori Kanr | This study | |
| pCS106 | PLlacO1::MCS Cola ori Kan | This study | |
| pCS127 | PLlacO1::terFS Cola ori Kanr | This study | |
| pCS128 | PLlacO1::terFJ Cola ori Kanr | This study | |
| pCS138 | PLlacO1:: fdhCB pSC101 ori Cmr | This study | |
| pEL11 | PLlacO1::atoBEC-adhE2CA-crtCA-hbdCA ColE1 ori Ampr | This study | |
| pEL12 | PLlacO1::atoBEC-crtCA-hbdCA ColE1 ori Ampr | This study | |
| pEL16 | PLlacO1::terTV Cola ori Kanr | This study | |
| pIM8 | PLlacO1::terTD Cola ori Kanr | This study | |
| pIM11 | PLlacO1::bcdCA-etfACA-etfBCA Cola ori Kanr | This study | |
| pHJ2 | PLlacO1::terFS(M11K/A397V) Cola ori Kanr | This study | |
| pHJ3 | PLlacO1::terFS(M11K/T325I) Cola ori Kanr | This study | |
| pHJ6 | PLlacO1::terFS(M11K) Cola ori Kanr | This study | |
| Primers | |||
| Fdhfacc65 | TCAGGTACCATGAAAATTGTGCTGGTGTTATATGATG | pCS138 | |
| Fdhrsal | TCAGTCGACTCACTTTTTATCATGTTTTCCGTAC | pCS138 | |
| Tdeterfacc65 | TCAGGTACCATGATTGTAAAACCAATGGTTAGGAACA | pIM8 | |
| Tdeterrsal | TCAGTCGACTAAATCCTGTCGAACCTTTCTACCTCG | pIM8 | |
| CacBcdfacc65 | TCAGGTACCATGGATTTTAATTTAACAAGAGAACAAGAAT | pIM11 | |
| CacEtfBrsal | TCAGTCGACTTAATTATTAGCAGCTTTAACTTGAGCTATTAATT | pIM11 | |
| Crtfsph | GGGAAAGCATGCAGGAGATATACCATGGAACTAAACAATGTCATCCTTGAAAAGG | pEL12 | |
| Crtfxba | GGGAAATCTAGAAGGAGATATACCATGGAACTAAACAATGTCATCCTTGAAAAGG | pEL11 | |
| CrtrSOE | CCTTTTTCATGGTATATCTCCTCTATCTATTTTTGAAGCCTTCAATTTTTCTTTTC | pEL11 | |
| HbdfSOE | AAATAGATAGAGGAGATATACCATGAAAAAGGTATGTGTTATAGGTGCAGG | pEL11 | |
| Hbdrxba | GGGAAATCTAGATTATTTTGAATAATCGTAGAAACCTTTTCCTGATT | pEL11 | |
| Tviterfacc65 | GGGAAAGGTACCATGAGTATGAAACCGATGCTGAGAA | pEL16 | |
| Tviterrsal | TCAGTCGACTTATATCCGGTCGAACCGGTCAATC | pEL16 | |
| Tviterrbam | GGGAAAGGATCCTTATATCCGGTCGAACCGGTCAATC | pEL16 | |
| Fsuterfacc65 | TCAGGTACCATGATTATCAAGCCGCTCATTCGT | pCS127 | |
| Fsuterrsal | ACGCAGTCGACTTAGATAGAGGTCAGGGTCTGAACATCT | pCS127 | |
| Fjoterfacc65 | TCAGGTACCATGATTATCGAACCGCGTATGCG | pCS128 | |
| Fjoterrsal | ACGCAGTCGACTTATTTGATAGATTCGATATTAACAACTTCGT | pCS128 |
In plasmid descriptions, subscripts indicate the source of the gene as follows: CA, Clostridium acetobutylicum; TD, Treponema denticola; TV, Treponema vincentii; FJ, Flavobacterium johnsoniae; FS, Fibrobacter succinogenes; CB, Candida boidinii. Primer sequences are shown 5′ → 3′.
Plasmid pEL11 was constructed by inserting the crt-hbd fragment into pJCL17 (2). The crt-hbd fragment was created by individually amplifying crt with primers crtfxba and crtrSOE and hbd with hbdfSOE and hbdrxba using pJCL60 (2) as a template. The two pieces were then connected together by splicing by overlap extension (SOE) and further amplified with crtfxba and hbdrxba. The resulting crt-hbd fragment and pJCL17 were both digested with XbaI and ligated to give pEL11. To make pEL12, pJCL17 was digested with SphI and XbaI and ligated with the crt-hbd fragment cut with the same pair of restriction enzymes amplified by crtfsph and hbdrxba using pEL11 as a template.
To clone individual ter genes from various organisms, the ter gene was amplified with PCR from the corresponding genomic DNA purchased from ATCC. For Treponema denticola, the resulting fragment of ter amplified with Tdeterfacc65 and Tdeterrsal was digested and ligated into pCS106 cut with Acc65I and SalI to yield pIM8. For Treponema vincentii, for which genomic DNA was not available, the ter was amplified by colony PCR using the commercially available polymerase kit ImmoMix-Red (Bioline) and the microorganism suspended in liquid. The primers used for this PCR were Tviterfacc65 and Tviterrbam. The resulting DNA fragment was double digested with Acc65I and BamHI and ligated into the corresponding sites of pCS106 to give pEL16. For Fibrobacter succinogenes and Flavobacterium johnsoniae, for which the genomic DNA or strain is not available, PCR-based gene synthesis was performed. Plasmids pCS127 and pCS128 were created by ligating the PCR-assembled ter genes into pCS106 cut with Acc65I and SalI.
C. acetobutylicum Bcd-EtfAB was cloned into pCS106 by amplification of the operon using primers CacBcdfacc65 and CacEtfBrsal from genomic DNA, followed by digestion with Acc65I and SalI, yielding pIM11.
PCR-based gene synthesis.
The ter genes from Fibrobacter succinogenes and Flavobacterium johnsoniae were synthesized by PCR assembly of 50-bp oligonucleotides. The oligonucleotide sequences were obtained by using automated oligonucleotide design from Helix Systems (NIH; http://helixweb.nih.gov/dnaworks/). The protein sequences of the two ter genes were entered into Helix systems. The default parameters were used. The codon frequency was selected for E. coli standard, and Acc65I and SalI restriction sites were added to the sequence for subsequent insertion into a vector plasmid. The oligonucleotides received were then mixed together into a supermix and used in PCRs.
Culture conditions for anaerobic growth rescue. (i) Liquid.
Cells of E. coli strain BW ΔadhE ΔldhA Δfrd (JCL166) and its derivatives were cultured overnight in LB with tetracycline and other appropriate antibiotics at 37°C. The next day, overnight cells were inoculated (usually 10 μl) into 5 ml of fresh LB with 1% glucose, 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and appropriate antibiotics to a starting optical density at 600 nm (OD600) of around 0.005 in the 10-ml BD (San Jose, CA) Vacutainer sealed tubes. A needle (20 G by 1 1/2 in.; BD) was inserted through the rubber cap of the glass tube with the other end attached to a Millipore PES (polyethersulfone) filter (0.22 μm). Oxygen in the headspace and medium were then evacuated through the needle by repeated vacuuming and refilling of nitrogen and hydrogen in the anaerobic transfer chamber. The needles were removed from the caps inside the anaerobic chamber. The sealed tubes were then taken out and wrapped with parafilm and tape to prevent bursting of the caps (when cells grow, pressure builds up due to CO2 release). The cultures were then incubated at 37°C in a rotary shaker (250 rpm) for a few days, and growth rates were examined.
(ii) Plates.
To perform the growth rescue experiment on plates (petri dish), transformed cells of the E. coli strain BW ΔadhE ΔldhA Δfrd (JCL166) were either directly picked from LB plates or cultured overnight in LB prior to streaking on fresh LB plates with 1% glucose, 0.1 mM IPTG, and appropriate antibiotics. The plates were then placed in a BD GasPak EZ anaerobic pouch, sealed, and incubated at 37°C for the next few days.
1-Butanol production media.
Unless specified otherwise, production of 1-butanol was performed in terrific broth (TB) (12 g tryptone, 24 g yeast extract, 2.31 g KH2PO4, 12.54 g K2HPO4, 4 ml glycerol per liter of water) supplemented with 2% glucose. For medium analysis, each component in the TB was subtracted out individually.
Culture conditions for 1-butanol production.
Single colonies of the strain transformed with the desired plasmids were cultured overnight in LB with appropriate antibiotics (ampicillin 100 μg/ml, kanamycin 50 μg/ml, and chloramphenicol 50 μg/ml). On the next day, the overnight culture was inoculated at 1% into 5 ml (test tube for anaerobic preculture) or 20 ml (250-ml screw cap flasks for microaerobic preculture) of fresh TB–2% glucose medium (unless otherwise noted). The cultures were grown at 37°C to an OD600 of 0.4 to 0.6 and then induced with 0.1 mM IPTG for another 1 to 2 h aerobically.
For microaerobic production, the 20-ml induced cultures were allowed to grow and produce at 37°C in the 250-ml screw cap flasks for a few days, and samples were taken under aerobic conditions.
For the anaerobic conditions, the 5-ml induced cultures were transferred from test tubes to the 10-ml BD Vacutainer sealed tubes. Oxygen was evacuated by the same method as described above for anaerobic growth rescue. Cultures were then incubated at 37°C in a rotary shaker (250 rpm), and daily samples were taken inside the anaerobic chamber to maintain anaerobicity. If a time course was taken, 1.5% glucose in 1× TB medium was fed to the cultures every day. Unless otherwise noted, the culture pH was adjusted to around 7 using 10 M NaOH on a daily basis.
Bioreactor production of 1-butanol.
Strain JCL299 bearing plasmids pEL11, pIM8, and pCS138 was used in the fermentation for bioreactor production of 1-butanol. The overnight preculture was inoculated in LB containing the appropriate antibiotics and allowed to grow at 37°C in a rotary shaker (250 rpm).
1-Butanol fermentation was performed in a 1-liter stirred-tank bioreactor (Applikon Biotechnology, Schiedam, Netherlands), using a working volume of 0.35 liters. The bioreactor was inoculated with 4 ml of overnight preculture, and 0.1 mM IPTG was added at the time of inoculation to induce the expression of the enzymes involved in the 1-butanol production pathway. Dissolved oxygen (DO) during the aerobic stage was maintained above 20% with respect to air saturation by raising the stirrer speed (from 200 to 600 rpm). The cells were grown at 30°C under aerobic conditions in batch mode until the optical density reached about 8. Then, 2 vvm (volume of gas per volume of liquid per minute) of nitrogen was bubbled through the bioreactor with two goals: (i) to switch to anaerobic conditions and (ii) to accomplish in situ gas stripping of 1-butanol. Upon the anaerobic switch, intermittent linear feeding of glucose solution (500 g/liter) was initiated to maintain a glucose concentration of between 10 and 20 g/liter. The evaporated 1-butanol was condensed using two Graham condensers connected in series. The exhaust gases from the fermentor were bubbled in a trap cooled with ice and then circulated through condenser 1 maintained at 4°C. After that, gas continued circulating through a second equal loop. The pH was controlled at 6.8 at all times by the automatic addition of 2 M NaOH solution. Fermentation samples were collected to determine cell growth, 1-butanol production, organic acids, and glucose concentrations.
Cell extract preparation.
To determine the in vitro activity of each 1-butanol synthetic enzyme in the production strain, E. coli strain JCL166 carrying pEL11 and pIM8 was cultured under conditions and in media identical to those described above for the anaerobic 1-butanol production method. To capture the enzyme activities before and during the production phase, crude cell extracts were prepared from 0.5 ml of the induced culture at the time of anaerobic switch or from the anaerobic culture in the sealed Vacutainer tubes after 20 h of fermentation.
Disruption of cells and preparation of crude extracts were performed under anaerobic conditions. Cells were harvested by centrifugation at 15,000 rpm at room temperature aerobically. The pellets were then resuspended with 0.2 ml of the lysis buffer (50 mM Tris-HCl at pH 7.5, 10× Bugbuster, and 1,000× Lysonase) inside the anaerobic chamber. Lysis was allowed to proceed for 10 to 20 min until the cell resuspension turned clear. The lysate was then centrifuged at 13,200 rpm for 20 min at 4°C. The supernatant was then retrieved for subsequent enzyme assays.
AtoB assay.
All spectrophotometric assays were performed using the Biotek microplate reader (model Powerwave XS) at 30°C under aerobic conditions. The reaction mixture volume was 0.2 ml. Protein concentrations were determined using the Bradford assay or a Nanodrop 2000C spectrophotometer from Thermo Scientific.
The AtoB activity was measured by monitoring the disappearance of acetoacetyl-CoA, corresponding to the thiolysis direction of the enzymatic reaction (22). The disappearance of acetoacetyl-CoA was monitored by the decrease in absorbance at 303 nm, which is the characteristic absorption band of an enolate complex (39) formed by acetoacetyl-CoA with Mg2+. The reaction mixture contained 100 mM Tris-HCl, pH 8.0, 10 mM MgSO4, 200 μM acetoacetyl-CoA, 200 μM CoA, and cell extract prepared as described above. A standard curve was constructed by measuring the absorbance of acetoacetyl-CoA at different concentrations with 10 mM Mg2+.
Hbd assay.
The Hbd activity was measured by monitoring the decrease of absorption at 340 nm, corresponding to consumption of NADH (22). The reaction mixture contained 100 mM 3-(N-morpholino) propanesulfonic acid (MOPS), pH 7.0, 200 μM NADH, 200 μM acetoacetyl-CoA, and crude cell extract. The reaction was initiated by the addition of the cell extract.
Crt assay.
The Crt activity was measured by the decrease of absorption at 263 nm, corresponding to disruption of the α-β unsaturation of crotonyl-CoA (22). The assay mixture contained 100 mM Tris-HCl pH 7.6, 100 μM crotonyl-CoA, and the crude extract. The reaction was initiated by the addition of the cell extract. The standard curves for crotonyl-CoA and 3-hydroxybutyryl-CoA were constructed by measuring the absorbance of the two compounds at 263 nm at different concentrations.
Ter assay.
The Ter activity for crotonyl-CoA was measured at 340 nm. The reaction mixture contained 100 mM potassium phosphate buffer, pH 6.2, 200 μM NADH, 200 μM crotonyl-CoA, and crude extract. The reaction was initiated by the addition of the extract.
To detect the Ter activity for butyryl-CoA, the reaction mixture contained 1 mM NAD+, 0.4 mM butyryl-CoA, and crude extract in 100 mM Tris HCl, pH 7.5. The absorbance was monitored at 340 nm at 30°C. The reaction was initiated by the addition of the extract.
AdhE2 assay.
The aldehyde and alcohol dehydrogenase activities of AdhE2 were measured by monitoring the decrease of absorbance at 340 nm corresponding to the consumption of NADH or NADPH. The reaction mixture contained 100 mM Tris-HCl, pH 7.5, 5 mM dithiothreitol (DTT), 300 μM NADH, and 1 mM butyryl-CoA for the butyraldehyde dehydrogenase (BYDH) reaction and 50 mM butyraldehyde for the butanol dehydrogenase (BDH) reaction. The reaction was initiated by the addition of the extract.
NADH assay.
A fluorescent NAD/NADH detection kit purchased from Cell Technology (Mountain View, CA) was used. Cells were harvested by centrifugation at 13,200 rpm at 4°C. The pellets were then resuspended with 0.2 ml of the NAD/NADH extraction buffer and 0.2 ml of the lysis buffer (provided). Lysis was allowed to proceed for 10 to 20 min at 60°C until the cell resuspension turned clear. The lysate was then centrifuged at 8,000 rpm for 5 min at 4°C. The supernatant was retrieved for subsequent NADH assays.
For the measurement of intracellular NADH levels, the cell lysates were mixed with the enzyme and the fluorescent detection reagent provided in the kit. The reaction was allowed to proceed for 1 to 1.5 h at room temperature in the dark, and then readings were taken with excitation at 530 to 570 nm and emission at 590 to 600 nm.
Ter mutation and selection.
Error-prone PCR was performed using a GenemorphII random mutagenesis kit (Strategene) with plasmids pEL16, pCS127, and pCS128 as templates to amplify the specific ter gene. The initial template concentration was maintained at around 300 to 500 ng for a cycle number of 30. The following primers were used: Tviterfacc65 and Tviterrsal for pEL16, Fjoterfacc65 and Fjoterrsal for pCS128, and Fsuterfacc65 and Fsuterrsal for pCS127. The resulting fragments were gel purified and digested with Acc65 and SalI, followed by DpnI to cut any remaining methylated DNA. The digested PCR products were then ligated into pCS106 cut with the same pair of enzymes. The ligation mixture was transformed into commercial E-Shot DH-10B T1R electrocompetent cells (Invitrogen). The transformed cells were rescued in 10 ml LB in 250-ml screw cap flasks for 1 h, and 10 μl of the cells were plated on LB plates with kanamycin to calculate the library size. At the same time, kanamycin (50 mg/liter) was added to the 10-ml rescued culture. A mutant library size of between 0.5 and 1 million was achieved.
On the next day, the mutant libraries were miniprepped and cotransformed with pEL11 into JCL166. The resulting cells were plated on LB–1% glucose with appropriate antibiotics and incubated at 37°C anaerobically for the next few days. Single colonies were picked based on size, restreaked on fresh LB-glucose plates, and then incubated anaerobically to isolate single colonies again. Plasmids were retrieved from the potential positive cells, and XbaI was used to cut pEL11, leaving behind only the plasmid containing the mutant ter. The digested mixture was again transformed into XL-1 Blue to obtain the pure mutant plasmid, followed by DNA sequencing. The production and growth rescue tests were then repeated to confirm the validity of the ter mutants. The three most successful mutant plasmids were digested with Acc65 and SalI to retrieve the mutagenized ter coding region, followed by ligation into the empty medium-copy-number plasmid pCS106 cut with the same enzymes. The resulting plasmids were sequenced and named pHJ2, pHJ3, and pHJ6.
Quantification of metabolites.
Alcohols were quantified by a gas chromatograph (GC) equipped with a flame ionization detector. The system consisted of a model 6890N GC and a model 6850 automatic injector, sampler, and controller (Hewlett-Packard). The supernatant of culture broth was injected in split injection mode (1:15 split ratio) using 2-methyl-1-pentanol as the internal standard. Detailed procedures were described previously (2).
Glucose was quantified with a YSI glucose analyzer 2700. For other secreted metabolites, filtered supernatant was applied (0.02 ml) to an Agilent 1200 high-pressure liquid chromatography (HPLC) system equipped with an autosampler (Agilent Technologies) and a Bio-Rad (Bio-Rad Laboratories, Hercules, CA) Aminex HPX87 column (5 mM H2SO4, 0.6 ml/min, column temperature at 35°C). Organic acids were detected by using a photodiode array detector at 210 nm. Concentrations were determined by extrapolation from standard curves.
RESULTS
Implementation of the strategy: T. denticola Ter rescued anaerobic growth.
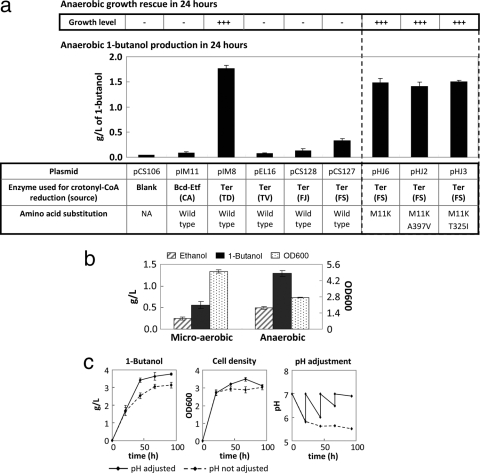
To implement the NADH driving force scheme for 1-butanol production, JCL166 (ΔadhE ΔldhA Δfrd) was used as the host. This strain cannot grow anaerobically without a heterologous NADH-consuming pathway being expressed. To rescue JCL166 for anaerobic growth, the modified CoA-dependent 1-butanol pathway was overexpressed. The four genes atoB (E. coli), adhE2 (C. acetobutylicum), crt (C. acetobutylicum), and hbd (C. acetobutylicum) were expressed as an artificial operon under PLlacO1 promoter control on the high-copy-number plasmid pEL11, and the T. denticola Ter was cloned onto the medium-copy-number plasmid pIM8 by itself for ease in replacing different Ter-equivalent genes, including Bcd-EtfAB, in and out of the system. The expression of E. coli atoB and the native Clostridium gene set (hbd, crt, bcd-etfAB, and adhE2) in JCL166 failed to rescue its anaerobic growth, suggesting that the 1-butanol pathway did not couple effectively to the NADH driving force. The replacement of Bcd-EtfAB with T. denticola Ter successfully restored anaerobic cell growth of JCL166 (Fig. 2a). In addition, with aerobic growth followed by anaerobic fermentation, JCL166 transformed with plasmids pEL11 and pIM8 yielded 1.8 g/liter of 1-butanol in 24 h compared to only 0.1 g/liter generated by an equivalent construct harboring Bcd-EtfAB (Fig. 2a). In contrast, the 1-butanol titer (550 mg/liter) achieved previously by our group (2) using the Bcd-EtfAB complex required semianaerobic conditions with precultures grown in minimal medium.
Fig. 2.
(a) Comparison of anaerobic 1-butanol production using different Ter homologues and mutants. Production level with C. acetobutylicum Bcd-EtfAB is also shown. Successful Ter mutants (dashed box) as a result of first-round growth selection and their corresponding 1-butanol titers are shown, with point mutations specified in the table at the bottom. Host strain JCL166 carrying plasmid pEL11 harboring the artificial operon atoB-adhE2-crt-hbd, along with a second plasmid containing Bcd-EtfAB (pIM11) or various Ter homologous and mutants, was used in all production assays. The extent of anaerobic growth rescue of each strain is indicated in the top panel, where one “+” refers to an OD600 of around 0.2 to 0.3 in liquid cultures. It is important to note that anaerobic growth rescue and 1-butanol production procedures were performed in separate experiments under different culturing conditions (Materials and Methods). CA, Clostridium acetobutylicum; TD, Treponema denticola; TV, Treponema vincentii; FJ, Flavobacterium johnsoniae; FS, Fibrobacter succinogenes; NA, not applicable. JCL166, BW ΔldhA ΔadhE ΔfrdBC. (b) Effect of aeration level on 1-butanol production. Fermentations of strain JCL166 harboring plasmids pEL11 and pIM8 were performed under two different oxygen conditions, as indicated. Detailed procedures for each condition are described in Materials and Methods. Samples were taken after 24 h. Cell densities are listed on the right y axis. JCL166, BW ΔldhA ΔadhE ΔfrdBC. (c) Effect of pH adjustment on anaerobic 1-butanol production. Fermentations of strain JCL166 harboring plasmids pEL11 and pIM8 were performed with or without pH adjustments. “Time” indicates time since inoculation. JCL166, BW ΔldhA ΔadhE ΔfrdBC. Error bars show standard deviations. L, liter.
The activities of all the enzymes in the synthetic 1-butanol pathway were detected by performing in vitro assays (Table 2). AdhE2 showed significantly lower activities than the other enzymes, which may be attributed to its oxygen sensitivity and/or insolubility. Interestingly, we found that Ter is irreversible, based on the in vitro enzyme assay results. Ter uses NADH directly as the electron donor, presumably through a hydride transfer mechanism, which typically is reversible (e.g., lactate dehydrogenase or alcohol dehydrogenase). However, we could not detect Ter activity in the reverse direction with surplus levels of butyryl-CoA and NAD+. The irreversibility of Ter may provide an additional driving force for the 1-butanol production.
Table 2.
In vitro activity of each enzyme in the 1-butanol synthetic pathway
| Enzyme, substratea | Activity (μmol/min/mg) with: |
||
|---|---|---|---|
| No plasmid | pEL11 + pIM8 atb: |
||
| 0 h | 20 h | ||
| AtoB | 0.13 | 10 ± 4 | 17 ± 2 |
| Hbd | 0.043 | 2.7 ± 1 | 4.6 ± 0.8 |
| Crt | 0.75 | 128.4 ± 7.1 | 97.9 ± 13.6 |
| Ter | ND | 1.2 ± 0.2 | 3.7 ± 0.5 |
| AdhE2 | |||
| BYDH | ND | ND | 0.014 ± 0.001 |
| BDH | ND | ND | 0.007 ± 0.001 |
The bifunctional AdhE2 activities were assayed with either substrate listed below. BYDH indicates AdhE2 activity toward butyryl-CoA, and BDH refers to the AdhE2 activity toward butyraldehyde.
JCL166 carrying no plasmid was used as the negative control. Crude extracts were prepared from the production strain (JCL166/pEL11/pIM8) at anaerobic switch (0 h) and in the middle of fermentation (20 h). The resulting in vitro activities were compared.
C. acetobutylicum AdhE2 is essential for 1-butanol production.
Like its E. coli counterpart, C. acetobutylicum AdhE2 also contains an iron-coupled motif, which makes it potentially oxygen sensitive. Currently, C. acetobutylicum AdhE2 is the only bifunctional aldehyde/alcohol dehydrogenase that has been clearly demonstrated to act on butyryl-CoA (20). Since the AdhE2 activity in E. coli remained inconclusive, we speculated that unspecific native alcohol dehydrogenase (in addition to the product of the deleted adhE) present in E. coli strain JCL166 could have contributed to the 1-butanol production. Plasmid pEL12 was constructed by deleting adhE2 on plasmid pEL11 while keeping the other genes. With only AtoB, Hbd, Crt, and Ter overexpressed on plasmids pEL12 and pIM8, 1-butanol production dropped below 0.05 g/liter in 24 h, and the strain failed to restore anaerobic growth of JCL166 (data not shown). Since no formation of butyrate was detected from various 1-butanol production strains, the indigenous butyrate synthetic enzymes may not be actively expressed. Thus, without AdhE2, the only carbon outlet for the 1-butanol pathway was interrupted, and the flux stopped due to backward inactivation of the pathway or metabolic toxicity from intracellular accumulation of acyl-CoA compounds. These results verified the activity of C. acetobutylicum AdhE2 in E. coli and its significant contribution to the production of 1-butanol in the recombinant strain.
Higher 1-butanol titer achieved under anaerobic conditions and neutral pH.
Upon the replacement of Bcd-EtfAB with Ter in the synthetic 1-butanol pathway, the effect of oxygen on 1-butanol productivity was determined by analyzing different 1-butanol production conditions, microaerobic and strictly anaerobic. Contrary to what was reported previously using the Bcd-EtfAB complex (2), where a small amount of oxygen was necessary to achieve higher 1-butanol productivity, the highest accumulation of 1-butanol was observed under fully anaerobic conditions in this case (Fig. 2b). This phenomenon may be an indication of tighter coupling of the 1-butanol pathway with the synthetic NADH driving force upon the replacement of Bcd-EtfAB with Ter and, thus, no need for respiration to recycle the excess NADH. This result also demonstrates the sufficiency of every essential enzyme under strictly anaerobic conditions for 1-butanol production. The presence of oxygen weakened the NADH driving force due to the activity of aerobic respiration. The divergence of carbon flux from 1-butanol into biomass was shown by the increase in cell density and decrease of the 1-butanol titer under the microaerobic conditions (Fig. 2b). The beneficial effect of oxygen as described previously (2) may be attributed to the inefficiency of Bcd-EtfAB for NAD+ regeneration.
Because solventogenesis in Clostridium species is triggered by the low pH resulting from the butyric and acetic acid secretion during the acidogenic phase (47), we set out to investigate whether low pH (around 5.5) is also critical for heterologous enzyme activities and 1-butanol production in the recombinant E. coli strain. Although no butyrate was produced and no solventogenic regulation occurred in the E. coli strain, the effect of pH change due to acetate and pyruvate production may alter the biochemical property of the enzymes. Time courses of 1-butanol fermentation using strain JCL166 transformed with plasmids pEL11 and pIM8 were performed for 4 days, where the pH for one set of cultures was adjusted to 7 daily while the other set was left unadjusted. As shown in Fig. 2c, neutral pH still appeared to be beneficial for E. coli growth and the 1-butanol production rate. This suggests that low pH is not required for the in vivo activity of the nonnative 1-butanol enzymes.
Increasing the NADH driving force by Fdh overexpression improved 1-butanol productivity.
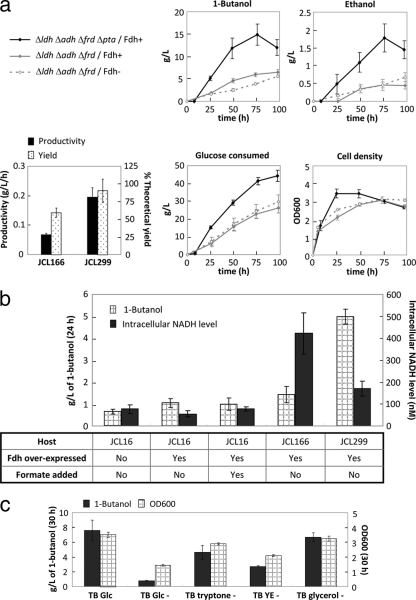
To further increase the intracellular NADH driving force, formate dehydrogenase (Fdh) from C. boidinii (1) carried on plasmid pCS138 was overexpressed to oxidize formate into CO2 and NADH (8, 28, 32). The production profiles of the strains (JCL166 transformed with plasmids pEL11 and pIM8) with and without C. boidinii Fdh overexpression are compared in Fig. 3a. The Fdh-overexpressing strain showed decreased formate secretion, while the strain without C. boidinii Fdh accumulated more pyruvate as a sink for the excess carbon flux due to the redox requirement of the 1-butanol pathway. This observation is consistent with the stoichiometric balance of the 1-butanol pathway, as shown in Fig. 4. The introduction of Fdh slightly improved the production rate and yield of 1-butanol in the first few days of fermentation; however, the Fdh− strain gradually reached the 6.5-g/liter cumulative titer, 60% of the theoretical yield (maximum theoretical yield, 0.41 g/g of glucose), and 0.07 g/liter/h productivity achieved by the Fdh+ strain (Fig. 3a). This result suggests that there may be other limiting factors in the 1-butanol pathway that are unrelated to the NADH/NAD+ ratio. Nevertheless, the stepwise improvement of the 1-butanol titer from that of the wild type is reflected by the increasing intracellular NADH levels in the corresponding strains, as summarized in Fig. 3b.
Fig. 3.
(a) Effects of Fdh overexpression and Pta deletion on anaerobic 1-butanol production. Time courses of alcohol production, cell growth, and glucose consumption are shown. A much higher yield and productivity of 1-butanol was achieved in JCL299 than in JCL166. It is important to note that other components present in the TB medium (such as yeast extracts) also contributed slightly to the 1-butanol titer, therefore affecting the yield. Solid black lines (Δldh Δadh Δfrd Δpta/Fdh+) refer to JCL299 transformed with plasmids pEL11, pIM8, and pCS138. Solid gray lines (Δldh Δadh Δfrd/Fdh+) represent JCL166 transformed with plasmids pEL11, pIM8, and pCS138. Dashed gray lines (Δldh Δadh Δfrd/Fdh−) refer to JCL166 transformed with plasmids pEL11 and pIM8. “Time” indicates time since inoculation. JCL166, BW ΔldhA ΔadhE ΔfrdBC; JCL299, BW ΔldhA ΔadhE ΔfrdBC Δpta. (b) Comparison of intracellular NADH levels and anaerobic 1-butanol production titers in the wild type and the engineered strains. All strains contained plasmids pEL11 and pIM8. Strains indicated as “Fdh over-expressed” also carry plasmid pCS138. A concentration of 20 mM formate was fed to the culture at the time of anaerobic switch where noted. The intracellular NADH level was measured using crude extracts prepared from the production culture after 24 h of fermentation. JCL16, wild type; JCL166, BW ΔldhA ΔadhE ΔfrdBC; JCL299, BW ΔldhA ΔadhE ΔfrdBC Δpta. (c) Medium analysis for anaerobic 1-butanol production. Fermentations of strain JCL299 harboring plasmids pEL11, pIM8, and pCS138 were performed in different medium compositions as indicated on the x axis (“−” indicates the absence of the particular component). The contribution of every element in the TB medium to 1-butanol production was analyzed by the subtraction of each component one by one (Glc, glucose; YE, yeast extract). Samples were taken after 30 h of fermentation. Cell densities are listed on the right y axis. Error bars show standard deviations. L, liter.
Fig. 4.
Stoichiometric balances for 1-butanol synthesis.
The modified 1-butanol pathway without the NADH driving force performed poorly.
Interestingly, when the same heterologous pathway genes carried on plasmids pEL11 and pIM8 were expressed in a wild-type host (JCL16) without the artificial NADH driving force, the anaerobic 1-butanol production was dramatically reduced, to less than 0.5 g/liter in 24 h (Fig. 3b), suggesting that the NADH driving force is more important than the irreversible Ter step. On the other hand, when plasmid pCS138 harboring Fdh was introduced to increase the NADH driving force, in addition to plasmids pEL11 and pIM8, the 1-butanol productivity in JCL16 doubled. The significance of the NADH driving force in 1-butanol production is in sharp contrast to the case of isobutanol, where an intrinsic driving force with CO2 evolution effectively drives the production. Thus, the same wild-type host (JCL16) was able to accumulate a considerable amount of isobutanol (40 g/liter in the wild type versus 50 g/liter in the engineered host) (5) when the synthetic genes were introduced. These results suggest that the artificial NADH driving force did play an important and specific role in channeling the carbon flux into the synthetic CoA-dependent 1-butanol pathway.
Increasing the acetyl-CoA driving force significantly elevated 1-butanol production.
To build the acetyl-CoA driving force, the phosphate acetyltransferase (pta) involved in acetate synthesis was deleted in strain JCL166. The resulting strain JCL299 (ΔadhE ΔldhA Δfrd Δpta), transformed with plasmids pEL11, pIM8, and pCS138, increased 1-butanol production significantly and reached a titer of 15 g/liter and a yield of about 88% of the theoretical in 3 days (Fig. 3a), comparable to the levels achieved by Clostridium species. Production of 1-butanol continued after cells entered the stationary phase. Medium analysis revealed that more than 85% of the 1-butanol synthesized resulted from glucose (Fig. 3c). Glycerol had no observable contribution, while yeast extract appeared to be an important nitrogen source that enhanced cell growth and helped lead to higher titers of 1-butanol (30). The significant elevation in the glucose consumption rate and, hence, the 1-butanol productivity might have resulted from a decreasing ATP/ADP ratio upon the elimination of Pta, similar to the effects observed upon the disruption of ATP synthase (12) and activation of a futile cycle (13, 14, 33). Thus, deletion of the pta gene might have played a dual beneficial role, building up the acetyl-CoA driving force and decreasing the ATP pool. Overall, we have achieved more than 10-fold increases in 1-butanol titers via the NADH and acetyl-CoA driving forces (Fig. 3a and b). While the NADH driving force was required to direct carbon flux into the reversible 1-butanol pathway, the acetyl-CoA driving force coupled with an increased glucose consumption rate was the major contributor to the high-flux production of 1-butanol (Fig. 3b).
Selection of Ter homologues and mutants.
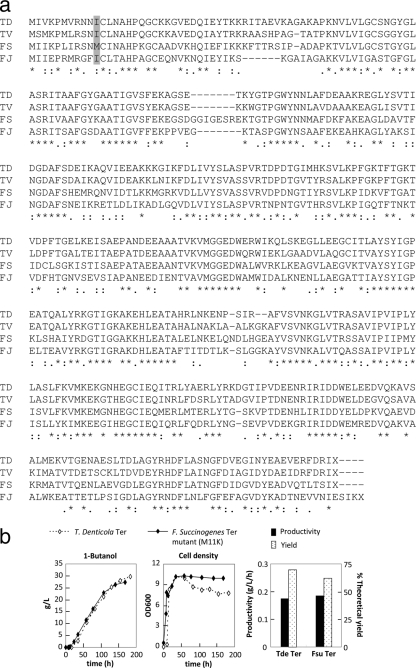
The NADH driving force created by a cell's inability to regenerate NAD+ anaerobically without an external electron acceptor can also be used as a selection platform to improve enzymes and pathways that consume NADH. On the basis of protein sequence homology with the T. denticola Ter, we identified three additional candidate Ter proteins from various organisms, including Treponema vincentii, Fibrobacter succinogenes, and Flavobacterium johnsoniae, in order of decreasing homology (Fig. 5a). Of the three Ter homologues tested, the enzyme from F. succinogenes yielded the highest level of 1-butanol (0.3 g/liter) with aerobic growth followed by anaerobic fermentation (Fig. 2a). However, none of the Ter homologues supported apparent growth of JCL166 under anaerobic conditions.
Fig. 5.
(a) Multiple sequence alignments of Ter homologues from various organisms using ClustalW. The M11K amino acid substitution found in the F. succinogenes Ter mutants is shaded. Fully conserved residues are noted with asterisks. TD, T. denticola; TV, T. vincentii; FS, F. succinogenes; FJ, F. johnsoniae. (b) Anaerobic 1-butanol production in the pH-controlled fed batch fermentor with gas-stripping using either the T. denticola (Tde) Ter or the F. succinogenes (Fsu) Ter M11K mutant. Strain JCL299 was transformed with plasmids pEL11 and pCS138, in addition to plasmid pIM8 carrying the T. denticola Ter or plasmid pHJ6 carrying the F. succinogenes Ter mutant. Similar performances were observed in both cases in terms of 1-butanol titer (30 g/liter), productivity (0.18 g/liter/h), and cumulative yield (around 70% of the theoretical). “Time” indicates time since inoculation. JCL299, BW ΔldhA ΔadhE ΔfrdBC Δpta. L, liter.
To demonstrate that the NADH surplus created in the host can be used as a selection pressure for optimizing the 1-butanol pathway, we subjected the three Ter homologues from T. vincentii, F. succinogenes, and F. johnsoniae to error-prone PCR mutagenesis followed by cotransformation with plasmid pEL11 into strain JCL166. Anaerobic growth was then examined on LB-glucose plates incubated at 37°C for the next 1 to 3 days. Potential positive mutants were isolated based on colony size, purified, sequenced, and confirmed to have 1-butanol production by retransformation with plasmid pEL11 into JCL166. As shown in Fig. 2a, a few first-round mutants of the F. succinogenes Ter successfully restored anaerobic growth of JCL166 and demonstrated an encouraging 1-butanol titer of 1.5 g/liter in 24 h with aerobic growth followed by anaerobic fermentation, comparable to the level achieved with T. denticola Ter. The three best mutants of F. succinogenes Ter carry the identical amino acid substitution Met11Lys, in addition to other silent mutations that may have contributed to the optimization of protein expression and stability in the heterologous system. The importance of the single Met11Lys substitution was confirmed by reintroducing this mutation, using site-directed mutagenesis, into the wild-type F. succinogenes ter gene, followed by anaerobic 1-butanol production using the resulting Ter variant. The success in the mutation and selection of Ter homologues established the basis for strain and/or enzyme evolution for NADH-consuming pathways using this platform, even under conditions of incomplete NADH balance.
Continuous product removal as a driving force successfully doubled the 1-butanol titer.
To further strengthen the driving force and to minimize the adverse effects of 1-butanol toxicity, fermentation was conducted using a 1-liter stirred-tank bioreactor with continuous gas stripping. Strain JCL299 harboring plasmids pEL11, pIM8, and pCS138 was grown aerobically at 30°C to an optical density close to 8 prior to switching to completely anaerobic conditions by means of bubbling nitrogen. Both the T. denticola Ter and the F. succinogenes Ter Met11Lys mutant were tested in individual fermentation experiments to compare their performance. Continuous removal of 1-butanol from the production medium in the pH-controlled fed batch fermentor led to nearly 30 g/liter of 1-butanol in 7 days (Fig. 5b) with intermittent linear feeding of glucose. The two Ter enzymes performed practically the same. Yield was maintained at approximately 70% of the theoretical throughout the fermentation period, with productivity of around 0.18 g/liter/h. In conclusion, gas-stripping effectively minimized the toxic effect of 1-butanol on the production culture and successfully created a stronger entropic driving force to channel carbon flux toward the end product 1-butanol.
DISCUSSION
This work demonstrates the importance of driving forces in high-titer 1-butanol synthesis in E. coli. With the NADH and acetyl-CoA driving forces coupled with Ter, the 1-butanol production in E. coli achieved a level comparable to the levels produced by Clostridium species in flasks and batch fermentors (15, 18, 21, 34, 35), with productivity of 0.2 g/liter/h, titers of 15 g/liter in flasks and 30 g/liter in the fermentor, and yields of approximately 88% of the theoretical in flasks and 70% of the theoretical in the fermentor. The artificial driving force created by NADH accumulation is similar to the transition from butyrate to 1-butanol production in Clostridium that is induced upon the addition of more reductive substrates (5, 36, 40). The deletion of pta showed a significant effect on 1-butanol production (Fig. 3b), suggesting that the acetyl-CoA driving force is also important. As shown in Fig. 3b, the use of the irreversible Ter reaction without any driving force (strain JCL16) produced only one-third the amount of 1-butanol produced by the strain with the NADH driving force (JCL166), which accumulated about one-third the amount of 1-butanol produced by the strain with both NADH and acetyl-CoA driving forces (JCL299). These results indicate the effectiveness of the NADH and acetyl-CoA driving forces in high-titer production systems.
The effectiveness of the NADH driving force as a selection pressure for optimizing the 1-butanol pathway was also demonstrated by our success in retrieving positive Ter mutants with improved catalytic activity or protein expression from the anaerobic-growth rescue platform. This general scheme of alleviating the anaerobic redox imbalance (22, 25, 38, 40, 42) caused by the inactivation of NADH-consuming reactions with target production pathways is applicable to systems where directed evolution is desired. Coupling of the desired reaction(s) as the primary NAD+ regeneration route to the growth phenotype allows the evolution of target enzymes for higher catalytic efficiency, alternative substrate utilization, change of cofactor specificity, and greater robustness.
The stoichiometric redox balances of the 1-butanol pathway in different production strains are summarized in Fig. 4. While 1-butanol synthesis (D) requires 4 mol of NADH for 1 mol of 1-butanol, glycolysis (A) can only provide 2 mol of NADH from 1 mol of glucose. Therefore, the maximum theoretical yield of 1-butanol relies on the host cell's ability to generate additional NADH from pyruvate. Under anaerobic conditions, pyruvate formate lyase (Pfl) converts pyruvate into acetyl-CoA and formate with no generation of NADH. As a result, 2 molecules of glucose are necessary to balance the NADH requirement of the synthetic 1-butanol pathway with pyruvate secretion (E), which was observed from the production strain without C. boidinii Fdh overexpression. On the other hand, overexpression of C. boidinii Fdh allows 2 additional mol of NADH to be generated from formate (C) and fulfills the redox demand of the 1-butanol synthesis pathway (F). The introduction of Fdh effectively balances NADH for the 1-butanol pathway with 1 molecule of glucose and raises the molar theoretical yield to 100%.
Whereas solventogenic acetone-butanol-ethanol (3:6:1) fermentation from the obligate anaerobe Clostridium has to occur after the acidogenic phase that is required for ATP synthesis and redox balance (37, 47), the synthesis of 1-butanol in E. coli allows higher flexibility in the culture conditions, with lower levels of by-products. The tractability of nonnative hosts compared to Clostridium species and the high 1-butanol titer demonstrated in this work present a promising alternative to the traditional acetone-butanol-ethanol fermentation for 1-butanol production. The efficiency of the CoA-dependent clostridial 1-butanol pathway under high-flux conditions as illustrated in this work also suggests its industrial potential for the synthesis of related alcohols and chemicals in nonnative systems. Understanding the role of driving forces would facilitate metabolic engineering efforts to achieve high-titer production in various organisms.
ACKNOWLEDGMENTS
This work was supported by The Kaiteki Institute, Japan. Y.D. was on leave from Mitsubishi Chemical Group Science and Technology Research Center, Inc., Japan.
We thank Iara Machado and Hyun-Jung Lim for their technical assistance.
Footnotes
Published ahead of print on 11 March 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Andreadeli A., Platis D., Tishkov V., Popov V., Labrou N. E. 2008. Structure-guided alteration of coenzyme specificity of formate dehydrogenase by saturation mutagenesis to enable efficient utilization of NADP+. FEBS J. 275:3859–3869 [DOI] [PubMed] [Google Scholar]
- 2. Atsumi S., et al. 2008. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10:305–311 [DOI] [PubMed] [Google Scholar]
- 3. Atsumi S., Hanai T., Liao J. C. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89 [DOI] [PubMed] [Google Scholar]
- 4. Atsumi S., et al. 2010. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 6:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baez A., Cho K. M., Liao J. C. 10 March 2011. High-titer isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. doi:10.1007/s00253-011-3173-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reference deleted.
- 7. Berezina O. V., et al. 2010. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl. Microbiol. Biotechnol. 87:635–646 [DOI] [PubMed] [Google Scholar]
- 8. Berrios-Rivera S. J., Bennett G. N., San K. Y. 2002. Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metab. Eng. 4:217–229 [DOI] [PubMed] [Google Scholar]
- 9. Berrios-Rivera S. J., Sanchez A. M., Bennett G. N., San K. Y. 2004. Effect of different levels of NADH availability on metabolite distribution in Escherichia coli fermentation in minimal and complex media. Appl. Microbiol. Biotechnol. 65:426–432 [DOI] [PubMed] [Google Scholar]
- 10. Boynton Z. L., Bennet G. N., Rudolph F. B. 1996. Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 178:3015–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buelter T., et al. June 2009. Engineered microorganisms for producing n-butanol and related methods. U.S. patent application 20090155869 A1
- 12. Causey T. B., Shanmugam K. T., Yomano L. P., Ingram L. O. 2004. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc. Natl. Acad. Sci. U. S. A. 101:2235–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chao Y. P., Liao J. C. 1993. Alteration of growth yield by overexpression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in Escherichia coli. Appl. Environ. Microbiol. 59:4261–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chao Y. P., Liao J. C. 1994. Metabolic responses to substrate futile cycling in Escherichia coli. J. Biol. Chem. 269:5122–5126 [PubMed] [Google Scholar]
- 15. Chen C. K., Blaschek H. P. 1999. Acetate enhances solvent production and prevents degeneration in Clostridium beijerinckii BA101. Appl. Microbiol. Biotechnol. 52:170–173 [DOI] [PubMed] [Google Scholar]
- 16. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duncombe G. R., Frerman F. E. 1976. Molecular and catalytic properties of the acetoacetyl-coenzyme A thiolase of Escherichia coli. Arch. Biochem. Biophys. 176:159–170 [DOI] [PubMed] [Google Scholar]
- 18. Ezeji T. C., Qureshi N., Blaschek H. P. 2004. Butanol fermentation research: upstream and downstream manipulations. Chem. Rec. 4:305–314 [DOI] [PubMed] [Google Scholar]
- 19. Fischer C. R., Tseng H. C., Tai M., Prather K. L., Stephanopoulos G. 2010. Assessment of heterologous butyrate and butanol pathway activity by measurement of intracellular pathway intermediates in recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 88:265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fontaine L., et al. 2002. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Formanek J., Mackie R., Blaschek H. P. 1997. Enhanced butanol production by Clostridium beijerinckii BA101 grown in semidefined P2 medium containing 6 percent maltodextrin or glucose. Appl. Environ. Microbiol. 63:2306–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hartmanis M. G., Gatenbeck S. 1984. Intermediary metabolism in Clostridium acetobutylicum: levels of enzymes involved in the formation of acetate and butyrate. Appl. Environ. Microbiol. 47:1277–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffmeister M., Piotrowski M., Nowitzki U., Martin W. 2005. Mitochondrial trans-2-enoyl-CoA reductase of wax ester fermentation from Euglena gracilis defines a new family of enzymes involved in lipid synthesis. J. Biol. Chem. 280:4329–4338 [DOI] [PubMed] [Google Scholar]
- 24. Inokuma K., Liao J. C., Okamoto M., Hanai T. 2010. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 110:696–701 [DOI] [PubMed] [Google Scholar]
- 25. Inui M., et al. 2008. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 77:1305–1316 [DOI] [PubMed] [Google Scholar]
- 26. Jantama K., et al. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99:1140–1153 [DOI] [PubMed] [Google Scholar]
- 27. Jarboe L. R., Grabar T. B., Yomano L. P., Shanmugan K. T., Ingram L. O. 2007. Development of ethanologenic bacteria. Adv. Biochem. Eng. Biotechnol. 108:237–261 [DOI] [PubMed] [Google Scholar]
- 28. Kaup B., Bringer-Meyer S., Sahm H. 2004. Metabolic engineering of Escherichia coli: construction of an efficient biocatalyst for D-mannitol formation in a whole-cell biotransformation. Appl. Microbiol. Biotechnol. 64:333–339 [DOI] [PubMed] [Google Scholar]
- 29. Kim Y., Ingram L. O., Shanmugam K. T. 2007. Construction of an Escherichia coli K-12 mutant for homoethanologenic fermentation of glucose or xylose without foreign genes. Appl. Environ. Microbiol. 73:1766–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee S. Y., et al. 2008. Fermentative butanol production by clostridia. Biotechnol. Bioeng. 101:209–228 [DOI] [PubMed] [Google Scholar]
- 31. Li F., et al. 2008. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nielsen D. R., et al. 2009. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 11:262–273 [DOI] [PubMed] [Google Scholar]
- 33. Patnaik R., Roof W. D., Young R. F., Liao J. C. 1992. Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. J. Bacteriol. 174:7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qureshi N., Lolas A., Blaschek H. P. 2001. Soy molasses as fermentation substrate for production of butanol using Clostridium beijerinckii BA101. J. Ind. Microbiol. Biotechnol. 26:290–295 [DOI] [PubMed] [Google Scholar]
- 35. Qureshi N., Saha B. C., Cotta M. A. 2007. Butanol production from wheat straw hydrolysate using Clostridium beijerinckii. Bioprocess. Biosyst. Eng. 30:419–427 [DOI] [PubMed] [Google Scholar]
- 36. Rao G., Mutharasan R. 1987. Altered electron flow in continuous cultures of Clostridium acetobutylicum induced by viologen dyes. Appl. Environ. Microbiol. 53:1232–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sillers R., Chow A., Tracy B., Papoutsakis E. T. 2008. Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab. Eng. 10:321–332 [DOI] [PubMed] [Google Scholar]
- 38. Steen E. J., et al. 2008. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell Fact. 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stern J. R. 1956. Optical properties of acetoacetyl-S-coenzyme A and its metal chelates. J. Biol. Chem. 221:33–44 [PubMed] [Google Scholar]
- 40. Tashiro Y., et al. 2007. Novel high-efficient butanol production from butyrate by non-growing Clostridium saccharoperbutylacetonicum N1-4 (ATCC 13564) with methyl viologen. J. Biosci. Bioeng. 104:238–240 [DOI] [PubMed] [Google Scholar]
- 41. Tucci S., Martin W. 2007. A novel prokaryotic trans-2-enoyl-CoA reductase from the spirochete Treponema denticola. FEBS Lett. 581:1561–1566 [DOI] [PubMed] [Google Scholar]
- 42. Vadali R. V., Bennett G. N., San K. Y. 2004. Cofactor engineering of intracellular CoA/acetyl-CoA and its effect on metabolic flux redistribution in Escherichia coli. Metab. Eng. 6:133–139 [DOI] [PubMed] [Google Scholar]
- 43. Wiesenborn D. P., Rudolph F. B., Papoutsakis E. T. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 54:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yomano L. P., York S. W., Zhou S., Shanmugam K. T., Ingram L. O. 2008. Re-engineering Escherichia coli for ethanol production. Biotechnol. Lett. 30:2097–2103 [DOI] [PubMed] [Google Scholar]
- 45. York S. W., Ingram L. O. 1996. Ethanol production by recombinant Escherichia coli KO11 using crude yeast autolysate as a nutrient supplement. Biotechnol. Lett. 18:683–688 [Google Scholar]
- 46. Zhang X., Jantama K., Moore J. C., Shanmugam K. T., Ingram L. O. 2007. Production of L-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:355–366 [DOI] [PubMed] [Google Scholar]
- 47. Zheng Y. N., et al. 2009. Problems with the microbial production of butanol. J. Ind. Microbiol. Biotechnol. 36:1127–1138 [DOI] [PubMed] [Google Scholar]
- 48. Zhou S., Causey T. B., Hasona A., Shanmugam K. T., Ingram L. O. 2003. Production of optically pure d-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl. Environ. Microbiol. 69:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou S., Shanmugam K. T., Yomano L. P., Grabar T. B., Ingram L. O. 2006. Fermentation of 12% (w/v) glucose to 1.2 M lactate by Escherichia coli strain SZ194 using mineral salts medium. Biotechnol. Lett. 28:663–670 [DOI] [PubMed] [Google Scholar]