Abstract
Alteration of nucleosomes by ATP-dependent remodeling complexes represents a critical step in the regulation of transcription. The human SWI/SNF (hSWI/SNF) family is composed of complexes that contain either Brg1 or hBrm as the central ATPase; however, these separate complexes have not been compared functionally. Here we describe the establishment of cell lines that express epitope-tagged Brg1 and hBrm and a characterization of the complexes associated with these two ATPases. We show that Brg1 fractionates into two complexes that differ in activity and subunit composition, whereas hBrm is found in one complex with lower activity than the Brg1 complexes. These three complexes can remodel nucleosomal arrays, increase restriction enzyme accessibility, and hydrolyze ATP in a DNA-dependent manner. The three complexes differ markedly in their ability to remodel mononucleosomal core particles. We also show that the hBrm complex and one of the Brg1 complexes contain components of the mammalian Sin3 (mSin3) complex. In addition, we have found that Brg1, hBrm, and BAF155 can interact specifically with mSin3A in vitro, showing a direct association of hSWI/SNF complexes with proteins involved in gene repression. These unexpected functional characteristics indicate that these hSWI/SNF complexes play diverse regulatory roles.
Keywords: Chromatin remodeling, ATPase activity, histone deacetylases, Sin3 repressor complex, human SWI/SNF complexes
The mechanisms by which genomic DNA is packaged into chromatin and is unwound during precise stages of cell growth and development have remained obscure. However, with the discovery of chromatin remodelers, histone acetyl-transferases and deacetylases, and DNA methylases and demethylases, it has become clear that cells use a variety of cellular machines to establish and maintain various programs of gene expression (for reviews, see Bird and Wolffe 1999; Kingston and Narlikar 1999; Knoepfler and Eisenman 1999; Kouzarides 1999; Wolffe et al. 1999). Among the activities that facilitate the transition of chromatin from a closed to a more open conformation are multisubunit complexes that use the energy of ATP to alter nucleosome structure. All chromatin remodeling complexes contain a central ATPase that has homology with the yeast SWI2/SNF2. There are three types of ATPases that can be grouped into different subfamilies depending on whether they contain either a bromodomain (SWI2/SNF2 subfamily), two copies of a chromodomain (Mi-2/CHD subfamily), or lack both domains (ISWI subfamily) (Eisen et al. 1995). Members of all three subfamilies have been shown to be part of multisubunit complexes that can remodel nucleosomes in an ATP-dependent manner and either enhance or alleviate the repressive effects of chromatin (for review, see Vignali et al. 2000).
In humans all chromatin remodelers purified to date contain different subunits, except the Brg1 and hBrm complexes, which have been shown to share several highly conserved subunits (Kwon et al. 1994; Wang et al. 1996a,b; Sif et al. 1998). Some of the shared subunits, such as p155 and p170, which are the homologs of yeast SWI3, have been shown to be highly related to each other (Wang et al. 1996b). In addition, three splice variants of p60, which is the human homolog of yeast SWP73, have been shown to be expressed in a tissue-specific manner and are part of different remodeling complexes (Wang et al. 1996b). This heterogeneity of the Brg1 and hBrm complexes has been proposed to be growth and developmentally relevant. The theme of similarity between subunits has also been found in the case of human nucleosome remodeling and deacetylase complex (NuRD), which contains Mi-2 as its central ATPase. NuRD also contains subunits that are highly related to each other, which include histone deacetylases (HDACs) 1 and 2 and retinoblastoma associated proteins (RbA) p46 and p48 (Tong et al. 1998; Wade et al. 1998; Xue et al. 1998; Zhang et al. 1998a). Presently, it is not clear why there are subunits that are highly related within each complex.
The second type of modification that has been shown to play an important role in the regulation of transcription and to have an effect on nucleosome stability is histone acetylation and deacetylation. Modification of histones by acetylation has been linked to gene activation, whereas histone deacetylation has been correlated with gene repression and silencing (for review, see Grunstein 1997). Recent studies have shown that HDAC1 and 2 can associate with mammalian homologs of the yeast corepressor Sin3, mSin3A and B (Alland et al. 1997; Hassig et al. 1997; Laherty et al. 1997; Nagy et al. 1997). This Sin3–histone deacetylase complex also contains RbAp46 and RbAp48, as well as the mSin3A-associated proteins, SAP18 and SAP30, whose functions are unknown (Zhang et al. 1997; Zhang et al. 1998b). Several reports have shown that the Sin3 complex can be targeted by specific transcription factors to repress gene expression. Therefore, it was proposed that repression can be achieved by recruiting histone deacetylase activity to specific promoters. However, it was not clear how HDACs might function in the context of chromatin. Purification of NuRD, which lacks mSin3A subunits, provided the first link between histone deacetylation and chromatin remodeling (Tong et al. 1998; Xue et al. 1998; Zhang et al. 1998a). This finding indicated that repressor proteins can inhibit transcription by targeting complexes with dual function, which can both alter nucleosome structure and deacetylate histones.
To begin to understand some of the biological properties of Brg1 and hBrm, we have examined the activity of these two ATPases across the cell cycle and found that they are regulated differently by phosphorylation during mitosis (Muchardt et al. 1996; Sif et al. 1998). Furthermore, Brg1 and hBrm have been implicated in the control of cell growth and proliferation through their interaction with the Rb family of tumor suppressor proteins (Dunaief et al. 1994; Strober et al. 1996; Zhang et al. 2000). Brg1 has also been shown to be involved in the repression of the c-fos gene through a pathway that is Rb-dependent (Murphy et al. 1999). These results were surprising because Brg1 and hBrm have been shown previously to be able to stimulate transcriptional activation by various transcription factors (Muchardt and Yaniv 1993; Chiba et al. 1994; Singh et al. 1995). Taken together, these results indicate that although Brg1 and hBrm are highly related and interact with similar subunits, their activities are regulated and targeted differently.
To shed more light on the subunit composition as well as the chromatin remodeling activity of the Brg1 and hBrm complexes, we have established cell lines that express either Flag-tagged Brg1 or hBrm. We report that there are two forms of the Brg1 complex that differ by their subunit composition and that can remodel chromatin more efficiently than the hBrm complex. We also present evidence that the Brg1 and hBrm complexes contain subunits of the human Sin3 complex. These differences in mechanism, composition, and associated proteins are likely to be important for the ability of Brg1 and hBrm complexes to participate in both activation and repression of transcription.
Results
Purification of Flag-tagged Brg1- and hBrm-based chromatin remodeling complexes
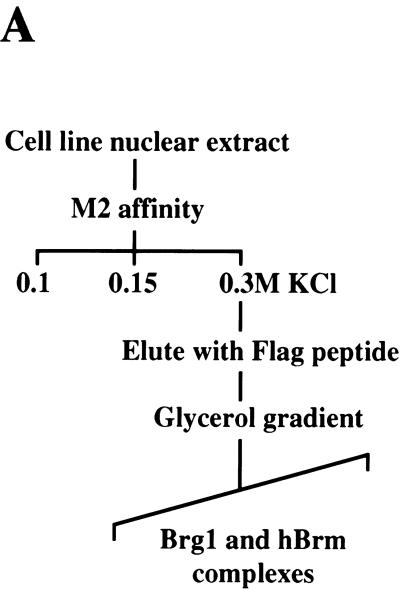
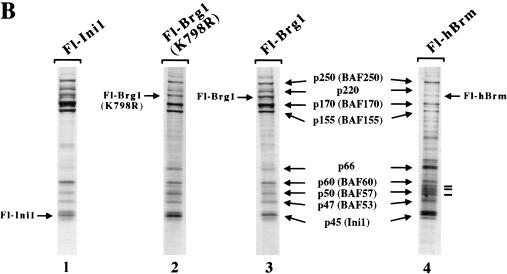
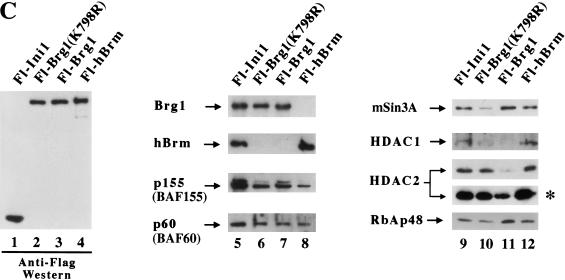
We have reported previously that we can isolate endogenous Brg1- and hBrm-based chromatin remodeling complexes using a two-step purification scheme (Sif et al. 1998). However, using various purification strategies, we have been unable to separate the Brg1 and hBrm complexes (Kwon et al. 1994; data not shown). To separate these two complexes, we created cell lines that express either Flag-tagged Brg1 or hBrm. Nuclear extracts from these cell lines were incubated with anti-Flag M2 affinity gel, and after several washes with buffer containing increasing salt concentrations, the proteins retained on the column were eluted with buffer containing Flag peptide (Fig. 1A). Analysis of equal amounts of wild-type and mutant Fl-Brg1 and Fl-hBrm fractions by SDS-PAGE and silver staining showed that they contain several prominent bands of the same size as the hSWI/SNF subunits (arrows, Fig. 1B, lanes 2–4). For comparison, Brg1 and hBrm complexes purified through the Ini1 subunit are shown (Fig. 1B, lane 1). Approximately 144 ng of each fraction was also analyzed by Western blotting using anti-Flag antibodies (Fig. 1C). There were similar amounts of epitope-tagged mutant and wild-type Brg1 as well as hBrm in the affinity-purified fractions (lanes 2–4), indicating that the concentration of the complexes that contain these proteins is similar in each fraction. Using antibodies specific to cloned hSWI/SNF subunits, we were able to determine that the Fl-Brg1 and Fl-hBrm complexes contain common subunits such as p155 and p60 (Fig. 1C, lanes 5–8).
Figure 1.
Purification of proteins associated with Fl-Ini1, wild-type and mutant Fl-Brg1, and Fl-hBrm. (A) Scheme for purification of Brg1 and hBrm complexes. See Materials and Methods for details. (B) SDS-PAGE analysis of Brg1 and hBrm complexes. Approximately 144 ng of affinity-purified fractions from cell lines that express Fl-Ini1 (lane 1), mutant Fl-Brg1 (lane 2), wild-type Fl-Brg1 (lane 3) and Fl-hBrm (lane 4) were analyzed by SDS-PAGE, and the proteins were visualized by silver staining. (Arrows) Shared subunits; (horizontal bars alongside the hBrm gel) novel subunits. (C) Brg1 and hBrm fractions contain components of the human Sin3 complex. The same amounts of proteins analyzed by silver staining were analyzed by Western blotting using affinity-purified complexes from Fl-Ini1, which contain endogenous Brg1 and hBrm (lanes 1,5,9), mutant Fl-Brg1 (lanes 2,6,10), wild-type Fl-Brg1 (lanes 3,7,11), and Fl-hBrm (lanes 4,8,12). (Asterisk) Long exposure of the HDAC2 Western blot.
Previously, endogenous Brg1 and hBrm have been shown to exist in two distinct complexes (Wang et al. 1996b). Therefore, we wished to determine whether Fl-Brg1 and Fl-hBrm can be found in the same complex. We used specific antibodies to show that Fl-Brg1 and Fl-hBrm reside in two separate complexes when isolated from the epitope-tagged cell lines (Fig. 1C, cf. lanes 6 and 7 with lane 8).
Affinity-purified Fl-Brg1 and Fl-hBrm fractions contain subunits of the human Sin3 complex
We noted that there were additional proteins associated with both the Fl-hBrm and Fl-Brg1 immunopurified complexes. We used antibodies to test these fractions for a wide variety of proteins that modify chromatin structure (see below). Surprisingly, we found that mSin3A, HDAC1, HDAC2, and RbAp48 all copurified with Fl-hBrm (Fig. 1C, lane 12). All of these subunits have been shown previously to coexist in the human Sin3 histone deacetylase complex, which has been implicated in the repression of transcription. Affinity-purified Fl-Brg1 fractions contained mSin3A, HDAC2, and RbAp48, but did not contain detectable levels of HDAC1 (Fig. 1C, lane 11). Mutant Fl-Brg1 fractions contained the same set of proteins; however, their ratios appear to be different (Fig. 1C, cf. lanes 10 and 11). We also determined that HDAC3, Mi-2α (CHD3), and Mi-2β (CHD4) were not present in the affinity-purified hSWI/SNF complexes (data not shown), indicating that the association of mSin3A, HDAC1, and HDAC2 with Brg1 and hBrm complexes is specific.
Fl-Brg1- and Fl-hBrm-based chromatin remodeling complexes are functionally different
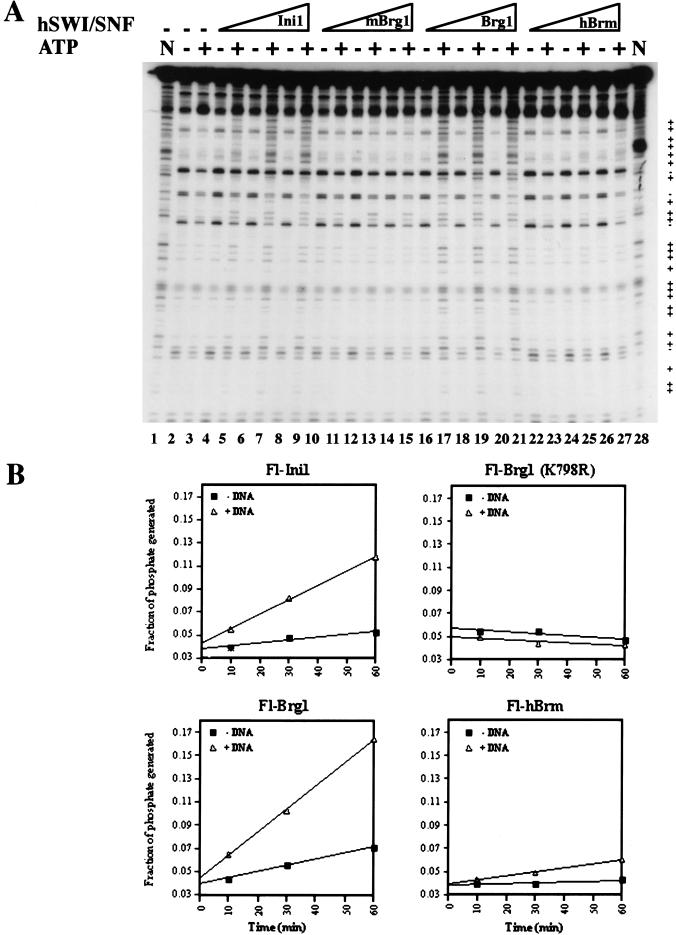
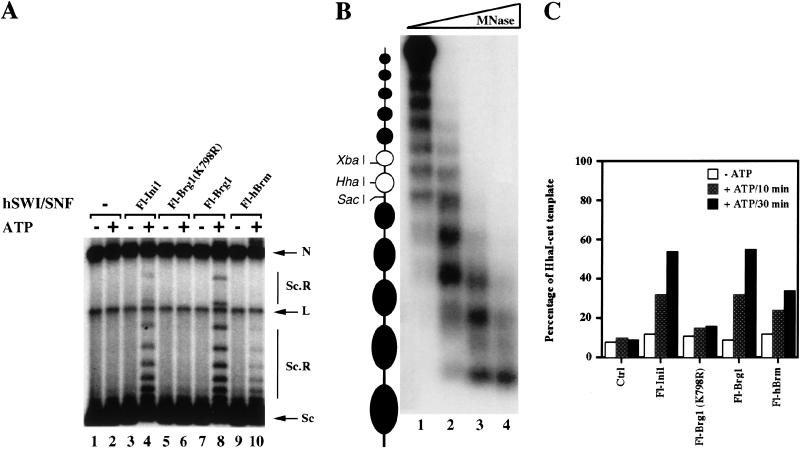
Brg1 and hBrm complexes were purified previously; however, their ability to remodel chromatin individually has not been tested. Therefore, we wanted to measure and compare the chromatin remodeling activity of these complexes by testing their ability to disrupt nucleosomal core particles. We titrated equivalent amounts of affinity-purified Fl-Brg1 and Fl-hBrm complexes into reactions that contained labeled mononucleosomes. The Fl-Brg1 fractions caused an ATP-dependent change in the DNase I digestion pattern (Fig. 2A, lanes 16–21). In contrast, when equal amounts of Fl-hBrm fractions were incubated with core nucleosomes in the presence of ATP, there was no noticeable change in cutting by DNase I (Fig. 2A, lanes 22–27). As a control, fractions containing Brg1 and hBrm complexes purified through the Ini1 subunit (Fig. 2A, lanes 4–9), or mutant Fl-Brg1 (K798R), which is unable to bind ATP (lanes 10–15), are shown.
Figure 2.
Biochemical characterization of Flag-tagged Brg1 and hBrm complexes. (A) Mononucleosome disruption by Brg1 and hBrm complexes. Equal amounts of affinity-purified fractions (in twofold increments from 36 ng [0.72nM] to 144 ng [2.88 nM]) containing either Fl-Ini1 complexes (lanes 4–9), mutant Fl-Brg1 (lanes 10–15), wild-type Fl-Brg1 (lanes 16–21), or Fl-hBrm (lanes 22–27) were incubated with nucleosome cores with or without ATP as indicated. As a control, naked DNA (N, lanes 1,28) and nucleosome cores with and without ATP (lanes 2,3) are shown. (+) Increased DNase I cleavage; (−) decreased DNase I cleavage. (B) ATPase activity of immunopurified Brg1 and hBrm complexes. Approximately 72 ng (3.6 nM) of either Fl-Ini1, mutant Fl-Brg1 (K798R), wild-type Fl-Brg1, or Fl-hBrm was incubated with 20 nM naked plasmid DNA and [γ-32P]ATP for the indicated times. The phosphate present at time zero is due to the presence of [γ-32P]phosphate in the [γ-32P]ATP stock. The ratio of inorganic phosphate to ATP was quantitated for each time point using a Molecular Dynamics PhosphorImager.
All SWI/SNF related complexes that have been studied have a DNA-stimulated ATPase activity. Therefore, we measured the ability of the Fl-Brg1 and Fl-hBrm complexes to hydrolyze ATP both in the presence and absence of DNA using equal amounts of protein (Fig. 2B). Under saturating conditions of ATP and DNA, the ATPase activity of affinity-purified Fl-Brg1, Fl-Ini1, and Fl-hBrm complexes was stimulated fourfold by DNA relative to ATPase activity in the absence of DNA. In contrast, affinity-purified Fl-Brg1 (K798R), which cannot bind ATP, was catalytically inactive. When we compared the DNA-stimulated rate of ATP hydrolysis, we found that Fl-Brg1 complexes have a 1.5- and 5-fold higher specific activity than the complexes purified through the Ini1 and hBrm, respectively.
Because the fractions that contain Fl-hBrm appear to hydrolyze ATP but were unable to remodel mononucleosomes, we tested their ability to remodel chromatin using polynucleosomal templates. First, we tested the ability of affinity-purified Fl-Brg1 and Fl-hBrm complexes to alter the topology of plasmid DNA that has been assembled into arrays of nucleosomes using Drosophila S190 extracts (Fig. 3A). When equal amounts of affinity-purified Fl-Brg1 and Fl-hBrm fractions were incubated with highly negatively supercoiled plasmid DNA in the absence of ATP, there was no change in topology of the DNA templates (Fig. 3A, lanes 7,9). However, when ATP was added to the reactions, Fl-Brg1 and Fl-hBrm complexes were able to remodel as shown by the increase in the amount of slowly migrating DNA species that contain fewer negative supercoils (Fig. 3A, cf. lanes 8 and 10 with lane 2). For comparison, fractions that contain a mixture of endogenous Brg1 and hBrm, or mutant Fl-Brg1 (K798R), which is catalytically inactive, are shown (Fig. 3A, lanes 3–6). In agreement with the ATPase assay, these results show that the Fl-hBrm complex can hydrolyze ATP and remodel chromatin.
Figure 3.
Disruption of polynucleosomal templates. (A) Brg1 and hBrm complexes can alter the topology of highly negatively supercoiled chromatin templates. Approximately 144 ng (2.88 nM) of affinity-purified Fl-Ini1 complexes (lanes 3,4), mutant Fl-Brg1 (lanes 5,6), wild-type Fl-Brg1 (lanes 7,8), and Fl-hBrm (lanes 9,10) was incubated with nucleosome-assembled plasmid DNA in the absence or presence of ATP as indicated. (Lanes 1,2) Assembled templates without Brg1 and hBrm complexes. (N) Nicked closed circular DNA; (L) linear DNA; (Sc) supercoiled DNA; (Sc.R) less supercoiled and relaxed DNA. (B) Analysis of the 5S polynucleosomal array. To determine the extent of assembly of the 5S array, increasing amounts of MNase (lane 1, 1 mU; lane 2, 10 mU; lane 3, 100 mU; lane 4, 1U) were added to 2 ng of assembled template, and the 5S DNA was detected by Southern blotting as described in Materials and Methods. (C) Brg1 and hBrm complexes can increase restriction enzyme accessibility. Equal amounts (144 ng, or 2.88 nM) of affinity-purified Brg1 and hBrm complexes were incubated with 6 ng (0.14 nM) of 5S nucleosomal arrays. After either 10 or 30 min at 30°C, HhaI was added, and the reactions were incubated for an additional 60 min at 30°C. The percentage of HhaI-cut template represents the ratio of cut DNA to total amount of template.
To rule out the possibility that there are additional factors in the Drosophila S190 extracts that might assist Fl-hBrm complexes in remodeling chromatin templates, we also tested Fl-Brg1 and Fl-hBrm complexes for their ability to remodel a linear DNA template that was assembled using purified components. The DNA fragment used in this assay contains an array of 12 nucleosomes. The middle two nucleosomes include unique restriction sites that are located either within the nucleosome (Fig. 3B, HhaI and XbaI sites) or in the linker region between nucleosomes (SacI site) (Neely et al. 1999). The assembly of this template was achieved by salt dialysis using purified DNA and H1-depleted HeLa core histones. The nucleosomal phasing was determined by restriction enzyme cutting (see below) and MNase/Southern blotting (Fig. 3B).
In this assay, the chromatin remodeling activity is measured by the ability of the immunopurified complexes to either increase or decrease accessibility to restriction enzyme sites. When equal amounts of Fl-Brg1 and Fl-hBrm complexes were incubated with the 5S array and accessibility to the otherwise occluded HhaI site was measured by incubating the template with restriction enzyme, we were able to see an increase in cutting only in the presence of ATP (Fig. 3C). Similar results were obtained when XbaI was used (data not shown). Quantification of the amount of template digested by HhaI after 30 min of chromatin remodeling revealed that the Fl-Brg1 complexes increased cutting by 6.9-fold, whereas the Fl-hBrm complex caused a 4.3-fold increase. Furthermore, when the Ini1 fractions were tested in this assay, there was a 6.8-fold stimulation of cutting by HhaI. Mutant Fl-Brg1 (K798R), which lacks ATPase activity, showed a twofold stimulation; there is no activity of this fraction following further purification (see below). We have also tested the ability of wild-type and mutant Fl-Brg1, Fl-hBrm, and Fl-Ini1 complexes to remodel 5S arrays using substoichiometric amounts of hSWI/SNF proteins and obtained similar results (data not shown). We conclude that the Fl-hBrm complexes have a 1.6-fold lower specific activity than the Fl-Brg1 complexes in the protocol that measures remodeling of arrays, but that there is a much more significant decrease in the relative ability of the Fl-Brm complex to remodel mononucleosomes.
Components of the Sin3 complex cofractionate with the Brg1 and hBrm complexes
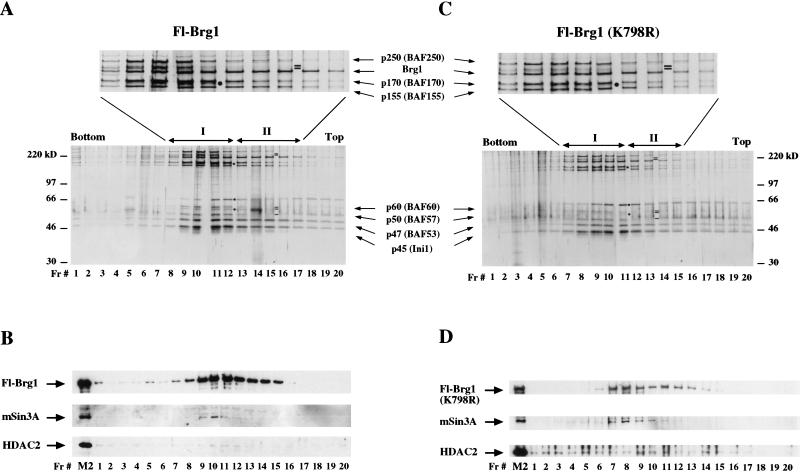
Affinity-purified Brg1 and hBrm fractions contain subunits of the Sin3 complex (Fig. 1C). To determine whether these subunits cofractionate with Brg1 and hBrm, we separated equivalent amounts of each complex using a glycerol gradient and analyzed each fraction by silver staining and Western blot analysis (Figs. 4,5). Fractionation of wild-type and mutant Fl-Brg1 complexes followed by silver stain analysis revealed two different types of complexes (labeled I and II, Fig. 4A,C). We show below that the location of these two peaks correlates with two different peaks of remodeling activity. Wild-type Fl-Brg1 complex I elutes in fractions 8 through 12, and complex II, which lacks the 50-kD subunit, and the 60-kD and 220-kD doublets, elutes in fractions 13 through 17 (Fig. 4A). The wild-type and mutant Fl-Brg1 complexes have similar polypeptide banding patterns (Fig. 4C). By Western blotting, Brg1 was present in complexes I and II; however, the mSin3A subunit was found only in complex I (Fig. 4B, fractions 9–11 and Fig. 4D, fractions 7–10). HDAC2 was detected in most fractions of mutant Fl-Brg1 (K798R), and was hardly detectable in the wild-type fractions because its levels were reduced relative to the Fl-Ini1, mutant Fl-Brg1, and Fl-hBrm fractions (Fig. 1C, lanes 9–12).
Figure 4.
SDS-PAGE analysis of wild-type and mutant Fl-Brg1 glycerol gradient fractions. (A) Ten microliters of each fraction derived from immunopurified Fl-Brg1 complexes was electrophoresed on an 8% SDS–polyacrylamide gel, and the proteins were visualized by silver staining. (B) Analysis of 10 μL of each fraction by Western blot analysis revealed that components of the Sin3 complex cosediment with Fl-Brg1 complex I. Equal amounts (10 μL) of glycerol gradient fractions from mutant Fl-Brg1 were analyzed as described for wild-type Fl-Brg1, and the proteins were visualized either by silver staining (C) or by Western blotting (D). (Filled circles) Novel subunits that cofractionate with the Brg1 complexes; (small black bars) missing subunits; (I and II) complexes that differ by their subunit composition; (Fr.#) fraction number; (M2) lanes containing 144 ng of affinity-purified fractions.
Figure 5.
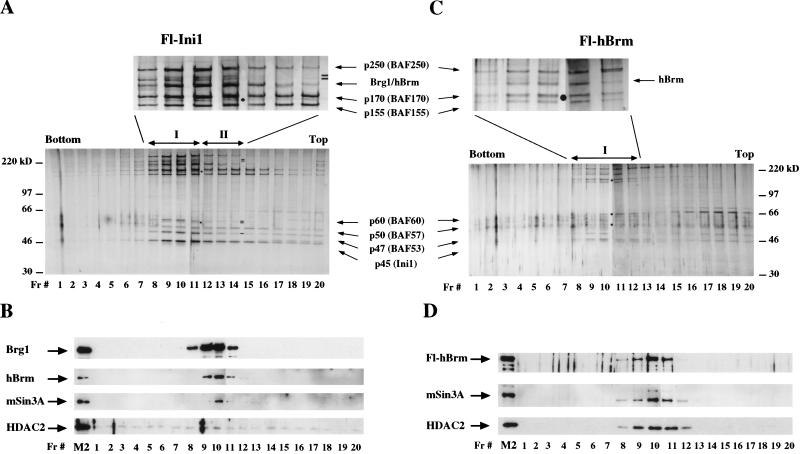
Fl-Ini1 and Fl-hBrm complexes contain components of the Sin3 complex. Ten microliters of glycerol gradient fractions derived from affinity-purified Fl-Ini1 complexes, which contain endogenous Brg1 and hBrm, was analyzed by SDS-PAGE, and hSWI/SNF subunits were visualized either by silver staining (A) or Western blot analysis (B). Equal amounts (10 μL) of each glycerol gradient fraction derived from immunopurified Fl-hBrm complexes were analyzed by either silver staining (C) or Western blotting (D). (Filled circles) Bands that cosediment with Brg1 and hBrm complexes; (small black bars) subunits that are missing; (Fr.#) fraction number; (M2) lanes containing approximately 144 ng of affinity-purified fractions.
There are at least two types of complexes in the affinity-purified Fl-Ini1 fractions, which differ by their subunit composition (Fig. 5A). The first type of complex coincides with the peak of anti-Brg1 and anti-hBrm activity (Fig. 5B, fractions 8–11), whereas complex II appears to lack the subunits that migrate as 60- and 220-kD doublets and contains substoichiometric amounts of the 47-kD subunit (Fig. 5A). There are also complexes that lack the upper high molecular weight subunits (Fig. 5B, fractions 15–19). Western blot analysis of the glycerol gradient fractions revealed that endogenous Brg1 and hBrm cofractionate in the complexes isolated using tagged Ini1 (Fig. 5B, fractions 9–11). HDAC1 and mSin3A cofractionate with the peak of Brg1 and hBrm complexes (fraction 10, and data not shown). HDAC2 was also detected in these gradient fractions; however, its elution profile appeared to be more widespread than the other components of the Sin3 complex.
Analysis of Fl-hBrm fractions showed that there is only one major complex that elutes in fractions 9 through 11 and correlates with the activity of anti-Fl-hBrm antibodies (Fig. 5C,D). This complex also correlates with the presence of remodeling activity (see below). By Western blotting, it appears that mSin3A, HDAC1, and HDAC2 cofractionate with Fl-hBrm and its associated hSWI/SNF subunits (Fig. 5C,D). It is important to note that unlike Fl-Brg1, which is found in at least two different complexes that appear to lack HDAC1 and have reduced levels of HDAC2 (Fig. 4B), Fl-hBrm is found in only one complex and is associated with both HDAC1 and HDAC2 (Fig. 5D). Furthermore, by silver staining it appears that the Fl-hBrm complex lacks only the 220-kD doublet (Fig. 5C, fractions 9–11). Taken together, these results show that although Fl-Brg1 and Fl-hBrm complexes contain highly conserved subunits, they interact differently with other proteins, and consequently this might affect their ability to remodel chromatin.
We have found that mSin3A cofractionates with Brg1 and with hBrm in all four cell lines that we have analyzed. In addition, there are Brg1 complexes that have remodeling activity but do not associate with mSin3A (complex II). Therefore, we conclude that there is a direct association of mSin3A, which is normally involved in gene repression, with specific Brg1- and hBrm-based chromatin remodeling complexes.
Interaction of mSin3A with hSWI/SNF subunits in vitro
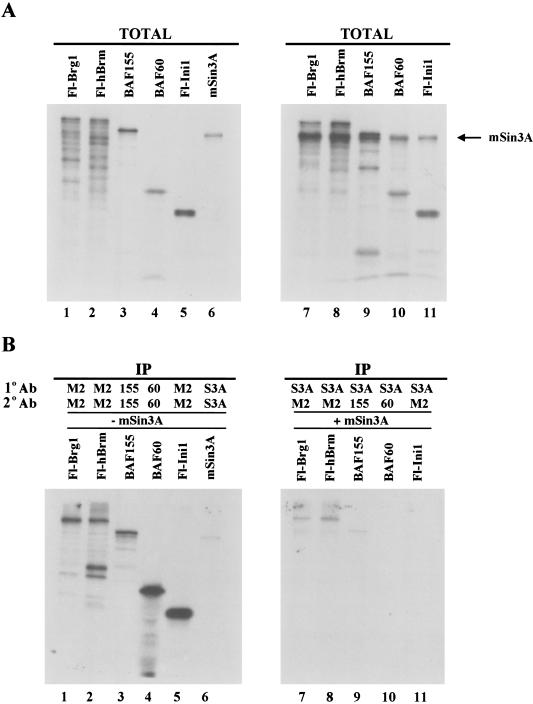
To further study which components of hSWI/SNF complexes mediate the interaction with the Sin3A complex, we performed immunoprecipitation experiments using in vitro translated proteins. Fl-Brg1, Fl-hBrm, BAF155, BAF60, and Fl-Ini1 were expressed either individually (Fig. 6A, lanes 1–5) or in combination with mSin3A (Fig. 6A, lanes 7–11) in the presence of [35S]methionine and were subjected to immunoprecipitation with an affinity-purified anti-mSin3A antibody (Fig. 6B, lanes 7–11). After extensive washing of the immunoprecipitates, hSWI/SNF subunits that interacted with mSin3A were released by heat denaturation and subjected to a second immunoprecipitation using specific antibodies. Fl-Brg1, F-hBrm, and BAF155 were the only hSWI/SNF subunits detected, indicating that these subunits can interact with mSin3A. As a control, immunoprecipitations of individually translated proteins are shown (Fig. 6B, lanes 1–6). When preimmune anti-Brg1, anti-hBrm, and anti-BAF155 antisera were used, no labeled proteins were detected (data not shown). In light of these results, we conclude that the association of mSin3A with the Brg1 and hBrm complexes is specific.
Figure 6.
mSin3A can associate specifically with hSWI/SNF subunits in vitro. (A) hSWI/SNF subunits were in vitro translated either individually (lanes 1–5) or in combination with mSin3A (lanes 7–11) using the Promega TNT coupled reticulocyte lysate system as described in Materials and Methods. TOTAL represents 33% of the total amount of protein used in the immunoprecipitation assays. (B) Individual proteins that were immunoprecipitated (IP) (lanes 1–6) were first immunoprecipitated (1° Ab) with either anti-Flag (M2), anti-BAF155 (155), anti-BAF60 (60), or anti-mSin3A (S3A) antibodies and then subjected to a second immunoprecipitation (2° Ab) as indicated. (Lanes 7–11) Double immunoprecipitations of hSWI/SNF subunits that were cotranslated with mSin3A.
Brg1 complexes fractionate into two functionally distinct complexes on glycerol gradients
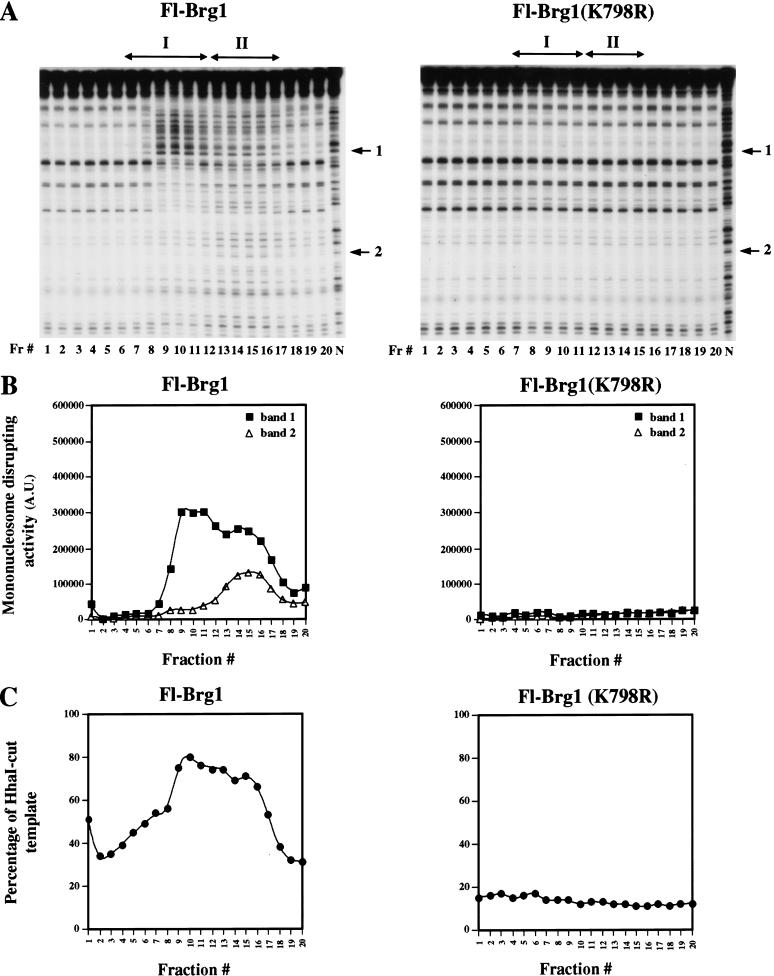
To characterize further the function of the Brg1 and hBrm complexes in chromatin remodeling, we separated equal amounts of these complexes using glycerol gradient sedimentation and tested the resultant fractions for remodeling activity. We assayed each fraction for its ability to disrupt the DNase I digestion pattern of mononucleosomes in the presence of ATP, and we also tested the ability of each fraction to enhance restriction enzyme access to a 5S nucleosomal array. We were surprised to find two different peaks of activity when we fractionated the affinity-purified Brg1 complexes (Fig. 7A,B, Fl-Brg1 panels). In the mononucleosome remodeling assay, fractions 9–11 displayed strong disruption of the DNase I digestion pattern in regions that correspond to the shoulder of the nucleosome, but showed less disruption near the dyad axis. In contrast, fractions 13–16 showed disruption of the DNase I digestion pattern throughout the mononucleosome. When these fractions were tested for activity on nucleosomal arrays using the restriction enzyme cleavage protocol, we found maximal activities in fractions 9 through 16 (Fig. 7C). Although all active fractions contain Brg1 (Fig. 4B), there are differences in associated proteins that correlate with the differences in the mononucleosome disruption assay. In all assays, mutant Fl-Brg1 complexes had no activity (Fig. 7A,C, Fl-Brg1/K798R panels), showing that the chromatin remodeling activity of these complexes depends on having an active Brg1 ATPase.
Figure 7.
Characterization of the chromatin remodeling activity of glycerol gradient-purified fractions. (A) Wild-type Fl-Brg1 complexes I and II can remodel nucleosomal core particles. DNase I disruption assays were performed as described in Materials and Methods. Mutant Fl-Brg1 (K798R) complexes were inactive. (Fr.#) Fraction number; (N) naked DNA; (arrows 1 and 2) bands used to quantify the nucleosome remodeling activity of each fraction in arbitrary units (A.U.). (B) Quantification of bands 1 (filled squares) and 2 (open triangles) revealed that the peaks of nucleosome remodeling activity correlate with the elution profile of active Brg1 complexes. (C) Fl-Brg1 complexes can remodel nucleosomal arrays and increase restriction enzyme accessibility. Equal amounts of each fraction (10 μL) were incubated with the 5S array and assayed for activity by incubating the remodeled template with HhaI. The percentage of cut template represents the ratio of cut template to total DNA.
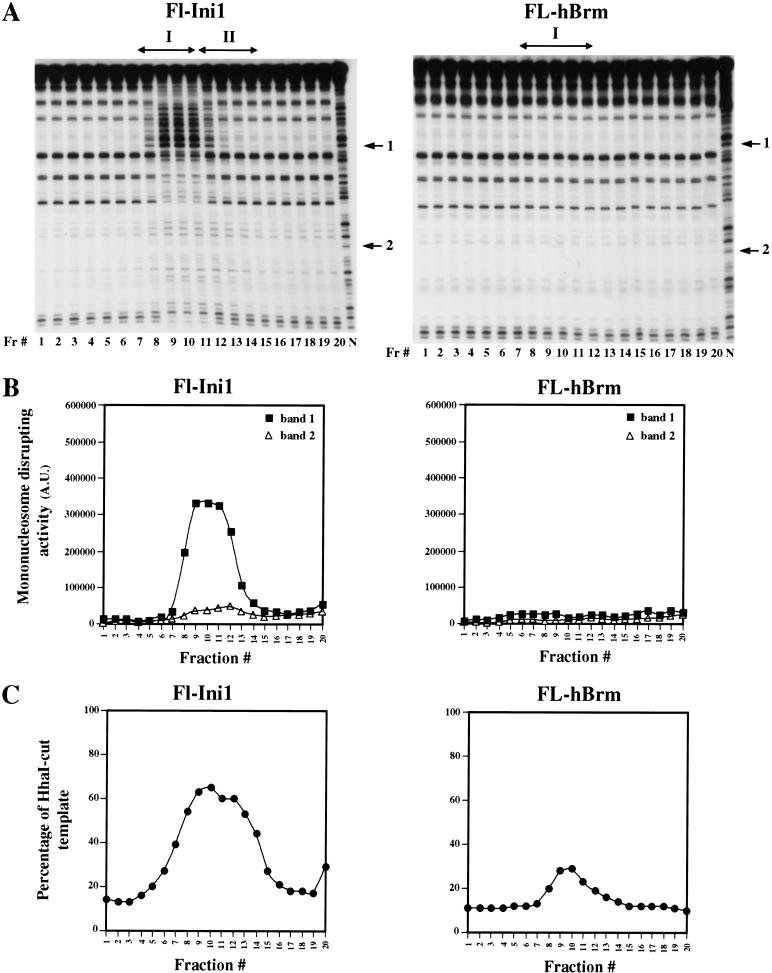
Fractionation of the Fl-Ini1 complexes, which contain a mixture of Brg1 and hBrm-based complexes, resulted in a broad peak of chromatin remodeling activity (Fig. 8). The activity in the DNase I disruption protocol correlated with the presence of Brg1 and hBrm (cf. Fig. 8, Fl-Ini1 panels, with Fig. 5B), and displayed a disruption pattern that was similar to the disruption observed with Brg1 complexes I and II (cf. Fig. 8A with 7A). Analysis of the Fl-hBrm complexes revealed that there is one peak of activity in the restriction enzyme accessibility protocol (Fig. 8C). These fractions had no activity in the DNase I disruption protocol, as expected (Fig. 8A,B). These results indicate that the hBrm complex has less activity than each of the two Brg1-based complexes.
Figure 8.
Chromatin remodeling activity of Fl-Ini1 and Fl-hBrm complexes. (A) Fl-Ini1 and Fl-hBrm glycerol gradient fractions (10 μL each) were incubated with nucleosome cores as described in Materials and Methods. Fl-Ini1 fractions show a single broad peak of nucleosome-disrupting activity, which correlates with the activity of anti-Brg1 and anti-hBrm antibodies. Fl-hBrm complex did not alter the mononucleosome 10-bp ladder. (Fr.#) Fraction number; (N) naked DNA; (arrows 1 and 2) bands used to generate the graphs shown in B. (C) Fl-Ini1 and Fl-hBrm complexes can remodel nucleosomal arrays. Equal amounts (10 μL) of each glycerol gradient fraction were assayed for activity as described for Fl-Brg1 complexes. The percentage of cut template represents the ratio of cut template to total DNA.
Discussion
The Brg1 and hBrm complexes that we have characterized display significant functional differences. The two Brg1 complexes can efficiently remodel nucleosomes as core particles as well as arrays, whereas the hBrm complex can remodel only arrays of nucleosomes. These complexes also differ in composition. One of the Brg1 complexes and the hBrm complex associate with mSin3A, a repressor that is involved in the mechanisms that regulate cell growth. It had been recognized previously that there are multiple members of the SWI/SNF family of remodeling complexes. One possibility is that these differences cause these complexes to be targeted differentially to different genes. The studies reported here show that these complexes will bring different remodeling capabilities when they are targeted to genes and also that they can bring, by association, proteins involved directly in repression. The hSWI/SNF family of complexes is therefore likely to contribute more broadly to regulation of gene expression than previously thought.
Brg1 and hBrm complexes are functionally different
Studies that examined the levels of Brg1 and hBrm under either growth-inhibiting or growth-inducing conditions indicated that the levels of hBrm increase in cells that have withdrawn from the cell cycle and decrease in cells that are actively dividing (Muchardt et al. 1998). Based on these results, it was proposed that hBrm negatively regulates cell growth (Muchardt et al. 1998; Reyes et al. 1998). Other evidence that supports the notion that Brg1 and hBrm are different stems from the findings that show that Brg1 and hBrm complexes are inactivated by phosphorylation in vivo; however, only the hBrm complex is targeted for degradation during mitosis (Muchardt et al. 1996; Sif et al. 1998).
We have now purified complexes that contain either Brg1 or hBrm from HeLa cells and showed that Brg1 is associated with at least two different complexes, which differ by their subunit composition, lack HDAC1, and contain low levels of HDAC2. In contrast, hBrm is found in one complex that contains both HDAC1 and HDAC2. We have also shown that mSin3A cofractionates with Brg1 complex I and the hBrm complex and can interact with Brg1, hBrm, and BAF155 in vitro. The differential ability of Brg1 and hBrm complexes to associate with different members of the Sin3 complex might be a key reason for the differences that these complexes play in biological regulation.
Using different assays, we have shown that the Brg1 complexes are more active than the hBrm complex, particularly in their ability to remodel mononucleosome templates. There are several possible explanations for why the hBrm complex does not remodel chromatin as efficiently as the Brg1 complexes. The possibility that we favor is that additional subunits repress the remodeling activity of the hBrm complex. Add-back experiments show that when immunopurified Fl-hBrm complex is added in stoichiometric amounts to Fl-Brg1 complexes, it can decrease the remodeling activity on nucleosomal arrays (data not shown). In contrast, experiments using baculovirus-expressed proteins show that Fl-Brg1 and Fl-hBrm have similar specific activities (G.J. Narlikar and R.E. Kingston, unpubl.), and addition of affinity-purified Fl-hBrm to Fl-Brg1 does not inhibit the ability of Brg1 to remodel nucleosomal arrays (Phelan et al. 1999). Thus, inhibition in these add-back experiments requires proteins other than the ATPase. One attractive possibility is that HDAC1 and HDAC2 directly affect the remodeling activity of the hBrm complex; these proteins are found associated predominantly with hBrm (Figs. 1C and 5D).
Brg1 is the ATPase of two distinct chromatin remodeling complexes
Purification of affinity-purified mutant and wild-type Fl-Brg1 complexes by glycerol gradient sedimentation revealed that there exist two different complexes that contain Brg1 as their central ATPase. Fl-Brg1 complex I contains all known hSWI/SNF subunits; however, Fl-Brg1 complex II lacks p50, and p60 and p220 doublets. It remains possible that Fl-Brg1 complex II is a partial version of complex I, but we believe that this is very unlikely because the missing subunits are not limiting (data not shown). Both complexes can remodel nucleosomal arrays and display different mononucleosomal disrupting activities. Complexes that were isolated through Fl-Brg1 (K798R) were inactive in all assays, indicating that Brg1 is the major ATPase in these complexes. Furthermore, we have noticed that the levels of mSin3A and RbAp48 are reduced in mutant Brg1 affinity-purified fractions. It is possible that these two genes might require Brg1 complexes for their expression. Therefore, when mutant Brg1, which is unable to remodel chromatin, is expressed, it partially interferes with their activation.
There are clear differences in the way Brg1 complexes I and II alter nucleosome structure, as shown by the mononucleosome disruption assay (Fig. 7A,B). Brg1 complex I disrupts the edge of the nucleosome and shows protection from DNase I cleavage near the dyad axis. In contrast, Brg1 complex II induces cutting throughout the nucleosome cores. It is likely that the lack of certain subunits in Brg1 complex II might contribute to the widespread disruption of the nucleosome ladder. Evidence that supports this hypothesis comes from studies by Wang et al. (1998), which show that BAF57 (p50 in our complexes) can avidly bind four-way junction DNA through its HMG-like DNA binding domain. Based on this result and the studies of Quinn et al. (1996), it was speculated that BAF57 might bind nucleosomes near the dyad axis, where DNA enters and exits the nucleosome, and therefore might adopt a structure that resembles a four-way junction. Our results show that Brg1 complex I, which contains p50 (BAF57), can induce protection over the region that spans the dyad axis. In contrast, Brg1 complex II, which lacks p50 as well as p60 and p220 subunits, shows a different pattern of disruption of the 10-bp ladder. Therefore, it is possible that the p50 subunit either alone or in combination with other hSWI/SNF subunits is responsible for blocking remodeling at the dyad axis.
There is ample evidence indicating that chromatin remodeling complexes are targeted by transcription factors to perform specific changes at specific loci (Cosma et al. 1999; Gregory et al. 1999; Krebs et al. 1999). The nature of the changes introduced by chromatin remodeling complexes is still not clear, although it is likely that these changes are going to be regulated and specific in nature. Recent studies have shown that the yeast SWI/SNF and RSC complexes can remodel nucleosomes by catalyzing the transfer of the core histones in cis or in trans (Lorch et al. 1999; Whitehouse et al. 1999; Jaskelioff et al. 2000). In contrast, members of the ISWI subfamily of chromatin remodeling complexes can alter nucleosome structure by promoting histone octamer sliding in cis, but not in trans (Hamiche et al. 1999; Längst et al. 1999). Therefore, different classes of transcription factors might exploit the intrinsic differences that exist between different forms of remodeling complexes to induce specific alterations of nucleosome structure. For example, Brg1 complex I might be recruited to induce changes in localized regions of the nucleosome, whereas Brg1 complex II might be targeted to cause a more widespread disruption of chromatin. It is also conceivable that the difference in activity between Brg1 and hBrm complexes might be used selectively by various transcription factors to achieve different levels of chromatin remodeling.
Interaction of Brg1 and hBrm complexes with components of the Sin3 complex
Several studies have shown that multisubunit chromatin remodeling complexes can act in concert with histone acetyl transferases to activate transcription (Cosma et al. 1999; Gregory et al. 1999; Krebs et al. 1999). More recently, members of the Mi-2 subfamily of ATPases have been found in association with HDAC1 and 2, providing a link between chromatin remodeling and histone deacetylation (Tong et al. 1998; Xue et al. 1998; Zhang et al. 1998a). We now show that Brg1 and hBrm complexes associate with components of the Sin3 complex, which has been shown to be involved in the repression of transcription by a wide variety of factors. Characterization of the chromatin remodeling activity of these complexes shows that they have different specific activities and can alter nucleosome structure in a different manner. Taken together, these results indicate that proteins that target the Sin3A/histone deacetylase complex might also indirectly target the Brg1 and hBrm complexes. Precedents for such a mechanism are provided by the NuRD complex, which has been shown to be targeted by factors that can repress transcription (Kim et al. 1999; O'Neill et al. 2000).
There are several questions that remain unanswered. For example, why is HDAC1 not present in the Brg1 complexes? Why is mSin3A found in association only with Brg1 complex I and not with complex II? Both complexes remodel nucleosomes; however, it appears that Brg1 complex I has a different pattern of disruption of the 10-bp ladder than complex II. Does this mean that the Sin3 complex can interact only with complexes that can induce localized changes in nucleosome structure? Using the Brg1 and hBrm epitope-tagged cell lines, we can begin to address these questions and elucidate the mechanisms by which Brg1 and hBrm complexes are regulated and targeted.
Materials and methods
Plasmid constructions
Plasmid pBabe/Fl-Ini1 for in vivo expression of C-terminally Flag-tagged Ini1 was described previously (Sif et al. 1998). Plasmid pBS(KS+)/Fl-Ini1 was generated by subcloning a 1.2-kb EcoRI fragment excised out of pBabe/Fl-Ini1 into the unique EcoRI site of Bluescript. Retroviral vectors used to express C-terminally Flag-tagged wild-type and mutant Brg1, and hBrm, were generated using inserts from pBS(KS+)/Fl-Brg1, pBS(KS+)/Fl-Brg1 (K798R), and pBS(KS+)/Fl-hBrm, respectively. To generate pBS(KS+)/Fl-Brg1, full-length Brg1 was excised out of pBJ5/Brg1 (Khavari et al. 1993) as an HpaI fragment and subcloned into EcoRV-linearized pBS(KS+). Two primers were then used to introduce a Flag tag before the stop codon followed by SalI and SpeI restriction sites. The 5′ primer (5′-GAC CAA GAC CCT GAT GAA CAC CAT CAT GCA GCT GCG GAA GAT CTG CAA CCA CCC CTA CAT GTT CCA GCAC-3′) spanned the BglII site at nucleotide position 3190, and the 3′ primer (5′-GAC TAG TCG CGT CGA CTT ATC ATT TGT CAT CGT CGT CCT TGT AGT CGT CTT CTT CGC TGC CAC TTC CTG AGC GGT CCT CCT CT-3′) contained the Flag-tag, SalI, and SpeI sequences. Flag sequences are indicated in boldface, and the restriction sites are underlined. The PCR fragment generated with these primers was then subcloned into pBS(KS+)/Brg1, which was digested with BglII–SpeI, thereby generating pBS(KS+)/Fl-Brg1. The full-length Flag-tagged Brg1 cDNA was then excised out of pBS(KS+)/Fl-Brg1 as a SalI fragment and subcloned into pBabe to generate pBabe/Fl-Brg1. To create Flag-tagged Brg1 containing a mutation in the ATP binding site, which substitutes a lysine at amino acid position 798 for an arginine, an NsiI–BglII restriction fragment was isolated from pBJ5/Brg1 (K798R) (Khavari et al. 1993) and subcloned into the respective sites of pBS(KS+)/Fl-Brg1. Plasmid pBabe/Fl-Brg1 (K798R) was generated by subcloning a SalI fragment from pBS(KS+)/Fl-Brg1 (K798R) into SalI-linearized pBabe. Plasmids pBS(KS+)/BAF155 and pBS(KS−)/BAF60 were described previously (Wang et al. 1996b; Phelan et al. 1999). Plasmid pBS(KS+)/Fl-hBrm was constructed as follows. First, two primers were designed so that the 5′ primer (5′-CGC AGC AAC AAC AGC AGC CGG CCC TTG TTA ACT ACA ACA GAC CAT CTG GCC CGG GGC CGG AGC TGA GCG G-3′) spanned the internal HpaI site at nucleotide position 1070, and the 3′ primer (5′-CGG GGT ACC CGG AAT TCT TAT CAT TTG TCA TCG TCG TCC TTG TAG TCC TCA TCA TCC GTC CCA CTT CCT TCT GAC TGT TCA CGT-3′) encoded the Flag tag followed by EcoRI and KpnI restriction sites. These two primers were used to generate a PCR fragment from pCG/hBrm (Muchardt and Yaniv 1993), which was digested with HpaI and KpnI and cloned back into pCG/hBrm. The resulting plasmid, pCG/Fl-hBrm, was digested subsequently with EcoRI, and the full-length Flag-tagged hBrm cDNA was cloned as an EcoRI fragment into pBabe. Plasmids pTPT and p2085S-G5E4 used to generate the DNA fragments to assemble mononucleosomal core particles and the 5S nucleosomal arrays, respectively, were described previously (Sif et al. 1998; Neely et al. 1999).
Establishment of cell lines
Cells were grown in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% fetal bovine serum (FBS). To create cell lines that express Flag-tagged wild-type and mutant Brg1, and hBrm, retroviral vectors for the expression of these proteins were transfected into the Bing packaging cell line essentially as described (Sif et al. 1998). Several individual colonies were isolated for each construct, expanded into cell lines, and screened for expression. Two clones for each construct were expanded into media containing puromycin and used to immunopurify Brg1 and hBrm complexes.
Purification of Flag-tagged hSWI/SNF complexes
Nuclear extracts were prepared from each cell line as described previously (Dignam et al. 1983). Affinity purification of epitope-tagged hSWI/SNF complexes was performed as described previously (Sif et al. 1998). To further purify hSWI/SNF complexes, approximately 4 μg of affinity-purified complexes was applied to a 2-mL 20%–40% glycerol gradient (glycerol, 50 mM Tris-HCl at pH 7.5, 1 mM EDTA) and centrifuged in a TLS55 rotor at 30,000 rpm for 24 h at 4°C. Fractions (∼120 μL) were collected and analyzed as described in the figure legends.
Mononucleosome disruption and ATPase assays
Mononucleosome disruption assays were performed as described previously (Imbalzano et al. 1996; Sif et al. 1998), except that the core particles were assembled by octamer transfer instead of by salt dilution (Rhodes and Laskey 1989). A 155-bp EcoRI–MluI DNA fragment derived from the pTPT plasmid was 32P-labeled at the EcoRI site by Klenow treatment and assembled into core particles in the presence of 80-fold molar excess of H1-depleted HeLa nucleosomes. Assembled cores were applied to a 5%–30% glycerol gradient and purified as described (Sif et al. 1998). Approximately 3.3 ng (0.44 nM) of nucleosomal core particles, which includes 0.3 ng (0.04 nM) labeled and 3 ng (0.4 nM) unlabeled DNA, were incubated with or without 2 mM ATP and affinity-purified Brg1 and hBrm complexes in a 25-μL reaction containing 12 mM HEPES (pH 7.9), 60 mM KCl, 6 mM MgCl2, 60 μM EDTA, 2 mM DTT, 13% glycerol for 30 min at 30°C. DNase I treatment and analysis were as described (Sif et al. 1998). To measure the ATPase activity of Brg1 and hBrm complexes, approximately 72 ng (3.6 nM) of immunopurified complexes was incubated with or without 20 nM plasmid DNA and 100 μM ATP in a 10-μL reaction as described (Phelan et al. 1999).
Chromatin remodeling assays
Nucleosome-assembled plasmid templates used in the supercoiling assay were prepared as described previously (Sif et al. 1998). Nucleosome reconstitution of Lytechinus variegatus 5S ribosomal DNA repeat sequences was performed using plasmid p2085S-G5E4 (Neely et al. 1999). The 5S array was gel purified following digestion of p2085S-G5E4 with Asp718, ClaI, and DdeI, 32P-labeled at the Asp 718 site by Klenow treatment, and used to assemble polynucleosomal templates as reported previously (Richmond et al. 1988; Carruthers et al. 1999). Briefly, a total of 10 μg of gel-purified 5S-G5E4 fragment was incubated with 13.1 μg of H1-depleted HeLa core histones in a 100-μL final reaction volume containing high salt buffer (2 M NaCl, 20 mM Tris-HCl at pH 7.8, 1 mM EDTA, 10 mM DTT, 0.5 mM benzamidine hydrochloride) supplemented with 100 μg/mL BSA. The reaction was transferred to a dialysis tubing and placed into 200 mL of high salt buffer (2 M NaCl, 20 mM Tris-HCl at pH 7.8, 1 mM EDTA, 10 mM DTT, 0.5 mM benzamidine hydrochloride). Assembly of the 5S array was performed by continuous dilution of the dialysis buffer using a low salt buffer (0.25 M NaCl, 20 mM Tris-HCl at pH 7.8, 1 mM EDTA, 10 mM DTT, 0.5 mM benzamidine hydrochloride) and keeping the volume of the dialysis reaction constant. Dialysis was allowed to proceed for 48 h until the salt concentration of the dialysis buffer was lower than 500 mM NaCl. The assembled 5S linear array was stabilized by a final dialysis in 200 mL of a buffer containing 10 mM Tris-HCl (pH 7.8) and 0.25 mM EDTA for 12–16 h at 4°C (Hansen et al. 1991). The amount of HeLa core histones added to the initial reaction was determined empirically as one that resulted in more than 50% saturated arrays as determined by EcoRI digestion analysis (Carruthers et al. 1999), concomitant increases in agarose gel electrophoretic mobility (Steger and Workman 1999), and micrococcal nuclease (MNase) digestion.
To analyze the extent of assembly of the 5S array, we treated 2 ng of unlabeled and assembled template with increasing amounts of MNase for 2 min at room temperature. The reactions were stopped with stop buffer (50 mM Tris-HCl at pH 8.0, 0.1 mM EDTA, 25% (v/v) glycerol, 3% (w/v) SDS, 0.04% (w/v) bromophenol blue, 0.04% (w/v) xylene cyanol), which was supplemented with 2 mg/mL proteinase K, and incubated for 45–60 min at 37°C. Samples were loaded on a 1% agarose gel and electrophoresed for 12–15 h at 60 V. The denatured DNA was transferred to Hybond-XL membrane (Amersham Pharmacia Biotech) overnight in 20× SSC (3M NaCl, 0.3M sodium citrate at pH 7.0). Following UV irradiation to cross-link the DNA, the membrane was prehybridized and hybridized using ExpressHyb hybridization solution (Clontech) according to the manufacturer's protocol. A 5S-specific oligonucleotide (5′-GGT ATG GTC GTA GGC TCT TGC TTG-3′) was 32P-labeled by kinase treatment and used at 2 × 106 cpm/mL of hybridization solution to detect the 5S DNA.
To assay for nucleosome remodeling,∼6 ng of either plasmid chromatin (0.12 nM) or 5S nucleosomal arrays (0.14 nM) was incubated with or without ATP and Brg1 and hBrm complexes in a 25-μL reaction as described for the mononucleosome disruption assay. Reactions that contained plasmid chromatin were supplemented with 0.1 unit of wheat germ topoisomerase I (Promega) and incubated for 60–90 min at 30°C. When the 5S nucleosomal array was added, samples were incubated with remodeling complexes for either 10 or 30 min at 30°C before adding apyrase (2.5 units per reaction). Next, HhaI was added (15 units per reaction), and the reactions were allowed to incubate for an additional 60 min at 30°C. All reactions were stopped with stop buffer containing 2 mg/mL proteinase K and were incubated for 45–60 min at 37°C. The samples that contained plasmid DNA were analyzed on a 2% agarose gel in 1× phosphate buffer (40 mM Tris-base, 30 mM NaPO4, 1 mM EDTA) for 42 h at 40 V, whereas samples that contained 5S arrays were analyzed on 1% agarose gel in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA) for 4–5 h at 140 V.
Antibodies and Western blot analysis
Western blot analysis and anti-Brg1, anti-hBrm, and anti-hSWI3 rabbit polyclonal antisera have been described previously (Sif et al. 1998). Anti-BAF60 rabbit polyclonal antibodies were raised against a GST-BAF60 (amino acids 10–84). Anti-mSin3A and anti-Flag M2 antibodies were purchased from Santa Cruz, anti-HDAC1 and 3 were purchased from Upstate Biotechnologies, and anti-HDAC2 and anti-RbAp48 antibodies were purchased from Zymed.
Immunoprecipitation of in vitro translated proteins
Proteins were synthesized by incubating 1–2 μg of plasmid DNA with 25 μL of the TNT-coupled reticulocyte lysate and 22 μCi of [35S]methionine and cysteine (NEN) according to the manufacturers recommendations (Promega). In vitro translated proteins were immunoprecipitated by incubating 40–50 × 103 cpm with antibodies in a 250-μL reaction containing 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 0.1% NP-40, 1% Aprotinin. After a 60 min incubation on ice, 75 μL of a 50% slurry of protein A sepharose beads (Pharmacia) was added, and the reactions were incubated overnight at 4°C with gentle mixing. When anti-Flag M2 affinity gel was used, protein A sepharose beads were not added. After the first immunoprecipitation, beads were washed three times with 0.5 mL of washing buffer (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 0.1% NP-40, 1% Aprotinin). The retained proteins were then released by heat denaturation for 5 min in the presence of 0.5% SDS, and the supernatant was used to perform a second immunoprecipitation using specific antibodies as described above. Beads were washed four times with washing buffer and analyzed by SDS-PAGE.
Acknowledgments
We thank R.N. Eisenman, J. Workman, and K. Neely for providing plasmids; W. Wang, A. Kuzmicher, and D. Reinberg for providing antibodies; H. Su for help generating cell lines; and J. Vandeusen and M. Caligiuri for help with the PhosphorImager. We thank C. Varughese and M. Hirschel from Cellex Biosciences, Inc., for growing cell lines. We are grateful to K. Lee, T. Gilmore, S. Ackerman, and C. Peterson for critical reading of the manuscript. A.J.S. is a Human Frontier Science Program fellow. This work is supported by NIH grant GM48405 to R.E.K and by the James Cancer Hospital and Solove Research Institute Research Fund and NCI grant 1 K01 CA89854–01 to S.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Sif.1@osu.edu; FAX (614) 292-4118.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.872801.
References
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Carruthers LM, Tse C, Walker KP, Hansen JC. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 1999;304:19–35. doi: 10.1016/s0076-6879(99)04004-5. [DOI] [PubMed] [Google Scholar]
- Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PD, Schmid A, Zavari M, Münsterkötter M, Hörz W. Chromatin remodelling at the PHO8 promoter requires SWI–SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Van Holde KE, Lohr D. The mechanism of nucleosome assembly onto oligomers of the sea urchin 5S DNA positioning sequence. J Biol Chem. 1991;266:4276–4282. [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Schnitzler GR, Kingston RE. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- Jaskelioff M, Gavin IM, Peterson CL, Logie C. SWI/SNF mediated nucleosome remodeling: Role of histone octamer mobility in the persistence of the remodeled state. Mol Cell Biol. 2000;20:3058–3068. doi: 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Kuo M-H, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes & Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang W-M, Sun J-M, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Längst G, Bonte EJ, Corona DFV, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg R D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Reyes J-C, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Bourachot B, Reyes J-C, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2α ATPase from the mammalian SWI-SNF complex. EMBO J. 1998;17:223–231. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Hardy S, Engel DA. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao H-Y, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig H-P, et al. An Ikaros-containing chromatin remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. DNA binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- Reyes J-C, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Laskey RA. Assembly of nucleosomes and chromatin in vitro. Methods Enzymol. 1989;170:575–585. doi: 10.1016/0076-6879(89)70065-3. [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Searles MA, Simpson RT. Crystals of a nucleosome core particle containing defined sequence DNA. J Mol Biol. 1988;199:161–170. doi: 10.1016/0022-2836(88)90386-5. [DOI] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes & Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Workman JL. Transcriptional analysis of purified histone acetyltransferase complexes. Methods: A companion to methods in enzymology. 1999;19:410–416. doi: 10.1006/meth.1999.0877. [DOI] [PubMed] [Google Scholar]
- Strober BE, Dunaief JL, Guha S, Goff SP. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wang W, Côté C, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996a;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes & Dev. 1996b;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang W, Chi T, Xue Y, Zhou S, Kuo A. Architectural DNA binding by a high mobility group/kinesin like subunit in mammalian SWI/SNF related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Jones PL, Wade PA. DNA demethylation. Proc Natl Acad Sci USA. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998a;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Cell. 1998b;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]