Abstract
Adherence of bacteria to epithelial cells is an important step in colonization and immune modulation in the large bowel. The aims of this study were to use a three-stage continuous-culture system (CCS) to investigate how environmental factors affect bacterial attachment to Caco-2 cells and modulation of cytokine expression by gut microorganisms, including a probiotic Bifidobacterium longum strain, DD2004. The CCS simulated environmental conditions in the proximal large intestine (vessel 1 [V1]) and distal colon (V2 and V3) at two different system retention times (R) within the range of normal colonic transits (20 and 60 h). The model was inoculated with human fecal material, and fluorescence in situ hybridization (FISH) was used to characterize microbial populations and to assess bacterial attachment to Caco-2 cells. Real-time quantitative PCR (qPCR) was employed to measure cytokine gene expression following challenge with bacteria from different components of the CCS in the presence and absence of B. longum. At an R of 60 h, bacterial adherence increased from V1 to V3, but this trend was reversed at an R of 20 h. Atopobia were the predominant adherent organisms detected at both system retention times in each culture vessel. Modulation of transforming growth factor β1 (TGF-β1), interleukin 6 (IL-6), and IL-18 gene expression by CCS bacteria was marked at an R of 60 h, while at an R of 20 h, IL-4, IL-10, TGF-β2, IL-1α, and tumor necrosis factor alpha (TNF-α) were significantly affected. The addition of B. longum affected cytokine expression significantly at both retention times. This study demonstrates that environmental determinants regulate the adherence properties of intestinal bacteria and their abilities to regulate cytokine synthesis.
INTRODUCTION
The human gastrointestinal tract is a complex bacterial ecosystem containing several hundred different species and strains that constitute the normal microbiota (11, 39). This commensal microbiota is predominantly anaerobic and plays a fundamental role in the health and well-being of the host (10, 35, 39, 41).
Marked interindividual variations in fecal bacterial community profiles have been shown, as well as in different parts of the gut; however, the dominant bacterial communities are generally host specific and remain stable over time (12, 45), with the Bacteroides-Prevotella group, eubacteria, and clostridia usually predominating (19, 23). Because many intestinal anaerobes are not readily amenable to culture, a variety of molecular techniques, largely based on analyses of the 16S RNA gene, have been developed (1, 2, 44, 45). However, ethical considerations have hindered study of the structure of the normal gut microbiota and its metabolic activities in different parts of the large bowel, and most investigations have been done on fecal material. To overcome this, an in vitro three-stage continuous-culture system (CCS) model was developed to investigate the behavior of the microbiota under environmental conditions simulating those in the proximal and distal gut (7, 24). This system provides a controlled operator-defined environment that can be maintained in a steady state and that simulates the complexity and diversity of the microbiota (7, 29, 37).
Probiotics, such as bifidobacteria and lactobacilli, are normal inhabitants of the intestinal tract (36, 38). Evidence suggests that some of these organisms can modulate the composition of the gut microbiota and affect gut immune regulation (15, 32, 43). Interactions between intestinal bacteria and the innate immune system are often manifested through differential expression of chemical mediators, such as cytokines (27, 28, 33). Direct contact with and adhesion to host cells or tissues may be essential for some organisms to exert their effects on the host. It is known that in human epithelial cell lines cytokine production can be modulated by pathogenic bacteria; however, the interactions of these bacteria with members of the normal commensal microbiota in relation to changes in cytokine profiles have been less studied.
The aim of this investigation was to use a three-stage gut model to investigate the adherence properties of bacteria grown under different environmental conditions in different culture vessels to Caco-2 cells at varying retention (transit) times (R). The abilities of CCS-derived bacteria to modulate cytokine production by colonic epithelial cells, with or without the addition of a known probiotic Bifidobacterium longum strain (13), were also assessed to investigate their immunomodulatory potentials.
MATERIALS AND METHODS
Three-stage continuous-culture system.
The CCS model was operated as described previously (24). The system comprised three continuous-culture vessels (V1, V2, and V3) and was operated at a total system retention time of either 20 or 60 h, which are in the range of normal human colonic transits (9). A headspace of O2-free N2 gas was maintained in each vessel, and the culture pH was maintained at 5.5 (V1), 6.2 (V2), and 6.8 (V3) to simulate environments in the proximal, transverse, and distal colons, respectively. The sterile culture medium was treated continuously with nitrogen and was introduced via a peristaltic pump to V1, which sequentially supplied V2 and V3 through a series of weirs. Several polymerized carbon sources were included in the growth medium to maximize species diversity in the fermentation system. The culture medium consisted of the following (g liter−1) in distilled water: Lintner's starch (BDH Ltd., Lutterworth, United Kingdom), 2.0; pectin (citrus), 0.5; Raftiline LS (Orafti, Tienen, Belgium), 0.5; xylan (oat spelt), 0.5; arabinogalactan (larch wood), 0.5; guar gum, 0.5; mucin (porcine gastric type III), 0.5; tryptone, 3.0; peptone water, 3.0; yeast extract, 4.5; bile salts no. 3, 0.4; cysteine, 0.8; hemin, 0.05; Tween 80, 1.0; NaCl, 4.5; KCl, 2.5; MgCl2 · 6H2O, 4.5; CaCl2 · 6H2O, 0.2; and KH2PO4, 0.4. Two milliliters of a trace element solution containing (g liter−1) MgSO4 · 7H2O, 3.0; MnCl2 · 4H2O, 0.32; FeSO4 · 7H2O, 0.1; CoSO4 · 7H2O, 0.18; CaCl2 · 2H2O, 0.1; ZnSO4 · 7H2O, 0.18; CuSO4 · 5H2O, 0.01; and NiCl2 · 6H2O, 0.092 was also added. The final medium was adjusted to pH 5.5 by adding 1 M HCl prior to autoclaving it. Fecal material from a healthy 30-year-old male volunteer who had not taken antibiotics for at least 12 months prior to commencement of the study was used to inoculate the CCS, using methods described previously (7, 24). Short-chain fatty acid (SCFA) analysis was used to confirm that the model was operating under steady-state growth conditions (7, 24).
Caco-2 cell assay preparation.
The Caco-2 cells (34) were obtained from the European Collection of Cell Cultures (ECACC) at the Public Health Laboratory Service (Porton Down, Wiltshire, United Kingdom). The cells were grown and maintained in high-glucose Dulbecco's modified Eagle medium (DMEM) (Sigma-Aldrich, Gillingham, United Kingdom) supplemented with 10% fetal calf serum (FCS) (Gibco-BRL, Life Technologies, Paisley, United Kingdom), 1% (vol/vol) HEPES (Gibco-BRL), 1% (vol/vol) penicillin-streptomycin (Gibco-BRL), and 2 mM l-glutamine (Gibco-BRL). All cell cultures were done in 5% CO2 in a humidified atmosphere at 37°C. Cells were seeded at high density in 24-multiwell tissue culture plates (Corning, Costar, High Wycombe, United Kingdom) and grown to 95 to 100% confluence. The cells were challenged with bacteria from the CCS, B. longum, or a combination of both. For adhesion assays, Caco-2 cells were grown to confluence on glass slides cut into square shapes (18 mm by 18 mm) placed in petri dishes. Before each experiment, the culture medium and dead cells were aspirated off, and the cells were washed three times with antibiotic-free culture medium.
Analysis of bacterial communities in the CCS.
Qualitative and quantitative fluorescence in situ hybridization (FISH) was used to demonstrate that the CCS supported a diverse range of bacteria. Planktonic samples from all three vessels were prepared and processed as described by Harmsen et al. (16), using a range of group- and species-specific 16S rRNA oligonucleotide probes to cover a large range of intestinal bacteria, as detailed by Child et al. (7). To improve cell attachment, the hybridization slides were immersed in a solution of 0.1% (wt/vol) gelatin and 0.01% (wt/vol) chromium potassium sulfate at 60°C and allowed to air dry for 3 h at room temperature. After hybridization, the samples were covered with 10 μl of Citifluor (Citifluor Ltd., London, United Kingdom) to prevent fading of fluorescence and were visualized using a Zeiss Axiophot epifluorescence microscope (Carl Zeiss, Welwyn Garden City, United Kingdom) connected to a Dell Optiplex GX110 PC, with C-Imaging System Simple PCI imaging software (Compix Inc., PA). Between 10 and 25 microscope fields were quantified, depending on the number of fluorescing bacteria (magnification, ×1,000). Results are given as percentages of the total DAPI (4′,6-diamidino-2-phenylindole) counts (7).
In vitro adhesion assays.
Samples were removed from each vessel of the CCS and centrifuged for 20 min at 3,000 × g. The supernatants were discarded, and the bacterial pellets were resuspended in antibiotic-free culture medium to 1 × 1010 bacteria ml−1 (to simulate the number of planktonic cells in the CCS system), as assessed by direct microscopic examination. One-milliliter aliquots of the CCS samples were then added to Caco-2 cells and incubated for 3 h in a 5% CO2 atmosphere at 37°C. The adhesion slides were subsequently washed twice with antibiotic-free culture medium, air dried, and fixed in fresh 4% (vol/vol) paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.0) for 1 h at room temperature, washed with PBS (10 min), and dried in a hybridization oven at 46°C for 15 min. FISH hybridization and bacterial attachment were quantified as for planktonic samples, and the number of adhering bacteria per Caco-2 cell was calculated.
Preparation of B. longum.
The probiotic B. longum strain DD2004 was initially isolated from the rectal mucosa of a healthy individual (26) and has been shown to be of therapeutic value, when combined as a symbiotic, in reducing symptoms in patients with active ulcerative colitis (13). The organism was maintained on Wilkins Chalgren (WC) agar under anaerobic conditions. For cell culture assays, 20 ml of an overnight culture of B. longum grown in anaerobic WC broth was transferred to a 50-ml centrifuge tube and centrifuged for 20 min at 3,000 × g. The supernatant was gently discarded, and the bacterial pellet was resuspended in 20 ml antibiotic-free tissue culture medium.
In vitro bacterial assays.
The resuspended B. longum cultures from each vessel of the CCS and combinations of equal numbers of B. longum and CCS bacteria were added to Caco-2 cells in cell culture medium for 3 h, with untreated cells as controls. One milliliter of a bacterial suspension containing 1 × 1010 bacteria ml−1 was applied to each well in duplicate.
Measurements of cytokine gene expression.
Expression profiles for nine cytokine genes by Caco-2 cells after stimulation by bacteria from the CCS and B. longum were determined relative to the hBD-1 housekeeping gene by extraction of RNA, cDNA synthesis, and quantification by real-time PCR, as described below.
RNA extraction and cDNA synthesis.
Total mRNA was extracted using liquid nitrogen snap-freezing and mechanical grinding (13). The RNA was purified using the RNeasy Mini Kit (Qiagen, Crawley, United Kingdom) according to the manufacturer's protocol, with an initial cleanup step, using a Qiashredder column (Qiagen) and an additional 15-min on-column DNase I digestion step (Qiagen), to ensure that there was no genomic-DNA contamination. RNA was eluted in 40 μl RNase-free water and stored at −83°C before being reverse transcribed using the Quick reverse transcription (RT) system (Promega Ltd., Southampton, United Kingdom), utilizing oligo(dT)15 primers. The reaction mixtures were incubated for 90 min at 42°C, followed by 5 min at 95°C to inactivate the reverse transcriptase enzyme. Negative controls included RT reactions without reverse transcriptase and/or template. The reaction mixtures were cooled on ice for 5 min before the DNA products were dispensed in 2-μl aliquots and stored at −83°C. The resulting cDNA was used as a template in real-time quantitative PCRs (qPCRs) to quantitate cytokine gene expression. Aliquots of the same cDNA sample were used with all primer sets.
Quantitative PCR.
Quantitative real-time PCR (SYBR green) analysis was done using an iCycler Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) with iCycler Optical System Interface software (version 3.0) as described previously (13). The PCR primers (interleukin 4 [IL-4], IL-10, transforming growth factor β1 [TGF-β1], TGF-β2, TGF-β3, IL-1α, IL-6, IL-18, and tumor necrosis factor alpha [TNF-α]), methods of standard development, and optimization used in these experiments were described in a previous report (3). The real-time PCR was done in duplicate in 96-well plates in a final volume of 20 μl. The reaction mixture comprised 10 μl iQ SYBR green Supermix (containing 3 mM MgCl2, 20 mM Tris-HCl [pH 8.4], 50 mM KCl, 200 mM each deoxynucleoside triphosphate, SYBR green I, 10 nM fluorescein, and 0.625 U iTaq DNA polymerase), 1 μl of each primer (0.5 μM), 6 μl PCR water, and 2 μl of the respective standardized cDNA as a template. The 96-well plate format included six 10-fold dilutions in duplicate of the plasmid DNA standards, starting at a concentration of 106 to 101 molecules μl−1, except for the housekeeping gene, which had 108 to 101 molecules μl−1. The standard amplification protocol consisted of an initial denaturation step (95°C; 3 min), followed by 38 cycles of denaturation (95°C; 30 s), annealing (30 s) at appropriate temperatures ranging from 55°C to 70°C, and one final denaturation cycle (95°C; 30 s). Samples were then renatured (50°C; 30 s), followed by consecutive data acquisition and melting-curve analysis consisting of 35 cycles in which the temperature was ramped up from 65°C to 96°C in 1°C increments for 10 s. All PCRs were routinely run with nontemplate control. Target gene copy numbers were subsequently extrapolated from appropriate standard curves and normalized against the housekeeping gene (13).
Statistical analysis.
Data analysis was done using GraphPad Prism, version 4 (GraphPad Software Inc., San Diego, CA). One-way analysis of variance (with Tukey's posttest) was used for comparison of different groups. P values of ≤0.05 were taken as statistically significant.
Chemicals.
Unless otherwise stated, all chemicals were obtained from Sigma (Poole, Dorset, United Kingdom) and biological culture media from Oxoid Ltd. (Basingstoke, Hamps, United Kingdom).
RESULTS
Bacterial communities in the colon model.
The CCS used in this study was operated at system retention times of 20 and 60 h. The predominant planktonic populations detected by FISH analysis in each of the culture vessels are shown in Tables 1 and 2. Bacterial populations changed markedly in different fermentors, while the community structure was strongly influenced by the system retention time. At both retention times, total eubacterial FISH numbers declined as a percentage of the DAPI (4′,6-diamidino-2-phenylindole) counts from V1 to V3. Organisms belonging to the Atopobium cluster and, to a lesser degree, segmented filamentous bacteria (SFB) were unaffected by this phenomenon. The Eubacterium rectale-Clostridium coccoides group always predominated in the fermentation system, irrespective of culture vessel or retention time. At an R of 60 h (Table 1), Bacteroides-Prevotella, Faecalibacterium prausnitzii, and bifidobacteria and lactic acid bacteria were major groups in V1 and V2, but atopobia and SFB became more significant in V3. At an R of 20 h, previously minor components of the microbiota at an R of 60 h, such as SFB, Eubacterium cylindroides, ruminococci, and Roseburia intestinalis, predominated, with the E. rectale-C. coccoides group, largely replacing bacteroides and bifidobacteria, particularly in V1 and V2 (Table 2).
Table 1.
Compositions of major bacterial populations in the gut model determined using FISH probes at a system retention time of 60 h
| Population | % of total DAPI counta |
||
|---|---|---|---|
| Vessel 1 | Vessel 2 | Vessel 3 | |
| Total eubacteria | 95.6 ± 1.0 | 87.8 ± 0.6 | 77.8 ± 1.5 |
| Bifidobacteria | 7.1 ± 0.5 | 2.0 ± 0.3 | 0.9 ± 0.1 |
| Bacteroides-Prevotella | 10.7 ± 0.3 | 5.8 ± 0.2 | 3.9 ± 0.2 |
| F. prausnitzii | 8.0 ± 0.3 | 6.4 ± 0.2 | 4.4 ± 0.2 |
| Ruminococcus spp. | 0.7 ± 0.3 | 0.6 ± 0.1 | NDb |
| E. cylindroides | 0.1 ± 0.0 | 0.2 ± 0.4 | —c |
| Lactic acid bacteria | 6.4 ± 0.4 | 5.1 ± 0.3 | 1.7 ± 0.1 |
| Atopobium cluster | 5.5 ± 0.5 | 4.5 ± 0.2 | 4.8 ± 0.2 |
| Enterobacteriaceae | 0.4 ± 0.1 | 0.2 ± 0.0 | 0.2 ± 0.0 |
| Segmented filamentous bacteria | 2.2 ± 0.6 | 4.6 ± 0.1 | 4.9 ± 0.2 |
| E. rectale-C. coccoides group | 16.5 ± 1.2 | 10.2 ± 0.5 | 6.5 ± 0.2 |
| R. intestinalis | 1.6 ± 0.1 | 0.8 ± 0.2 | 0.5 ± 0.1 |
The results are means of three separate measurements ± standard errors of the mean.
ND, not detected.
−, bacteria were detectable, but at levels of <0.01% of the total DAPI count.
Table 2.
Compositions of major bacterial populations in the gut model determined using FISH probes at a system retention time of 20 h
| Population | % of total DAPI counta |
||
|---|---|---|---|
| Vessel 1 | Vessel 2 | Vessel 3 | |
| Total eubacteria | 92.4 ± 0.7 | 83.9 ± 1.7 | 81.3 ± 2.1 |
| Bifidobacteria | 3.4 ± 0.3 | 2.7 ± 0.1 | 2.2 ± 0.3 |
| Bacteroides-Prevotella | 5.7 ± 0.2 | 5.1 ± 0.2 | 3.2 ± 0.2 |
| F. prausnitzii | 7.2 ± 0.3 | 8.2 ± 0.2 | 3.1 ± 1.6 |
| Ruminococcus spp. | 7.8 ± 0.3 | 2.6 ± 0.4 | 1.7 ± 0.3 |
| E. cylindroides | 10.0 ± 0.5 | 7.8 ± 0.1 | 6.1 ± 0.3 |
| Lactic acid bacteria | 5.9 ± 0.2 | 5.9 ± 0.2 | 2.9 ± 0.4 |
| Atopobium cluster | 3.2 ± 0.3 | 3.6 ± 0.4 | 3.8 ± 0.3 |
| Enterobacteria | 0.6 ± 0.1 | 0.3 ± 0.2 | 0.2 ± 0 |
| Segmented filamentous bacteria | 15.6 ± 0.5 | 14.4 ± 1.5 | 7.7 ± 0.6 |
| E. rectale-C. coccoides group | 27.2 ± 1.9 | 20.3 ± 0.6 | 16.4 ± 1.2 |
| R. intestinalis | 10.7 ± 0.9 | 8.2 ± 0.4 | 2.1 ± 0.2 |
The results are means of three separate measurements ± standard errors of the mean.
Adherence assays.
Adherence of CCS bacteria to Caco-2 cells is shown in Tables 3 and 4. In general terms, at an R of 60 h, bacterial attachment increased from V1 to V3, with the Atopobium group being the predominant adhering organisms. Bifidobacteria were also strongly adherent at an R of 60 h. However, their attachment decreased from V1 to V3. At an R of 20 h, the abilities of CCS bacteria to attach to Caco-2 cells were considerably greater (Table 4). Again, atopobia were the predominant attaching species, although R. intestinalis and the E. rectale-C. coccoides group from V1 were also strongly adherent.
Table 3.
Adhesion of bacteria taken from each fermentation vessel to Caco-2 cells at a system retention time of 60 h
| Population | No. of bacteria per Caco-2 cella |
||
|---|---|---|---|
| Vessel 1 | Vessel 2 | Vessel 3 | |
| Total eubacteria | 5.5 ± 0.4 | 5.7 ± 0.5 | 7.2 ± 0.9 |
| Bifidobacteria | 0.8 ± 0.1 | 0.7 ± 0.1 | 0.4 ± 0.1 |
| Bacteroides-Prevotella | 0.3 ± 0.0 | 0.4 ± 0.0 | 0.3 ± 0.1 |
| F. prausnitzii | NDb | ND | ND |
| Ruminococcus spp. | ND | ND | ND |
| E. cylindroides | ND | ND | ND |
| Lactic acid bacteria | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 |
| Atopobium cluster | 3.2 ± 0.2 | 4.8 ± 0.4 | 5.6 ± 0.7 |
| Enterobacteria | ND | ND | ND |
| Segmented filamentous bacteria | ND | ND | ND |
| E. rectale-C. coccoides group | 0.5 ± 0.1 | 0.5 ± 0.0 | 0.5 ± 0.0 |
| R. intestinalis | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.0 |
The results are means ± standard deviations. Bacteria were quantified in 10 to 25 microscope fields.
ND, not detected.
Table 4.
Adhesion of bacteria taken from each fermentation vessel to Caco-2 cells at a system retention time of 20 h
| Population | No. of bacteria per Caco-2 cella |
||
|---|---|---|---|
| Vessel 1 | Vessel 2 | Vessel 3 | |
| Total eubacteria | 36.0 ± 4.6 | 19.4 ± 1.9 | 17.2 ± 1.6 |
| Bifidobacteria | 3.0 ± 0.4 | 2.5 ± 0.3 | 1.0 ± 0.1 |
| Bacteroides-Prevotella | 1.5 ± 0.2 | NDb | ND |
| F. prausnitzii | ND | ND | ND |
| Ruminococcus spp. | ND | ND | ND |
| E. cylindroides | ND | ND | ND |
| Lactic acid bacteria | ND | ND | ND |
| Atopobium cluster | 34.2 ± 3.7 | 11.4 ± 1.5 | 14.8 ± 1.5 |
| Enterobacteria | ND | ND | ND |
| Segmented filamentous bacteria | ND | ND | ND |
| E. rectale-C. coccoides group | 6.0 ± 0.1 | 1.3 ± 0 | 1.5 ± 0.2 |
| R. intestinalis | 5.3 ± 1.0 | 1.0 ± 0.2 | 1.2 ± 0.1 |
The results are means ± standard deviations. Bacteria were quantified in 10 to 25 microscope fields.
ND, not detected.
Effects of CCS bacteria on anti-inflammatory cytokine gene expression.
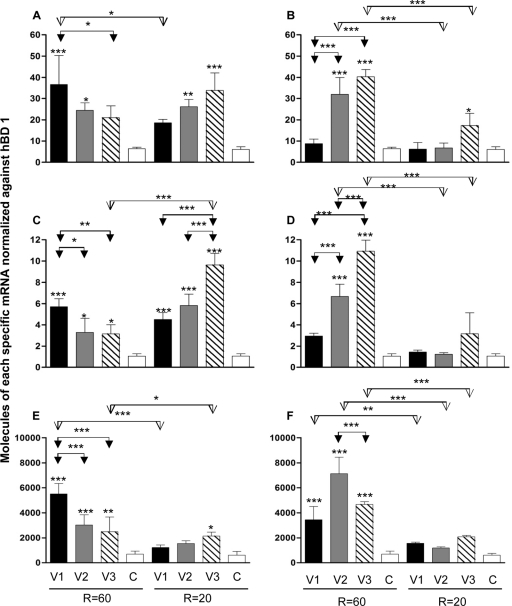
Figures 1 and 2 show the physiological effects of adding bacteria from different CCS vessels to Caco-2 cells, with or without the probiotic B. longum. Figure 1A shows IL-4 mRNA expression levels at the two retention times. The cytokine was significantly increased at an R of 60 h after the introduction of bacteria from V1 and V2 (P ≤ 0.001 and P ≤ 0.05, respectively). While at an R of 20 h the expression level with V2 and V3 CCS bacteria was significantly increased (P ≤ 0.01 and P ≤ 0.001, respectively), retention time was found to have a significant effect only with bacteria from V1 (P ≤ 0.05). The addition of B. longum (Fig. 2B) had a stimulatory effect on the expression of IL-4 at an R of 60 h when added to bacteria from V2 and V3 (P ≤ 0.001). At an R of 20 h, there was no significant increase, except with V3 (P ≤ 0.05). The retention time had a marked effect on expression of IL-4 by V2 and V3 CCS bacteria (P ≤ 0.001).
Fig. 1.
Cytokine profiles induced by bacteria taken from each vessel of the CCS (with or without B. longum) after addition to Caco-2 cells at retention times of 60 and 20 h. (A) IL-4 and CCS. (B) IL-4 and CCS with B. longum. (C) IL-10 and CCS. (D) IL-10 and CCS with B. longum. (E) TGF-β1 and CCS. (F) CCS with B. longum. The results are mean values from three separate experiments plus standard deviations and are normalized against hBD-1. The asterisks denote significant differences from the control (C). Significant differences between vessels and retention times are indicated by arrows connecting bars. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Fig. 2.
Cytokine profiles induced by bacteria taken from each vessel of the CCS (with or without B. longum) after addition to Caco-2 cells at retention times of 60 and 20 h. (A) TGF-β2 and CCS. (B) TGF-β2 and CCS with B. longum. (C) TGF-β3 and CCS. (D) TGF-β3 and CCS with B. longum. (E) IL-1α and CCS. (F) IL-1α and CCS with B. longum. The results are mean values from three separate experiments plus standard deviations. The values are normalized against hBD-1. The asterisks denote significant differences from the control (C). Significant differences between vessels and retention times are indicated by arrows connecting bars. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
IL-10 expression was greatly increased at both retention times with the CCS microbiota from all culture vessels: at an R of 60 h, V1, P ≤ 0.001, and V2 and V3, P ≤ 0.05; at an R of 20 h, V1 to V3, P ≤ 0.001 (Fig. 1C). The effect of the retention time on IL-10 expression was significant only with V3 bacteria (P ≤ 0.001). Addition of B. longum to V2 and V3 CCS bacteria had a significant stimulatory effect on the expression of IL-10 (P ≤ 0.001) at an R of 60 h (Fig. 1D). However, there was no significant increase with the probiotic at an R of 20 h (Fig. 1D). The retention time had a significant effect on expression of IL-10 with B. longum and bacteria from V2 and V3 (P ≤ 0.001).
TGF-β1 expression was greatly increased at an R of 60 h, when samples from V1 and V2 (P ≤ 0.001) and V3 (P ≤ 0.01) microbiotas were added to Caco-2 cells (Fig. 1E), while at an R of 20 h, the level was significantly increased only by V3 bacteria (P ≤ 0.05). The effect of the retention time on TGF-β1 expression was found to be significant only with organisms from V1 (P ≤ 0.001) and V3 (P ≤ 0.05). Addition of B. longum to CCS bacteria had a stimulatory effect on the expression of TGF-β1 at an R of 60 in V1 to V3 (P ≤ 0.001), while there was no significant increase at an R of 20 h (Fig. 1F). Retention time had a significant effect on the expression of TGF-β1, when V1 (P ≤ 0.01) and V2 and V3 (P ≤ 0.001) CCS bacteria were combined with B. longum.
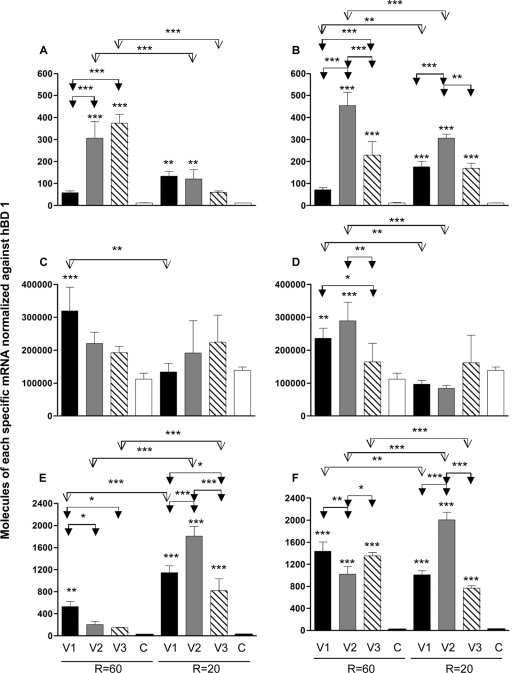
TGF-β2 expression was initially stimulated at an R of 60 h with bacteria from V1 (P ≤ 0.05) and subsequently reduced with bacteria from V2 and V3; however, only cytokine modulation withV3 CCS bacteria was significant (P ≤ 0.001) (Fig. 2A). TGF-β2 expression at an R of 20 h was initially downregulated with bacteria from V1 (P ≤ 0.001) and then increased withV2 CCS and subsequently reduced with V3 CCS (P ≤ 0.001). The effect of the retention time on TGF-β2 expression was found to be significant in cells from V1 (P ≤ 0.001). In Fig. 2B, it can be seen that the addition of B. longum at both retention times significantly reduced expression of TGF-β2 (P ≤ 0.001).
Figure 2C shows that no significant changes in TGF-β3 expression were observed at either retention time, except for V1 bacteria at an R of 20 h (P ≤ 0.05). The effect of the system retention time was significant with V3 bacteria (P ≤ 0.001). Addition of B. longum to V1 to V3 microbiotas at either an R of 20 h or an R of 60 h elicited no significant changes in TGF-β3 expression (Fig. 2D). IL-1α expression at an R of 60 h increased marginally with CCS bacteria from V1 to V3 (Fig. 2E). In contrast, at an R of 20 h, IL-1β increased significantly with bacteria from V1 to V3 (P ≤ 0.05, P ≤ 0.001, and P ≤ 0.001, respectively) (Fig. 2F). B. longum had significant effects on the expression of IL-1α at an R of 20 h when added to bacteria from V1 and V3 (P ≤ 0.001). The effect of retention time was significant with bacteria from V1 (P ≤ 0.001) and V3 (P ≤ 0.05).
Effects of CCS bacteria on proinflammatory cytokine gene expression in the presence and absence of B. longum.
IL-6 expression at an R of 60 h was significantly increased after the addition of bacteria from V2 and V3 to Caco-2 cells (P < 0.001) (Fig. 3A). At an R of 20 h, significant stimulation was observed with V1 and V2 bacteria (P ≤ 0.01). The addition of B. longum (Fig. 3B) to CCS bacteria at both CCS retention times significantly increased IL-6 expression, especially with V2 and V3 bacteria at an R of 60 h (P ≤ 0.001) and V1 to V3 bacteria at an R of 20 h (P ≤ 0.001). The retention time markedly affected gene expression with the probiotic and V1 and V2 bacteria (P ≤ 0.01 and P ≤ 0.001). As can be seen in Fig. 3C, IL-18 gene expression increased after the addition of bacterial samples from CCS vessels at both retention times but was significant only at an R of 60 h for V1 (P ≤ 0.001). The retention time markedly affected gene expression with bacteria from V1 (P ≤ 0.01). B. longum (Fig. 3D) at an R of 60 h significantly increased IL-18 with bacteria fromV1 and V2 (P ≤ 0.01 and P ≤ 0.001, respectively).
Fig. 3.
Cytokine profiles induced by bacteria taken from each vessel of the CCS (with or without B. longum) after addition to Caco-2 cells at retention times of 60 and 20 h. (A) IL-6 and CCS. (B) IL-6 and CCS with B. longum. (C) IL-18 and CCS. (D) IL-18 and CCS with B. longum. (E) TNF-α and CCS. (F) TNF-α and CCS with B. longum. The results are mean values from three separate experiments plus standard deviations and are normalized against hBD-1. The asterisks denote significant differences from the control (C). Significant differences between vessels and retention times are indicated by arrows connecting bars. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
TNF-α expression increased at both retention times when bacteria from V1 to V3 were added to Caco-2 cells, with significant increases at an R of 60 h for V1 (P ≤ 0.01) and at an R of 20 h for all vessels (P ≤ 0.001) (Fig. 3E). The introduction of B. longum resulted in significant increases in TNF-α expression levels at both retention times (P ≤ 0.001), with the retention time having a significant effect on gene expression: V1, P ≤ 0.01, and V2 and V3, P ≤ 0.001.
DISCUSSION
The health effects of commensal microorganisms in the colon are generally not mediated by the presence or absence of individual bacterial species, but rather by the global composition of the microbiota and its metabolic properties. Due to the fact that bacterial community structure and metabolism vary throughout the large intestine (1, 23, 24), an artificial gut simulator was used in this study to generate complex bacterial communities growing under environmental conditions similar to those found in different parts of the large bowel to assess their effects on cytokine expression by Caco-2 cell lines. Because colonic transit time is important in regulating the microbiota and its activities, the significance of changing system retention times was also investigated.
Bacterial adherence to intestinal epithelial cells and long colonic transit times have been linked to intestinal putrefaction, bowel cancer, ulcerative colitis (UC), and antibiotic-associated colitis, as well as constipation (1, 14, 20, 25, 26). Furthermore, adherence and colonization are prerequisite attributes for bacterial functionality, particularly with respect to their interactions with intestinal enterocytes. In this investigation, the adherence abilities of colonic microorganisms increased from V1 to V3 at an R of 60 h, but the reverse occurred at an R of 20 h. These results demonstrate that bacterial attachment to Caco-2 cells was strongly dependent on the environmental conditions under which the organisms were growing. Interestingly, the greater adherence abilities of bacteria during slow, nutrient-limited growth might indicate a possible correlation between slow gut transit and colonic disease.
FISH studies showed that overall bacterial numbers in the gut model were generally similar at both retention times. However, cell counts for individual groups of organisms as a percentage of the total detectable microbiota, such as Bifidobacterium spp., Bacteroides-Prevotella, and F. prausnitzii, were higher at an R of 60 h. Thus, it might be expected that these organisms would represent a higher proportion of the adherent microbiota, but results showed that this was not the case and that the principal adherent organisms belonged to the genus Atopobium, together with R. intestinalis. This could be due to a number of factors; for example, bacteria cultured in vitro often differ from freshly isolated organisms in relation to their adherence properties. Studies by Smith (40) showed that bacterial cell adhesion in vitro is influenced by the composition of the culture medium and the growth environment. Moreover, in a previous investigation in this laboratory, it was observed that a number of species underwent changes in cellular morphology under different culture conditions in the CCS (7); these included bacteria belonging to the segmented filamentous group, whose morphology was transformed from large irregular filaments, which formed the dominant component in V1 but were supplemented and then replaced by smaller, regular filaments and rod-shaped organisms in V2 and V3. In that study, a large number of adhering butyrate-producing organisms, including E. rectale and R. intestinalis (both Clostridium cluster XIVa) were able to attach to the Caco-2 cells. The fact that butyrate is the major fuel for the colonic epithelium, particularly in the distal large bowel, may reflect a closer symbiotic role for these bacteria with colonocytes than was previously thought (9, 18, 19).
The absence of attachment of lactic acid bacteria (LAB) at an R of 20 h may be explained by environmental regulation of cell surface protein structures involved in adhesion processes (8), differential expression of lipoteichoic acids (5), or carbohydrate structures (4). LAB are heterotrophic but lack many biosynthetic capabilities, and most species have multiple requirements for complex nutrients, such as amino acids and vitamins. These needs might not be met during rapid growth at an R of 20 h, with the result that the organisms could not be sustained in the ecosystem. It should also be noted that the LAB probe used in this study is a broad probe that targets lactobacilli, enterococci, and similar species.
The effects of environmental and physiological determinants on cytokine expression in human colonic epithelial cells has not previously been studied. Thus, in this investigation, colonic epithelial cells were exposed to bacteria from vessels simulating the proximal and distal colons. In a previous study, environmental conditions, such as pH, were shown to be able to affect NF-κB, which regulates the transcription of a large number of genes involved in inflammation and immune responses, including those coding for gamma interferon (IFN-γ), IL-1α, IL-2, IL-6, IL-8, TNF-α, and granulocyte/macrophage or granulocyte/colony-stimulating factor (22). Thus, if the environmental location was solely responsible for cytokine induction, as indicated in studies with samples from the CCS at an R of 60 h, the same trends would be expected at an R of 20 h, but at that retention time, the cytokine expression trend was reversed. In addition, it was seen that, although cytokine expression patterns could not be directly related to patterns of adherence, at an R of 60 h, bacterial attachment increased from V1 to V3, while at an R of 20 h, adherence was reduced.
Although there was an increase in expression of the proinflammatory cytokine TNF-α with B. longum combined with CCS bacteria on Caco 2 cells, previous work with the HT29 cell line indicated that there was no stimulation of TNF-α with the probiotic (3). Other investigations have also pointed to the induction of proinflammatory cytokines, such as IL-18, IFN-γ, and TNF-α, by putatively probiotic bacteria (6, 17, 30, 31), and the results seem to be dependent on the type of cell line used. In vivo measurements often give information different from that obtained using cell line models, for example, the synbiotic combination of B. longum and the prebiotic Synergy 1 was shown to significantly reduce levels of mucosal TNF-α in human feeding studies with ulcerative colitis and Crohn's disease patients (13, 42).
The inability to observe clear induction/modulation of pro- and anti-inflammatory cytokines by mixed communities of bacteria from the CCS and B. longum may be due to a number of factors. First, these organisms form part of the commensal gut microbiota, to which there is a high degree of immune tolerance, which contributes to immune homeostasis (21). Second, heterogeneous microbial populations from the CCS expressing a multitude of antigenic structures are likely to modify any interactions between the probiotic and Caco-2 cells.
In conclusion, it has been demonstrated that the in vitro model system is a useful tool for studying interactions that occur between commensal organisms and probiotic bacteria in relation to cytokine production in human epithelial cells. Cytokine profiles were determined by direct addition of B. longum and the chemostat microbiotas to Caco-2 cells, and if added to the gut model, the probiotic could also act by outcompeting other microorganisms in the ecosystem, thereby changing the microbiota community structure and its effects on the immune response. Although the amount of the probiotic added to the cell lines may be in excess of what could be delivered in vivo, synbiotic feeding studies with this organism show that it proliferates in the gut and can become a major component of mucosal microbiotas in Crohn's disease patients (42). A limitation of this study is that fecal material was obtained from one healthy individual, and other parameters, such as disease and differences in individual microbiotas, are certain to affect host immune responses. Further work is therefore needed to investigate how probiotics, such as B. longum, and commensal species interact to enable predictions to be made regarding their effects on gut microecology and metabolic processes within the microbiota.
Footnotes
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Ahmed S., et al. 2007. Mucosal-associated bacterial diversity associated with human terminal ileum and colonic biopsy samples. Appl. Environ. Microbiol. 73:7435–7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amann R. I., Ludwig W., Schleifer K. H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bahrami B., Macfarlane S., Macfarlane G. T. 2011. Induction of cytokine formation by human intestinal bacteria in gut epithelial cell lines. J. Appl. Microbiol. 110:353–363 [DOI] [PubMed] [Google Scholar]
- 4. Brooker B. E., Fuller R. 1975. Adhesion of lactobacilli to the chicken crop epithelium. J. Ultrastruct. Res. 52:21–31 [DOI] [PubMed] [Google Scholar]
- 5. Chan R. C. Y., Reid G., Irvin R. T., Bruce A. W., Costerton J. W. 1985. Competitive exclusion of uropathogens from human uroepithelial cells by Lactobacillus whole cells and cell wall fragments. Infect. Immun. 47:84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen T., Isomaki P., Rimpilainen M., Toivanen P. 1999. Human cytokine responses induced by gram-positive cell walls of normal intestinal microbiota. Clin. Exp. Immunol. 118:261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Child M. W., et al. 2006. Studies on the effect of system retention time on bacterial populations colonizing a three-stage continuous culture model of the human large gut using FISH techniques. FEMS Microbiol. Ecol. 55:299–310 [DOI] [PubMed] [Google Scholar]
- 8. Conway P. L., Kjelleberg S. 1989. Protein-mediated adhesion of Lactobacillus fermentum strain 737 to mouse stomach squamous epithelium. J. Gen. Microbiol. 135:1175–1186 [DOI] [PubMed] [Google Scholar]
- 9. Cummings J. H. 1981. Short chain fatty acids in the human colon. Gut 22:763–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Falk P. G., Hooper L. V., Midtvedt T., Gordon J. I. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finegold S. M., Attebery H. R., Sutter V. L. 1974. Effect of diet on human fecal flora: Comparison of Japanese and American diets. Am. J. Clin. Nutr. 27:1546–1569 [DOI] [PubMed] [Google Scholar]
- 12. Franks A. H., et al. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furrie E., et al. 2005. Synbiotic therapy (Bifidobacterium longum/Synergy 1TM) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 54:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibson G. R., Macfarlane G. T. 1994. Intestinal bacteria and disease, p. 53–62 In Gibson S. A.W. (ed.), Human health: the contribution of microorganisms. Springer-Verlag, London, United Kingdom [Google Scholar]
- 15. Haller D., et al. 2000. Non-pathogenic bacteria elicit a different cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut 47:79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harmsen H. J. M., et al. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hessle C., Hanson L. A., Wold A. E. 1999. Lactobacilli from human gastrointestinal mucosa are strong stimulators of IL-12 production. Clin. Exp. Immunol. 116:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hold G. L., Pryde S. E., Russell V. J., Furrie E., Flint H. J. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33–39 [DOI] [PubMed] [Google Scholar]
- 19. Hold G. L., Schwiertz A., Aminov R. I., Blaut M., Flint H. J. 2003. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69:4320–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hughes R., Rowland I. R. 2003. Nutritional and microbial modulation of carcinogenesis, p. 208–220 In Fuller R., Perdigon G. (ed.), Gut flora, nutrition, immunity and health. Blackwell Publishing, Oxford, United Kingdom [Google Scholar]
- 21. Kelly D., et al. 2003. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat. Immunol. 5:104–112 [DOI] [PubMed] [Google Scholar]
- 22. Kopp E. B., Ghosh S. 1995. NF-kappa B and Rel proteins in innate immunity. Adv. Immunol. 58:1–27 [DOI] [PubMed] [Google Scholar]
- 23. Macfarlane G. T., Macfarlane S. 1997. Human colonic microbiota: ecology, physiology and metabolic potential of intestinal bacteria. Scand. J. Gastroenterol. 222:S3–S9 [DOI] [PubMed] [Google Scholar]
- 24. Macfarlane G. T., Macfarlane S., Gibson G. R. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 35:180–187 [DOI] [PubMed] [Google Scholar]
- 25. Macfarlane G. T., McBain A. J. 1999. The human colonic microbiota, p. 1–25 In Gibson G. R., Roberfroid M. B. (ed.), Colonic microbiota, nutrition and health. Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 26. Macfarlane S., Furrie E., Cummings J. H., Macfarlane G. T. 2004. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 38:1690–1699 [DOI] [PubMed] [Google Scholar]
- 27. Mahida Y. R., Rolfe V. E. 2004. Host-bacterial interactions in inflammatory bowel disease. Clin. Sci. 107:331–341 [DOI] [PubMed] [Google Scholar]
- 28. Marin M. L., Lee J. H., Murtha J., Ustunol Z., Pestka J. J. 1997. Differential cytokine production in clonal macrophage and T-cell lines cultured with bifidobacteria. J. Dairy Sci. 80:2713–2720 [DOI] [PubMed] [Google Scholar]
- 29. Marsh P. D. 1995. The role of continuous culture in modeling the human microflora. J. Chem. Technol. Biotechnol. 64:1–9 [Google Scholar]
- 30. Miettinen M., Vuopio-Varkila J., Varkila K. 1996. Production of human tumor necrosis factor alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect. Immun. 64:5403–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miettinen M., et al. 1998. Lactobacilli and streptococci induce interleukin-12 (IL-12), IL-18, and gamma interferon production in human peripheral blood mononuclear cells. Infect. Immun. 66:6058–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perdigon G., Alvarez S., Rachid M., Aguero G., Gobbato N. 1995. Immune system stimulation by probiotics. J. Dairy Sci. 78:1597–1606 [DOI] [PubMed] [Google Scholar]
- 33. Pessi T., Sutas Y., Hurme M., Isolauri E. 2000. Interleukin-10 generation in atopic children following oral Lactobacillus rhamnosus GG. Clin. Exp. Med. 30:1804–1808 [DOI] [PubMed] [Google Scholar]
- 34. Pinto M., et al. 1983. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 47:323–330 [Google Scholar]
- 35. Pitcher M. C. L., Cummings J. H. 1996. Hydrogen sulphide: a bacterial toxin in ulcerative colitis? Gut 39:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reid G., et al. 2003. New scientific paradigms for probiotics and probiotics. J. Clin. Gastroenterol. 37:105–118 [DOI] [PubMed] [Google Scholar]
- 37. Rumney C. J., Rowland I. R. 1992. In vivo and in vitro models of the human colonic flora. Crit. Rev. Food Sci. Nutr. 31:299–331 [DOI] [PubMed] [Google Scholar]
- 38. Salminen S., et al. 1998. Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80:147–171 [DOI] [PubMed] [Google Scholar]
- 39. Savage D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107–133 [DOI] [PubMed] [Google Scholar]
- 40. Smith H. 1977. Microbial surfaces in relation to pathogenicity. Bacteriol. Rev. 41:475–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sonnenburg J. L., Angenent L. T., Gordon J. I. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5:569–573 [DOI] [PubMed] [Google Scholar]
- 42. Steed H., et al. 2010. Clinical trial: The microbiological and immunological effects of synbiotic consumption—a randomized double-blind placebo-controlled study in active Crohn's disease. Aliment. Pharmacol. Ther. 32:872–883 [DOI] [PubMed] [Google Scholar]
- 43. Tannock G. W. 1998. Studies of the intestinal microflora: a prerequisite for the development of probiotics. Int. Dairy J. 8:527–533 [Google Scholar]
- 44. Tannock G. W. 2002. Molecular methods for exploring the intestinal ecosystem. Br. J. Nutr. 87:S199–S201 [DOI] [PubMed] [Google Scholar]
- 45. Zoentendal E. G., Akkermans A. D. L., De Vos W. M. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]