Abstract
To identify genes targeted by the tobacco KNOX homeodomain protein, Nicotiana tabacum homeobox 15 (NTH15), we have generated an inducible system using the human glucocorticoid receptor. In this system, steroid treatment strictly induced NTH15 function and immediately suppressed the expression of a gibberellin (GA) biosynthetic gene encoding GA 20-oxidase (Ntc12) and also resulted in a decrease in bioactive GA levels. The repression of Ntc12 was observed even when indirect effects were blocked by cycloheximide. NTH15 mRNA was present in corpus cells of the shoot apical meristem (SAM), whereas Ntc12 mRNA was observed in leaf primordia and rib meristem but not in the corpus. Recombinant NTH15 protein strongly bound to a 5-bp dyadsymmetric sequence, GTGAC, in the first intron of Ntc12 in vitro. Mutation of this sequence in the Ntc12 gene abolished the NTH15-dependent suppression of Ntc12 in the corpus of the SAM. Our results indicate that NTH15 directly represses Ntc12 expression in the corpus of the wild-type SAM to maintain the indeterminate state of corpus cells. The suppression of NTH15 within cells at the flanks of the SAM permits GA biosynthesis, which promotes organized cell proliferation and consequently induces the determination of cell fate.
Keywords: DNA binding, gibberellin 20-oxidase, in situ hybridization, KNOX homeodomain protein, shoot apical meristem
The shoot apical meristem (SAM) successively produces leaves, internodes, and axillary buds, keeping itself as a collection of indeterminate stem-like cells throughout plant development (Steeves and Sussex 1989). KNOX homeodomain proteins encoded by knotted1-like homeobox (knox) genes are preferentially accumulated in the indeterminate cells around the SAM, but not in the determinate lateral organs, such as leaves (Jackson et al. 1994; Lincoln et al. 1994; Nishimura et al. 1999; Sentoku et al. 1999). Loss-of-function mutations in the Arabidopsis knox gene SHOOTMERISTEMLESS (STM) and the maize knotted1 (kn1) gene result in defects in shoot meristem development or maintenance (Long et al. 1996; Kerstetter et al. 1997). The opposite phenotype, namely, formation of adventitious meristems on leaves, has been observed in transgenic plants that constitutively and ectopically express knox genes (Matsuoka et al. 1993; Sinha et al. 1993; Chuck et al. 1996; Nishimura et al. 2000). On the basis of this evidence, KNOX homeodomain proteins are considered to play a role in the maintenance of the indeterminate properties of cells in the SAM (Reiser et al. 2000); however their direct function is still unresolved.
To understand the mechanism of KNOX homeodomain protein function, it is necessary to characterize genes that are targeted by these proteins. In this study, we attempted to identify a target gene of the tobacco KNOX homeodomain protein, Nicotiana tabacum homeobox 15 (NTH15). Previously, we reported that ectopic expression of NTH15 in transgenic tobacco plants under control of the cauliflower mosaic virus (CaMV) 35S promoter causes alterations in leaf morphology, including formation of adventitious meristems with a drastic decrease in the level of endogenous bioactive gibberellin (GA), GA1 (Tamaoki et al. 1997a). A similar result has also been reported in transgenic tobacco plants that express a rice knox gene, OSH1 (Kusaba et al. 1998b). The decrease in GA1 levels is probably caused by the suppression of a GA 20-oxidase gene, Ntc12, which encodes a key enzyme in GA biosynthesis and is mainly expressed in leaf primordia and young leaves (Kusaba et al. 1998a; Tanaka-Ueguchi et al. 1998). Based on these observations, we hypothesized that ectopic expression of NTH15 inhibits the expression of Ntc12 in leaf primordia, resulting in abnormal leaf morphologies in transgenic plants through the reduction in GA1 levels.
Because a number of physiological changes, such as abnormal leaves with adventitious meristems, dwarfism, and loss of apical dominance, occur in transgenic plants that constitutively express knox genes, it has not been clear whether a direct interaction between NTH15 and Ntc12 expression occurs or not. The levels of endogenous auxin and cytokinin also change in transgenic plants that constitutively express knox genes (Tamaoki et al. 1997a; Kusaba et al. 1998b; Frugis et al. 1999; Ori et al. 1999; Hewelt et al. 2000). In addition, many photosynthesis-related genes, which are induced by exogenous cytokinin treatment, are up-regulated in transgenic tobacco plants that express OSH1 (Tamaoki et al. 1997b). These results highlight the difficulty in identifying the primary effects of KNOX homeodomain protein function in transgenic plants that constitutively express knox genes.
Recently, an inducible system was developed for plant cells using the regulatory mechanism of the glucocorticoid receptor (GR; Schena et al. 1991; Lloyd et al. 1994; Aoyama et al. 1995; Aoyama and Chua 1997). The steroid binding domain of the GR inactivates the function of a neighboring domain in the chimeric protein molecule in the absence of steroid ligand, but the function is restored in the presence of ligand, dexamethasone (DEX) even in plants (Schena et al. 1991). This system may provide us with an important advantage in understanding the function of KNOX homeodomain proteins.
In this study, we expressed NTH15–GR fusion protein in transgenic tobacco plants. These transformants showed DEX-dependent induction of NTH15 function. Using this inducible system, we found that NTH15 directly suppresses the Ntc12 expression. Further analyses revealed that the NTH15 binding sequence in the first intron of Ntc12 acts as cis-motif in the SAM. Based on these results, we propose that NTH15 maintains the indeterminate properties of cells in the corpus of the SAM through the direct suppression of Ntc12 expression and GA biosynthesis.
Results
NTH15 function is inducible by artificial steroid treatment
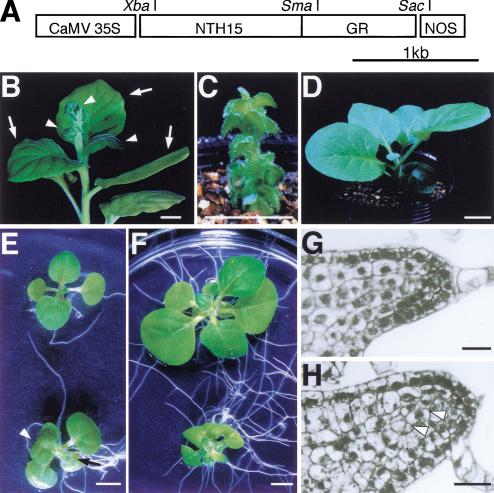
To artificially regulate NTH15 activity in vivo, the translational stop codon of NTH15 was removed and replaced with a 278-amino-acid segment of the human GR that contains the ligand-binding domain. The chimeric protein was expressed under control of the CaMV 35S promoter (35S::NTH15–GR; Fig. 1A) and was introduced into tobacco plants by Agrobacterium-mediated transformation.
Figure 1.
Diagram of NTH15–GR fusion construct and represented phenotype. (A) The chimeric gene consists of an in-frame fusion of the entire NTH15 cDNA and the steroid-binding domain of the human glucocorticoid receptor (GR). This gene was driven by the cauliflower mosaic virus 35S promoter. (B) Transformants carrying the fusion construct produced normal leaves in the absence of dexamethasone (DEX; arrows) and developed severely malformed leaves after a drop of DEX was applied on the shoot apical meristem (arrowheads). (C,D) Transgenic seedlings grown with (C) or without (D) 10 μM DEX. (E,F) Morphological changes in wild-type (upper) and 35S::NTH15–GR transgenic (lower) seedlings at 7 d (E) and 14 d (F) after DEX treatment. Arrowhead and arrow indicate the third and fourth true leaves, respectively. (G,H) Blade initiation in a leaf primordium of wild-type (G) and 35S::NTH15–GR transgenic (H) seedlings at 3 d after DEX treatment. Open arrowheads indicate the site of abnormal mesophyll cell division. Bars, 1 cm (B–D); 5 mm (E,F); 20 μm (G,H).
All primary transformants (11 independent lines) developed normal leaves without the steroid (Fig. 1B, arrows), but they produced malformed leaves after treatment with 10 μM DEX (Fig. 1B, arrowheads). Seeds from self-pollinated transformants were sown with or without 10 μM DEX and grown for 1 mo. Compared to wild-type seedlings, DEX-treated seedlings were severely dwarfed, and their leaves were very small and thick (Fig. 1C), as found in the severe phenotype of 35S::NTH15 transformants (Tamaoki et al. 1997a). This phenotype was not observed in either 35S::NTH15–GR seedlings grown in the absence of DEX (Fig. 1D) or in wild-type seedlings treated with the same concentration of DEX (Fig. 1E,F, upper plants), which confirms that NTH15 function was successfully induced in a ligand-dependent manner by DEX treatment.
Abnormal cell divisions result in the aberrant leaf development
To clarify the kinetics of NTH15 action, we followed the process of leaf development after induction of NTH15 function. 35S::NTH15–GR seedlings were grown without DEX for 14 d and then treated with 10 μM DEX. At this time, the third true leaf had just reached a visible size. The first indication of abnormal morphology was apparent on the third true leaf; namely, there was a small reduction in the length of midvein and leaf blade was slightly wrinkled (Fig. 1E, arrowhead). The fourth true leaf reached a visible size at 7 d after DEX treatment and was poorly developed, similar to the leaves that had been constitutively treated with 10 μM DEX (Fig. 1E, arrow). After emergence of the fourth true leaf, gross morphology of 35S::NTH15–GR seedlings was clearly distinguishable from that of wild-type seedlings (Fig. 1F, cf. lower plant to upper plant).
Because the abnormality in phenotype was first recognized as malformed leaves that emerged from the SAM, we investigated the anatomy of leaf primordia of DEX-treated 35S::NTH15–GR seedlings. In wild-type tobacco, leaf blade consists of six to seven cell layers that are arranged from the adaxial to abaxial side as: The upper epidermis, the palisade mesophyll, the middle spongy mesophyll, the lower spongy mesophyll, and the lower epidermis (Fig. 1G; McHale 1993). The cell layers are maintained during lateral expansion of leaf blade by a strict pattern of anticlinal divisions. We observed the morphology in leaf primordia every 24 h and found that abnormal cell divisions first became apparent at 3 d after DEX treatment. Although the anticlinal cell divisions were maintained in wild-type leaves (Fig. 1G), abnormal periclinal divisions in the mesophyll cell layers occurred sporadically in leaf blades of 35S::NTH15–GR seedlings (Fig. 1H). A similar phenomenon has also been observed in PR1a::OSH1 transgenic tobacco plants, which showed wrinkled and/or lobed leaves (Sato et al. 1996). These abnormal divisions cause the intercalation of cells into the cell layers, which disrupts the lineage of these layers and may result in the formation of abnormally shaped, thicker leaves.
Activation of NTH15 immediately suppresses Ntc12 expression
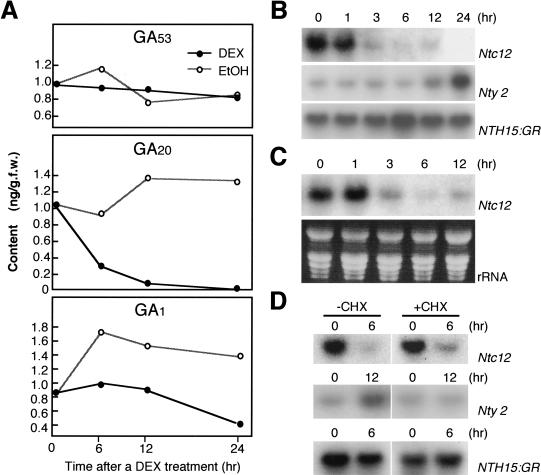
To follow the changes in endogenous GA biosynthesis, the levels of three GAs were analyzed. The bioactive GA1 is synthesized from GA53 through GA44, GA19, and GA20. GA 20-oxidase catalyzes the whole series of oxidation reactions at C-20, the steps involved in conversion from GA53 to GA20, and GA 3β-hydroxylase catalyzes the 3β-hydroxylation of GA20 to form GA1 (Hedden and Kamiya 1997). We found that the level of GA20 was drastically decreased within 6 h of DEX treatment (Fig. 2A). GA1 levels decreased gradually from 6 h to 24 h, whereas the level of GA53 was not significantly changed in either the presence or absence of DEX. These results show that activation of NTH15 interferes within 6 h with a particular step in GA biosynthesis catalyzed by GA 20-oxidase, causing a gradual decrease in bioactive GA1 levels.
Figure 2.
Analysis of the endogenous levels of gibberellins (GAs) and Ntc12 expression after dexamethasone (DEX) treatment. (A) Changes in the level of endogenous GAs. 35S::NTH15–GR seedlings were harvested at 0 h, 6 h, 12 h, and 24 h after treatment with (closed circle) or without (open circle) DEX. (B–D) RNA gel blot analysis of Ntc12 and Nty2. Total RNAs (20 μg) at 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h after DEX treatment without (B) or with (C) α-amanitin were probed with labeled Ntc12 or Nty2. α-Amanitin and DEX were applied at the same time. For blocking of protein synthesis, CHX was treated 1 h before DEX treatment (D). Numbers indicate times after DEX treatment. Expression of NTH15–GR (B,D) or rRNA (C) is shown as a loading control.
To investigate whether Ntc12 mRNA levels were also reduced ≤6 h after DEX treatment, RNA gel blot analysis was performed. The level of Ntc12 mRNA was drastically reduced from 1 h to 3 h after the treatment (Fig. 2B). In contrast, mRNA levels of Nty2, which encodes a GA 3β-hydroxylase gene (Itoh et al. 1999), remained unchanged until 6 h, after which time the mRNA levels increased, probably because of a feedback up-regulation triggered by the decrease in GA1 levels (Fig. 2B). These results strongly suggest that the immediate suppression of Ntc12 within 3 h of DEX treatment caused the change in the level of bioactive GA1 via a rapid decrease in GA20 levels within 6 h.
The kinetics of the decrease in Ntc12 mRNA levels was the same even under treatment with α-amanitin, an inhibitor of RNA polymerase II activity (Fig. 2C). This indicates that reduction in the level of Ntc12 mRNA after DEX treatment reflects the degradation kinetics of Ntc12 mRNA in vivo. To examine whether the NTH15-dependent suppression of Ntc12 expression requires prior synthesis of other proteins, 35S::NTH15–GR seedlings were treated with a protein synthesis inhibitor, cycloheximide (CHX), and changes in Ntc12 mRNA levels were monitored by RNA gel blot analysis. CHX treatment had no effect on the decrease in Ntc12 expression (Fig. 2D), but it completely abolished the increase in Nty2 mRNA levels that occurred in response to DEX treatment (Fig. 2D). The effect of CHX on the incorporation of [35S]methionine confirmed that CHX treatment successfully inhibited protein synthesis (<5%). Taken together, these data indicate that NTH15 directly suppresses the Ntc12 expression without prior synthesis of any other proteins.
NTH15 binds to the 5′ flanking and intron sequences of the Ntc12 gene
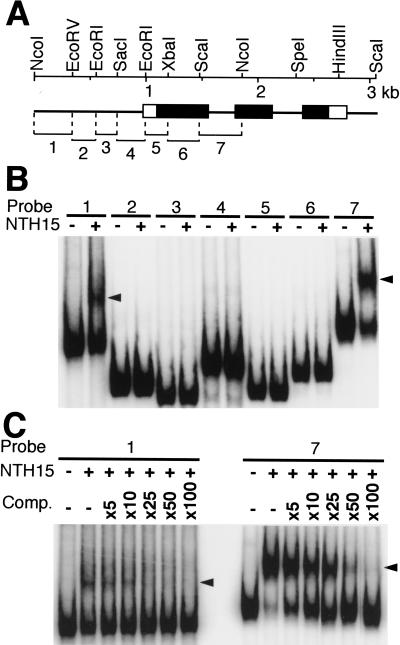
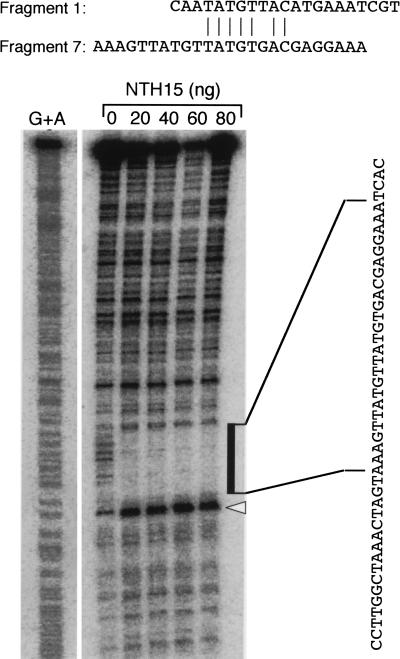
The Ntc12 gene consists of three exons and two introns (Fig. 3A). A 1.9-kb segment of Ntc12, which includes 1.0 kb of the 5′ flanking region, the first exon, and the entire first intron, is sufficient to achieve correct GUS reporter expression in transgenic tobacco plants (M. Ueguchi-Tanaka, T. Sakamoto, and M. Matsuoka, unpubl.). To investigate the possibility of a direct interaction between NTH15 and the Ntc12 sequence, this 1.9-kb region was cleaved into seven ∼350-bp fragments (Fig. 3A) and analyzed with electrophoresis mobility shift assay (EMSA; Ausubel et al. 1995). Recombinant NTH15 protein expressed in Escherichia coli cells bound to fragments 1 and 7, resulting in bands with lower mobility than free fragments (Fig. 3B). Bacterial lysate prepared from E. coli cells that carry only the vector did not generate complex (data not shown), which indicates that the retardation was specifically caused by an interaction with NTH15. The retarded complex was specifically competed by increasing concentrations of nonlabeled fragment 7 (Fig. 3C). The nonlabeled fragment 7 competed more strongly with the fragment-1 complex than with the fragment-7 complex. This shows that fragment 7, which encompasses the entire sequence of the first intron, has a higher binding affinity with NTH15 than does fragment 1, which is located furthest upstream of the 5′ flanking region that we used. The precise NTH15-binding sites in these fragments were identified by DNase I protection assay, which revealed that NTH15 interacted with a sequence TATGTTAC in fragment 1 and with TATGTGAC in fragment 7 (Fig. 4).
Figure 3.
Binding of NTH15 to the 5′-flanking and first intron sequences in the Ntc12 gene. (A) Genomic structure of Ntc12. Lines, open, and closed boxes indicate introns, noncoding, and coding sequences, respectively. Shown at the top is a restriction map of Ntc12. Fragments used as probes for electrophoresis mobility shift assay (EMSA) are numbered from 1 to 7. (B) EMSA of the seven probes with recombinant NTH15 protein. DNA fragments were incubated without (−) or with (+) NTH15. Fragments 1 and 7 were shifted by incubation with NTH15 as shown by arrowheads. (C) Fragment 7 binds to NTH15 much more strongly than fragment 1. Binding affinity was estimated using nonlabeled fragment 7 as a competitor, which was added in excess over the probe as indicated (Comp.). The NTH15–DNA complexes are indicated by arrowheads.
Figure 4.
DNase I protection assay of the NTH15-binding sites. An alignment of the regions protected from DNase I digestion by NTH15 in fragments 1 and 7 is shown at top. The reaction of the labeled fragment 7 is presented below. The region protected from DNase I digestion by NTH15 is marked by filled box. Open arrowhead indicates the NTH15-induced DNase I hypersensitive site. The scale to the left shows the reference ladders generated by the A + G sequence reaction of the same DNA fragments performed by the Maxam-Gilbert method.
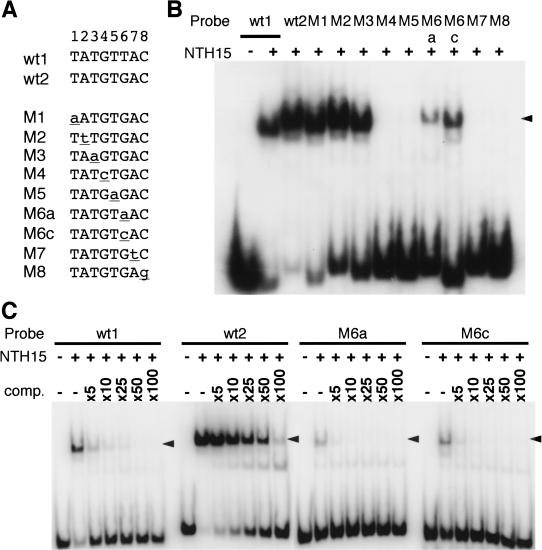
To confirm that these binding sequences were specifically recognized by NTH15, DNA sequence-specific binding of NTH15 was analyzed by EMSA using a series of mutagenized probes. Nucleotide positions within the TATGT(G/T)AC element were defined as 1–8 from the 5′ end to 3′ end, and corresponding mutations were defined as M1–M8 (Fig. 5A). Single nucleotide substitutions at positions 1–3 (M1–M3) had no effect on the affinity of binding with NTH15; however, substitutions at positions 4, 5, 7, and 8 (M4, M5, M7, and M8) dramatically reduced binding to undetectable levels (Fig. 5B). In contrast, substitutions at position 6 led to a reduction in the NTH15-binding affinity, but did not completely eliminate it (Fig. 5B). Competition experiments using four interactive oligonucleotides (wt1, wt2, M6a, and M6c) as probes and nonlabeled wt2 fragment as a competitor revealed that NTH15 preferably interacted with the sequence containing G at position 6 (Fig. 5C). These findings led us to conclude that NTH15 recognizes the 5-bp dyadsymmetric sequence GTNAC at positions 4–8 with a preference for G at position 6.
Figure 5.
Preference of the NTH15-binding sequence. (A) Core sequences of the NTH15-binding site found in fragments 1 and 7 (wt1 and wt2) and its derivative sequences used for the experiments (M1 to M8). Numbers at the top indicate the nucleotide positions. (B) electrophoresis mobility shift assay (EMSA) of each DNA fragment. Synthetic oligonucleotides containing the wild-type or mutagenized binding site were used as probes in EMSA. (C) NTH15 preferably interacted with the sequence containing G at position 6. The binding affinity was estimated with nonlabeled wt2 as a competitor (Comp.). The NTH15–DNA complexes are indicated by arrowheads.
The NTH15-binding sequence in the first intron of Ntc12 is required for the NTH15-dependent suppression of Ntc12
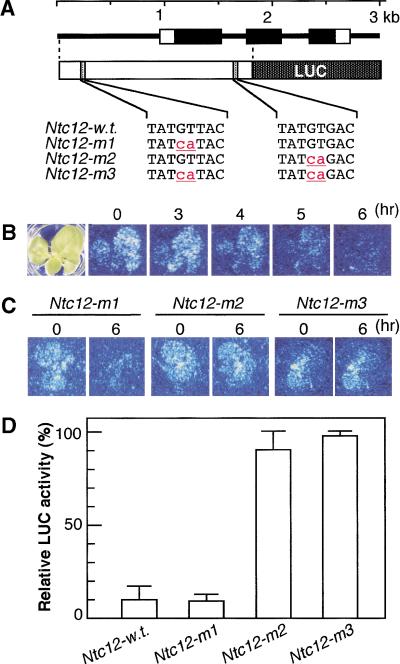
Because NTH15 binds to two different sites on Ntc12 in vitro, these two sites may act as cis-motives for transcriptional repression of Ntc12 by NTH15 in vivo. To examine this possibility, we introduced nucleotide substitutions independently at these two sites (Fig. 6A), generated a fusion construct with luciferase (LUC) reporter gene, and then transferred into 35S::NTH15–GR transformants. LUC luminescence was ubiquitously found at a low level in young leaves and around the SAM grown in the absence of DEX (Fig. 6B, time indicated as 0). This low luminescence activity is consistent with previous observations from RNA gel blot and in situ hybridization analyses of Ntc12 expression in wild type (Tanaka-Ueguchi et al. 1998). Following DEX treatment, in vivo luminescence was steeply decreased in transformants that carry the wild-type promoter (Ntc12-w.t.; Fig. 6B), such that LUC activity was reduced to 11.4% of initial levels 6 h after the treatment (Fig. 6D). Similar repression was observed in transformants that contained the mutant promoter at the distal site (Ntc12-m1; Fig. 6C,D). However, this suppression of luminescence was not observed in transformants that carry the mutant promoter at either the proximal site or at both sites (Ntc12-m2 or Ntc12-m3, respectively) in which the relative LUC activity remained at 90.2% and 98.3%, respectively (Fig. 6C,D). Thus, the NTH15-binding sequence TATGTGAC in the first intron of Ntc12 is essential for the suppression of Ntc12 expression through the activation of NTH15.
Figure 6.
NTH15-dependent repression of Ntc12 expression and the effect of two-base substitutions on Ntc12 repression. (A) Schematic structure of the promoter–luciferase (LUC) construct and nucleotide substitutions in the three mutants (Ntc12-m1 to Ntc12-m3) compared to the wild type (Ntc12-w.t.). (B) In vivo luminescence of LUC reporter was imaged using a high-sensitivity camera system. Ntc12-w.t. seedling was measured at 0 h, 3 h, 4 h, 5 h, and 6 h after dexamethasone (DEX) treatment. The appearance of treated seedling is shown in the left panel. (C) Ntc12-m1, Ntc12-m2, and Ntc12-m3 seedlings were measured 0 h and 6 h after DEX treatment. (D) Relative LUC activity 6 h after DEX treatment. The value obtained before DEX treatment was arbitrarily set at 100%. Each data point is the average of five plants.
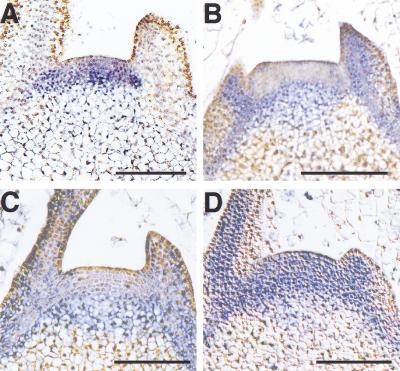
In situ hybridization of the tobacco SAM was performed to confirm that NTH15 suppresses Ntc12 expression in wild type. NTH15 mRNA was present in corpus cells, the inner layers of the SAM, but was not seen in the outermost layers (L1), rib meristem or leaf primordia (Fig. 7A), as previously reported by Tamaoki et al. (1997a). In contrast, Ntc12 transcript was localized to leaf primordia and rib meristem, areas in which NTH15 mRNA was not detected, and repressed in the corpus of the SAM (Fig. 7B).
Figure 7.
NTH15 directly represses Ntc12 expression in vivo around the shoot apical meristem (SAM). Probes were labeled with digoxygenin-UTP and the transcript-specific hybridization signal is visible as purple staining. (A,B) In situ hybridization of NTH15 (A) and Ntc12 (B) mRNA, respectively, in the wild-type tobacco SAM. (C,D) Localization of luciferase (LUC) reporter gene transcript in the SAM of transformants carrying the wild-type (Ntc12-w.t.; C) and mutant promoter–reporter construct (Ntc12-m2.; D), respectively. Bars, 100 μm.
We also determined the in situ mRNA localization of LUC reporter gene driven by the wild-type and mutant Ntc12 promoter. As for endogenous Ntc12 mRNA expression, LUC mRNA was localized to leaf primordia and rib meristem but not in the corpus of the SAM in Ntc12-w.t. transformants (Fig. 7C). However, in Ntc12-m2 transformants, LUC mRNA was expanded in the corpus of the SAM (Fig. 7D), in which NTH15 mRNA was detected and Ntc12 mRNA was not.
Discussion
It has often been reported that ectopic expression of knox genes from various plants causes drastic alterations in leaf and flower morphogenesis. However, such phenomena should be the result of a complex combination of direct and/or indirect effects of KNOX homeodomain protein function. Indeed, numerous kinds of genes are up-regulated in transgenic tobacco plants that carry a rice knox gene, OSH1, and most of them are considered to be induced indirectly by physiological stresses and changes in hormonal levels triggered by OSH1 expression (Tamaoki et al. 1997b).
To distinguish the direct effects of KNOX homeodomain proteins from the various changes observed in transgenic plants, we generated an artificial inducible system of NTH15 function using the human GR. This induction system has been applied to a number of genes that are involved in plant development, including: The Arabidopsis homeobox gene, Athb1; the flowering-time gene, CONSTANS; the floral homeotic genes, APETALA3/PISTILLATA; and the maize transcriptional regulator, R (Lloyd et al. 1994; Aoyama et al. 1995; Simon et al. 1996; Robert et al. 1998).
Using this inducible system, we found that the inhibition of GA biosynthesis via the specific suppression of Ntc12 expression was one of the earliest events caused by the activation of NTH15. The fact that this NTH15-dependent suppression of Ntc12 expression did not require new protein synthesis indicates that Ntc12 may be a member of target genes in the genetic systems regulated by NTH15. Indeed, recombinant NTH15 protein bound to the specific intron sequence of Ntc12 in vitro.
Biochemical studies with EMSA showed that NTH15 recognized the 5-bp dyadsymmetric sequence GTNAC and that it favored G relative to other nucleotides at the third position. In contrast to the typical homeodomain proteins, such as Antennapedia and Ultrabithorax of Drosophila, which recognize the core sequence TAAT, TALE-atypical (for three amino acid loop extension between helices I and II) homeodomain proteins interact with a non-TAAT sequence (plant KNOX homeodomain proteins belong to this superclass). For example, the human TALE homeodomain protein, TGIF, binds to the TGTCA core sequence (Bertolino et al. 1995), and the barley KNOX homeodomain protein, Hooded (an ortholog of maize KN1), binds to the core sequence GTCA (Krusell et al. 1997). The NTH15 binding sequence, GTNAC, is very similar to these core sequences but not identical. NTH15 preferentially binds to GTGAC rather than GTCAC, which contains the core sequence of TGIF and Hooded. Conversely, the binding affinity of TGIF is significantly reduced by a single nucleotide substitution from C to G within its core sequence TGTCA (Bertolino et al. 1995). Taken together, these data indicate that the binding property of NTH15 is slightly different from that of other TALE homeodomain proteins. Such minor differences between TALE homeodomain proteins may enable them to target different genes and consequently cause a unique function of each homeodomain protein.
Analysis of the NTH15-binding sites revealed that the binding sequence in the first intron of Ntc12 was a functional target of NTH15 for negative regulation of Ntc12 expression. Mutation in this sequence abolished the suppression of Ntc12 by the activation of NTH15 function in vivo. In addition, NTH15 and Ntc12 showed an exclusive pattern of in situ mRNA localization in the wild-type SAM. This inverse relationship between NTH15 and Ntc12 expression strongly suggests that the suppression of Ntc12 caused by NTH15 also occurs in the wild-type SAM. This possibility is also supported by the observation that mutation in the NTH15-binding sequence resulted in the expanded expression of Ntc12 in the corpus of the SAM of Ntc12-m2 transformants.
It is noteworthy that cells divide randomly in the inner layers of the SAM, where NTH15 is expressed and Ntc12 is not. However, the orientation of cell division is regulated in leaf primordia and rib meristem, where Ntc12 mRNA is present and NTH15 is suppressed. In addition, activation of NTH15 induces abnormal periclinal divisions in the mesophyll cell layers in leaves, where Ntc12 and GA biosynthesis are suppressed. Bioactive GA is known to enhance transverse cell division and longitudinal cell expansion by affecting the arrangement of newly deposited cellulose microfibrils in the cell wall (Gunning 1982). Therefore it is possible that NTH15, through the repression of Ntc12 and GA biosynthesis, plays a regulatory role in maintaining the flexibility of the orientation of cell division to preserve the indeterminate properties of cells in the inner layers of the SAM. We propose that suppression of NTH15 within cells at the flanks of the SAM permits the expression of Ntc12 and enables them to increase their bioactive GA content, which promotes the organized cell division and consequently induces the determination of cell fate.
Materials and methods
Plasmid constructs
To create NTH15–GR fusion protein, the stop codon of NTH15 was replaced with SmaI site (CCCGGG) by polymerase chain reaction (PCR) as reported previously (Sakamoto et al. 1999). In the same way, we amplified DNA fragments that encode the steroid-binding domain of the human GR (Hollenberg et al. 1985). The chimeric protein sequence was (NTH15 aa 1–342)-Pro-Gly-(GR aa 500–777). NTH15–GR was then cloned between the CaMV 35S promoter and the nopaline synthase (NOS) polyadenylation signal of pBI121 (Fig. 1A; Clonetech).
To create a chimeric construct with a reporter gene under control of the promoter of Ntc12, 1.0 kb of the 5′-flanking sequence plus the sequences of entire first exon, first intron, and part of second exon of Ntc12 were introduced at the front of Luc cDNA from pSP-luc+ vector (Promega). Nucleotide substitutions were introduced by PCR as previously reported (Sakamoto et al. 1999). The resulting chimeric genes (Ntc12-w.t., Ntc12-m1, Ntc12-m2, and Ntc12-m3) were inserted into the 5′ end of the NOS polyadenylation signal of the hygromycin resistant binary vector pBI-H1 (Ohta et al. 1990).
Plant transformation and growth conditions
Constructs were introduced into Agrobacterium tumefaciens LBA4404 by electroporation. Agrobacterium-mediated transformation of Nicotiana tabacum cv Samsun NN was performed with leaf discs as reported previously (Matsuoka and Sanada 1991). Transgenic plants were selected on media containing 100 mg/L kanamycin or 50 mg/L hygromycin. Antibiotic-resistant plants and seedlings from self-pollinated transformants were adapted to hydroponic growth conditions and grown at 25°C under continuous light.
Chemical treatments
DEX (Wako Pure Chemical Industries) was dissolved in ethanol to 30 mM before use and diluted with growth medium to 10 μM. The same volume of ethanol was added to the control medium. For treatment of whole plants, roots were submerged in growth medium containing 10 μM DEX. To inhibit RNA synthesis, 5 μM α-amanitin (Sigma) was added to growth medium. For inhibition of protein synthesis, seedlings were first soaked in growth medium containing 10 μM CHX (Sigma) for 1 h and then incubated in medium containing 10 μM CHX and 10 μM DEX or 10 μM CHX and ethanol as a control. The effect of CHX on protein synthesis was determined according to the method of Lam et al. (1989).
Histological analysis
Plant material was fixed overnight at 4°C in 4% paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4), dehydrated through a graded ethanol series followed by a t-butanol series (Sass 1958), and finally embedded in Paraplast Plus (Sherwood Medical). Microtome sections of 7–10 μm in thickness were applied to silan-coated glass slides (Matsunami Glass). The sections were deparaffinized in xylene, rehydrated through a graded ethanol series, and dried overnight before staining with hematoxylin.
GA analysis
Whole above-ground portion of 35S::NTH15–GR seedlings was harvested and used for the analysis of endogenous GA levels. Samples of plant material were extracted with methanol. After addition of the radio-labeled internal standards, methanol extracts were purified as described previously (Nakayama et al. 1989). The purified and methylated samples were then analyzed by GC-SIM with a Hewlett-Packard 5989 B GC-MS system fitted with a DB-1 fused-silica chemically bonded capillary column (J & W Scientific) as described previously (Tanaka-Ueguchi et al. 1998).
RNA gel blot analysis
Total RNA from whole above-ground portion of 35S::NTH15–GR seedlings was extracted and 20 μg of each RNA sample was electrophoresed on a 1.2% agarose gel, then transferred onto Hybond N+ membrane (Amersham Pharmacia Biotech). Hybridization was performed in 5× SSC, 5× Denhardt's solution, 0.5% SDS, 10% dextran sulphate, and 20 mg/L salmon sperm DNA at 65°C for 14 h. The filter was washed in 2× SSC containing 0.1% SDS at 65°C and then further washed in 0.2× SSC containing 0.1% SDS at 65°C.
EMSA and DNase I protection assay
To produce NTH15 protein, the full-length NTH15 cDNA was inserted in the sense orientation as a translational fusion into the pET-32a expression vector (Novagen) and expressed in BL21 (DE3) E. coli cells (Stratagene). The Ntc12 DNA fragments of ∼350 bp were excised with restriction endonucleases, purified on 8% polyacrylamide gels, labeled with [32P]dATP using Klenow fragment and purified on Sephadex G-50 columns. DNA-binding reactions were performed at 4°C for 30 min in 10 mM Tris (pH 7.5), 50 mM NaCl, 1 mM EDTA, 500 μM DTT, 10% glycerol, 0.05% IGEPAL CA-630 (Sigma), and 50 mg/L poly(dI-dC) · poly(dI-dC) (Amersham Pharmacia Biotech), and subjected to EMSA with 8% polyacrylamide gels in 0.25 × Tris-borate-EDTA buffer.
For DNase I protection experiment, labeled DNA fragments and NTH15 protein were incubated at 4°C for 30 min in 50 μL of the same solution described above. After incubation, 5 μL of diluent (10 mM MgCl2, 5 mM CaCl2) was added, followed immediately by 1 unit of DNase I (Amersham Pharmacia Biotech). After 1 min, reactions were terminated by addition of 150 μL of a stop buffer (192 mM CH3COONa, 32 mM EDTA, 0.14% SDS, 64 mg/L yeast DNA). Following extraction with 0.2 mL of phenol:chloroform (1:1), nucleic acids were concentrated by ethanol precipitation, electrophoresed in 8% polyacrylamide gels containing 8 M urea, and detected by autoradiography. Reference ladders were generated by the A + G sequence reaction of the same DNA fragments performed by the Maxam-Gilbert method (Maxam and Gilbert 1977).
Luciferase assays
To image LUC luminescence, roots of transgenic seedlings were submerged in a solution of 10 mM potassium luciferin (Bio-Rad Laboratories) for 10 h before DEX treatment. LUC luminescence from transgenic seedlings was visualized using an image-intensifying camera and photon-counting image processors (Hamamatsu Photonic Systems ARGUS-50). Exposure time was 15 min for each assay. Luminescence was quantified from images captured in centroid-processing mode (this records a single pixel of unit depth for each photoelectron detected by the camera) and corrected for background counts from thermal electrons.
In situ hybridization analysis
Plant materials were fixed and embedded as described above. Microtome sections (7–10 μm in thickness) were applied to glass slides treated with Vectabond (Vector Laboratory). Hybridization with digoxygenin-labeled sense or antisense RNA and immunological detection of the hybridized probe were conducted according to the method described by Kouchi and Hata (1993).
Acknowledgments
We thank Takashi Aoyama and Nam-Hai Chua for providing plasmid containing the GR receptor domain, Akemi Tagiri for assistance with the in situ hybridization, and Naomi Oyama and Masaji Koshioka for their help with the GA analysis. This study was supported by the Research for the Future Program of the Japan Society for the Promotion of Science (JSPS) and Grant-in-Aid for Scientific Research on Priority Areas (Molecular Mechanisms Controlling Multicellar Organization of Plant) from the Ministry of Education, Science, Sports, and Culture of Japan. T.S. is supported by a research fellowship from JSPS and Grant-in-Aid for JSPS Fellows from the Ministry of Education, Science, Sports and Culture of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL j45751a@nucc.cc.nagoya-u.ac.jp; FAX 81-52-789-5226.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.867901.
References
- Aoyama T, Dong C-H, Wu Y, Carabelli M, Sessa G, Ruberti I, Morelli G, Chua N-H. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell. 1995;7:1773–1785. doi: 10.1105/tpc.7.11.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: Wiley; 1995. [Google Scholar]
- Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes a retinoid-responsive motif. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugis G, Giannino D, Mele G, Nicolodi C, Innocenti AM, Chiappetta A, Bitonti MB, Dewitte W, Van Onckelen H, Mariotti D. Are homeobox knotted-like genes and cytokinins the leaf architects? Plant Physiol. 1999;199:371–374. doi: 10.1104/pp.119.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning BES. The cytokinetic apparatus: Its development and spatial regulation. In: Lloyd CW, editor. The cytoskeleton in plant growth and development. New York: Academic Press; 1982. pp. 229–292. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hewelt A, Prinsen E, Thomas M, Onckelen HV, Meins F. Ectopic expression of maize knotted1 results in the cytokinin-autotrophic growth of cultured tobacco tissues. Planta. 2000;210:884–889. doi: 10.1007/s004250050693. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development. 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Krusell L, Rasmussen I, Gausing K. DNA binding sites recognized in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare) FEBS Lett. 1997;408:25–29. doi: 10.1016/s0014-5793(97)00382-7. [DOI] [PubMed] [Google Scholar]
- Kusaba S, Fukumoto M, Honda C, Yamaguchi I, Sakamoto T, Kano-Murakami Y. Decreased GA1 content caused by the overexpression of OSH1 is accompanied by suppression of GA 20-oxidase gene expression. Plant Physiol. 1998a;117:1179–1184. doi: 10.1104/pp.117.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba S, Kano-Murakami Y, Matsuoka M, Tamaoki M, Sakamoto T, Yamaguchi I, Fukumoto M. Alteration of hormone levels in transgenic tobacco plants overexpressing a rice homeobox gene OSH1. Plant Physiol. 1998b;116:471–476. doi: 10.1104/pp.116.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Green PJ, Wong M, Chua N-H. Phytochrome activation of two nuclear genes requires cytoplasmic protein synthesis. EMBO J. 1989;8:2777–2783. doi: 10.1002/j.1460-2075.1989.tb08423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Sanada Y. Expression of photosynthetic genes from the C4 plant, maize, in tobacco. Mol Gen Genet. 1991;225:411–419. doi: 10.1007/BF00261681. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano-Murakami Y. Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell. 1993;5:1039–1048. doi: 10.1105/tpc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale NA. LAM-1 and FAT genes control development of the leaf blade in Nicotiana sylvestris. Plant Cell. 1993;5:1029–1038. doi: 10.1105/tpc.5.9.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Yamane H, Yamaguchi I, Murofushi N, Takahashi N, Katsumi M. Endogenous gibberellins in the vegetative shoots of tall and dwarf cultivars of Phaseolus vulgaris L. J Plant Growth Regul. 1989;8:237–247. [Google Scholar]
- Nishimura A, Tamaoki M, Sato Y, Matsuoka M. The expression of tobacco knotted1-type class 1 homeobox genes correspond to regions predicted by the cytohistological zonation model. Plant J. 1999;18:337–347. doi: 10.1046/j.1365-313x.1999.00457.x. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sakamoto T, Matsuoka M. Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol. 2000;41:583–590. doi: 10.1093/pcp/41.5.583. [DOI] [PubMed] [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K. Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol. 1990;31:805–813. [Google Scholar]
- Ori N, Juarez MT, Jackson D, Yamaguchi J, Banowetz GM, Hake S. Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell. 1999;11:1073–1080. [PMC free article] [PubMed] [Google Scholar]
- Reiser L, Sanchez-Baracaldo P, Hake S. Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol Biol. 2000;42:151–166. [PubMed] [Google Scholar]
- Robert W, Sablowski M, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Nishimura A, Tamaoki M, Kuba M, Tanaka H, Iwahori S, Matsuoka M. The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell. 1999;11:1419–1431. doi: 10.1105/tpc.11.8.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass AE. Botanical micro technique. 3rd ed. Ames, IA: Iowa State University Press; 1958. [Google Scholar]
- Sato Y, Tamaoki M, Murakami T, Yamamoto N, Kano-Murakami Y, Matsuoka M. Abnormal cell divisions in leaf primordia caused by the expression of the rice homeobox gene OSH1 lead to altered morphology of leaves in transgenic tobacco. Mol Gen Genet. 1996;251:13–22. doi: 10.1007/BF02174339. [DOI] [PubMed] [Google Scholar]
- Schena M, Lloyd AM, Davis RW. A steroid-inducible gene expression system for plant cells. Proc Natl Acad Sci. 1991;88:10421–10425. doi: 10.1073/pnas.88.23.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M. Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell. 1999;11:1651–1664. doi: 10.1105/tpc.11.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Igeño MI, Coupland G. Activation of floral meristem identity genes in Arabidopsis. Nature. 1996;384:59–62. doi: 10.1038/384059a0. [DOI] [PubMed] [Google Scholar]
- Sinha NR, Williams RE, Hake S. Overexpression of the maize homeo box gene, Knotted-1, causes a switch from determinate to indeterminate cell fates. Genes & Dev. 1993;7:787–795. doi: 10.1101/gad.7.5.787. [DOI] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in plant development. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Tamaoki M, Kusaba S, Kano-Murakami Y, Matsuoka M. Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol. 1997a;38:917–927. doi: 10.1093/oxfordjournals.pcp.a029252. [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Yamamoto N, Matsuoka M. Isolation and characterization of cDNA clones with enhanced expression in tobacco plants expressing the rice homeobox gene, OSH1. Plant Cell Physiol. 1997b;38:638–642. doi: 10.1093/oxfordjournals.pcp.a029215. [DOI] [PubMed] [Google Scholar]
- Tanaka-Ueguchi M, Itoh H, Oyama N, Koshioka M, Matsuoka M. Over-expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J. 1998;15:391–400. doi: 10.1046/j.1365-313x.1998.00217.x. [DOI] [PubMed] [Google Scholar]