Abstract
Molecular transport is a key process in cellular metabolism. This step is often limiting when using a nonnative carbon source, as exemplified by xylose catabolism in Saccharomyces cerevisiae. As a step toward addressing this limitation, this study seeks to characterize monosaccharide transport preference and efficiency. A group of 26 known and putative monosaccharide transport proteins was expressed in a recombinant Saccharomyces cerevisiae host unable to transport several monosaccharides. A growth-based assay was used to detect transport capacity across six different carbon sources (glucose, xylose, galactose, fructose, mannose, and ribose). A mixed glucose-and-xylose cofermentation was performed to determine substrate preference. These experiments identified 10 transporter proteins that function as transporters of one or more of these sugars. Most of these proteins exhibited broad substrate ranges, and glucose was preferred in all cases. The broadest transporters confer the highest growth rates and strongly prefer glucose. This study reports the first molecular characterization of the annotated XUT genes of Scheffersomyces stipitis and open reading frames from the yeasts Yarrowia lipolytica and Debaryomyces hansenii. Finally, a phylogenetic analysis demonstrates that transporter function clusters into three distinct groups. One particular group comprised of D. hansenii XylHP and S. stipitis XUT1 and XUT3 demonstrated moderate transport efficiency and higher xylose preferences.
INTRODUCTION
Lignocellulosic biomass is an attractive industrial feedstock that can be converted into liquid transportation fuels and other small molecule bioproducts via microbial and fungal fermentations (6). However, this material is quite recalcitrant to enzymatic digestion and contains a significant fraction of pentose sugars (especially d-xylose and l-arabinose). These pentose sugars cannot be readily metabolized by nonrecombinant versions of common fermentative host organisms such as the bacterium Zymomonas mobilis or the yeast Saccharomyces cerevisiae. While native pentose-utilizing organisms exist, a lack of well-developed genetic tools and low product tolerances (13) limit their utility as hosts for industrial scale lignocellulosic conversion processes. As a result, a significant effort has focused on the metabolic engineering of pentose catabolic pathways in the yeast S. cerevisiae to enable xylose and arabinose fermentation (12, 18). Despite these efforts, the transport of these exogenous sugars is still limited and can often be the rate-limiting step in metabolism (8). It has been demonstrated that xylose transport in recombinant yeast is facilitated by native glucose transporters (14, 27, 37). The lack of a dedicated xylose transport system in recombinant S. cerevisiae thus limits the capacity for dual xylose and glucose fermentation as well as high xylose catabolic pathway flux (17). This limitation highlights the need to identify and/or engineer efficient, heterologous xylose transport proteins in yeast.
Initial work to enable xylose utilization in yeast focused on establishing the essential, heterologous catabolic pathways. To this end, several metabolic engineering strategies for enabling the recombinant fermentation of pentoses in the yeast S. cerevisiae have been investigated (1, 16, 23, 41). The vast majority of work has focused on the most abundant pentose sugar, d-xylose, through a combination of heterologous pathway engineering and native pentose phosphate pathway (PPP) optimization (12, 42). The introduction of one of two basic heterologous xylose catabolic pathways, an oxidoreductase pathway commonly found in fungi (20) and an isomerase pathway commonly found in bacteria (25), can confer growth on xylose as a sole carbon source. Both of these pathways convert xylose to the natively fermentable ketose sugar xylulose. However, the oxidoreductase pathway suffers from cofactor imbalance (18), whereas the isomerase pathway has proved difficult to actively express (21) and suffers from lower throughput. Further overexpression and complementation of the native PPP enzymes xylulokinase (XKS) (5), transaldolase (TAL) (19), and transketolase (TKT) (38) have improved xylose catabolic rates. Together, these metabolic engineering efforts have led to increased xylose catabolic flux and improved ethanol yields. However, independently of the pathway used, xylose flux in recombinant yeast has been shown to be limited by transport (8, 14, 26, 35). As a result, further pathway and metabolic engineering efforts aimed at improving intracellular pathways will only increase this limitation (42).
Heterologous xylose transporter expression to alleviate this limitation has been explored (14, 15, 28, 35). These studies suggest that heterologous transporters can improve S. cerevisiae xylose fermentation characteristics (27). However, only a few proteins have been experimentally identified for enabling xylose transport in Saccharomyces cerevisiae, and all of these have been shown to favor glucose over xylose in a mixed-sugar culture. Moreover, while these proteins show affinity toward two structurally different monosaccharides (glucose and xylose), no work has examined other monosaccharides as potential substrates. Such a characterization would expand our understanding of molecular transporter function as well as suggest potentially useful classes of transport proteins for improving recombinant xylose utilization in yeast.
In this study, we pursue a functional survey and characterization of 23 heterologous and 3 native S. cerevisiae yeast proteins expressed in a recombinant xylose-utilizing S. cerevisiae host devoid of glucose and xylose transporters (40). These proteins represent both putative and known transporters capable of xylose transport spanning the organisms Arabidopsis thaliana, Candida intermedia, Cryptococcus neoformans, Debaryomyces hansenii, Escherichia coli, Scheffersomyces stipitis (formerly Pichia stipitis [24]), and Yarrowia lipolytica. We present growth-based assays using a variety of monosaccharides as sole carbon sources (glucose, xylose, galactose, fructose, mannose, and ribose) in an effort to characterize the substrate acceptance profiles of these transporters. Furthermore, we characterize the preference ratio of xylose to glucose using a competitive preference assay in order to measure the degree to which xylose transport is inhibited by glucose in a cofermentation. No prior study has evaluated putative transporters from several organisms, using a consistent strain background, in an effort to characterize carbon source profiles and preferences. Thus, these results present the largest-scale characterization of sugar transporter properties to date and suggest a path forward for improving xylose transport in recombinant Saccharomyces cerevisiae for biofuel applications.
MATERIALS AND METHODS
Strains and plasmids.
The microbial strains and plasmids used in this study are listed in Table 1. S. cerevisiae EBY.VW4000 (40) was obtained as a gift from Eckhard Boles of the Institute of Molecular Biosciences, Goethe-Universität, Frankfurt, Germany. D. hansenii CBS 767, E. coli K-12 MG1655, S. stipitis CBS 6054, and Y. lipolytica ATCC 8662 were purchased from the American Type Culture Collection (ATCC; Manassas, VA). S. cerevisiae BY4741 was obtained from EUROSCARF (Goethe-Universität, Frankfurt, Germany). C. intermedia NCYC 2504 was obtained from the National Collection of Yeast Cultures (NCYC; Colney, Norwich, United Kingdom). Escherichia coli 10-beta (New England Biolabs, Ipswich, MA) was routinely used for gene cloning. E. coli strains containing the Mumberg et al. yeast shuttle vectors for gene cloning (32) (ATCC 87669) were obtained from the ATCC. Vectors were isolated using a plasmid miniprep kit (Qiagen, Valencia, CA). A complete list of all vectors used in this study is given in Table 2.
Table 1.
Strains used in this study
| Straina | Description | Source or reference |
|---|---|---|
| Candida intermedia NCYC 2504 | Wild type | NCYC |
| Debaryomyces hansenii CBS 767 | Wild type | ATCC (ATCC 36239) |
| Escherichia coli 10-beta | araD139Δ(ara-leu)7697 fhuA laxX74 galK [φ80 Δ(lacZ)M15] mcrA galU recA1 end A1 nupG rpsL (Strr) Δ(mrr-hsdRMS-mcrBC) | New England Biolabs |
| Escherichia coli K-12 MG1655 | Sequenced E. coli strain | ATCC (ATCC 700926) |
| Scheffersomyces stipitis CBS 6054 | Wild type | ATCC (ATCC 58785) |
| Yarrowia lipolytica | Wild type | ATCC (ATCC 8662) |
| S. cerevisiae BY4741 | Standard laboratory yeast | EUROSCARF (accession no. Y00000) |
| S. cerevisiae EBY.VW4000 | MATα leu2-3,112 ura3-52 trp1-289 his3-Δ1 Mal2-8c SUC2hxt17Δ hxt13Δ::loxPhxt15Δ::loxPhxt16Δ::loxP hxt14Δ::loxP hxt12Δ::loxP hxt9Δ::loxP hxt11Δ::loxP hxt10Δ::loxP hxt8Δ::loxP hxt514::loxP hxt2Δ::loxP hxt367Δ::loxP gal2Δ stl1Δ::loxP agt1Δ::loxP ydl247wΔ::loxP yjr160cΔ::loxP | 41 |
| S. cerevisiae EY1 | S. cerevisiae EBY.VW4000(p16T.XYL1) | This study |
| S. cerevisiae EY12 | S. cerevisiae EBY.VW4000(p16T.XYL1)(p25G.XYL2) | This study |
| S. cerevisiae EY12.XX | S. cerevisiae EY12(p14T.XX) | This study |
This includes the strains from which genomic DNA was isolated, as well as the recombinant host EY12, which was used as a host for the experiments conducted in this study.
Table 2.
Plasmids constructeda
| Plasmid | Description | Reference or source |
|---|---|---|
| p416-TEF | URA3, CEN6/ARSH4 origin, TEFp | 33 |
| p425-GPD | LEU2, 2μm origin, GPDp | 33 |
| p414-TEF | TRP, CEN6/ARSH4 origin, TEFp | 33 |
| p16T.X1 | p416-TEF-SsXyl1 | This study |
| p25G.X2 | p425-GPD-SsXyl2 | This study |
| p14T.02 | p414-TEF-AtXYLL3 | This study |
| p14T.03 | p414-TEF-AtXYLL2 | This study |
| p14T.04 | p414-TEF-ScHXT3 | This study |
| p14T.05 | p414-TEF-CiGXF1 | This study |
| p14T.06 | p414-TEF-CiGXS1 | This study |
| p14T.07 | p414-TEF-DEHA0D02167 | This study |
| p14T.08 | p414-TEF-DEHA2B14278 | This study |
| p14T.09 | p414-TEF-DEHA2A14300 | This study |
| p14T.10 | p414-TEF-DEHA2F19140 | This study |
| p14T.12 | p414-TEF-DhXylHP | This study |
| p14T.13 | p414-TEF-EcXylE | This study |
| p14T.14 | p414-TEF-SsXUT1 | This study |
| p14T.15 | p414-TEF-SsXUT2 | This study |
| p14T.16 | p414-TEF-SsXUT3 | This study |
| p14T.17 | p414-TEF-SsXUT4 | This study |
| p14T.18 | p414-TEF-SsXUT5 | This study |
| p14T.19 | p414-TEF-SsXUT6 | This study |
| p14T.20 | p414-TEF-SsXUT7 | This study |
| p14T.21 | p414-TEF-YALI0B06391 | This study |
| p14T.22 | p414-TEF-YALI0B01342 | This study |
| p14T.23 | p414-TEF-YALI0F06776 | This study |
| p14T.24 | p414-TEF-YALI0C06424 | This study |
| p14T.25 | p414-TEF-YALI0C08943 | This study |
| p14T.26 | p414-TEF-CNBC3990 | This study |
| p14T.27 | p414-TEF-ScHXT7 | This study |
| p14T.35 | p414-TEF-ScHXT13 | This study |
| p14T.36 | p414-TEF-ScGAL2 | This study |
The xylose metabolic genes XYL1 and XYL2 from Scheffersomyces stipitis were expressed in Mumberg et al. shuttle vectors (32), along with all known and putative transporter ORFs included in this study.
Media and culture conditions.
Yeast and bacterial strains were stored at −80°C in 15% glycerol. E. coli strains were grown in LB-Miller broth and supplemented with 50 μg/ml of ampicillin for plasmid propagation when necessary. C. intermedia and S. stipitis were cultivated at 30°C in YM broth (3 g/liter yeast extract, 3 g/liter malt extract, 5 g/liter Bacto peptone, 10 g/liter glucose). D. hansenii, S. cerevisiae BY4741, and Y. lipolytica were cultivated at 30°C in YP medium (10 g/liter yeast extract, 20 g/liter Bacto peptone) with 20 g/liter glucose. S. cerevisiae EBY.VW4000 was cultivated at 30°C in YP medium with 20 g/liter maltose (YPM) (40). All strains were cultivated with 225-rpm orbital shaking. To select transformants, yeast synthetic complete (YSC) medium composed of 6.7 g/liter yeast nitrogen base, 15 g/liter agar, and either complete supplement mixture (CSM)-Ura, CSM-Leu-Ura, or CSM-Leu-Trp-Ura (MP Biomedicals, Solon, OH) were added, depending on the required auxotrophic selection. The carbon source used for selection, propagation, and preculturing of EBY.VW4000-derived strains was 20 g/liter maltose. Growth characterization experiments used YSC medium with CSM-Leu-Trp-Ura and various carbon sources, detailed below.
Identifying known and putative transporter genes.
The collection of 23 heterologous and 3 native S. cerevisiae transporters listed in Table 3 was chosen in order to characterize a more extensive carbon source profile and to compare novel open reading frames (ORFs) with native and literature-identified transporters. To this end, the S. cerevisiae HXT7 and GAL2 genes were included as positive xylose transport controls, and S. cerevisiae HXT13 was included as a negative xylose transport control based on prior literature evidence (3, 14, 35, 37). The two C. intermedia genes, GXF1 and GXS1 (9, 35), were included to assay for additional monosaccharide substrates and to provide another benchmark from which to evaluate novel ORFs. Two ORFs from Arabidopsis thaliana, At5g59250 and At5g17010, were included to investigate a disagreement in the literature as to the ability of these transporters to confer improved xylose uptake characteristics (14, 15). The Escherichia coli xylE transporter was also included on the basis of its exclusive specificity for xylose (4), despite previous unsuccessful expression attempts in S. cerevisiae (14). The seven annotated XUT genes from S. stipitis were chosen in an attempt to identify the high-affinity xylose transporter(s) hypothesized to exist in this yeast. The National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) (2) was then used to identify genes with a high degree of similarity to those transporters reported, yet those with over 90% homology were generally discarded. C. intermedia GXS1, Trichoderma reesei xlt1, and S. stipitis XUT3 and XUT4 served as the reference sequences for the BLAST search. ORFs from the sequenced yeasts Y. lipolytica and D. hansenii occurred frequently over multiple BLAST searches, which resulted in the inclusion of five ORFs from each organism in this study. D. hansenii xylHP, in addition to recovery by BLAST, has been mentioned as a potential xylose transporter (27). In addition, one gene from C. neoformans was included due to its homology to S. stipitis XUT4. Combined, these ORFs comprise the 23 heterologous transporter genes surveyed in this work. Since the SUT1-3 genes of S. stipitis (22, 39) are so similar to each other and to C. intermedia GXF1, they were not included in this survey.
Table 3.
ORFs cloneda
| Organism | Gene/locus tag | UniProt accession no. | Source or reference | Primer | RE(s) used | Sequence |
|---|---|---|---|---|---|---|
| S. stipitis | XYL1 | P31867 | 24 | EY037 | XbaI | GCTCTAGAATGCCTTCTATTAAGTTGAACTCTGGTTAC |
| EY038 | ClaI | CCATCGATTTATTTCCTCTCTATAAAGCAACCTTCTTAG | ||||
| XYL2 | P22144 | 24 | EY103 | BamHI-HF | GGCGCGGATCCATGACTGCTAACCCTTCCTTGGT | |
| EY104 | XmaI | TCCCCCGGGGGATTACTCAGGGCCGTCAATGAGAC | ||||
| A. thaliana | At5g59250 | Q0WWW9 | 16 | EY052 | EcoRI | CGGAATTCATGGCTTTCGCTGTCTCGGT |
| EY004 | ClaI | CCATCGATTCACTTCAAGATTTTTGATTCAATTTCTTCC | ||||
| At5g17010 | Q6AWX0 | 16 | EY177 | BamHI-HF | GGCGCGGATCCATGGCGCTTGATCCTGAGCA | |
| EY178 | ClaI | CCGTATCCATCGATTTAGAGACATTTGGCTTCAATTTCCTCA | ||||
| C. intermedia | GXF1 | Q2MDH1 | 29 | EY175 | BamHI-HF | GGCGCGGATCCATGTCACAAGATTCGCATTCTTCT |
| EY176 | ClaI | CCGTATCCATCGATTTAAACCTGTTCGTCGGTGGCC | ||||
| GXS1 | Q2MEV7 | 29 | EY083 | BamHI-HF | CGGGATCCATGGGTTTGGAGGACAATAGAATGG | |
| EY082 | ClaI | CCATCGATTTAAACAGAAGCTTCTTCAGACATAATAGC | ||||
| D. hansenii | DEHA0D02167 | Q6BTD8 | BLAST | EY163 | XmaI | TATTCCCCCGGGATGGGTTTAGAAGATAATGCGCTTAT |
| EY164 | XhoI | CCGTGGCTCGAGTTAGACTGAAGTGGTTTCAATTTCAGT | ||||
| DEHA2B14278 | Q6BW54 | BLAST | EY087 | BamHI-HF | CCATCGATCTACGAACTAGAGTCCTTGGTATCAAC | |
| EY086 | ClaI | CGGGATCCGAGAGAAGGAGAAAAATAATACGAAATGTGG | ||||
| DEHA2A14300 | B5RSN0 | BLAST | EY181 | EcoRI | GAGTGGCGGAATTCATGTTCAATAAAATCAGATTTGGTTTCTGCA | |
| EY182 | ClaI | CCGTATCCATCGATCTACTTACTTATGCTATTTGATTCCACTTGTTC | ||||
| DEHA2F19140 | B5RUJ3 | BLAST | EY084 | BamHI-HF | CGGGATCCTTGATGTCTTCGTTATTGACCAACA | |
| EY085 | ClaI | CCATCGATTCAATTATTCGAGAGCAAAGTACGTTC | ||||
| xylHP | Q64L87 | 28 | EY167 | XmaI | TATTCCCCCGGGATGACTACTGCTGTTGGATTAGAAGATAATTCC | |
| EY168 | XhoI | CCGTGGCTCGAGTTAATCAGAATAATGTGCTTCCGAAATATCTGTTG | ||||
| E. coli | xylE | P0AGF4 | 15 | EY051 | BamHI-HF | CGGGATCCATGAATACCCAGTATAATTCCAGTTATATATTTTCG |
| EY001 | ClaI | CCATCGATTTACAGCGTAGCAGTTTGTTGTG | ||||
| S. stipitis | XUT1 | A3LY10 | NCBI | EY073 | BamHI-HF | CGGGATCCATGCACGGTGGTGGTGACGG |
| EY072 | ClaI | CCATCGATTTATTTTTCAACGTGGTAGACATCAGCCTTGC | ||||
| XUT2 | A3GIE8 | NCBI | EY061 | BamHI-HF | CGGGATCCATGAAGTATTTTCAAATCTGGAAATCAGGC | |
| EY032 | ClaI | CCATCGATCACTCAACTTCAATATGCTCGATTATTG | ||||
| XUT3 | A3GHU5 | NCBI | EY169 | XmaI | TATTCCCCCGGGATGAGAGAAGTTGGTATTCTTGATGTTGC | |
| EY170 | XhoI | CCGTGGCTCGAGTTATTCTGACATTTCAATCGAGTTGCG | ||||
| XUT4 | A3M0B9 | NCBI; intron-free version synthesized by Blue Heron Biotechnology | ||||
| XUT5 | A3LY79 | NCBI | EY068 | EcoRI | CGGAATTCATGACGGAAAGAAGCATTGGACCTT | |
| EY069 | ClaI | CCATCGATTTACTTCTTTGTATTAACAACAAAACCTTGTCTG | ||||
| XUT6 | A3M0N4 | NCBI | EY035 | EcoRI | CGGAATTCATGTCCAGTGTTGAAAAAAGTGCTGAAA | |
| EY036 | ClaI | CCATCGATTTAGCTGATGTTTTCGACATGCTCTAT | ||||
| XUT7 | A3GHF2 | NCBI | EY207 | XmaI | TATTCCCCCGGGATGATATCATCGCTTTTGGTAGC | |
| EY174 | ClaI | CCGTATCCATCGATCTAGAGTAATGTTCTTCTTGGAGACTCG | ||||
| Y. lipolytica | YALI0B06391 | Q6CFJ6 | BLAST | EY059 | EcoRI | CGGAATTCATGATTGGAAACGCTCAAATTAACCA |
| EY027 | ClaI | CCATCGATTTACAATTGAGAGGGAGGGGCG | ||||
| YALI0B01342 | Q6CG30 | BLAST | EY058 | EcoRI | CGGAATTCATGTACAAGGTCCATAACCCCTACC | |
| EY026 | ClaI | CCATCGATTTAGACATGCTCAGTTCCAGGATACT | ||||
| YALI0F06776 | Q6C2L7 | BLAST | EY023 | BamHI-HF | CGGGATCCATGTTTTCGTTAACGGGCAAACCG | |
| EY024 | ClaI | CCATCGATTTATACCGGAGGTTGAGGGAAGTC | ||||
| YALI0C06424 | Q6CCU6 | BLAST | EY171 | XmaI | TATTCCCCCGGGATGGGACTCGCTAACATCATCAACC | |
| EY172 | ClaI | CCGTATCCATCGATCTAGACAGACTCAATGTAGACGTGCTGTC | ||||
| YALI0C08943 | Q6CCJ1 | BLAST | EY210 | XmaI | TATTCCCCCGGGATGGCCATTATTGTGGCTG | |
| EY211 | ClaI | CGGTATCCATCGATCTAATCCGAATCAAATCCAGAAT | ||||
| C. neoformans | CNBC3990 | Q55VT8 | BLAST | Introns, Synthesized by Blue Heron Biotechnology | ||
| S. cerevisiae | GAL2 | P13181 | 3, 38 | EY220 | EcoRI | TATTCCGAATTCATGGCAGTTGAGGAGAA |
| EY221 | ClaI | CGGTATCCATCGATTTATTCTAGCATGGCCTTG | ||||
| HXT4 | P32466 | 15, 36 | EY194 | XmaI | TATTCCCCCGGGATGAATTCAACTCCAGATTTAATATCTCC | |
| EY208 | ClaI | CGGTATCCATCGATTTATTTCTTGCCGAACATTTTCTT | ||||
| HXT7 | P39004 | 15, 36 | EY159 | BamHI-HF | GGCGCGGATCCATGTCACAAGACGCTGCTATTGC | |
| EY180 | ClaI | CCGTATCCATCGATTTATTTGGTGCTGAACATTCTCTTGTACAATGG | ||||
| HXT13 | P39924 | 15, 36 | EY196 | XmaI | TATTCCCCCGGGATGTCTAGTGCGCAATCCTC | |
| EY209 | ClaI | CGGTATCCATCGATTCAATCAGAATTCTTTGAGAACTTC |
Many genes were cloned using PCR and restriction enzyme (RE) cloning. The complete list is given here, including the restriction enzymes and primers used.
Cloning heterologous genes.
PCR protocols utilizing Phusion DNA polymerase (Thermo Fisher Scientific, Waltham, MA) were performed using standard protocols. The PCR primers used to amplify all genes as well as the restriction enzymes used in this study are listed in Table 3. Standard restriction enzyme cloning and bacterial transformations were performed according to the work by Sambrook and Russell (36). Yeast transformations were conducted according to the protocol described by Gietz and Schiestl (10). Genomic DNA was isolated from C. intermedia, D. hansenii, E. coli, S. stipitis, S. cerevisiae, and Y. lipolytica using the Wizard genomic DNA isolation kit (Promega, Madison, WI). Arabidopsis thaliana cDNA originally isolated from the CD4-30 library was provided as a gift from Alan Lloyd at The University of Texas at Austin. Since both S. stipitis XUT4 and C. neoformans CNBC3990 possessed one or more introns, the ORFs were synthesized as an intron-free gene by Blue Heron Biotechnology (Bothell, WA). The native ORF was used; no yeast codon optimization was selected. All heterologous ORFs were cloned into the multiple cloning site of the Mumberg et al. plasmid p414-TEF (Table 2) and then sequence verified to ensure correct cloning. To create a xylose utilization pathway, S. stipitis XYL1 was cloned into p416-TEF and S. stipitis XYL2 was cloned into p425-GPD. All cloned genes were sequence confirmed after cloning and prior to yeast transformations. All primers were purchased from Integrated DNA Technologies (Coralville, IA), restriction enzymes were purchased from New England Biolabs (Ipswich, MA), and the remaining chemicals were purchased through Thermo Fisher Scientific (Waltham, MA).
Growth rate measurements.
Growth rates of the transformed yeast strains were measured with a Bioscreen C system (Growth Curves USA, Piscataway, NJ) with biological triplicates using the wide-band filter (420 to 580 nm) recommended for optical density (OD) measurements. A 1-μl inoculum of fully grown culture was added to each well with 250 μl of YSC medium plus 20 g/liter of the carbon source under investigation (glucose, xylose, galactose, fructose, mannose, or ribose) as well as an additional condition that contained only 5 g/liter xylose. The experiment was run for 48 h with high continuous shaking and sampling every 10 min. Exponential growth rates were calculated using a solution algorithm written in MATLAB. Strains with average growth rates that were below that of the empty vector control are reported as bc, rates statistically equal to that of the empty vector control are labeled ec, and strains that demonstrated growth rates higher than that of the control but did not double in optical density in 48 h are reported as dnd. All other values are reported as calculated, and the growth rates that were most significant (with a Z test value that rounded to 1) were considered top performers.
Competitive fermentation assays.
Glucose-and-xylose mixed-sugar fermentations were used to assay sugar preference by these transporters. These glucose-xylose cofermentations were performed at a high yeast optical density in biological duplicates. A 2-day preculture was used to inoculate 250 ml of YSC plus 20 g/liter maltose. Once this culture exceeded an optical density (OD) of 4 (∼24 h), the cells were pelleted, resuspended, and inoculated into 40 ml YSC plus 20 g/liter glucose and 20 g/liter xylose at an initial approximate OD of 20. Sample time points were taken at 0, 1, 3, 4, 6, 8, 12, and 24 h. At each time point, a 500-μl sample was taken and pelleted for 2 min at full speed in a microcentrifuge. The supernatant was diluted 1:10, and the glucose and xylose concentrations were measured using the YSI Life Sciences bioanalyzer 7100 MBS. Initial glucose and xylose consumption rates were calculated based on the change in concentration of the sugars over time. The linear range of the glucose consumption curve was found, and the slope was calculated. Over the same time interval, a xylose consumption slope was also calculated. The ratio of xylose consumption rate to glucose consumption rate was then computed and converted to a mole ratio to give the X/G preference ratio. High values of the preference ratio would be interpreted as a xylose selective transporter, although no ORFs in this study exhibited such behavior. In addition to the preference ratio, the percentage of sugar transported was computed by subtracting the total sugar remaining after 24 h from the initial total amount of sugars and dividing that by the initial total amount of sugars. This metric provided an assessment of overall throughput of the transporter under study.
RESULTS
Cloning and selection of known and putative xylose transporters.
A total of 26 transporters were evaluated in this study, consisting of 23 heterologous and 3 native S. cerevisiae transporter proteins, which comprise the largest survey of transporter ORFs for xylose transport function to date (Table 3). These transporters were selected based on homology searches and literature evidence of xylose transport capacity, as described above in Materials and Methods. This collection of transporters encompasses a wide array of phylogenetic diversity (Fig. 1). Once cloned into the shuttle vector p414-TEF, the transporters were transformed into a hexose transporter-null strain of yeast (EBY.VW4000) complemented with a S. stipitis-based oxidoreductase pathway (the genes XYL1 and XYL2 are expressed by plasmids). This strain, named EY12, was used throughout this study. This host background is unable to support substantial growth on glucose or xylose due to the transporter deletions, validated by our growth rate assays in which the strain was unable to double in OD in the 48-h duration of the experiment. Similar growth kinetics (lack of doubling within 100 h or more) have been observed for similarly constructed yeast strains (14). S. cerevisiae EY12 thus provides a suitable host for comparing specific functions of transporter proteins.
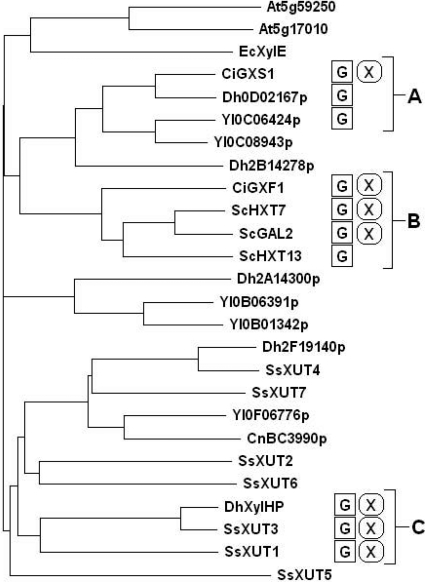
Fig. 1.
Phylogenetic analysis of transporter proteins used in this study. Cladogram of protein sequences included in this study assembled by ClustalW and visualized in TreeView X. The functional ORFs show a great deal of clustering. Group A consists of ORFs with low growth rates, and only C. intermedia GXS1 confers growth on xylose. Group B consists of nonspecific but efficient transporters. Members of group C confer xylose growth and exhibit moderate to high xylose-to-glucose preference ratios yet still prefer glucose. Prefixes: At, A. thaliana; Ec, E. coli, Ci, C. intermedia; Dh, D. hansenii; Yl, Y. lipolytica; Sc, S. cerevisiae; Ss, S. stipitis; Cn, C. neoformans. X, xylose; G, glucose.
Growth phenotypes of recombinant strains using different carbon sources.
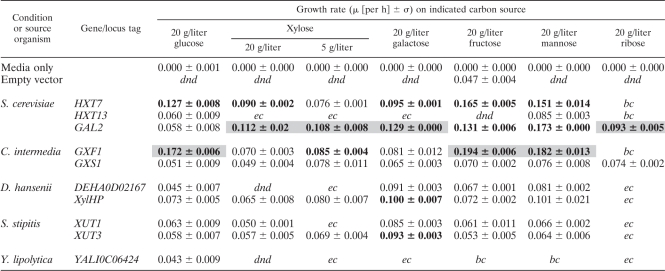
In order to characterize sugar utilization profiles, the growth rates of S. cerevisiae EY12 strains harboring these 26 transporters were measured using glucose, xylose, low xylose (5 g/liter), galactose, fructose, mannose, or ribose as the sole carbon source in minimal media. With the exception of fructose, the host strain is unable to support growth under experimental conditions on any of these sugars without the aid of a heterologous transporter protein. Fructose supports low growth rates in the control strain (an OD doubling occurred within 48 h). Of the 26 transporters evaluated, 10 ORFs conferred statistically significant growth phenotypes on one or more of these tested carbon sources. Table 4 depicts the carbon source-associated growth rates for these 10 transporters compared with that for an empty vector control. The remaining 16 transporters did not enable growth at a rate that was significantly above the control (denoted bc or ec) or did not double in optical density in 48 h (dnd) (Table 5). It is important to note that the EY12 strain used to express these transporters is a highly modified strain of yeast (with over 20 gene knockouts and three plasmids) and thus is not expected to exhibit absolute growth rates that match those of industrial or unmodified strains of yeast. However, the qualitative trends and rank order analysis presented here provide a means of comparing the performance of isolated heterologous transporters.
Table 4.
Growth rates conferred by transporter expression in S. cerevisiae EY12a
Here the growth rate (μ) is reported for the 10 genes that conferred growth on any of the carbon sources assayed. Standard deviations are reported, and the genes with significant Z test scores for each carbon source are shown in boldface. The top performer in each category is shaded. Generally, the transporters assayed show a trade-off between efficiency and specificity. There are three abbreviations used to indicate a slow-growth phenotype. The measured growth rate was either below that of the control (bc), indicating detrimental effects on the host, equal to that of the control (ec), showing performance statistically similar to that of the empty vector control, or did not produce a doubling in optical density in 48 h (dnd).
Table 5.
ORFs that did not confer growth in S. cerevisiae EY12a
| Organism | Gene/locus tag/promoter |
|---|---|
| A. thaliana | At5g59250 |
| At5g17010 | |
| C. neoformans | CNBC3990 |
| D. hansenii | DEHA2A14300p |
| DEHA2B14278p | |
| DEHA2F19140p | |
| E. coli | xylE |
| S. stipitis | XUT2 |
| XUT4 | |
| XUT5 | |
| XUT6 | |
| XUT7 | |
| Y. lipolytica | YALI0B06391p |
| YALI0B01342p | |
| YALI0C08943p | |
| YALI0F06776p |
This list of 16 transporters exhibited ec, bc, or dnd growth phenotypes for all carbon sources used in this study.
This growth rate assay enables several observations about the behavior of molecular transport proteins. First, our results generally show both broad and narrow specificities for the carbon sources assayed. Of the 10 ORFs conferring growth phenotypes in S. cerevisiae EY12, all of them restored growth on glucose, whereas only 7 also conferred growth on xylose. These 7 transporters all conferred growth on the other hexoses assayed. The other 3 transporters showed various specificities for hexoses. Y. lipolytica YALI0C06424 conferred growth on glucose only, S. cerevisiae HXT13 conferred growth on glucose and mannose, and D. hansenii DEHA0D02167 was specific for hexoses. Growth on ribose was conferred only by S. cerevisiae GAL2 and C. intermedia GXS1, making this phenotype the rarest among the ORFs assayed. No strain assayed was found to grow solely on xylose; only strains that grew solely on glucose were observed.
Second, these data show a clear relationship between substrate specificity and transporter efficiency. Highly specific transporters conferred the lowest growth rates for those carbon sources on which they grew. S. cerevisiae GAL2, C. intermedia GXF1, and S. cerevisiae HXT7 were the least specific transporters and consistently conferred the highest growth rates. An exception to this trend is the broad transporter C. intermedia GXS1, which conferred modest to low growth on most carbon sources. This phenomenon may be due to the H+ symport mechanism of Gxs1p (28), which would reduce its efficiency compared to facilitated diffusion transport proteins when protons are scarce, unlike with xylose. However, C. intermedia GXS1 expression provided a 60% increase in the conferred growth rate when grown on 5 g/liter xylose compared to that when grown on 20 g/liter xylose. This improvement is a nearly 3-fold percent increase compared to those of C. intermedia GXF1, D. hansenii xylHP, and S. stipitis XUT3. Again, this improvement may be due to the proton symport mechanism of Gxs1p. In contrast, the native S. cerevisiae proteins all demonstrated decreased growth rates in reduced xylose concentrations.
Third, this study enables rank order analysis of the magnitude of growth rates for a given carbon source but not across carbon sources due to factors such as differing metabolic pathways. C. intermedia GXF1 conferred the highest growth rate on glucose, fructose, and mannose in addition to its high growth rate on 5 g/liter xylose. S. cerevisiae GAL2 was the most efficient transporter for xylose, galactose, and ribose in addition to high growth rates conferred when grown on fructose and mannose. Including the results obtained by this study, S. cerevisiae GAL2 has now been found to facilitate the transport of the following three different pentoses: xylose (14) (this study), ribose (this study), and l-arabinose (3). S. cerevisiae HXT7 also conferred high growth rates on all carbon sources except ribose and 5 g/liter xylose. D. hansenii xylHP and S. stipitis XUT3 expression enabled high growth rates on galactose.
Fourth, these experiments demonstrate that a high fraction of proteins putatively annotated as xylose transporters do not confer growth on any monosaccharide carbon source when heterologously expressed in recombinant S. cerevisiae. These 16 transporters are listed in Table 5. Most of the S. stipitis XUT family and E. coli and A. thaliana ORFs are included. The results for the plant and bacterial genes support the findings of Hamacher et al. (14). As for the S. stipitis transporters, it is not clear from our study why so many of them conferred no significant growth phenotype.
Future work is required to determine whether these proteins are not actually transporters, may transport other carbon sources not tested, or are not suited for heterologous expression in recombinant S. cerevisiae due to folding or membrane compatibility differences. There is also the possibility that one or more of these proteins acts as a membrane sensor protein that does not transfer substrates across the cellular membrane, like the RGT2 glucose sensor (7). Nevertheless, no observations of significant growth rates for a majority of the transporters assayed indicates a high failure rate for BLAST-identified ORFs. Also, the low number of ORFs able to confer growth on xylose may indicate that specific xylose transporters are structurally different from the reference genes used for the BLAST algorithm or are inefficient when expressed individually.
Collectively, these results upgrade our understanding of monosaccharide transport in several organisms. To our knowledge, this study presents the first experimental characterization of individual monosaccharide transporters from the yeasts D. hansenii and Y. lipolytica and of the S. stipitis XUT family. Within these organisms, it is evident that S. stipitis XUT1 and XUT3 and D. hansenii xylHP enable the transport of hexoses and xylose, while D. hansenii DEHA0D02167 and Y. lipolytica YALI0C06424 appear to be hexose transporters. These results also present the first characterization of S. cerevisiae HXT7, HXT13, and GAL2 and C. intermedia GXF1 and GXS1 on hexose and pentose sugars other than glucose and xylose.
Competitive preference of xylose in the presence of glucose.
Most industrial applications with engineered yeasts will be in an environment where multiple sugars are present. Thus, it is also important to evaluate the relative preference exhibited by transporter proteins for sugars as a means of qualitatively assessing Michaelis constant (Km) values for these transporters. It is inadvisable to compare growth rates across carbon sources in Table 4 to understand preference, since competitive inhibition may interfere with the ability to consume the sugar at the rates occurring in the sole carbon source growth assay. Thus, to address this issue, a competitive fermentation assay was conducted to test substrate preference in a more direct manner. This glucose-xylose cofermentation experiment was conducted with a high-cell-density culture of yeast cells with 20 g/liter glucose and 20 g/liter xylose present at the same time. The extracellular concentration of these sugars was monitored over time, and the consumption rate of each sugar was measured. The ratio of these consumption rates was computed to give the xylose to glucose preference ratio (termed the X/G preference ratio). These preference ratios are provided in Table 6. All of the transporters tested had low X/G preference ratios; the top performer imported only one molecule of xylose per two molecules of glucose. Thus, this experiment demonstrates that these transporter proteins possess a stronger preference for glucose over xylose. In addition, the percentage of total sugar consumed was calculated by determining the total amount of the 40 g/liter of sugars that had been consumed after 24 h of culturing and dividing that by 40 g/liter of sugars (Table 6). The percent total sugar consumed is a facile measure of collective transporter efficiency.
Table 6.
Xylose-to-glucose preference ratios and percent total sugar consumed during the competitive preference assaya
| Organism | Gene | X/G preference ratio | % sugars consumed |
|---|---|---|---|
| S. cerevisiae | HXT7 | 0.11 | 75 |
| GAL2 | 0.09 | 68 | |
| C. intermedia | GXF1 | 0.10 | 78 |
| GXS1 | 0.51 | 14 | |
| D. hansenii | xylHP | 0.17 | 58 |
| S. stipitis | XUT1 | 0.69 | 16 |
| XUT3 | 0.11 | 17 |
A cofermentation assay was conducted to determine transporter preference. The X/G preference ratio is calculated by first measuring the molar consumption rates of each sugar over the same time period from a stationary-phase yeast culture and then dividing the xylose molar consumption rate by the glucose molar consumption rate. This metric approximates how many xylose molecules per glucose molecule are imported. As a measure of transporter throughput, the total sugar consumption over 24 h was measured and taken as a mass percentage of total sugars. Several cultures exhibit diauxic sugar consumption, in which glucose is consumed first, resulting in low X/G preference ratios and high percent sugars consumed.
These results demonstrate that glucose preference and transport efficiency are linked. In particular, the most efficient transporters typically exhibited the lowest preference ratios, whereas the opposite was true for the least efficient transporters. S. stipitis XUT1 demonstrated the highest preference for xylose over glucose among the ORFs studied. However, this transporter was capable of importing only 16% of the sugars present in 24 h. The transporter with the second highest preference ratio, C. intermedia GXS1, also transported only 14% of total sugars. Moreover, the native transporters S. cerevisiae GAL2 and S. cerevisiae HXT7 exhibit a preference ratio below 0.1, indicating that these transporters mediate glucose uptake almost to the exclusion of xylose. Nevertheless, these transporters are quite efficient at total utilization, as over 65% of the sugars present were transported in 24 h. Therefore, the expression of these transporters creates a strong diauxic growth phenotype. Likewise, C. intermedia GXF1 and D. hansenii xylHP had low preference ratios (0.10 and 0.17, respectively) yet consumed 78% and 58% of the total sugars, respectively. Finally, S. stipitis XUT3 had low performance with both metrics. In all of these cases, glucose preference is directly linked with transport efficiency.
DISCUSSION
This study provides the first large-scale analysis of substrate specificity and preference across a varied group of putative and known transporter proteins. By studying 26 transporters from seven different organisms, we evaluated a diverse collection of transporters and identified 10 that function as monosaccharide transporters in the absence of the native hexose transport system of S. cerevisiae. Our results reveal that efficiency trends inversely with specificity and directly with glucose preference. These results are expected when considering the diauxic growth phenomena observed for a large number of microbes, including S. stipitis (30), and that broad substrate efficiency is evolutionarily advantageous, since naturally available carbon sources are somewhat unpredictable. Therefore, expression of an efficient, broad transporter allows a great deal of flexibility without requiring the induction and expression of new transporters for different carbon sources.
The most specific transporters in this study (S. cerevisiae HXT13 and Y. lipolytica YALI0C06424) exhibited low overall efficiency. Therefore, it is presumed that loss of transporter efficiency is the fitness trade-off associated with improved specificity. This trade-off does not bode well for discovering an efficient, xylose-specific yeast transporter in organisms that have evolutionarily been exposed to a broad range of carbon sources. While xylose-preferring transporters may exist in yeast, they are likely to exhibit low net transport rates and efficiencies based on the evidence described here. Exclusive xylose transporters are also likely be rare, since glucose exclusivity is shown to be rare in this study. Yet, it is known that the E. coli xylE transporter functions as an exclusive xylose transporter (4), so these transporters exist in bacteria. The unsuccessful expression of xylE in S. cerevisiae (14) (this study) could be due to membrane incompatibility, expression, and folding difficulties experienced with bacterial proteins. A similar problem exists when the bacterial xylose isomerase pathway is imported (31), which suggests a protein engineering approach could enable the use of this transporter in yeast.
Collectively, this study provides a sugar preference profiling of native and heterologous transporters. Sixteen of these proteins conferred no growth on any of the carbon sources assayed. Host incompatibility may partially explain these observations, although there are many possible explanations. Nevertheless, there are few explanations for observed functionality other than that the transporter is expressed and naturally accepts the specific carbon source. Therefore, this study reconfirms that S. cerevisiae HXT7 transports glucose and xylose (14, 35, 37) and provides evidence that the transporter accepts additional hexoses. S. cerevisiae HXT13 is reconfirmed as being able to transport glucose yet not xylose (14), and evidence here demonstrates an affinity for mannose. This study also supports the previous research involving S. cerevisiae GAL2 (14, 37) and, further, finds that the transporter accepts all carbon sources studied, in addition to arabinose (3). S. cerevisiae GAL2 thus encodes a very broad transporter.
Of the heterologous transport proteins, the two C. intermedia transporters are probably the most extensively studied (28, 29, 34). Previous reports focus on only the glucose and xylose transport capabilities of these proteins. The findings shown here indicate that these ORFs encode broad hexose transporters that have additional affinity for pentoses—xylose for GXF1 and both xylose and ribose for GXS1. This study also shows that D. hansenii xylHP, previously indicated as a potential candidate for heterologous expression (27), is also a hexose transporter with affinity for xylose. The only other functional ORF from D. hansenii, DEHA0D02167, appears to encode a hexose transporter only. While previous studies were unable to identify functional transporters from S. stipitis (14), this study reports that S. stipitis XUT1 and XUT3 encode hexose transporters with affinity for xylose. Previously uninvestigated Y. lipolytica YALI0C06424 is an exclusive glucose transporter; therefore, we propose that the annotation of this ORF be reclassified as a glucose transporter and be renamed as a HXT protein. Most of these characterizations demonstrate previously unknown substrates for these transporter proteins. This includes the first experimental validation for the XUT family. All other ORFs assayed have unknown functions, as they did not support growth on any of the carbon sources used in this study.
Of all the carbon sources assayed, ribose was seen as a substrate for only two proteins, S. cerevisiae GAL2 and C. intermedia GXS1. It remains to be explained why these hexose transporters should have such affinity for pentoses. Ribose transport activity may be a concomitant result of other adaptations rather than an advantage, since free ribose in nature is rare. Also, the rarity of ribose acceptance implies that certain limits to broad substrate specificity exist. The permeases investigated are not merely open pores but engage in some type of protein-mediated substrate recognition. The ubiquity of glucose transport among these proteins illustrates a potential evolutionary path taken among these proteins. Moreover, these findings agree with what is already known about the metabolism of most microorganisms—hexoses are preferred carbon sources. These data also provide evidence that ribose transport across the cell membrane is almost unnecessary for most organisms or is carried out by a different class of proteins.
Our results also show that in a cofermentation, glucose transport is preferred over xylose; all ORFs investigated imported, at the most, one molecule of xylose for every two of glucose. While this phenomenon was observed only within the limited search space of this study, it may be the result of natural selection to produce sugar transporters that prefer hexoses. Even so, it remains possible that xylose fermenters such as S. stipitis and D. hansenii have dedicated xylose transport systems similar in scope to the hexose transport systems of S. cerevisiae based on their respective whole-cell transport phenotypes (11, 33). However, even S. stipitis demonstrates a diauxic growth phenotype in glucose-xylose cofermentation (11). Therefore, if xylose-preferring transport systems do exist in organisms such as S. stipitis, they are likely to have very low throughput and be quite distinct in sequence space compared to those of the glucose transporters.
As a final analysis to link protein sequences and observed phenotype, a phylogenetic tree was created based on the multiple protein sequence alignment of these 26 transporters generated by ClustalW (Fig. 1). As is evident from this figure, all of the transporters that confer growth on xylose cluster into three distinct functional groups. The first group (denoted group A on the tree) is characterized by the permissive proton symporter C. intermedia GXS1, which also demonstrates one of the highest xylose-to-glucose preference ratios. However, the other members of this group are specific for hexoses. Group B is composed of C. intermedia GXF1 and several native S. cerevisiae transporters, which have very efficient transport characteristics yet very low preference ratios. Group C is composed of transporters from S. stipitis and D. hansenii xylHP, which all demonstrate moderate efficiencies and high preference ratios. Therefore, group C presents favorable candidates for further bioprospecting.
In conclusion, this survey revealed a subset of heterologous transporters that, when expressed in a hexose transporter-null strain of S. cerevisiae, permit growth on glucose and on xylose as well as on other hexose and pentose sugars. These transporters cluster into three well-defined groups, one of which is worthy of further investigation for lignocellulosic biomass fermentation. This study presents the first molecular characterization of ORFs from several organisms of industrial interest across multiple carbon sources. As a result, novel ORFs from the yeasts S. stipitis and D. hansenii that are able to confer growth on xylose in recombinant S. cerevisiae were identified. In addition, we have demonstrated that transporters in nature exhibit a trade-off between specificity and efficiency. As a result, the solution to xylose transport limitations in yeast may require the development of new genetic approaches.
ACKNOWLEDGMENTS
This work was supported by the Camille and Henry Dreyfus New Faculty Award and the National Science Foundation Graduate Research Fellowship Program.
We recognize the generosity of Eckhard Boles for the contribution of S. cerevisiae EBY.VW4000. We also acknowledge the technical contributions of Erik Quandt, Jared Ellefson, John Blazeck, and Amanda Lanza to this work.
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Alper H., Stephanopoulos G. 2009. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7:715–723 [DOI] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Becker J., Boles E. 2003. A modified Saccharomyces cerevisiae strain that consumes l-arabinose and produces ethanol. Appl. Environ. Microbiol. 69:4144–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis E. O., Henderson P. J. F. 1987. The cloning and DNA-sequence of the gene xylE for xylose-proton symport in Escherichia coli K-12. J. Biol. Chem. 262:13928–13932 [PubMed] [Google Scholar]
- 5. Eliasson A., Christensson C., Wahlbom C. F., Hahn-Hagerdal B. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farrell A. E., et al. 2006. Ethanol can contribute to energy and environmental goals. Science 311:506–508 [DOI] [PubMed] [Google Scholar]
- 7. Forsberg H., Ljungdahl P. O. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91–109 [DOI] [PubMed] [Google Scholar]
- 8. Gardonyi M., Jeppsson M., Liden G., Gorwa-Grausland M. F., Hahn-Hagerdal B. 2003. Control of xylose consumption by xylose transport in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 82:818–824 [DOI] [PubMed] [Google Scholar]
- 9. Gardonyi M., Osterberg M., Rodrigues C., Spencer-Martins I., Hahn-Hagerdal B. 2003. High capacity xylose transport in Candida intermedia PYCC 4715. FEMS Yeast Res. 3:45–52 [DOI] [PubMed] [Google Scholar]
- 10. Gietz R. D., Schiestl R. H. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2:31–34 [DOI] [PubMed] [Google Scholar]
- 11. Grootjen D. R. J., Vanderlans R., Luyben K. 1991. Conversion of glucose-xylose mixtures by Pichia stipitis under oxygen-limited conditions. Enzyme Microb. Technol. 13:648–654 [Google Scholar]
- 12. Hahn-Hagerdal B., Karhumaa K., Jeppsson M., Gorwa-Grauslund M. F. 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:147–177 [DOI] [PubMed] [Google Scholar]
- 13. Hahn-Hagerdal B., Linden T., Senac T., Skoog K. 1991. Ethanolic fermentation of pentoses in lignocellulose hydrolysates. Appl. Biochem. Biotechnol. 28–29:131–144 [DOI] [PubMed] [Google Scholar]
- 14. Hamacher T., Becker J., Gardonyi M., Hahn-Hagerdal B., Boles E. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783–2788 [DOI] [PubMed] [Google Scholar]
- 15. Hector R. E., Qureshi N., Hughes S. R., Cotta M. A. 2008. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl. Microbiol. Biotechnol. 80:675–684 [DOI] [PubMed] [Google Scholar]
- 16. Ho N. W., Chen Z., Brainard A. P. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeffries T. W. 2006. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 17:320–326 [DOI] [PubMed] [Google Scholar]
- 18. Jeffries T. W., Jin Y. S. 2004. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 63:495–509 [DOI] [PubMed] [Google Scholar]
- 19. Jin Y. S., Alper H., Yang Y. T., Stephanopoulos G. 2005. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl. Environ. Microbiol. 71:8249–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karhumaa K., Fromanger R., Hahn-Hagerdal B., Gorwa-Grauslund M. F. 2007. High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 73:1039–1046 [DOI] [PubMed] [Google Scholar]
- 21. Karhumaa K., Garcia Sanchez R., Hahn-Hagerdal B., Gorwa-Grauslund M. F. 2007. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katahira S., et al. 2008. Improvement of ethanol productivity during xylose and glucose co-fermentation by xylose-assimilating S. cerevisiae via expression of glucose transporter SUT1. Enzyme Microb. Technol. 43:115–119 [Google Scholar]
- 23. Kotter P., Amore R., Hollenberg C. P., Ciriacy M. 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr. Genet. 18:493–500 [DOI] [PubMed] [Google Scholar]
- 24. Kurtzman C. P., Suzuki M. 2010. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q.-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience 51:2–14 [Google Scholar]
- 25. Kuyper M., et al. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5:399–409 [DOI] [PubMed] [Google Scholar]
- 26. Kuyper M., et al. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5:925–934 [DOI] [PubMed] [Google Scholar]
- 27. Leandro M. J., Fonseca C., Goncalves P. 2009. Hexose and pentose transport in ascomycetous yeasts: an overview. FEMS Yeast Res. 9:511–525 [DOI] [PubMed] [Google Scholar]
- 28. Leandro M. J., Goncalves P., Spencer-Martins I. 2006. Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H+ symporter. Biochem. J. 395:543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leandro M. J., Spencer-Martins I., Goncalves P. 2008. The expression in Saccharomyces cerevisiae of a glucose/xylose symporter from Candida intermedia is affected by the presence of a glucose/xylose facilitator. Microbiology 154:1646–1655 [DOI] [PubMed] [Google Scholar]
- 30. Ligthelm M. E., Prior B. A., Dupreez J. C., Brandt V. 1988. An investigation of D-(1-C-13) xylose metabolism in Pichia stipitis under aerobic and anaerobic conditions. Appl. Microbiol. Biotechnol. 28:293–296 [Google Scholar]
- 31. Madhavan A., et al. 2009. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl. Microbiol. Biotechnol. 82:1067–1078 [DOI] [PubMed] [Google Scholar]
- 32. Mumberg D., Muller R., Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122 [DOI] [PubMed] [Google Scholar]
- 33. Nobre A., Lucas C., Leao C. 1999. Transport and utilization of hexoses and pentoses in the halotolerant yeast Debaryomyces hansenii. Appl. Environ. Microbiol. 65:3594–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Runquist D., Fonseca C., Radstrom P., Spencer-Martins I., Hahn-Hagerdal B. 2009. Expression of the Gxf1 transporter from Candida intermedia improves fermentation performance in recombinant xylose-utilizing Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 82:123–130 [DOI] [PubMed] [Google Scholar]
- 35. Saloheimo A., et al. 2007. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl. Microbiol. Biotechnol. 74:1041–1052 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Sedlak M., Ho N. W. Y. 2004. Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast 21:671–684 [DOI] [PubMed] [Google Scholar]
- 38. Walfridsson M., Hallborn J., Penttila M., Keranen S., Hahnhagerdal B. 1995. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose-phosphate pathway enzymes transketolase and transaldolase Appl. Environ. Microbiol. 61:4184–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weierstall T., Hollenberg C. P., Boles E. 1999. Cloning and characterization of three genes (SUT1-3) encoding glucose transporters of the yeast Pichia stipitis. Mol. Microbiol. 31:871–883 [DOI] [PubMed] [Google Scholar]
- 40. Wieczorke R., et al. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123–128 [DOI] [PubMed] [Google Scholar]
- 41. Wiedemann B., Boles E. 2008. Codon-optimized bacterial genes improve l-arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Young E., Lee S. M., Alper H. 2010. Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol. Biofuels 3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]