Abstract
Cyclopropane fatty acids (CFAs) are synthetized in situ by the transfer of a methylene group from S-adenosyl-l-methionine to a double bond of unsaturated fatty acid chains of membrane phospholipids. This conversion, catalyzed by the Cfa synthase enzyme, occurs in many bacteria and is recognized to play a key role in the adaptation of bacteria in response to a drastic perturbation of the environment. The role of CFAs in the acid tolerance response was investigated in the lactic acid bacterium Lactococcus lactis MG1363. A mutant of the cfa gene was constructed by allelic exchange. The cfa gene encoding the Cfa synthase was cloned and introduced into the mutant to obtain the complemented strain for homologous system studies. Data obtained by gas chromatography (GC) and GC-mass spectrometry (GC-MS) validated that the mutant could not produce CFA. The CFA levels in both the wild-type and complemented strains increased upon their entry to stationary phase, especially with acid-adapted cells or, more surprisingly, with ethanol-adapted cells. The results obtained by performing quantitative reverse transcription-PCR (qRT-PCR) experiments showed that transcription of the cfa gene was highly induced by acidity (by 10-fold with cells grown at pH 5.0) and by ethanol (by 9-fold with cells grown with 6% ethanol) in comparison with that in stationary phase. Cell viability experiments were performed after an acidic shock on the mutant strain, the wild-type strain, and the complemented strain, as a control. The higher viability level of the acid-adapted cells of the three strains after 3 h of shock proved that the cyclopropanation of unsaturated fatty acids is not essential for L. lactis subsp. cremoris survival under acidic conditions. Moreover, fluorescence anisotropy data showed that CFA itself could not maintain the membrane fluidity level, particularly with ethanol-grown cells.
INTRODUCTION
Cyclopropane fatty acids (CFAs) are synthetized in situ by the transfer of a methylene group from S-adenosyl-l-methionine to a double bond of unsaturated fatty acid (UFA) chains of membrane phospholipids. This conversion, catalyzed by the Cfa synthase enzyme, occurs in many Gram-negative (4, 9, 26, 32) and Gram-positive (22, 24, 42–44, 47) bacteria. It is recognized nowadays that this energetically expensive conversion (CFA synthesis requires three ATP molecules per cyclopropane ring formed [46]) plays a major role in the adaptation of bacteria in response to a drastic perturbation of the environment. In Escherichia coli, a correlation between the level of CFAs in membrane and the resistance of the bacterium to a lethal pH was reported (6). The strategies, including the construction and the physiological study of cfa-deficient cells in E. coli (9) or Salmonella enterica serovar Typhimurium (33), likewise demonstrated the high sensitivity of the deficient cells to acid stress. An overexpression of the cfa gene in Clostridium acetobutylicum increased the CFA content of cells as well as acid and butanol resistance (47).
When focusing on stress tolerance of lactic acid bacteria, the cyclopropane fatty acids are regarded as key fatty acids. Lactic acid bacteria often used as starters in food fermentation (including probiotics) are confronted with various kinds of environmental perturbations, such as starvation and osmotic, thermal, acid, and ethanol stresses, from their production in industry to their implementation. It has been reported that the cyclopropanation of the monounsaturated oleic acid increased after acid and cold stresses in Lactobacillus helveticus (37). In the lactic acid bacterium of wine, Oenococcus oeni, cells respond to culture in the presence of ethanol by increasing their CFA content (44). The data obtained by Grandvalet et al. (22) demonstrated that O. oeni acid-grown cells or cells harvested in stationary growth phase increased their CFA content at the expense of oleic acid, similar to that increased by ethanol-grown cells. In the case of Lactococcus lactis subsp. cremoris, the predominance of CFAs over other unsaturated and saturated fatty acids is observed in cells submitted to osmotic stress or high temperature (5, 24). The proteomic strategy developed to characterize the acid tolerance response in L. lactis subsp. cremoris underlines that Cfa synthase can be considered not only a key effector of the lactococcal acid tolerance but also a general stress protein (8).
It is unanimously agreed that the extent of the cyclopropanation of the monounsaturated fatty acids represents one of the major adaptive responses of the bacterial cells in order to stabilize the membrane fluidity known as “homeoviscous adaptation” (42). However, the role of CFAs in membrane fluidity adjustments remains unclear. According to the hypothesis of Härtig et al. (25), the presence of CFAs could make the membrane more rigid because of their higher lipid melting points and their poorer ability to pack into the acyl chain array of the phospholipid bilayer in comparison those of with unsaturated fatty acids (23). But contrary effects were obtained for measurements of membrane physical changes due to cyclopropane formation. For example, a cyclopropane fatty acid-deficient strain of Pseudomonas putida and the wild-type strain could not be differentiated by the fluidity levels of their membranes (38). However, the results obtained from lipid extracts from the same bacterium by using two independent methods (fluorescence polarization and Fourier transform infrared spectroscopy [FTIR] measurement) indicate that CFAs definitely render lipid bilayers more rigid (36).
The purpose of this study was to investigate the role of CFAs in the stress response of the lactic acid bacterium Lactococcus lactis subsp. cremoris by physiological studies of a cfa-deficient mutant. Strains were exposed to acid and ethanol sublethal stresses which are associated with cyclopropanation of membrane fatty acids in lactic acid bacteria. The impact of the mutation on the survival of strains under acidic conditions was discussed as the relationship of membrane fluidity variations and changes with membrane fatty acid components.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and their phenotypes are listed in Table 1. Lactococcus lactis subsp. cremoris MG1363 was grown at 30°C in M17/7 medium (7). M17/7 medium corresponded to M17 containing MOPS (3-N-morpholino propane sulfonic acid) at 200 mmol/liter rather than β-disodium glycerophosphate and supplemented with glucose (5 g/liter). The pH was adjusted to 7.2 with NaOH. M17/5 and M17/3 media corresponded to M17/7 acidified with HCl to pH 5 or pH 3, respectively. For M17/7 medium plus 6% (vol/vol) ethanol, absolute ethanol was added to the medium.
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Genotype/marker/descriptiona | Reference/source |
|---|---|---|
| L. lactis subsp. cremoris | ||
| MG1363 | Plasmid-free derivative of L. lactis NCDO 712 cured of ΦT712 prophage | 20 |
| MG1363 Δcfa | cfa mutant derivative of MG1363 | This study |
| MG1363 pDL278 | MG1363 harboring pDL278, Spr | This study |
| MG1363 Δcfa pDL278 | MG1363 Δcfa harboring pDL278, Spr | This study |
| MG1363 Δcfa pDLcfa12A3 | MG1363 Δcfa harboring pDLcfa12A3, Spr | This study |
| E. coli | ||
| K-12 and ER2738 | General cloning strains | Invitrogen |
| EC101 | E. coli JM101 with repA from pWV01 integrated into the chromosome; host strain used for construction of pGID023-derivative plasmids | 35 |
| Plasmids | ||
| pDL278 | E. coli-L. lactis shuttle vector (6.6 kbp), Spr | 11 |
| pDLcfa12A3 | pDL278-derivative vector used to express the cfa gene under an endogenous promoter, Spr | This study |
| pGID023 | Shuttle vector (8.2 kbp) for E. coli and L. lactis; derivative of pJDC9 containing the pE194 replication functions (thermosensitive ori); used as an unstable integration vector, Emr | 28 |
| pTMH1 | Integration plasmid and pGID023 derivative containing the cfa gene carrying a 28-bp internal deletion (nucleotides 514 to 541 of the coding sequence), Emr | This study |
Emr, erythromycin resistance (10 μg ml−1 for L. lactis subsp. cremoris; 250 μg ml−1 for E. coli); Spr, spectinomycin resistance (1,000 μg ml−1 for L. lactis subsp. cremoris; 50 μg ml−1 for E. coli).
For recombinant strains, the medium was appropriately supplemented with spectinomycin (1,000 μg/ml for E. coli and 15 μg/ml for L. lactis subsp. cremoris) or erythromycin (10 μg/ml for L. lactis subsp. cremoris and 250 μg/ml for E. coli). E. coli strain ER2738 was used for DNA cloning and overexpression and was grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C.
Growth under sublethal stress conditions or acid shock.
Overnight cultures of L. lactis subsp. cremoris strains were used to inoculate M17/7 medium at an initial optical density at 600 nm (OD600) of 0.1. Stationary-phase cells were harvested after 5.5 h of incubation (OD600 = 2.6) at 30°C. For ethanol- and acid-adapted and grown cells, L. lactis subsp. cremoris was inoculated at the same initial OD600 as that used previously in M17/7 medium with 6% (vol/vol) ethanol or in M17/5 medium. Adapted and grown cells were harvested by centrifugation (6,500 × g, 10 min, and 25°C) from early stationary phase after 7 h (OD600 = 0.6) and 6 h (OD600 = 1.1) of growth with 6% ethanol and at pH 5, respectively. The specific growth rate values were similar for the tested strains and depended only on the growth conditions used. In M17/7 medium, the mean value of μ was 1.2 h−1, whereas that of cells cultured at pH 5 or in the presence of 6% (vol/vol) ethanol decreased to 0.7 h−1 or 0.3 h−1, respectively. The cells were resuspended in M17/3 medium for acid shock. Aliquots were collected at timed intervals (0 min, 90 min, and 180 min), and viable cells were measured by serial dilutions, being plated on M17/7 agar supplemented with spectinomycin, and counting colonies after 24 h of incubation. The strain survival was defined as the ratio of colonies (number of CFU) formed on M17/7 agar medium at the different sampling times versus T0. Each experiment was performed in triplicate.
DNA manipulation and transformation.
Standard procedures such as purification, ligation, restriction analysis, and gel electrophoresis were carried out as described by Sambrook and Pollack (40). L. lactis subsp. cremoris chromosomal DNA was prepared by the classical method described by Posno et al. (39). PCR products and restriction products were purified with the GenElute PCR kits (Sigma, France). Conventional electroporation was used for E. coli. L. lactis subsp. cremoris was transformed by electroporation, as described by Dower (18).
Construction of the L. lactis subsp. cremoris Δcfa strain and homologous complementation of the mutant.
In order to delete an internal part of the cfa gene (28 bp deleted from nucleotides 514 to 541 of the coding sequence), two fragments generated by PCR were cloned into pGID023 in order to construct the integration plasmid pTMH1. The primers olcg100 (5′CCCCGAATTCTTGGTATTTGAAAAAACAGTCTTG-3′) and olcg101 (CCCCGGATCCCGGCCACCAGGTTTGCTAT), which include EcoRI and BamHI restriction sites, respectively (restriction sites are underlined), were used to amplify the 5′-end region of the coding sequence from nucleotides 1 to 512. The primer olcg102 (CCCCGGATCCACTTTGATGTTTAAAGCCGTGC), which includes the BamHI site (underlined), and the primer olcg103 (TCCAGCTTCAAAACTTGCGG) were used to amplify the 3′-end region of the coding sequence from nucleotides 541 to 1104. Both PCR products were digested with EcoRI-BamHI and BamHI-HindIII using the cleavage site HindIII at position 1078 of the coding sequence. Purified digested PCR products were ligated using the BamHI site. The 1,056-bp fragment obtained was then subcloned into the EcoRI/HindIII sites of pGID023 in order to obtain the thermosensitive suicide vector pTMH1 for cfa gene replacement in L. lactis subsp. cremoris. The 8.2-kb cloning vector pGID023 is stable in E. coli and unstable in Gram-positive bacteria under nonselective conditions. Mutagenesis by allelic replacement is therefore possible in L. lactis subsp. cremoris by the following two successive homologous recombinations: an insertion followed by an excision (28). Erythromycin resistance was the selective marker in E. coli and in Gram-positive bacteria. Mutagenesis by allelic replacement at the cfa locus was performed in L. lactis subsp. cremoris. Plasmid pTHM1 was electroporated into L. lactis subsp. cremoris. Stable erythromycin-resistant transformants were selected for plasmid pTHM1 integration at the cfa locus. After growth in the presence of erythromycin, erythromycin-sensitive excisants were isolated. To confirm the deletion of the cfa gene in these excisants, their genomic DNA was tested by PCR amplification using primers olcg104 (AATTGCTAAAGTTCATCATATCTT) and olcg105 (CATCTGCTGAAAGTCCGTATT). The 28-bp deletion and the cleavage BamHI site were confirmed, demonstrating the presence of the mutation in the tested excisants. One excisant, named the L. lactis subsp. cremoris Δcfa strain, was used for physiological analysis in this study.
Homologous complementation of the L. lactis subsp. cremoris Δcfa strain was performed as the control. The cfa gene of L. lactis subsp. cremoris wild-type MG1363 with its promoter was amplified by PCR with high-fidelity Taq DNA polymerase (Roche), using primers MG1363cfa1 (ATGGAATTCTTTTTTATTATATCAAAAAACTAT) and MG1363cfa2 (AGCGGATCCTAAAAATGTCCCCCAATCTAAAT), which include EcoRI and BamHI sites (underlined), respectively. The 1,894-bp fragment obtained was digested with EcoRI and BamHI and cloned into pDL278 in order to yield the pDLcfa12A3 vector. This vector was electroporated into the L. lactis subsp. cremoris Δcfa strain. The complemented strain was named L. lactis subsp. cremoris Δcfa + pDLcfa.
RNA extraction and analysis.
A bacterial culture in stationary phase of growth was centrifuged and washed with water-DMPC (dimethyl pyrocarbonate; 0.1% DMPC-treated water before being autoclaved to inactivate RNases). The cell pellet was taken in 1 ml TRI reagent (Sigma-Aldrich), and cells were broken in Precellys disruptor by 2 cycles of 30 s at 6,000 rotations per minute in the presence of 200 mg of glass beads (diameter, 70/100 μm). Total RNAs were then extracted with phenol-chloroform, precipitated with isopropanol, and resuspended in RNase-free water. Reverse transcription-PCR was carried out using the reverse transcriptase iScript (Bio-Rad), as recommended by the supplier.
The real-time PCR (quantitative reverse transcription-PCR [qRT-PCR]) was carried out on a Bio-Rad iCycler with the iQ SYBR Green supermix (Bio-Rad) in 96-well plates. For each measurement, a threshold cycle (CT) value was determined. Quantification of mRNA was calculated by the method of threshold (ΔΔCT) in which the quantity of target cDNA is adjusted relative to that of a reference made by the amount of cDNA as described by Desroche et al. (17). The results were normalized by using the butB gene coding for 2,3-butanediol dehydrogenase as an internal control. We have previously verified that the level of transcription of the butB gene was independent of the conditions of growth tested in this study.
Fatty acid determination.
The total lipids were extracted from cells, cultured under adapted or unadapted conditions, and harvested in stationary phase of growth. The extraction was conducted with a chloroform-methanol (1:2) mixture, according to the method described by Bligh and Dyer (2). The fatty acids were directly transesterified with 3% methanol in sulfuric acid for 2 h at 82°C. The separation and determination of the fatty acid methyl esters were performed with a Chrompack CP 9002 gas chromatograph equipped with an injector and flame ionization detector (GC-FID). The separation was carried out in a capillary Varian FactorFour column (30 m by 0.32 mm inside diameter [ID]; film thickness, 0.25 μm). The temperature detector and injector used were 230°C and 220°C, respectively. The initial oven temperature used was 160°C for 10 min and then was increased to 180°C at 2°C/min. The fatty acid esters were designated by comparing their retention times with those of the standard compounds (Nu-Chek Prep, Elysian, MN). The molar percentage of each component was calculated from the peak area obtained (ratio of peak area/total area of all peaks).
Fatty acid assignments were completed by GC-mass spectrometry (GC-MS). 4,4-Dimethyloxazoline (DMOX) derivatives were prepared in a simple one-pot reaction. Total lipids were converted to their DMOX derivatives by treatment with 2-amino-2-methylpropanol in a sealed ampoule at 180°C for 18 h. The reaction mixture was cooled, dissolved in dichloromethane, and washed twice with 1 ml of water. After the organic phase was dried, the solvent was removed under a stream of nitrogen, and the sample was dissolved in hexane. The sample was applied to a short column of Florisil, which was subsequently washed with hexane prior to elution of the DMOX derivatives with a mixture of hexane-acetone (96:4, vol/vol).
Cell membrane labeling and fluorescence anisotropy measurements.
Before each measurement, bacterial cells were energized in 50 mM MES [2-(N-morpholino)ethanesulfonic acid]-KOH buffer (pH 7.2) containing 10 mM glucose to prevent a decrease in membrane fluidity caused by bacterial death independent of the stress nature (45). Membrane fluidity was determined continuously by measuring fluorescence anisotropy using hydrophobic 1,6-diphenyl-1,3,5-hexatriene (DPH) (Molecular Probes, Oregon) as a probe, as previously described in reference 13. Anisotropy values were automatically calculated by a spectrofluorometer, according to Shinitzky (41). Fluorescence anisotropy (inversely proportional to membrane fluidity) was measured at 30°C, with determinations made every 8 s. To ensure probe stabilization within cell membranes for optimal anisotropy determination, control and stress-grown and adapted cells were incubated with a DPH probe in 50 mM MES-KOH buffer and 10 mM glucose, pH 7.2, at 30°C for 15 min. Each measurement was repeated at least five times.
Statistical analysis.
A one-way analysis of variance (ANOVA) was performed to test whether there was a significant difference among cell viability percentages, membrane fluidity values, and membrane fatty acid compositions. The confidence interval for a difference in the means was set at 95% (P < 0.05), and the Tukey test was used to locate these significant differences. SigmaStat software was used to carry out these statistical analyses.
RESULTS
Influence of culture conditions on the fatty acid composition.
The fatty acid composition of the L. lactis subsp. cremoris MG1363 strain was compared to those of the mutant (L. lactis subsp. cremoris Δcfa strain) and the complemented strain (L. lactis subsp. cremoris Δcfa + pDLcfa) grown under different culture conditions. This analysis was performed with stationary-phase-grown cells cultured in optimal medium or under sublethal conditions of growth (pH 5 or with 6% ethanol) (Table 2). We have previously verified that the deletion of the cfa gene had no influence on the specific growth rate (data not shown). Main fatty acids were identified as follows: C14:0, iso-C15, anteiso-C15, C16:0, C16:1n-7, C18:0, C18:1n-7, and C19:0n-7 cyc. Two unknown fatty acids, A4 and A5, were detected. In order to identify these fatty acids, a preparation of ester derivatives of fatty acids by the basic method was conducted. In this case, no fatty acid corresponding to the same retention times was detected by chromatographic analysis (data not shown). A4 and A5 probably resulted from C18:1n-7 and C19:0n-7 cyc as transient compounds generated by transesterification under the acidic condition.
Table 2.
Fatty acid compositions of L. lactis subsp. cremoris MG1363 strains grown at 30°C on M17/7 medium or under sublethal stress conditions
| Fatty acidd | Fatty acid composition (molar %)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| M17/7 medium |
M17/5 medium |
M17/7 medium plus 6% ethanol |
|||||||
| Wild type | Δcfa strain | Δcfa + pDLcfa | Wild type | Δcfa strain | Δcfa + pDLcfa | Wild type | Δcfa strain | Δcfa + pDLcfa | |
| Iso-C15:0 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 6.84 ± 8.44 | 6.46 ± 3.19 | 13.33 ± 5.92 |
| C14:0 | 9.21 ± 0.35 | 9.91 ± 0.75 | 11.07 ± 1.45 | 7.46 ± 1.09 | 6.86 ± 0.97 | 8.82 ± 1.08 | 10.45 ± 2.13 | 8.92 ± 0.73 | 9.95 ± 1.21 |
| Anteiso-C15:0 | 5.15 ± 1.43 | 3.65 ± 0.28 | 3.77 ± 0.78 | 7.22 ± 0.50 | 5.71 ± 0.16 | 4.66 ± 3.58 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| C16:0 | 22.63 ± 2.08 | 24.04 ± 1.32 | 23.59 ± 1.92 | 29.53 ± 1.47 | 28.34 ± 0.95 | 33.22 ± 1.82 | 23.12 ± 3.27 | 26.20 ± 2.93 | 30.34 ± 1.70 |
| C16:1n-7 | 2.42 ± 0.53 | 2.16 ± 0.18 | 2.95 ± 1.31 | 2.13 ± 1.58 | 2.60 ± 1.84 | 0.00 ± 0.00 | 17.36 ± 5.76 | 16.45 ± 5.15 | 0.00 ± 0.00 |
| A4b | 7.59 ± 0.60 | 8.33 ± 0.46 | 3.96 ± 2.15 | 1.76 ± 1.44 | 16.23 ± 1.57 | 0.00 ± 0.00 | 0.40 ± 0.41 | 2.06 ± 0.81 | 0.00 ± 0.00 |
| C18:0 | 1.33 ± 0.51 | 1.63 ± 0.79 | 1.54 ± 0.40 | 3.44 ± 0.71 | 3.93 ± 0.64 | 5.11 ± 0.99 | 6.31 ± 1.62 | 8.14 ± 3.29 | 10.11 ± 0.58 |
| C18:1n-7 | 32.29 ± 5.67 | 50.28 ± 1.69 | 19.42 ± 4.86 | 3.60 ± 0.95 | 36.32 ± 1.91 | 0.00 ± 0.00 | 9.93 ± 1.72 | 31.77 ± 7.47 | 0.00 ± 0.00 |
| A5b | 1.23 ± 0.71 | 0.00 ± 0.00 | 4.70 ± 0.17 | 13.74 ± 0.67 | 0.00 ± 0.00 | 4.60 ± 2.86 | 0.27 ± 0.47 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| C19:0n-7 cyc | 18.15 ± 3.23 | 0.00 ± 0.00 | 29.00 ± 3.69 | 31.11 ± 0.43 | 0.00 ± 0.00 | 43.58 ± 2.89 | 25.32 ± 2.41 | 0.00 ± 0.00 | 36.28 ± 3.61 |
| CFA conversion (%)c | 35.99 | 0.00 | 59.89 | 89.62 | 0.00 | 100.00 | 71.83 | 0.00 | 100.00 |
Samples were taken from stationary phase of growth. Each value is the mean obtained from three separate analyses. The summed data presented is shown as the mean value ± standard deviation of three replicate values.
Unidentified fatty acids.
CFA conversion = 100 × [Σ(C19:0n-7 cyc)/Σ(C19:0n-7 cyc + C18:1n-7)].
C14:0, myristic acid; C16:0, palmitic acid; C16:1n-7, palmitoleic acid; C18:0, stearic acid; C18:1n-7, cis-vaccenic acid; C19:0n-7 cyc, lactobacillic acid.
Under all growth conditions, the L. lactis subsp. cremoris Δcfa mutant strain could not produce lactobacillic acid, and a high level of the CFA precursor (C18:1n-7) was detected, compared to those of the wild-type and complemented strains. Thanks to the copy number of the pDLcfa12A3 vector, overexpression of the cfa gene in the complemented strain L. lactis subsp. cremoris Δcfa + pDLcfa allowed an increase in the production of lactobacillic acid from cis-vaccenic acid compared to that allowed by the wild-type strain. These data validate our strategy to obtain a cfa-deficient strain and a homologous recombinant strain as a control.
Under optimal growth conditions (M17/7), the amount of C16:0 represented around 24% (molar percent) in the three strains. A stable quantity of anteiso-C15:0 was detected for all strains. There was 36% of CFA conversion for the wild-type strain versus 60% for the complemented strain, thanks to the multicopy vector. In cells grown under acid conditions (M17/5), we observed that 90% of cis-vaccenic acid was converted into lactobacillic acid for the wild-type strain and 100% for the complemented strain. On cells grown with ethanol (M17/7 plus 6% ethanol), except for the control, we noted a large amount of C16:1n-7 (17% [molar percent] versus 2.5% in M17/7). For all strains, the growth in the presence of ethanol induced an increase in the stearic acid molar percentage (8% versus 1.5% in M17/7). Moreover, no anteiso-C15:0 was detected, and we observed the apparition of C15:0. Inversely, the conversion of cis-vaccenic acid into lactobacillic acid was not affected by the growth condition with 71% and 100% of CFA conversion in the wild-type and control strains, respectively.
Expression of the cfa gene was induced by acidity and ethanol growth conditions.
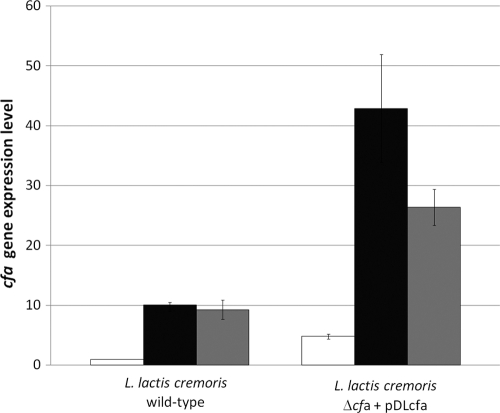
The differential rates of conversion of C18:1n-7 to C19:0n-7 cyc observed previously between the wild-type and control strains led us to study the level of transcription of the cfa gene under tested growth conditions. The results obtained by qRT-PCR are presented in Fig. 1. By using the comparative critical threshold (ΔΔCT) method with the butB gene of L. lactis subsp. cremoris as an internal control, we found that the cfa mRNA level increased 10 times when the wild-type strain cells were cultured at pH 5 and increased 9 times when cultured in the presence of 6% ethanol. Thanks to the multicopy plasmid and compared to the wild-type strain, the complemented strain showed an increase in the cfa mRNA levels by 5-, 42-, and 24-fold in optimal, acid-grown, and ethanol-grown cells, respectively. This overexpression allowed this strain to convert all C18:1n-7 to C19:0n-7 cyc. Budin-Verneuil et al. (7) previously showed the induction of cfa gene expression by moderate acidity by using a reporter system. Our results agreed with the results found by those authors and demonstrate that the cfa promoter of L. lactis subsp. cremoris is highly regulated at the transcriptional level by both acidity and ethanol.
Fig. 1.
Relative mRNA levels of the L. lactis subsp. cremoris cfa gene in response to various conditions of growth, as determined by qRT-PCR. Total RNA was extracted from the stationary-phase cells growing in M17/7 medium (open bars), in M17/5 medium (black bars), or in M17/7 medium plus 6% ethanol (gray bars). The expression level was normalized to that of the butB gene encoding 2,3-butanediol dehydrogenase.
Is the presence of cyclopropane fatty acids a key factor to maintain membrane rigidity in L. lactis subsp. cremoris strains?
The DPH probe is widely used for the membrane fluidity state evaluation. Indeed, it partitions favorably to the membrane interior, it has intense fluorescence, it does not appear to bind to proteins, and it is sensitive to the physical state of the membrane (13, 21, 45). Fluorescence anisotropy values of DPH obtained in stationary-phase-grown cells are shown in Table 3. When the strains were grown under optimal conditions (M17/7 medium), the initial value of anisotropy was not significantly different for the three strains (mean R value = 0.138). It means that there was no difference in the membrane fluidity levels among the strains, independent of the percentage of unsaturated fatty acid (UFA) cyclopropanation. In cells grown under acidic conditions (M17/5 medium), the value of anisotropy was maintained for the L. lactis subsp. cremoris Δcfa mutant strain. Membranes appeared to be significantly more rigid (as noted by an increasing of R values) for the wild-type and complemented strains (mean R value = 0.144). We could correlate these data with the higher levels of the CFA conversion (90% and 100% for the wild-type strain and the complemented strain, respectively). In cells grown with ethanol (M17/7 plus 6% ethanol), despite of the fact that the membrane appeared more fluid in the mutant strain, all fluidity R values dropped in comparison with values obtained under an optimal or acidic condition of growth, meaning that a fluidization of the membrane was induced by growth in the presence of ethanol.
Table 3.
Anisotropy R values of DPH in L. lactis subsp. cremoris strains grown at 30°C on M17/7 medium or under sublethal stress conditionsa
| Strain/genotype | Anisotropy valueb |
||
|---|---|---|---|
| M17/7 medium | M17/5 medium | M17/7 medium plus 6% ethanol | |
| Wild type | 0.141 ± 0.012 A,D | 0.139 ± 0.011 B,D | 0.122 ± 0.003 B,C |
| Δcfa | 0.136 ± 0.016 A,D | 0.125 ± 0.006 A,D | 0.116 ± 0.006 A,C |
| Δcfa + pDLcfa | 0.139 ± 0.011 A,C | 0.150 ± 0.008 B,D | 0.127 ± 0.006 B,C |
Samples were taken from stationary phase of growth. Each value is the mean obtained from at least five separate analyses. The summed data presented is shown as the mean value ± standard deviation of replicate values. Anisotropy R values of DPH were continuously monitored for a period of 15 min.
Statistical analysis was done by using the Tukey test (P < 0.05). Comparison done by column, with value A significantly less than value B; comparison done by line, with value C significantly less than value D.
Is the presence of cyclopropane fatty acids a key factor for the survival of L. lactis subsp. cremoris strains after an acidic challenge?
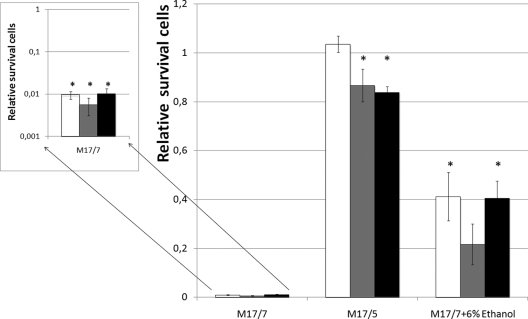
In order to test the impact of the mutation on the acid resistance of L. lactis subsp. cremoris, the three strains were cultivated under optimal conditions (pH 7), acid-adapted conditions (M17/5), or ethanol-adapted conditions (M17/7 plus 6% ethanol) until early stationary phase before being shocked. The results of survival are reported in Fig. 2. With cells grown under optimal conditions (M17/7), a decrease of 2 logs of viable cells was observed after 3 h of incubation at pH 3.0. There was no significant difference of viability among the three strains. The presence of CFA could not confer a better survival to the wild type nor to the complemented strains, which contained a larger amount of CFA. With acid-adapted cells, we observed an increase in the survival of all strains, including the mutant strain. After 3 h of incubation at pH 3.0, more than 80% cells were still alive. Surprisingly, with ethanol-adapted cells, an opposite effect was observed. The L. lactis subsp. cremoris Δcfa mutant strain was significantly more affected by the acid shock, with 80% mortality after 3 h of incubation at pH 3.0. No significant difference was observed between the wild-type and control strains, but a better survival level was obtained in comparison with that of the mutant strain, with 40% of cells viable after 3 h of treatment.
Fig. 2.
Survival of the L. lactis cremoris strains during acid challenge (pH 3.0) as a function of the growth conditions. L. lactis cremoris wild type, open bars; L. lactis cremoris Δcfa strain, gray bars; L. lactis cremoris Δcfa + pDLcfa, black bars. Cells were grown until stationary phase, harvested, and resuspended in M17/3. Viability cells were counted after 3 h of acid shock. The strain survival was defined as the ratio of colonies (number of CFU) formed on M17/7 agar medium at the sampling time versus T0. Each value is the mean from three determinations. The summed data presented are the mean values ± standard deviations of replicate values. An asterisk means that there is no significant difference between these values. A value of 1 for the relative survival corresponds to 3.108 cells/ml.
DISCUSSION
Modifications in fatty acid composition leading to an adjustment of the degree of saturation of the membrane lipids are well-known reactions of bacteria in response to a drastic modification of the environment (29, 32). Among these modifications, the synthesis of cyclopropane fatty acids is recognized to favor the stress tolerance, particularly in lactic acid bacteria, as well as the response to acid, ethanol, nutritional, and osmotic stresses (1, 22, 24, 36, 37, 44, 45). The lactobacillic acid C19:0n-7 cyc detected in the membranes of the L. lactis subsp. cremoris wild type and the control strain derives from the precursor cis-vaccenic acid C18:1n-7. The conversion ratio reached a value of 36% when the cells entered stationary phase of growth on an optimal medium such as M17/7. The acid growth condition induced CFA synthesis, with a conversion ratio of around 90%. This result correlates with increasing amounts of cfa mRNA transcripts with the number of acid-adapted cells grown and confirms that the gene cfa is regulated at the transcriptional level, according to the data of Budin-Verneuil et al. (7).
We have demonstrated previously using the wine lactic acid bacterium Oenococcus oeni that an increase in the CFA content was obtained with ethanol-grown cells (22). No data are available concerning the effect of ethanol on the membrane composition of L. lactis subsp. cremoris. Our results demonstrated that the presence of ethanol in the medium induced not only an increase in cyclopropanation (2-fold conversion ratio in comparison with that of cells grown under optimal conditions) but also an increase in the level of transcripts of cfa mRNA.
In E. coli, two promoters are involved in cfa transcription. P1 is reported to be transcribed by the σ70 RNA polymerase, and P2 transcription depends on σS (46). The transcriptional upregulation of cfa was shown to be dependent on ppGpp via the ppGpp-dependent growth phase activation of the σS factor (19). The decrease in growth rates observed with acid- or ethanol-grown cells in L. lactis strains in relation with high CFA content could suggest a similar mechanism of transcriptional regulation. However, L. lactis lacks global stress sigma factors homologous to σS, and a unique promoter with a sequence closely resembling the −10 and −35 sequences recognized by σ70 RNA polymerase was identified (7). The mechanism involved in the control of cfa expression has not yet been elucidated in this bacterium. The work of Budin-Verneuil et al. (7) underlined that the cfa gene transcription was upregulated by growth phase and acidity. However, the induction of cfa transcription by acidity was independent of ppGpp in this bacterium.
More interesting is the comparison of the fatty acid profiles between cells adapted to acid and those adapted to ethanol. First, we observed a disappearance of anteiso-C15:0 in favor of iso-C15:0 for the three strains. The growth-dependent changes in the ratio of terminally branched iso and anteiso fatty acids were suggested in Bacillus subtilis as mechanisms to adjust fluidity (25, 31, 34) without a change in the saturation level of membrane fatty acids. Second, an accumulation of palmitoleic acid in the wild-type and mutant strains cultivated in the presence of ethanol was obtained (17% [molar percentage] versus 2.5% with acid-grown cells). Another adaptive strategy to adjust membrane fluidity consists of a growth-dependent elongation in fatty acid chain length (15, 25). The gene fabF encoding a 3-ketoacyl-acyl carrier protein synthase was found in the L. lactis subsp. cremoris genome (3). This protein, annotated FabF, is responsible for the elongation of C16:1n-7 (palmitoleic acid) to C18:1n-7 (cis-vaccenic acid). Basing on the proteomic characterization of the acid tolerance response (ATR) in L. lactis subsp. cremoris MG1363 (8), FabF was not shown to be overproduced when the cells were incubated at pH 5.0. However, the synthesis of this protein was induced in both of the acid stress-resistant mutants of L. lactis subsp. cremoris (7), suggesting to those authors that this mechanism must be implied in the acid adaptation of the bacterium. If we postulate that in L. lactis subsp. cremoris, ATR implies a mechanism of elongation of palmitoleic acid to cis-vaccenic acid, followed by a cyclopropanation of cis-vaccenic acid into lactobacillic acid, we would logically have observed an accumulation of cis-vaccenic acid for the mutant strain grown at pH 5.0. However, this fatty acid represented 36% of the total membrane fatty acid when the mutant cells were grown under acid conditions versus 50% of that when the mutant cells were grown at pH 7.0. This result casts doubt on the involvement of FabF in the ATR. An analysis of the level of expression of the fabF gene, correlated with the level of expression of the cfa gene, under different stress conditions should be able to clarify the role of FabF in stress response in the future. Moreover, the accumulation of palmitoleic acid observed in wild-type and mutant strains grown in the presence of ethanol suggests a repression of the transcription of the fabF gene.
The role of structural classes of fatty acids in bacterial membrane homeoviscous adaptation is very well documented (25, 27, 36). Concerning cyclopropane fatty acids, although the molecular mechanism controlling their formation was the subject of numerous data (10), the role of these acids is not yet fully understood. Compared to those of cis unsaturated fatty acids, the presence of CFAs should make the membrane more rigid because of their higher lipid melting points (25). The determination of membrane anisotropy R values with stationary-phase cells grown on M17/7 did not verify the link between the presence of CFAs and a less fluid membrane. In fact, the absence of cyclopropanation had no influence on the R value. In this case, the partial cyclopropanation of palmitoleic acid would not allow an adjustment of the membrane fluidity. An induction by the acidity of the synthesis of CFAs led to a significant increase in R, particularly for the control strain, which overproduced lactobacillic acid. Surprisingly, the induction of cyclopropanation by ethanol led to a significant decrease in R for the wild-type and control strains, reflecting a fluidization of the membrane. These results are in contrast with those of main studies of the CFA role in membrane fluidity (23, 25). Moreover, previous studies of the lactic acid bacterium O. oeni have shown significant increases in membrane anisotropy with cells harvested in stationary growth phase as well as with acid- or ethanol-grown cells (12, 16). In ethanol-grown cells (wild-type and mutant strains), we observed larger amounts of C16:1n-7 than C18:1n-7. Shorter chains of fatty acids are characterized by a lower melting point. These data could explain the membrane fluidization observed in both strains but does not explain the value of R observed in the complemented strain. In E. coli, one of the modifications that occurs in membrane lipid composition in response to ethanol is an increase in the amount of unsaturated fatty acids (29). The same results were observed in Acinetobacter calcoaceticus in the presence of short-chained alcohols in the medium (30). In these cases, the cell-adaptive responses lead to more fluid membranes. Further investigation is needed in order to understand the contradictory effects of ethanol on the adapted cell membrane fluidity.
With unadapted cells, the presence of cyclopropane fatty acids could not confer better survival after an acid shock to the wild-type or complemented strains, which contained a larger amount of CFA. This result was consistent with fluidity data, suggesting an absence of homeoviscous adaptation despite the synthesis of CFA. The increase in survival observed with acid-adapted cells, independent of the cyclopropanation of the cis-vaccenic acid, supports the fact that the synthesis of CFA is not essential in the acid tolerance response.
In comparison with data obtained from acid-grown cells, the lower survival values obtained with ethanol-adapted cells could be explained by a more fluid state of the membrane affecting the cellular integrity. In order to understand the different behaviors observed between acid-adapted and ethanol-adapted cells, we compared 10 gene expression levels in the three strains cultured under three conditions (M17/7, M17/5, and M17/7 plus 6% ethanol) by qRT-PCR. The chosen genes encode stress proteins highly induced by acidity in L. lactis subsp. cremoris MG1363 (8). The comparison of gene expression levels demonstrated that the ATR is strongly induced by acidity in the three strains and slightly induced by ethanol (data not shown). The ATR implies that L. lactis subsp. cremoris reorganizes its metabolisms in response to acid stress. The Cfa synthase has been demonstrated to be part of the 10 most overexpressed proteins in L. lactis subsp. cremoris MG1363 incubated at pH 5.0 (8). However, other stress proteins must predominate to maintain the membrane integrity against acid stress. In O. oeni, induction of expression of hsp18, encoding the lipochaperone protein Lo18, was previously shown (12). Lo18 was found to be peripherally associated with the cytoplasmic membrane. An involvement of this small Hsp in the maintenance of membrane integrity has been demonstrated (14). No gene encoding such a protein has been identified in the genome of L. lactis subsp. cremoris. The role of ATR-overproduced proteins in the stabilization of membrane fluidity remains to be elucidated in this bacterium.
Growth in the presence of ethanol induces an overproduction of CFA independent of the ATR. In this particular case, acid shock induced a mortality lower than that observed in cells grown under optimal conditions. The accumulation of the shorter-chain fatty acid C16:1n-7, known to decrease the hydrophobic interaction between the free acyl chains and the proteins, could explain the restriction of the deleterious effect of an acid treatment on cellular integrity.
We conclude that the mechanism of cyclopropanation of the membrane unsaturated fatty acids, even if it could contribute to better survival of L. lactis subsp. cremoris MG1363 in response to an acid stress, is not a key factor for acid stress resistance. The actual function of CFAs on the adaptation of bacteria to environmental stresses remains a nonelucidated question in the physiology of stress adaptation in bacteria.
ACKNOWLEDGMENTS
This study was supported by a grant from the Embassy of France in Vietnam, by the Regional Council of Burgundy, and by AgroSup Dijon, France.
The GC-MS analyses were performed by the Plate-Forme Lipides-Arômes (UMR-1324, Centre des Sciences du Goût et de l'Alimentation, INRA Dijon). We thank Olivier Berdeaux and Stéphane Grégoire for their critical interpretation of the fatty acid profiles. We thank Dominique Garmyn for the gift of the pGID023 vector and for his constant interest in this work.
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Beal C., Fonseca F., Corrieu G. 2001. Resistance to freezing and frozen storage of Streptococcus thermophilus is related to membrane fatty acid composition. J. Dairy Sci. 84:2347–2356 [DOI] [PubMed] [Google Scholar]
- 2. Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 3. Bolotin A., et al. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brian B. L., Gardner E. W. 1968. A simple procedure for detecting the presence of cyclopropane fatty acids in bacterial lipids. Appl. Microbiol. 16:549–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Broadbent J. R., Lin C. 1999. Effect of heat shock or cold shock treatment on the resistance of Lactococcus lactis to freezing and lyophilization. Cryobiology 39:88–102 [DOI] [PubMed] [Google Scholar]
- 6. Brown J. L., Ross T., McMeekin T. A., Nichols P. D. 1997. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int. J. Food Microbiol. 37:163–173 [DOI] [PubMed] [Google Scholar]
- 7. Budin-Verneuil A., Maguin E., Auffray Y., Ehrlich S. D., Pichereau V. 2005. Transcriptional analysis of the cyclopropane fatty acid synthase gene of Lactococcus lactis MG1363 at low pH. FEMS Microbiol. Lett. 250:189–194 [DOI] [PubMed] [Google Scholar]
- 8. Budin-Verneuil A., Pichereau V., Auffray Y., Ehrlich D., Maguin E. 2007. Proteome phenotyping of acid stress-resistant mutants of Lactococcus lactis MG1363. Proteomics 7:2038–2046 [DOI] [PubMed] [Google Scholar]
- 9. Chang Y. Y., Cronan J. E., Jr 1999. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33:249–259 [DOI] [PubMed] [Google Scholar]
- 10. Chang Y. Y., Eichel J., Cronan J. E., Jr 2000. Metabolic instability of Escherichia coli cyclopropane fatty acid synthase is due to RpoH-dependent proteolysis. J. Bacteriol. 182:4288–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y. Y., LeBlanc D. J. 1992. Genetic analysis of scrA and scrB from Streptococcus sobrinus 6715. Infect. Immun. 60:3739–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu-Ky S. 2005. Fluidité membranaire en relation avec l'adaptation de la bacterie lactique Oenococcus oeni au milieu vin. Ph.D. dissertation. Université de Bourgogne, Dijon, France [Google Scholar]
- 13. Chu-Ky S., Tourdot-Marechal R., Marechal P. A., Guzzo J. 2005. Combined cold, acid, ethanol shocks in Oenococcus oeni: effects on membrane fluidity and cell viability. Biochim. Biophys. Acta 1717:118–124 [DOI] [PubMed] [Google Scholar]
- 14. Coucheney F., et al. 2005. A small HSP, Lo18, interacts with the cell membrane and modulates lipid physical state under heat shock conditions in a lactic acid bacterium. Biochim. Biophys. Acta 1720:92–98 [DOI] [PubMed] [Google Scholar]
- 15. Cronan J. E., Jr., Gelmann E. P. 1975. Physical properties of membrane lipids: biological relevance and regulation. Bacteriol. Rev. 39:232–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Da Silveira M. G., Golovina E. A., Hoekstra F. A., Rombouts F. M., Abee T. 2003. Membrane fluidity adjustments in ethanol-stressed Oenococcus oeni cells. Appl. Environ. Microbiol. 69:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desroche N., Beltramo C., Guzzo J. 2005. Determination of an internal control to apply reverse transcription quantitative PCR to study stress response in the lactic acid bacterium Oenococcus oeni. J. Microbiol. Methods 60:325–333 [DOI] [PubMed] [Google Scholar]
- 18. Dower W. J. 1990. Electroporation of bacteria: a general approach to genetic transformation. Genet. Eng. (N. Y.) 12:275–295 [DOI] [PubMed] [Google Scholar]
- 19. Eichel J., Chang Y. Y., Riesenberg D., Cronan J. E., Jr 1999. Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (sigmaS). J. Bacteriol. 181:572–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gasson M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gennis P., Gallagher J., Falvo C., Baker S., Than W. 1989. Clinical criteria for the detection of pneumonia in adults: guidelines for ordering chest roentgenograms in the emergency department. J. Emerg. Med. 7:263–268 [DOI] [PubMed] [Google Scholar]
- 22. Grandvalet C., et al. 2008. Changes in membrane lipid composition in ethanol- and acid-adapted Oenococcus oeni cells: characterization of the cfa gene by heterologous complementation. Microbiology 154:2611–2619 [DOI] [PubMed] [Google Scholar]
- 23. Grogan D. W., Cronan J. E., Jr 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61:429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guillot A., Obis D., Mistou M. Y. 2000. Fatty acid membrane composition and activation of glycine-betaine transport in Lactococcus lactis subjected to osmotic stress. Int. J. Food Microbiol. 55:47–51 [DOI] [PubMed] [Google Scholar]
- 25. Härtig C., Loffhagen N., Harms H. 2005. Formation of trans fatty acids is not involved in growth-linked membrane adaptation of Pseudomonas putida. Appl. Environ. Microbiol. 71:1915–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heipieper H. J., de Bont J. A. 1994. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty acid composition of membranes. Appl. Environ. Microbiol. 60:4440–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heipieper H. J., Meinhardt F., Segura A. 2003. The cis-trans isomerase of unsaturated fatty acids in Pseudomonas and Vibrio: biochemistry, molecular biology and physiological function of a unique stress adaptive mechanism. FEMS Microbiol. Lett. 229:1–7 [DOI] [PubMed] [Google Scholar]
- 28. Hols P., Ferain T., Garmyn D., Bernard N., Delcour J. 1994. Use of homologous expression-secretion signals and vector-free stable chromosomal integration in engineering of Lactobacillus plantarum for alpha-amylase and levanase expression. Appl. Environ. Microbiol. 60:1401–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ingram L. O. 1976. Adaptation of membrane lipids to alcohols. J. Bacteriol. 125:670–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabelitz N., Santos P. M., Heipieper H. J. 2003. Effect of aliphatic alcohols on growth and degree of saturation of membrane lipids in Acinetobacter calcoaceticus. FEMS Microbiol. Lett. 220:223–227 [DOI] [PubMed] [Google Scholar]
- 31. Kaneda K., Yoshioka Y., Makita K., Toyooka H., Amaha K. 1997. Effects of carboxy-PTIO on systemic hemodynamics, liver energetics, and concentration of liver metabolites during endotoxic shock in rabbits: a 31P and 1H magnetic resonance spectroscopic study. Crit. Care Med. 25:1019–1029 [DOI] [PubMed] [Google Scholar]
- 32. Keweloh H., Heipieper H. J. 1996. Trans unsaturated fatty acids in bacteria. Lipids 31:129–137 [DOI] [PubMed] [Google Scholar]
- 33. Kim B. H., et al. 2005. The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology 151:209–218 [DOI] [PubMed] [Google Scholar]
- 34. Konopasek I., Strzalka K., Svobodova J. 2000. Cold shock in Bacillus subtilis: different effects of benzyl alcohol and ethanol on the membrane organisation and cell adaptation. Biochim. Biophys. Acta 1464:18–26 [DOI] [PubMed] [Google Scholar]
- 35. Law J., et al. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loffhagen N., Härtig C., Geyer W., Voyevoda M., Harms H. 2007. Competition between cis, trans and cyclopropane fatty acid formation and its impact on membrane fluidity. Eng. Life Sci. 7:67–74 [Google Scholar]
- 37. Montanari C., Sado Kamdem S. L., Serrazanetti D. I., Etoa F. X., Guerzoni M. E. 2010. Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol. 27:493–502 [DOI] [PubMed] [Google Scholar]
- 38. Munoz-Rojas J., et al. 2006. Involvement of cyclopropane fatty acids in the response of Pseudomonas putida KT2440 to freeze-drying. Appl. Environ. Microbiol. 72:472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Posno M., Verweij W. R., Dekker I. C., de Waard P. M., Groot G. S. 1986. The genes encoding chloroplast ribosomal proteins S7 and S12 are located in the inverted repeat of Spirodela oligorhiza chloroplast DNA. Curr. Genet. 11:25–34 [DOI] [PubMed] [Google Scholar]
- 40. Sambrook J., Pollack R. 1974. Basic methodology for cell culture-cell transformation. Methods Enzymol. 32(Part B):583–592 [DOI] [PubMed] [Google Scholar]
- 41. Shinitzky M. 1984. Membrane fluidity in malignancy. Adversative and recuperative. Biochim. Biophys. Acta 738:251–261 [DOI] [PubMed] [Google Scholar]
- 42. Sinensky M. 1974. Homeoviscous adaptation: a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 71:522–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suutari M., Laakso S. 1992. Temperature adaptation in Lactobacillus fermentum: interconversions of oleic, vaccenic and dihydrosterulic acids. J. Gen. Microbiol. 138:445–450 [DOI] [PubMed] [Google Scholar]
- 44. Teixeira H., Goncalves M. G., Rozes N., Ramos A., San Romao M. V. 2002. Lactobacillic acid accumulation in the plasma membrane of Oenococcus oeni: a response to ethanol stress? Microb. Ecol. 43:146–153 [DOI] [PubMed] [Google Scholar]
- 45. Tourdot-Maréchal R., Gaboriau D., Beney L., Diviès C. 2000. Membrane fluidity of stressed cells of Oenococcus oeni. Int. J. Food Microbiol. 55:269–273 [DOI] [PubMed] [Google Scholar]
- 46. Wang A. Y., Cronan J. E., Jr 1994. The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an RpoS(KatF)-dependent promoter plus enzyme instability. Mol. Microbiol. 11:1009–1017 [DOI] [PubMed] [Google Scholar]
- 47. Zhao Y., et al. 2003. Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 69:2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]