Abstract
Polyether ionophores are a unique class of polyketides with broad-spectrum activity and outstanding potency for the control of drug-resistant bacteria and parasites, and they are produced exclusively by actinomycetes. A special epoxidase gene encoding a critical tailoring enzyme involved in the biosynthesis of these compounds has been found in all five of the complete gene clusters of polyether ionophores published so far. To detect potential producer strains of these antibiotics, a pair of degenerate primers was designed according to the conserved regions of the five known polyether epoxidases. A total of 44 putative polyether epoxidase gene-positive strains were obtained by the PCR-based screening of 1,068 actinomycetes isolated from eight different habitats and 236 reference strains encompassing eight major families of Actinomycetales. The isolates spanned a wide taxonomic diversity based on 16S rRNA gene analysis, and actinomycetes isolated from acidic soils seemed to be a promising source of polyether ionophores. Four genera were detected to contain putative polyether epoxidases, including Micromonospora, which has not previously been reported to produce polyether ionophores. The designed primers also detected putative epoxidase genes from diverse known producer strains that produce polyether ionophores unrelated to the five published gene clusters. Moreover, phylogenetic and chemical analyses showed a strong correlation between the sequence of polyether epoxidases and the structure of encoded polyethers. Thirteen positive isolates were proven to be polyether ionophore producers as expected, and two new analogues were found. These results demonstrate the feasibility of using this epoxidase gene screening strategy to aid the rapid identification of known products and the discovery of unknown polyethers in actinomycetes.
INTRODUCTION
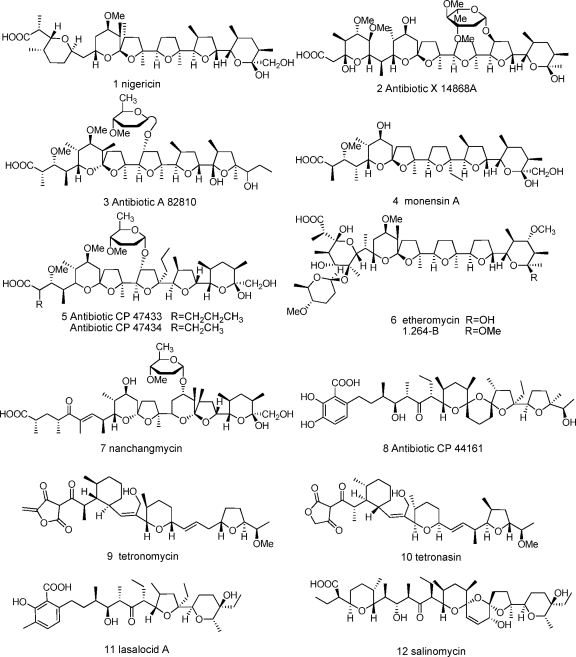
Programs aimed at the discovery of new secondary metabolites with interesting biological activities from microbial sources have found an impressive number of compounds during the past 50 years. Actinomycetes represent one of the most prolific microbial sources for the discovery of bioactive metabolites (5, 6), many of which are produced by polyketide synthase (PKS) systems (6, 42). Polyether ionophores (Fig. 1) are a unique class of type I polyketides with a high degree of promise for the control of drug-resistant bacteria and parasites, and they have been widely used as effective veterinary drugs and food additives in animal husbandry (13). These molecules also show a broad spectrum of bioactivity, including antifungal, antiviral, antitumor, herbicidal, and anti-inflammatory activity, as well as effects on the central nervous system (CNS) and immunoregulatory systems (24). The ability of polyethers to transport cations across plasma membranes leads to depolarization and succedent cell death. To date, more than 120 polyether ionophore structures have been characterized (16) of nearly 10,000 polyketides found so far (26). Without exception, all known polyether ionophores are produced by actinomycetes, with the vast majority being derived from the genera Streptomyces and Actinomadura (16) and the rest from Actinomyces, Dactylosporangium, Nocardia, and Nocardiopsis.
Fig. 1.
Structures of typical polyether ionophores. Structures 1, 4, 7, 9, and 11, compounds with complete polyether gene clusters already published; structure 6, etheromycin and its new analogue.
Despite the long history of the use of this class of drugs, until recently the biosynthetic pathway remained relatively obscure. However, five complete polyether ionophore gene clusters now have been cloned and fully sequenced: monensin (31), nanchangmycin (43), nigericin (20), tetronomycin (12), and lasalocid (34, 41). The complete polyether ionophore gene clusters contain a conserved tailoring enzyme gene encoding a flavin-linked epoxidase, which is responsible for the epoxidation of unsaturated polyether intermediates. The disruption of MonCI, an epoxidase involved in the biosynthesis of monensin, led to the isolation and characterization of a triene shunt metabolite, confirming its role (7). Given that genes involved in the same biosynthetic pathway tend to cluster together in a chromosome and that secondary metabolites with similar structures are likely to be biosynthesized by gene clusters that harbor certain homologous genes (32, 47), such genes could serve as markers in the identification and cloning of related gene clusters. KS domain-based PCR approaches are well established in the identification and cloning of type I PKS gene clusters in bacteria (18, 36). However, KS-based primers may identify all modular PKS, while the genome sequencing of actinomycetes has revealed that a single genome generally houses a large number of such clusters, hence more-selective primers targeting the polyether epoxidase gene are needed in the search for polyether ionophore gene clusters and products.
Here, we describe a PCR-based screening strategy for detecting polyether ionophore producers in actinomycetes. Designed primers enabled the cloning of an approximately 700-bp PCR fragment of the polyether epoxidase gene. The distribution of this gene among actinomycetes from different habitats and taxa were represented, and several polyether ionophores were identified from the producer strains. The results further suggest that the phylogenetic analyses of putative polyether epoxidases of the positive strains can provide a guide for the discovery of novel polyether ionophores.
MATERIALS AND METHODS
Strains and culture conditions.
A total of 1,068 actinomycetes previously isolated from samples collected from eight different habitats were screened. The first four in Table 1 were considered to be terrestrial samples and the latter four to be marine samples. A total of 236 reference strains of 35 genera and eight major families of Actinomycetales that represented the most productive producers of polyketides (2, 3, 8) also were screened. The majority of strains were incubated on GYM agar (JCM medium 43) plates for 7 to 14 days at 28°C, and the rest were incubated on appropriate medium, such as oatmeal agar (JCM medium 51), yeast-starch agar (JCM medium 42), and Bennett's agar (JCM medium 44). Most strains were cultivated at pH 7.3, except that acidic soil isolates and Streptacidiphilus strains were resuscitated at pH 5.0.
Table 1.
Epoxidase gene screening results and taxonomic diversity of the isolates from different environmental samples
| Strain category and source | No. of strains screened | No. (%) of positive strains | % Identity range of 16S rRNA genes | No. of OTU singletons | No. of OTU clusters | Total no. of OTUs |
|---|---|---|---|---|---|---|
| Terrestrial actinomycetes | ||||||
| Acidic soils from Jiangxi | 105 | 10 (9.5) | 84∼100 | 5 | 8 | 13 |
| Medicinal plants | 101 | 3 (3.0) | 83∼100 | 3 | 5 | 8 |
| Alpine soils from Qinghai-Tibet Plateau | 207 | 0 (0) | 83∼100 | 5 | 5 | 10 |
| Soils from lakeside of Kanas in Xinjiang | 70 | 1 (1.4) | 82∼100 | 5 | 4 | 9 |
| Marine-origin actinomycetes | ||||||
| Mangrove sediments from Hainan | 95 | 1 (1.1) | 88∼100 | 3 | 3 | 6 |
| Sponges from South China Sea | 220 | 5 (2.3) | 83∼100 | 7 | 6 | 13 |
| Coastal sediments from Bohai Bay, Dalian | 227 | 3 (1.3) | 81∼100 | 3 | 8 | 11 |
| Deep sea samples from southeast Indian Ocean | 43 | 0 (0) | 82∼100 | 2 | 2 | 4 |
| Total | 1,068 | 23 (2.2) | 33 | 41 | 74 |
Primer design and PCR amplifications of putative polyether epoxidase genes and 16S rRNA genes.
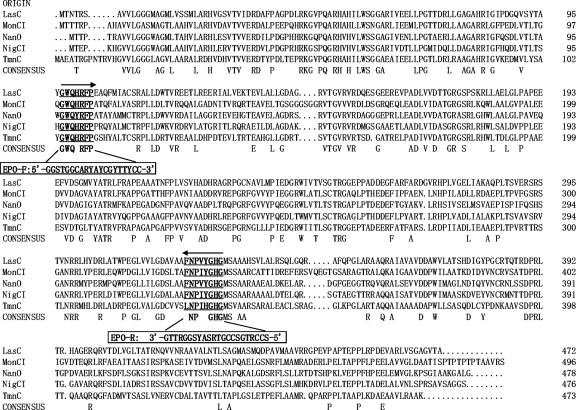
Amino acid and DNA sequences of the five known polyether epoxidases (lasalocid, monensin, nanchangmycin, nigericin, and tetronomycin) and other nonpolyether epoxidases, such as PimD, MycG, OleP, and ChmPI (1), and flavin-dependent epoxidases were retrieved from GenBank for primer design. Sequence alignments were carried out using the multiple alignment program Clustal W (30). A pair of degenerate primers, EPO-F (5′-GGSTGGCARYAYCGYTTYCC-3′) and EPO-R (5′-SCCRTGSCCGTRSAYSGGRTTG-3′), was designed according to the conserved regions of the five known polyether epoxidases (Fig. 2). Universal primers 27F and 1492R (29) were used to amplify the 16S rRNA gene.
Fig. 2.
Multiple amino acid sequence alignment of the five known polyether epoxidases (LasC, GenBank accession no. CAQ64694; MonCI, AAO65803; NanO, AAP42870; NigCI, ABC84466; TmnC, BAE93732). Conserved regions (underlined and marked with boldface) were used for polyether epoxidase gene-specific primer pair design. The boxed sequences indicate designed primers. The directions of arrows represent the directions of primers.
Total genomic DNAs from actinomycetes used in this study were extracted and purified as previously described by Hopwood et al. (21). PCR amplifications of polyether epoxidase and 16S rRNA genes were performed in a final volume of 50 μl containing 0.4 μmol of each primer, 0.2 mmol of each of the four deoxynucleoside triphosphates (dNTPs), 1 μl of extracted DNA, 5 U of Taq polymerase (with its recommended reaction buffer), and 3 μl of dimethylsulfoxide (DMSO). The thermal cycler (SensoQuest Labcycler) for the amplification of epoxidase genes was programmed according to the following parameters: 95°C for 8 min; 32 cycles at 95°C for 45 s, 59°C for 45 s, and 72°C for 1 min; and 72°C for 10 min, followed by cooling to 4°C. The PCR amplification of 16S rRNA genes was performed at 95°C for 4 min; 30 cycles at 95°C for 45 s, 55°C for 45 s, and 72°C for 1.5 min; and 72°C for 10 min. Fragments with the expected size of approximately 700 bp for epoxidase genes were purified, cloned, and sequenced using standard methods. PCR products of 16S rRNA genes were purified and sequenced directly.
Phylogenetic analysis.
The sequencing results were analyzed using BLASTP and BLASTN, which were accessed through the National Center for Biotechnology Information (NCBI) website. Sequences showing >40% amino acid identity to known polyether epoxidases were considered target genes. The phylogenetic analyses of amino acid sequences of the target epoxidases and 16S rRNA gene sequences of strains identified as positive for the polyether epoxidase gene were conducted using MEGA 4.0 (45), and neighbor-joining trees (39) were constructed with 2,000 bootstrap replicates. Epoxidase AmbJ served as the outgroup in the phylogenetic tree of polyether epoxidases. The nucleotide sequences that encoded putative polyether epoxidases and 16S rRNA genes (>1,350 bp) of strains identified as positive for the polyether epoxidase gene were deposited in the GenBank database under the accession numbers listed in Table S1 in the supplemental material.
Taxonomic diversity analysis of isolates from different habitats.
About 30% of the isolates from each of the eight habitats were randomly selected for 16S rRNA gene sequencing. Partial 16S rRNA gene sequences (600 bp) containing variable regions V1 to V4 (positions 94 to 694 in the Escherichia coli numbering system) (37) were aligned using Clustal X (30) and binned into groups of related sequences using the web-based tool Clusterer (25) with a distance parameter setting of 30. Given that the length of 16S rRNA gene sequences used for this purpose was 600 bp, the distribution of the distance setting 30 created sequence clusters that shared at least 95% identity. Each singleton and cluster was defined as an operational taxonomic unit (OTU) (23). One sequence from each OTU was used to construct a phylogenetic tree with related reference sequences as described above.
Fermentation, extraction, and chemical analysis.
Strains identified as positive for the polyether epoxidase gene were fermented either in liquid broth in 500-ml shake flasks (170 to 180 rpm) or on solid agar in petri dishes at 28°C for 7 days using two media, GYM (JCM medium 43) and SGG (1% starch, 1% glucose, 1% glycerol, 0.25% corn steep powder, 0.5% peptone, 0.2% yeast extract, 0.1% NaCl, 0.3% CaCO3 in tap water) (38). Liquid fermentation samples (100 ml) were centrifuged to separate the mycelium and supernatant; the mycelium was extracted once with 50 ml acetone, and the supernatant was extracted three times with 100 ml ethyl acetate. Solid fermentation samples (100 ml) were mashed and extracted three times with 100 ml ethanol. Different organic phases for each sample were pooled, and the entire organic layer was concentrated to dryness in a vacuum, and then the residue was redissolved in 2 ml DMSO. Two μl dissolved residues was used to test the activity against Staphylococcus aureus subsp. aureus CGMCC 1.2386 and Bacillus subtilis CGMCC 1.2428. Twenty μl of each active extract was subjected to high-performance liquid chromatography-UV-mass spectrometry/mass spectrometry (HPLC-UV-MS/MS) analysis (Shimadzu SPD-M20A and Thermo-Finnigan LCQ DECA XP) with a linear gradient of 50 to 100% aqueous methanol for 25 min (4.6- by 150-mm column; flow, 1.0 ml/min; photodiode array detector, 190 to 800 nm; Xbridge ODS). Mass spectra were collected (scanning 200 to 2,000 atomic mass units) in both positive and negative modes (electrospray ionization [ESI] voltage, 6.0 kV; capillary temperature, 275°C; sheath gas pressure, 12 U and 150 lb/in2). Compounds were identified by the comparison of molecular weights, UV spectra, and retention times with published chemical data from standard databases (e.g., DNP 2008 and SciFinder 2007) and references. Extracts showing no UV absorbance were submitted to thin-layer chromatography (TLC)-MS analysis using Silica-gel 60 F254 (Merck).
Large-scale solid fermentation was carried out for strains DSM 41766T (12 liters), FXJ1.076 (12 liters), and FXJ1.264 (25 liters), respectively. The fermentation samples were extracted three times with ethanol, and the combined extracts then were subjected to repeated silica gel (100 to 200 mesh; Qingdao Haiyang Chemical) and Sephadex LH-20 column chromatographies. The chemical structures of the purified compounds were determined by MS and nuclear magnetic resonance (NMR) (Bruker 400 MHz or Bruker 600 MHz) spectroscopic analyses.
Antimicrobial assays.
The antimicrobial activities of etheromycin and compound 1.264-B against bacteria and fungi were determined using liquid cultures in 96-well plates (Costar 3599; Corning Inc.) according to the method described by Liu et al. (33). Indicator strains Bacillus subtilis CGMCC 1.2428, E. coli CGMCC 1.2385, Staphylococcus aureus CGMCC 1.2386, Candidia albicans CGMCC 2.538, and Candida pseudorugosa CGMCC 2.3107 were obtained from the China General Microbiological Culture Collection Center; drug-resistant indicator strains E. coli EMBL 4-1, Klebsiella pneumoniae 5-1, Pseudomonas aeruginosa 6-1, and Staphylococcus aureus MRSA 1-1 were obtained from Weifang Medical University, China. All indicator strains were incubated in LB broth at 37°C for 24 h and diluted to 105 to 106 cells/ml using LB broth thereafter. Two hundred μl of each of the dilutions and 2 μl compound solutions (4 mg/ml as a stock solution in DMSO and serial dilutions) were added in triplicate into the 96-well plates, followed by incubation at 37°C overnight. The optical density at 600 nm (OD600) was measured by BioTek Synerge H4 to determine the MICs.
Nucleotide sequence accession numbers.
The nucleotide sequences that encoded putative polyether epoxidases and 16S rRNA genes (>1,350 bp) of strains identified as positive for the polyether epoxidase gene were deposited in the GenBank database under the accession numbers listed in Table S1 in the supplemental material.
RESULTS
Suitability of the primers to amplify polyether epoxidase genes.
To evaluate the utility of these primers, three polyether-producing strains, “Streptomyces nanchangensis” NS3226 (nanchangmycin), Streptomyces violaceusniger CGMCC 4.1423T (nigericin), and Streptomyces cinnamonensis CGMCC 4.1619T (monensin) were used as positive controls, and Streptomyces coelicolor A3(2) served as a negative control. Positive PCR products were obtained from the three polyether producers but not from S. coelicolor A3(2), and they were proven to be polyether epoxidase gene fragments nigCI, monCI, and nanO, respectively, by sequencing and BLAST analysis.
Furthermore, putative epoxidase gene fragments were identified from another five known producer strains that produce polyether ionophores unrelated to the five published gene clusters: Streptomyces albus subsp. albus JCM 4703 (salinomycin) (35), Actinomadura yumaensis NRRL 12515T (antibiotic X-14868) (27), Actinomadura macra CGMCC 4.1513T (antibiotics CP-47433 and CP-47434) (9), Actinomadura fibrosa NRRL 18348T (antibiotics A-82810) (19), and Dactylosporangium salmoneum ATCC 31222T (antibiotic CP-44161) (10).
PCR-based screening of putative polyether epoxidase genes from actinomycetes.
The PCR-based screening of 1,068 actinomycetes isolated from eight different habitats (Table 1) identified 23 (2.2%) putative polyether epoxidase gene-positive strains. Samples from alpine soils collected from the Qinghai-Tibet Plateau yielded no positive strains, although more than 200 isolates were screened. No positive strain was detected from deep-sea samples either. Few putative polyether epoxidase genes were detected in isolates from lakeside soils of Kanas Lake in Xinjiang Province (1.4%), from mangrove sediments in Hainan Province (1.1%), or from coastal sediments in Bohai Bay (1.3%). A comparatively high occurrence of this gene was observed in isolates from sponges in the South China Sea (2.3%) and from medicinal plants in Yunnan Province (3.0%). The highest occurrence of putative polyether epoxidase genes was observed in isolates from acidic soils collected in Jiangxi Province (9.5%). A further 21 positive strains were obtained from 236 reference actinomycetes, encompassing eight major families and 35 genera of Actinomycetales (see Table S1 and List S1 in the supplemental material). The positive strains belong to only three families and four genera: Streptomycetaceae (Streptomyces), Thermomonosporaceae (Actinomadura), and Micromonosporaceae (Dactylosporangium and Micromonospora).
Phylogenetic analysis of putative polyether epoxidase genes and 16S rRNA genes.
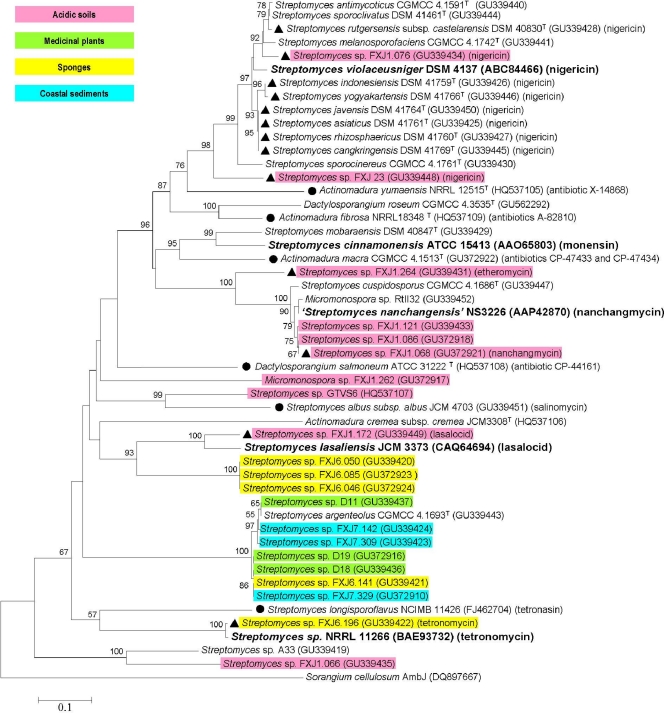
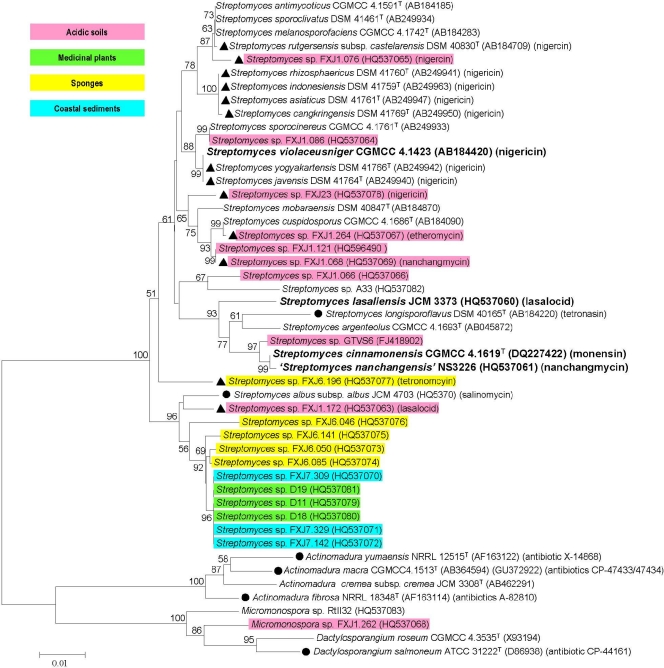
Phylogenetic analysis based on protein sequences revealed a large sequence diversity and novelty of the putative polyether epoxidases (Fig. 3), with identities ranging from 40 to 97%. Nearly half of the putative polyether epoxidase sequences clustered closely with known ones, and these results were supported by high bootstrap values. For instance, sequences from 11 reference strains and two isolates, FXJ1.076 and FXJ23, fell within a large and stable clade, with NigCI involved in the biosynthesis of nigericin in Streptomyces violaceusniger, supported by a bootstrap value of 98% and sharing relatively high sequence identities (>80%). Similarly, sequences from one reference strain and four isolates grouped into another stable clade, with NanO being involved in the biosynthesis of nanchangmycin in Streptomyces nanchangensis, and strains DSM 40847T, FXJ1.172, and FXJ6.196 grouped with MonCI, LasC, and TmnC, respectively, which are involved in the biosynthesis of monensin, lasalocid, and tetronomycin, respectively. Sequences from the other reported polyether producer strains (circles in Fig. 3) showed significant similarities to the five published epoxidases (with identities of 50 to 70%) and were located in well-separated clades. It was notable that all of the sequences obtained from coastal sediment samples and medicinal plant samples (blue and green in Fig. 3) fell into a well-circumscribed clade distantly related to the known epoxidases; in contrast, sequences from acidic soil isolates formed diverse branches interspersed in the whole tree. The topology of the epoxidase tree, where strains of different families and genera were mixed with each other, was completely incongruent with that of the 16S rRNA gene tree (Fig. 4), which clearly separated the four genera of the three families from each other.
Fig. 3.
Neighbor-joining tree of putative polyether epoxidases and five known polyether epoxidases (marked with boldface). Symbols: ▴, strains from which polyether products were identified in this study; •, strains producing known polyethers different from the five known ones; corresponding products are listed. Different colors represent different major sample sources of the positive strains. AmbJ, homolog of polyether epoxidases (serves as an outgroup). Significant bootstrap values (>50%) are indicated at the nodes. The scale bar represents 0.1 mutational events per site.
Fig. 4.
Neighbor-joining tree of 16S rRNA genes from polyether epoxidase gene-positive strains and four known polyether-producing strains (marked with boldface). Symbols: ▴, strains from which polyether products were identified in this study; •, strains producing known polyethers different from the five known ones; corresponding products are listed. Different colors represent different major sample sources of the positive strains. Significant bootstrap values (>50%) are indicated at the nodes. The scale bar represents 0.01 mutational events per site. The tetronomycin-producing strain and its 16S rRNA gene sequence were unavailable.
Taxonomic diversity of isolates from different habitats.
Using 95% sequence identity, more than half of the 16S rRNA gene sequences from deep-sea and mangrove sediments fell into one cluster, and fewer OTUs were obtained from these two sources than from the others. More OTUs were obtained for sequences from acidic soils and sponges (Table 1). A total of 74 OTUs were obtained, and the phylogenetic analysis (see Fig. S1 in the supplemental material) indicated that these OTUs covered 29 genera, 14 families, and 9 suborders of Actinomycetales. OTUs from each of the terrestrial habitats, from sponges, and from coastal sediments were interspersed among almost all of these suborders, showing wide taxonomic diversity. Streptomycetaceae, Streptosporangiaceae, Micromonosporaceae, Pseudonocardiaceae, and Nocardiaceae represented the majority of OTUs.
Chemical identification of polyether ionophores from strains identified as positive for the polyether epoxidase gene.
A total of 13 positive strains were examined and shown to produce polyether ionophores (see Table S2 in the supplemental material). The compound produced by strain FXJ6.196, with a molecular mass of 586 Da and maximum UV absorbances at 250 and 295 nm, was identified to be tetronomycin by comparison with the data in DNP 2008. The compound produced by strain FXJ1.172 had a molecular mass of 590 Da and a maximum UV absorbance at 304 nm, and it showed an MS2 fragmentation pattern identical to that of lasalocid A from the producer strain JCM 3373 (see Table S2); thus, it was designated lasalocid A. Nine strains of the large NigCI epoxidase clade all produced very similar active compounds, showing no UV absorbance and the same TLC behavior, with a molecular mass of 724 Da. 13C-NMR data for the compounds purified from large-scale fermentation extracts of strains DSM 41766T and FXJ1.076 (with a high yield of about 0.8 g) were the same (CDCl3, 150 MHz): 10.8(q, C-38), 13.0(q, C-31), 13.1(q, C-37), 13.2(q, C-36), 15.6(q, C-39), 16.3(q, C-33), 17.3(q, C-32), 22.7(q, C-34), 23.3(t, C-6), 25.8(t, C-18), 26.0(t, C-5), 27.4(q, C-35), 27.8(d, C-4), 30.9(t, C-19), 31.8(d, C-26), 32.3(t, C-23), 32.6(t, C-10), 35.2(d, C-22), 35.7(t, C-8), 36.7(d, C-12), 37.1(d, C-28), 37.4(t, C-27), 39.0(d, C-14), 42.3(t, C-15), 44.2(d, C-2), 57.4(q, C-40), 60.2(d, C-9), 68.2(t, C-30), 69.0(d, C-7), 72.9(d, C-3), 74.6(d, C-24), 77.2(d, C-25), 78.1(d, C-11), 81.7(d, C-17), 82.4(s, C-16), 83.5(s, C-21), 85.8(d, C-22), 97.0(s, C-29), 108.2(s, C-13), 177.5(s, C-1). According to the molecular mass and 13C-NMR results, these compounds were determined to be nigericin (11).
Strain FXJ1.068 produced two polyether-type compounds, 1.068-A and 1.068-B, bearing the same maximum UV absorbance at 234 nm and molecular masses of 866 and 880 Da, respectively. The multistage tandem mass spectrometry (MSn) fragmentation patterns of compound 1.068-A (see Table S3 in the supplemental material) exhibited high similarity to those of nanchangmycin (44), which strongly indicated this compound to be nanchangmycin or its stereoisomer. The MSn fragmentation patterns of compound 1.068-B (see Table S3) showed remarkable consistency with that of compound 1.068-A, with many fragment ions being 14 m/z higher than their counterparts in compound 1.068-A and some others identical to those of compound 1.068-A. These facts led to the designation of compound 1.068-B as a new analogue of nanchangmycin. However, the NMR of compound 1.068-B could not be done because of the low yield.
Two purified compounds (1.264-A [23.6 mg] and 1.264-B [23.2 mg]) were obtained from the large-scale fermentation of strain FXJ1.264. Compound 1.264-A was determined to be etheromycin by analyses of its ESI-MS and NMR data (Table 2) (40). Compound 1.264-B had a 13C-NMR spectrum similar to that of 1.264-A, and its molecular formula was determined to be C49H84O16 by analyses of its HRFT-ICRMS spectrum (m/z 927.5659 [M-H]−) and NMR data (Table 2). The 1H, 13C, DEPT, and HSQC spectra of 1.264-B showed 49 carbon signals for 11 methyl groups, 4 methoxy groups, 9 methylenes, 6 methines, 11 oxymethines, 1 dioxymethine, and 7 quaternary carbons (including one carboxyl carbon [δC 181.35]). The planar structure of 1.264-B was established by the comparison of the 13C-NMR spectrum to that of etheromycin (40) and by further two-dimensional NMR (2D-NMR) analysis (KEY 1H-1H COSY, 1H-13C HMBC; see Fig. S2 in the supplemental material). The relative configuration of 1.264-B was established by comparing the NMR data to that of etheromycin and 2D-ROESY analysis (see Fig. S2). The 1H- and 13C-NMR data of rings A to E and G were nearly identical to those of etheromycin, revealing their similar configurations. In the 2D-ROESY spectrum, the correlations of the C-4-methoxy group with H-5, the C-6-methoxy group with H-4, and the coupling constant of H4 and H5 (J4,5 = 11.4) revealed that the C-4-methoxy group and H-5 were on one side of ring A, while the C-6-methoxy group and H-4 were on the opposite side. The ROE correlation of the C-16-methoxy group with H-17, H-17 with the C-20-methoxy group, and the C-20-methoxy group with H-21 revealed that they were on the same side of rings C, D, and E. Although 1H- and 13C-NMR data of C-25 were not identical to those of etheromycin, the coupling constants (J24,25 and J25,26) were similar to those of etheromycin, and a careful analysis of 2D-ROESY signals of ring F revealed that 1.264-B and etheromycin shared identical relative configurations of ring F. The coupling constants of H-4′ and H-5′(J4′,5′ = 9.0 or 10.8) of ring G inferred that they were on opposite sides of the ring, like etheromycin. Due to the existence of the C-8-methoxy group, the configuration of ring A of 1.264-B should be the same as that of etheromycin (40). In addition, from a biogenetic perspective, the configuration of 1.264-B is considered identical to that of the cooccurring etheromycin, and its structure is shown in Fig. 1.
Table 2.
NMR (600 MHz, CDCl3) data for the free acid of 1.264-A (etheromycin) and 1.264-B
| Carbon | NMR data for: |
|||
|---|---|---|---|---|
| 1.264-Aa (etheromycin) |
1.264-B |
|||
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 COOH | 180.8 | 181.35b | ||
| 2 CH | 45.09 | 2.52, q(7.2) | 45.04 | 2.51, q(7.2) |
| 3 O—C—O | 99.5 | 99.5 | ||
| 4 CH | 38.9 | 1.64, m | 38.96 | 1.66, m |
| 5 O—CH | 76.56 | 3.66, d(11.4) | 76.58 | 3.67, d(11.4) |
| 6 C—O | 82.64 | 82.65 | ||
| 7 O—CH | 71.34 | 3.69, d(10.2) | 71.33 | 3.68, d(11.4) |
| 8 CH | 40.1 | 1.36, m | 40.1 | 1.38, m |
| 9 O—CH | 62.9 | 4.19, m | 63.5 | 4.21, m |
| 10 CH2 | 32.33 | 1.38, 1.63, m | 32.2 | 1.39, 1.63, m |
| 11 O—CH | 79.8 | 3.30, m | 79.81 | 3.30, m |
| 12 CH | 39.64 | 1.76, m | 40.2 | 1.76, m |
| 13 O—C—O | 109.1 | 109.05 | ||
| 14 CH2 | 36.75 | 1.86, 1.96, m | 37 | 1.93, 1.98, m |
| 15 CH2 | 32.77 | 1.61, 1.84, m | 32.2 | 1.70, 1.78, m |
| 16 C—O | 86.04 | 86.8 | ||
| 17 O—CH | 82.93 | 3.76, dd(8.1, 8.1) | 82.6 | 3.90, dd(11.4, 11.4) |
| 18 CH2 | 24.26 | 1.73, m | 24.8 | 1.78, 1.86, m |
| 19 CH2 | 28.6 | 1.42, 1.92, m | 28.63 | 1.56, m |
| 20 C—O | 85.2 | 85.83 | ||
| 21 O—CH | 83.87 | 4.20, m | 83.81 | 4.34, m |
| 22 CH2 | 29.74 | 1.39, 1.98, m | 29.65 | 1.23, 2.14, m |
| 23 CH2 | 24.36 | 1.73, 2.12, m | 24.82 | 2.00, m |
| 24 O—CH | 80.6 | 4.30, m | 80.69 | 4.34, m |
| 25 O—CH | 73.5 | 3.93, dd(10.5, 2.1) | 76.77 | 3.57, dd(10.2, 0) |
| 26 CH | 39.3 | 1.26, m | 39.56 | 1.28, m |
| 27 O—CH | 84.55 | 2.95, dd(9.6, 9.6) | 84.37 | 2.90, dd(10.2, 10.2) |
| 28 CH | 46.2 | 1.40, m | 46.89 | 1.45, dq(6.6, 9.6) |
| 29 O—C—O | 98.4 | 100.87 | ||
| 2-Me | 11.75 | 1.04, d(7.2) | 11.61 | 1.05, d(7.2) |
| 4-Me | 11.67 | 0.97, d(6.6) | 11.68 | 0.97, d(6.6) |
| 6-Me | 8.04 | 1.14, s | 8.05 | 1.15, s |
| 8-Me | 10.92 | 0.75, d(7.2) | 10.84 | 0.76, d(7.2) |
| 11-OMe | 58.74 | 3.47, s | 58.7 | 3.41, s |
| 12-Me | 13.54 | 0.98, d(7.2) | 13.32 | 0.96, d(7.2) |
| 16-Me | 29.2 | 1.22, s | 27.97 | 1.53, s |
| 20-Me | 22.55 | 1.06, s | 21.72 | 1.10, s |
| 26-Me | 13.2 | 0.91, d(6.6) | 13 | 0.94, d(6.6) |
| 27-OMe | 59.8 | 3.39, s | 60.19 | 3.40, s |
| 28-Me | 12.37 | 1.04, d(6.0) | 12.17 | 0.98, d(6.6) |
| 29-Me | 26.53 | 1.28, s | 21.9 | 1.22, s |
| 29-OMe | 48.15 | 3.13, s | ||
| Deoxysugar | ||||
| 1′ O—CH—O | 95.1 | 4.58, dd(0, 9.0) | 95.11 | 4.59, dd(1.5, 9.6) |
| 2′ CH2 | 30.44 | 1.53, 1.78, m | 30.5 | 1.55, 1.78, m |
| 3′ CH2 | 27.25 | 1.31, 2.13, m | 27.3 | 1.33, 2.15, m |
| 4′ O—CH | 79.8 | 2.78, ddd(4.2, 10.8, 13.2) | 79.88 | 2.79, ddd(4.2, 9.0, 10.8) |
| 5′ O—CH | 74.66 | 3.36, m | 74.72 | 3.37, m |
| 4′ OMe | 56.9 | 3.32, s | 56.91 | 3.32, s |
| 5′ Me | 18.16 | 1.26, d(6.0) | 18.18 | 1.26, d(6.0) |
Assignments based on reference 40.
No carboxyl signal in CDCl3; data were obtained from pyridine-d5.
Among the nine indicator strains, compound 1.264-B showed antimicrobial activity against only the three Gram-positive strains, i.e., Staphylococcus aureus CGMCC 1.2386, Staphylococcus aureus 1-1, and Bacillus subtilis CGMCC 1.2428, with MICs at 1.25, 2.5, and 10 μg/ml, respectively. The corresponding positive-control etheromycin (1.264-A) exhibited the same antimicrobial spectrum, with MICs at 0.625 to ∼1.25, 0.625, and 2.5 μg/ml, respectively.
1.264-B.
Pale yellow oil; IR (KBr) νmax 3425, 2972, 2879, 2935, 2831, 1595, 1458, 1377, 11111, 1095, 1072, 1004, 990, 959 cm−1; for 1H- and 13C-NMR data, see Table 2; HRFT-ICRMS m/z 927.5659 ([M-H]− calculated 927.5686).
DISCUSSION
Enormous efforts have been devoted to the discovery of novel bioactive secondary metabolites in the past 50 years, although only a few actinomycetes and their antibiotic products have been sampled (4). One of the most important steps for drug discovery from microbial resources is the careful selection of microorganisms. Genetic screening strategies provide a rapid method for cataloguing the biosynthetic potential of microorganisms. In this contribution, the gene of polyether epoxidase, which has been found to be a key enzyme for polyether biosynthesis, was chosen as a genetic marker for the discovery of polyether ionophores from actinomycetes. Although the degenerate primers were designed on the basis of only five polyether epoxidase genes from streptomycetes, these primers also detected epoxidase genes from another five known producer strains that produce polyethers different from the ones used for primer design, including four rare actinomycetes, thus demonstrating a good spectrum of the primers.
The PCR screening results showed that detection rates of putative polyether epoxidase genes were a little low, ranging from 0 to 9.5% in isolates from different habitats. Given that only about 120 of more than 10,000 actinomycete bioactive metabolites are polyether ionophores and that the incidence of type I PKS in actinomycetes ranges from 30 to 80% (2, 3), the low frequency of putative polyether epoxidase genes is considered normal. Obviously, higher occurrences of putative polyether epoxidase genes were observed in isolates from acidic soils with greater sequence diversity and novelty than those from other habitats. For the isolates from marine samples, only nine putative polyether epoxidases with low sequence diversity were identified out of more than 500 strains that covered as wide a taxonomic diversity as the acidic soil isolates according to 16S rRNA gene analysis, and no positive strain was obtained from deep-sea samples. Consequently, marine actinomycetes, which have been a research focus for discovering new bioactive metabolites (14, 15, 28), may not be the preferred option for polyether ionophore screening, whereas acidophilic actinomycetes are a better choice.
Eight positive rare actinomycetes were identified, four Actinomadura reference strains, two Dactylosporangium reference strains, and two Micromonospora isolates (RtII32 and FXJ1.262), four of which had been described to produce polyether ionophores according to the literature or patents. Micromonospora species have not, to our knowledge, been reported previously to be producers of polyether ionophores, thus the further characterization of polyether production from these putative epoxidase gene-positive strains will be of great interest. The rest of the 44 positive strains all were found to be Streptomyces spp. Moreover, among the 236 reference strains, occurrences of putative polyether epoxidases were much higher in the Streptomyces genus (15 out of 108 strains, 13.9%) than in rare actinomycete genera as a whole (6 out of 128 strains, 4.7%). It therefore is reasonable to propose that streptomycetes still are the main source of polyether ionophores, while the rare actinomycetes mentioned above also are worth consideration.
The results of our phylogenetic and chemical analyses showed a strong evolutionary correlation between putative polyether epoxidase genes and the corresponding polyether gene clusters. Four known polyethers were identified (see Table S2 in the supplemental material) from positive strains that grouped closely with the known producers in the epoxidase phylogeny (Fig. 3): nigericin, lasalocid A, nanchangmycins, and tetronomycin were found from strains (FXJ1.076 and eight others, FXJ1.172, FXJ1.068, and FXJ6.196) in the clades of NigCI, LasC, NanO, and TmnC, respectively. Etheromycin was found from strain FXJ1.264, which is located loosely at the periphery of the NanO clade. Further evidence of this correlation is that the main-chain structures of nigericin, monensin, nanchangmycin, antibiotic X-14868, antibiotic CP-47433/47434, antibiotic A-82810, and etheromycin are quite similar (Fig. 1), and the three clades of NigCI, MonCI, and NanO and the putative epoxidase from the other four producer strains were clustered together into a stable superclade (Fig. 3), while other epoxidases with distinct product structures were clustered discretely, such as LasC and TmnC. These findings suggested that strains containing homologous polyether epoxidases produce polyethers with similar structures and vice versa, indicating that the chemical structures of polyether ionophores could be predicted preliminarily by the phylogenetic analysis of the epoxidases involved, as long as there are reference sequences and structures. The screening strategy developed here paves the way for this, although the sequence data might not ensure that the strain contains a full suite of polyether ionophore biosynthetic genes or that the polyether gene cluster is expressed by the strain.
By comparing the epoxidase and 16S rRNA gene analyses, it was easy to find that several isolates producing known polyethers showed distant phylogenetic relationships with reference strains producing the same polyethers; e.g., strains FXJ23 (nigericin), FXJ1.068 (nanchangmycin), and FXJ1.172 (lasalocid) did not cluster with the respective producer strains for nigericin, nanchangmycin, and lasalocid in the 16S rRNA gene tree. Entirely different branch positions were observed for the producer reference strains of rare actinomycetes in the 16S rRNA gene and epoxidase phylogenies. These findings indicate that epoxidase genes, possibly accompanied by the corresponding polyether biosynthetic gene clusters, have undergone widespread horizontal transfer within the actinomycetes. On the other hand, although the horizontal gene transfer of epoxidases was evident, some taxonomically closely related strains also carried genes for highly homologous epoxidases, as exemplified by the clade of nigericin producer strains and strains from coastal sediments and medicinal plants, indicating the vertical inheritance of epoxidase genes within actinomycetes as well.
Polyketides often establish biological activity by constraints and modifications introduced by tailoring enzymes, which are encoded by genes clustered with the assembly line genes (46). Therefore, tailoring enzyme genes may contain sufficient phylogenetic information for the evolutionary history of corresponding gene clusters, and recent studies on these genes are beginning to give insights into the correlation between tailoring enzymes and corresponding chemical structures of products (17, 22). Our study indicates that a strong correlation does exist between polyether epoxidases and polyether ionophores and thus establishes a feasible genetic screening strategy that is useful for the rapid identification of known and the discovery of unknown polyether products in actinomycetes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to X. Guo and J. Gong (Institute of Microbiology, CAS) for their assistance in strain cultivation and DNA preparation and to Y. Luo (Institute of Microbiology, CAS) and J. Wang (Hisun Pharmaceutical Co., Ltd.) for their help in MS and NMR analyses. We also appreciate the international culture collections CGMCC, DSMZ, JCM, NBRC, and NRRL for providing reference strains.
This publication was supported by the Natural Science Foundation of China (NSFC; no. 30770002) and the National Hi-Tech Research and Development Program of China (grant no. 2007AA09Z420).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Anzai Y., et al. 2008. Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics. Chem. Biol. 15:950–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ayuso-Sacido A., Genilloud O. 2005. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 49:10–24 [DOI] [PubMed] [Google Scholar]
- 3. Ayuso A., et al. 2005. A novel actinomycete strain de-replication approach based on the diversity of polyketide synthase and nonribosomal peptide synthetase biosynthetic pathways. Appl. Microbiol. Biotechnol. 67:795–806 [DOI] [PubMed] [Google Scholar]
- 4. Baltz R. 2008. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharm. 8:557–563 [DOI] [PubMed] [Google Scholar]
- 5. Baltz R. H. 2007. Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131 [Google Scholar]
- 6. Bérdy J. 2005. Bioactive microbial metabolites. J. Antibiot. 58:1–26 [DOI] [PubMed] [Google Scholar]
- 7. Bhatt A., et al. 2005. Accumulation of an E, E, E-triene by the monensin-producing polyketide synthase when oxidative cyclization is blocked. Angew. Chem. Int. Ed. Engl. 44:7075–7078 [DOI] [PubMed] [Google Scholar]
- 8. Busti E., et al. 2006. Antibiotic-producing ability by representatives of a newly discovered lineage of actinomycetes. Microbiology 152:675–683 [DOI] [PubMed] [Google Scholar]
- 9. Celmer W., et al. April 1979. Polycyclic ether antibiotics produced by new species of actinomycete. U.S. patent 4,148,882. [Google Scholar]
- 10. Comai K., Sullivan A., Westley J. November 1981. Polyether ionophores as antiobesity and hypotriglyceridemic agents. U.S. patent 4,302,450. [Google Scholar]
- 11. David L., Ayala L. 1985. Abierixin, a new polyether antibiotic production, structrual determination and biological activities. J. Antibiot. 38:1655–1663 [DOI] [PubMed] [Google Scholar]
- 12. Demydchuk Y., et al. 2008. Analysis of the tetronomycin gene cluster: insights into the biosynthesis of a polyether tetronate antibiotic. ChemBioChem. 9:1136–1145 [DOI] [PubMed] [Google Scholar]
- 13. Dutton C., Banks B., Cooper C. 1995. Polyether ionophores. Nat. Prod. Rep. 12:165–181 [DOI] [PubMed] [Google Scholar]
- 14. Fenical W., Jensen P. 2006. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2:666–673 [DOI] [PubMed] [Google Scholar]
- 15. Fiedler H., et al. 2005. Marine actinomycetes as a source of novel secondary metabolites. Antonie Van Leeuwenhoek 87:37–42 [DOI] [PubMed] [Google Scholar]
- 16. Gallimore A. 2009. The biosynthesis of polyketide-derived polycyclic ethers. Nat. Prod. Rep. 26:266–280 [DOI] [PubMed] [Google Scholar]
- 17. Gao P., Huang Y. 2009. Detection and distribution of FADH2-dependent halogenase gene in major filamentous actinomycete taxonomic groups and its phylogenetic implication in organohalogen compound discovery. Appl. Environ. Microbiol. 75:4813–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ginolhac A., et al. 2005. Type I polyketide synthases may have evolved through horizontal gene transfer. J. Mol. Evol. 60:716–725 [DOI] [PubMed] [Google Scholar]
- 19. Hamill R., Yao R. March 1992. Process for producing antibiotic A8210 which comprises cultivating Actinomadura Fibrosa sp. nov. NRRL 18348, or an A82810-producing mutant thereof. U.S. patent 5,098,834. [Google Scholar]
- 20. Harvey B. M., et al. 2007. Insights into polyether biosynthesis from analysis of the nigericin biosynthetic gene cluster in Streptomyces sp. DSM 4137. Chem. Biol. 14:703–714 [DOI] [PubMed] [Google Scholar]
- 21. Hopwood D., et al. 1985. Genetic manipulation of Streptomyces: a laboratory manual. John Innes Foundation, Norwich, CT [Google Scholar]
- 22. Huitu Z., et al. 2009. PCR screening of 3-amino-5-hydroxybenzoic acid synthase gene leads to identification of ansamycins and AHBA-related antibiotic producers in actinomycetes. J. Appl. Microbiol. 106:755–763 [DOI] [PubMed] [Google Scholar]
- 23. Jensen P., Lauro F. 2008. An assessment of actinobacterial diversity in the marine environment. Antonie Van Leeuwenhoek 94:51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kevin I. I., Meujo D. D., Hamann M. 2009. Polyether ionophores: broad-spectrum and promising biologically active molecules for the control of drug-resistant bacteria and parasites. Expert Opin. Drug Discov. 4:109–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klepac-Ceraj V., Ceraj I., Polz M. 2006. Clusterer: extendable java application for sequence grouping and cluster analyses. Online J. Bioinf. 7:15–21 [Google Scholar]
- 26. Koskinen A. M., Karisalmi K. 2005. Polyketide stereotetrads in natural products. Chem. Soc. Rev. 34:677–690 [DOI] [PubMed] [Google Scholar]
- 27. Labeda D., Testa R., Lechevalier M., Lechevalier H. 1985. Actinomadura yumaensis sp. nov. Int. J. Syst. Bacteriol. 35:333–336 [Google Scholar]
- 28. Lam K. 2006. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 9:245–251 [DOI] [PubMed] [Google Scholar]
- 29. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 30. Larkin M., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 31. Leadlay P. F., et al. 2001. Engineering of complex polyketide biosynthesis-insights from sequencing of the monensin biosynthetic gene cluster. J. Ind. Microbiol. Biotechnol. 27:360–367 [DOI] [PubMed] [Google Scholar]
- 32. Liu W., et al. 2003. Rapid PCR amplification of minimal enediyne polyketide synthase cassettes leads to a predictive familial classification model. Proc. Natl. Acad. Sci. U. S. A. 100:11959–11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X. T., et al. 2006. ent-Rosane and labdane diterpenoids from Sagittaria sagittifolia and their antibacterial activity against three oral pathogens. J. Nat. Prod. 69:255–260 [DOI] [PubMed] [Google Scholar]
- 34. Migita A., et al. 2009. Identification of a gene cluster of polyether antibiotic lasalocid from Streptomyces lasaliensis. Biosci. Biotechnol. Biochem. 73:169–176 [DOI] [PubMed] [Google Scholar]
- 35. Miyazaki Y., et al. 1974. Salinomycin, a new polyether antibiotic. J. Antibiot. 27:814–821 [DOI] [PubMed] [Google Scholar]
- 36. Moffitt M., Neilan B. 2003. Evolutionary affiliations within the superfamily of ketosynthases reflect complex pathway associations. J. Mol. Evol. 56:446–457 [DOI] [PubMed] [Google Scholar]
- 37. Neefs J., Van de Peer Y., De Rijk P., Chapelle S., De Wachter R. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riedlinger J., et al. 2004. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. 57:271–279 [DOI] [PubMed] [Google Scholar]
- 39. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 40. Seto H., et al. 1979. Studies on the ionophorous antibiotics. XX. Some emperical rules for structural elucidation of polyether antibiotics by 13C-NMR spectroscopy. J. Antibiot. 32:239–243 [DOI] [PubMed] [Google Scholar]
- 41. Smith L., Hong H., Spencer J., Leadlay P. 2008. Analysis of specific mutants in the lasalocid gene cluster: evidence for enzymatic catalysis of a disfavoured polyether ring closure. ChemBioChem 9:2967–2975 [DOI] [PubMed] [Google Scholar]
- 42. Staunton J., Weissman K. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380–416 [DOI] [PubMed] [Google Scholar]
- 43. Sun Y., et al. 2003. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 10:431–441 [DOI] [PubMed] [Google Scholar]
- 44. Sun Y. H., Deng Z. X. 2003. Genetic and molecular basis for the biosynthesis of antibiotics in streptomyces nanchangensis. Ph.D. thesis. Shanghai Jiao Tong University, Shanghai, China [Google Scholar]
- 45. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 46. Walsh C. 2004. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303:1805–1810 [DOI] [PubMed] [Google Scholar]
- 47. Zazopoulos, et al. 2003. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat. Biotechnol. 21:187–190 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.