Abstract
gsiB, coding for glucose starvation-inducible protein B, is a characteristic member of the σΒ stress regulon of Bacillus subtilis and several other Gram-positive bacteria. Here we provide in silico evidence for the horizontal transfer of gsiB in lactic acid bacteria that are devoid of the σΒ factor.
INTRODUCTION
In Bacillus subtilis and many other Gram-positive species, the alternative sigma factor σΒ is responsible for redirecting RNA polymerase under stress conditions to transcribe a set of genes known as the general stress regulon (41). In contrast, lactic acid bacteria (LAB) are devoid of a σΒ ortholog and they have evolved other types of regulatory networks (36, 40, 44). Included among the genes of the σΒ regulon is the gene coding for the glucose starvation-inducible protein B (gsiB) (27). It is well established that gsiB is activated under different stress conditions, including starvation and exposure of cells to heat, acid, ethanol, and high osmolality, etc. (9, 25, 27). GsiB is of particular interest, since it belongs to the late embryogenesis abundant (LEA) family of proteins. LEA proteins were originally characterized in plants, where they were found to play an important role in the desiccation tolerance of maturing seeds and in vegetative organs under water deficit conditions (5, 42). In fact, B. subtilis GsiB was the first prokaryotic group 1 LEA protein to be described (35).
During our investigation of the plasmid content of Pediococcus pentosaceus ACA-DC 3431, isolated from traditional Formaela cheese, we sequenced and characterized plasmid pPS1. The protocols and the bioinformatic tools used have been described previously (4). Based on its features, pPS1 is a new member of the pC194/pUB110 family of rolling-circle replicating plasmids (data not shown) and it carries two open reading frames (ORFs). orf1 encodes a replication initiation protein (Rep) which exhibits 93% similarity (E value, 1.0e−159; 100% query coverage) to the respective protein encoded by the pLTK2 plasmid isolated from Lactobacillus plantarum (23). BLASTP searches for the orf2 product (128 amino acids) revealed an interesting similarity pattern. The most significant matches before the first nonbacterial protein could be classified into two categories. The first three hits were LAB proteins, i.e., a general stress protein (Gsp, corresponding to GenBank accession no. BAC99042 [direct submission]) encoded by plasmid pLS141-1 from Lactobacillus sakei LK141 (94% similarity; E value, 8.0e−44; 94% query coverage) and two identical GsiB proteins (corresponding to RefSeq accession no. ZP_06197568 and ZP_07367445 [direct submissions]) encoded on chromosomal contigs in the unfinished genomes of Pediococcus acidilactici strains 7_4 and DSM 20284 (93% similarity; E value, 8.0e−43; 98% query coverage). Remaining hits were also GsiB proteins, mainly from several Bacillales species (in all cases, similarity was ≥84%, the E value was ≤5.0e−17, and query coverage was ≥80%).
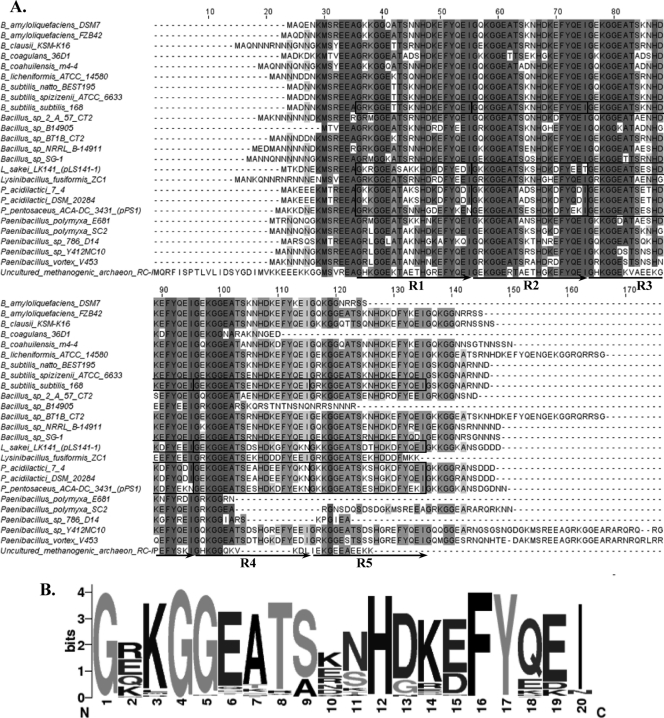
The multiple-sequence alignment of pPS1 GsiB with the related proteins mentioned above was generated by MUSCLE (17) and revealed a significant degree of conservation among these proteins (Fig. 1A). It should be noted that in the case of more than one BLAST match from the same species, the best hit was selected for the multiple-sequence alignment in order to exclude putative paralogs. Detailed inspection of the LAB GsiBs identified five tandem repeats of 20 amino acids in each sequence, an organization very similar to that previously reported for the GsiB of B. subtilis (Fig. 1A) (35). In fact, all GsiBs exhibited 20-mer tandem repeats in various numbers. The consensus sequence created by the WebLogo tool (14) for all repeats present in the multiple-sequence alignment revealed several highly conserved positions (Fig. 1B). ProDom (29) under default settings recognized several protein family domains corresponding to two LEA_5 (PF00477) and up to five KGG (PF10685) pfam motifs in each of the LAB GsiB sequences. The LEA_5 motif is characteristic of the group 1 LEA proteins (5, 21, 39), while the KGG motif is found in bacterial stress-induced proteins (33), as well as in eukaryotic LEA proteins (42). Furthermore, ProDom analysis of the sequence comprising the most conserved amino acid positions in the logo of the GsiB tandem repeats (i.e., GX1KGGEATSX2NHDKEFYQEI, where X1 is R, E, K, Q, or H and X2 is K, E, R, N, D, Q, or S) demonstrated that each tandem repeat is essentially part of the LEA_5 motif and includes the KGG motif (data not shown). In addition, the four LAB GsiB molecules each exhibited a significant hydrophilic index between −1 and −3 over its entire length, as revealed by the Kyte-Doolittle hydropathy analysis (performed at http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm) (16), and a high glycine content ranging from 15.2 to 15.8%. Our findings clearly suggest that LAB GsiBs belong to the hydrophilin-like superfamily that today contains diverse proteins (including all LEA groups) whose putative assigned function is to protect cells under conditions of dehydration (18). The in silico prediction of the existence of the gsiB gene in the pPS1 plasmid was further verified by reverse transcription-PCR (RT-PCR) using primers 5′-ATGGCTAAGAAAGATAACGA-3′ and 5′-GAATTGGCTTTTCCGCCT-3′ (data not shown) as described previously (4). Predictions concerning the secondary structures of the LAB GsiBs were inconclusive. Different predictors (e.g., PSIPRED [22] and Jpred 3 [12]) returned contradicting results, supporting both unstructured and highly structured organizations for these proteins (data not shown), which coincide with the current debate on the actual structure of hydrophilins (5, 10, 18, 19, 42).
Fig. 1.
Analysis of the sequences of LAB and non-LAB GsiBs. (A) Multiple-sequence alignment of pPS1 GsiB and related proteins. Gray shading in the alignment reflects the degree of amino acid conservation. Boxes indicate the 20-amino-acid tandem repeats identified manually in the LAB and B. subtilis GsiBs. (B) Consensus sequence created by the WebLogo tool (14) for all 20-mer repeats present in the multiple-sequence alignment. Regions R1 to R5 in the multiple-sequence alignment were used for this analysis.
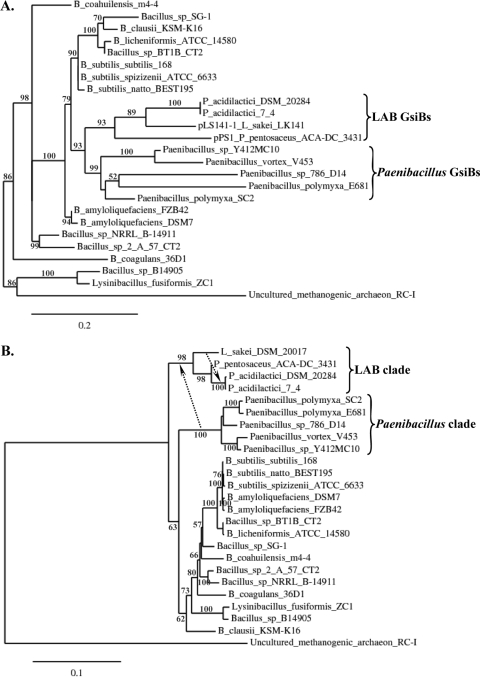
To gain more insight into the origin of GsiBs in LAB, their evolutionary relationship to other GsiBs was investigated. The multiple-sequence alignment shown in Fig. 1A was further curated with Gblocks using default parameters (11), and the phylogenetic tree of GsiBs was calculated by PhyML (20) with the WAG substitution model and the χ2-based parametric approximate likelihood-ratio test (aLRT) for branch support (3). The analysis was performed with the Phylogeny.fr pipeline (16). LAB GsiBs formed a separate clade within the phylogenetic tree that was fully surrounded by Bacillales-derived sequences (Fig. 2A). No other sequences belonging to LAB could be placed within this phylogenetic tree, since even a PSI-BLAST search for GsiB did not return any additional LAB homolog, even distantly related. While the phylogenetic distance between the LAB and the Bacillales species carrying GsiBs is obvious, the close phylogenetic relatedness of the GsiBs suggested some type of horizontal gene transfer (HGT). To assess HGT, we employed T-REX, a program that identifies topological violations in a gene/protein tree in relation to the respective species tree and calculates the possibility of HGT events during the evolution of the considered organisms (7, 26). For this reason, we constructed the 16S rRNA gene phylogenetic tree of all strains presented in Fig. 2A (Fig. 2B). Full or partial 16S rRNA gene sequences were retrieved from the Ribosomal Database Project website (13) or GenBank (6). A partial 16S rRNA gene sequence of P. pentosaceus ACA-DC 3431 was determined and deposited in the EMBL database. In the case of L. sakei, we used the 16S rRNA gene sequence of the L. sakei type strain (DSM 20017), since the 16S rRNA gene sequence of L. sakei LK141 is not available. Multiple-sequence alignment of the 16S rRNA gene sequences was performed using ClustalW (38), and the alignment was curated with Gblocks under settings for a less stringent selection (11). The phylogenetic tree was constructed with the neighbor-joining method (34) and the Kimura 2-parameter substitution model using the Phylogeny.fr pipeline (16). Branch support was estimated by bootstrapping (1,000 replicates). T-REX analysis using the detection mode of several HGTs by iteration and the optimization criterion of bipartition dissimilarity (8) predicted HGT routes that could resolve the differences between the GsiB phylogenetic tree and the relevant 16S rRNA gene sequence species tree. Among these routes, we identified one that could mediate the transfer of gsiB from the Paenibacillus clade to LAB and the dispersion of this gene from the pLS141-1 L. sakei plasmid to the chromosomes of the two strains of P. acidilactici (represented by arrows in Fig. 2B). A number of scenarios for the gsiB HGT among Bacillales were also predicted, including the transfer of this gene from Bacillus to Paenibacillus species (data not shown).
Fig. 2.
HGT route for the acquisition of GsiB by LAB. (A) Maximum-likelihood phylogenetic tree of pPS1 GsiB and related proteins. (B) Neighbor-joining phylogenetic tree of the 16S rRNA gene sequences of the species carrying GsiBs presented in panel A. Dotted arrows indicate HGT routes predicted by T-REX (26). Phylogenetic analysis was performed at the Phylogeny.fr pipeline (16), as described in the text. The lengths of the curated multiple-sequence alignments used for the construction of phylogenetic trees presented in panels A and B were 73 and 1,449 positions, respectively. Branch support values above 50% are indicated. Brackets highlight the positions of important clades in the phylogenetic trees that are further discussed in the text.
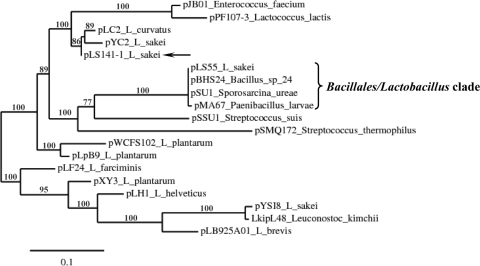
The HGT between Paenibacillus and LAB was also supported by further analysis of pLS141-1. The most significant matches in the BLASTP analysis for the pLS141-1 Rep protein (in all cases, similarity was ≥88%, the E value was ≤4.0e−81, and query coverage was ≥94%) were used to construct the phylogenetic tree shown in Fig. 3 with the same methodology used for the pPS1 GsiB tree. Among the evolutionary partners of the pLS141-1 Rep, the majority of which were of LAB origin, the Rep proteins encoded by the L. sakei plasmid pLS55 (1), the Paenibacillus larvae plasmid pMA67 (30), the Sporosarcina ureae plasmid pSU1 (corresponding to RefSeq accession no. YP_003560375 [direct submission]), and the Bacillus sp. strain #24 plasmid pBHS24 (32) could be identified (in all cases, similarity was ≥89%, the E value was ≤8.0e−99, and query coverage was 99%) (Fig. 3). The aforementioned plasmids, i.e., pLS55, pMA67, pSU1, and pBHS24, are practically identical (with fewer than 10 nucleotides differing over their 5-kb lengths), and they carry the tetracycline resistance gene tetL. Importantly, the partaking of the pLS55/pMA67 replicon by both P. larvae and L. sakei has been suggested to account for the HGT of the tetL gene in these species (30). Since the replication backbone of pLS141-1 is similar to the pLS55/pMA67/pSU1/pBHS24 replicon (data not shown), the plasmid is a perfect candidate as a Bacillales/Lactobacillus vehicle. Such an intraspecies vehicle, able to overcome the species barrier, is a prerequisite when HGT is mediated by plasmids in bacteria (37). pLS141-1 could have acted as an acceptor of the ancestral gsiB in Paenibacillus species. Transmission of pLS141-1 to LAB may account for their acquisition of gsiB, which could have further moved by recombination events to the chromosome (e.g., in the case of the gsiB genes of P. acidilactici strains 7_4 and DSM 20284) or to plasmids (e.g., in the case of gsiB of pPS1). In fact, Paenibacillus and LAB species coexist in several ecological niches, including food matrices like milk or dairy products (15), and thus HGT among these bacteria is feasible. Furthermore, it has been suggested previously that gsiB was transferred to B. subtilis by HGT from plants (24). In our opinion, the acquisition of gsiB by B. subtilis through HGT is also supported by the fact that the gsiB gene is absent in the species of the Bacillus cereus group (2). Consequently, LAB GsiBs seem to be the endpoint of a domino of HGT events that started from plants.
Fig. 3.
Relatedness of the pLS141-1 replicon with the pLS55/pMA67/pSU1/pBHS24 replicon. The phylogenetic tree of pLS141-1 Rep and related proteins was constructed using maximum likelihood at the Phylogeny.fr pipeline (16), as described in the text. The length of the curated multiple-sequence alignment used for the construction of the phylogenetic tree was 214 positions. The bracket and the solid arrow highlight the positions of the Bacillales/Lactobacillus clade and the pLS141-1 protein, respectively, which are further discussed in the text.
Finally, inspection of the LAB gsiB sequences revealed that no σΒ promoter (31) could be identified (Fig. 4). This finding shows that irrespective of the underlying evolutionary process of gsiB acquisition by LAB, the σΒ promoter was rejected, since it would have been useless for regulating the expression of the gene in these bacteria that are devoid of a σΒ ortholog.
Fig. 4.
Promoter regions of the LAB and B. subtilis strain 168 gsiB genes. In LAB, no σΒ promoters were detected that would satisfy the requirements of the consensus σΒ sequences GTTTAA (−35 region), and GGG(A/T)A(A/T) (−10 region) determined previously (31). The σΒ promoter was detected only for the gsiB gene of B. subtilis. In all other cases, the non-σΒ promoters and ribosome binding site (RBS) sequences were predicted as described before (4). The right arrow indicates the start codons of the genes.
It should be emphasized that no phenotype is as yet associated with the gsiB B. subtilis mutant (28) and heterologous expression of plant LEA proteins in Escherichia coli results in only a moderate improvement of its ability to grow under salt or low-temperature stress conditions (43). To the best of our knowledge, this is the first report concerning the identification of a putative GsiB in LAB, providing in silico evidence for the existence of group 1 LEA hydrophilins in these bacteria. We are now investigating the functional role of GsiB in LAB stress physiology.
Nucleotide sequence accession numbers.
The annotated nucleotide sequence of pPS1 (2,721 bp) was deposited in the EMBL database under accession no. FN869858. The partial 16S rRNA gene sequence of P. pentosaceus ACA-DC 3431 was deposited in the EMBL database under accession no. FR714835.
Acknowledgments
Ioanna-Areti Asteri was financially supported by the State Scholarships Foundation of Greece (IKY-Idryma Kratikon Ypotrofion).
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Ammor M. S., et al. 2008. Two different tetracycline resistance mechanisms, plasmid-carried tet(L) and chromosomally located transposon-associated tet(M), coexist in Lactobacillus sakei Rits 9. Appl. Environ. Microbiol. 74:1394–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson I., et al. 2005. Comparative genome analysis of Bacillus cereus group genomes with Bacillus subtilis. FEMS Microbiol. Lett. 250:175–184 [DOI] [PubMed] [Google Scholar]
- 3. Anisimova M., Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 4. Asteri I. A., et al. 2010. Characterization of pLAC1, a cryptic plasmid isolated from Lactobacillus acidipiscis and comparative analysis with its related plasmids. Int. J. Food Microbiol. 141:222–228 [DOI] [PubMed] [Google Scholar]
- 5. Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148:6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. 2008. GenBank. Nucleic Acids Res. 36:D25–D30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boc A., Makarenkov V. 2003. New efficient algorithm for detection of horizontal gene transfer events, p. 190–201In Benson G., Page R. (ed.), Algorithms in bioinformatics, vol. 2812 Springer, Berlin, Germany [Google Scholar]
- 8. Boc A., Philippe H., Makarenkov V. 2010. Inferring and validating horizontal gene transfer events using bipartition dissimilarity. Syst. Biol. 59:195–211 [DOI] [PubMed] [Google Scholar]
- 9. Brigulla M., et al. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Browne J. A., et al. 2004. Dehydration-specific induction of hydrophilic protein genes in the anhydrobiotic nematode Aphelenchus avenae. Eukaryot. Cell 3:966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 12. Cole C., Barber J. D., Barton G. J. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cole J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Jonghe V., et al. 2010. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int. J. Food Microbiol. 136:318–325 [DOI] [PubMed] [Google Scholar]
- 16. Dereeper A., et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garay-Arroyo A., Colmenero-Flores J. M., Garciarrubio A., Covarrubias A. A. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275:5668–5674 [DOI] [PubMed] [Google Scholar]
- 19. Goyal K., et al. 2003. Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J. Biol. Chem. 278:12977–12984 [DOI] [PubMed] [Google Scholar]
- 20. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 21. Hundertmark M., Hincha D. K. 2008. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202 [DOI] [PubMed] [Google Scholar]
- 23. Kaneko Y., et al. 2000. Development of a host-vector system for Lactobacillus plantarum L137 isolated from a traditional fermented food produced in the Philippines. J. Biosci. Bioeng. 89:62–67 [DOI] [PubMed] [Google Scholar]
- 24. Koonin E. V., Makarova K. S., Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kovacs T., Hargitai A., Kovacs K. L., Mecs I. 1998. pH-dependent activation of the alternative transcriptional factor sigmaB in Bacillus subtilis. FEMS Microbiol. Lett. 165:323–328 [DOI] [PubMed] [Google Scholar]
- 26. Makarenkov V. 2001. T-REX: reconstructing and visualizing phylogenetic trees and reticulation networks. Bioinformatics 17:664–668 [DOI] [PubMed] [Google Scholar]
- 27. Maul B., Volker U., Riethdorf S., Engelmann S., Hecker M. 1995. Sigma B-dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis. Mol. Gen. Genet. 248:114–120 [DOI] [PubMed] [Google Scholar]
- 28. Mueller J. P., Bukusoglu G., Sonenshein A. L. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J. Bacteriol. 174:4361–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mulder N., Apweiler R. 2007. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol. 396:59–70 [DOI] [PubMed] [Google Scholar]
- 30. Murray K. D., Aronstein K. A., de Leon J. H. 2007. Analysis of pMA67, a predicted rolling-circle replicating, mobilizable, tetracycline-resistance plasmid from the honey bee pathogen, Paenibacillus larvae. Plasmid 58:89–100 [DOI] [PubMed] [Google Scholar]
- 31. Petersohn A., et al. 1999. Identification of σB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phelan R. W., et al. 2011. Tetracycline resistance-encoding plasmid from Bacillus sp. strain #24, isolated from the marine sponge Haliclona simulans. Appl. Environ. Microbiol. 77:327–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robbe-Saule V., Lopes M. D., Kolb A., Norel F. 2007. Physiological effects of Crl in Salmonella are modulated by σS level and promoter specificity. J. Bacteriol. 189:2976–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 35. Stacy R. A., Aalen R. B. 1998. Identification of sequence homology between the internal hydrophilic repeated motifs of group 1 late-embryogenesis-abundant proteins in plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis. Planta 206:476–478 [DOI] [PubMed] [Google Scholar]
- 36. Sugimoto S., Abdullah-Al-Mahin, Sonomoto K. 2008. Molecular chaperones in lactic acid bacteria: physiological consequences and biochemical properties. J. Biosci. Bioeng. 106:324–336 [DOI] [PubMed] [Google Scholar]
- 37. Thomas C. M., Nielsen K. M. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 [DOI] [PubMed] [Google Scholar]
- 38. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tunnacliffe A., Wise M. J. 2007. The continuing conundrum of the LEA proteins. Naturwissenschaften 94:791–812 [DOI] [PubMed] [Google Scholar]
- 40. van de Guchte M., et al. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187–216 [PubMed] [Google Scholar]
- 41. van Schaik W., Abee T. 2005. The role of sigmaB in the stress response of Gram-positive bacteria—targets for food preservation and safety. Curr. Opin. Biotechnol. 16:218–224 [DOI] [PubMed] [Google Scholar]
- 42. Wise M. J., Tunnacliffe A. 2004. POPP the question: what do LEA proteins do? Trends Plant Sci. 9:13–17 [DOI] [PubMed] [Google Scholar]
- 43. Ying L. A. N., Dan C. A. I., Yi-Zhi Z. 2005. Expression in Escherichia coli of three different soybean late embryogenesis abundant (LEA) genes to investigate enhanced stress tolerance. J. Integr. Plant Biol. 47:613–621 [Google Scholar]
- 44. Yother J., Trieu-Cuot P., Klaenhammer T. R., de Vos W. M. 2002. Genetics of streptococci, lactococci, and enterococci: review of the sixth international conference. J. Bacteriol. 184:6085–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]