Abstract
The virulence determinants of uropathogenic Escherichia coli have been studied extensively over the years, but relatively little is known about what differentiates isolates causing various types of urinary tract infections. In this study, we compared the genomic profiles of 45 strains from a range of different clinical backgrounds, i.e., urosepsis, pyelonephritis, cystitis, and asymptomatic bacteriuria (ABU), using comparative genomic hybridization analysis. A microarray based on 31 complete E. coli sequences was used. It emerged that there is little correlation between the genotypes of the strains and their disease categories but strong correlation between the genotype and the phylogenetic group association. Also, very few genetic differences may exist between isolates causing symptomatic and asymptomatic infections. Only relatively few genes that could potentially differentiate between the individual disease categories were identified. Among these were two genomic islands, namely, pathogenicity island (PAI)-CFT073-serU and PAI-CFT073-pheU, which were significantly more associated with the pyelonephritis and urosepsis isolates than with the ABU and cystitis isolates. These two islands harbor genes encoding virulence factors, such as P fimbriae (pyelonephritis-associated fimbriae) and an important immunomodulatory protein, TcpC. It seems that both urovirulence and growth fitness can be attributed to an assortment of genes rather than to a specific gene set. Taken together, urovirulence and fitness are the results of the interplay of a mixture of factors taken from a rich menu of genes.
INTRODUCTION
Urinary tract infection (UTI) is one of the most common bacterial infections of humans and a major cause of morbidity. It is estimated that there are more than 10 million cases in Western Europe per year. UTI also accounts for 25 to 40% of all nosocomial infections, making these infections an important medical and financial burden on health care systems (16, 37). UTI usually starts as a bladder infection but can, depending on the bacterial strain, ascend to the kidneys and may ultimately can result in renal failure or dissemination to the blood. UTI is classified into disease categories according to the focal point and the severity of infection: bacteriuria (the urine), cystitis (the bladder), pyelonephritis (the kidneys), and urosepsis (the blood). Growth in urine in the absence of symptoms is called asymptomatic bacteriuria (ABU). ABU resembles a commensal-like carrier state, which often goes unnoticed by the patient. UTIs primarily affect women and girls; 40 to 50% of adult women will experience at least one UTI episode during their lifetime (8, 34).
Escherichia coli is the most important etiological agent of UTIs and is associated with more than 80% of all such infections (36). E. coli strains are usually associated with a commensal lifestyle in the gastrointestinal tract. However, some strains have acquired the ability to cause disease. Intestinal pathogenic E. coli can cause a range of intestinal disorders, while extraintestinal pathogenic E. coli (ExPEC) causes a variety of extraintestinal infections, such as urinary tract infections, septicemia, neonatal meningitis, and infections of the respiratory tract (19). Unlike diarrheagenic strains, ExPEC strains do not cause disease in the intestinal tract; however, they are normally excellent long-term colonizers of the intestine (17). ExPEC strains constitute about 20% of the E. coli strains of the human fecal flora in healthy individuals (35). It is generally believed that strains infecting the urinary tract originate from the fecal flora. E. coli is also found in the vagina in about 20% of healthy women (8, 25).
The phenotypic profile of a strain is reflected in the genome content. The genomes of E. coli strains vary in size from about 4.6 to 5.6 Mb. They have mosaic structures and consist of a conserved core of genes plus a flexible gene pool that is strain specific. The flexible gene pools typically comprise genes encoding niche-specific fitness factors and also, in the case of pathogenic strains, virulence factors (6). The genetic determinants of uropathogenic E. coli (UPEC) have been studied extensively, and a range of virulence and fitness factors, including various types of adhesins, toxins, and iron uptake systems, have been implicated in pathogenesis. However, the specific factors that differentiate strains of E. coli that cause different types of UTI remain unclear. While bacterial strain variation most likely plays an important part in determining the outcome of any initial colonization, individual patient parameters are also important. Understanding the genetic basis for pathogenicity in ExPEC, especially with regard to genetic determinants associated with different types of UTIs, will be important in the development of preventive measures.
Recently, we designed a multigenome E. coli microarray for comparative genomic hybridization (CGH) of E. coli isolates from diverse origins (40). The array was based on 31 complete E. coli genomic sequences representing a wide range of pathogenic, as well as commensal, isolates. The diverse nature of the CGH array makes it an excellent tool for studying the genomic contents of unsequenced E. coli isolates. Here, we employed this array to compare a range of E. coli strains causing urinary tract infections in an attempt to further discriminate between isolates causing different types of UTI.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains included in the study are listed in Table 1. The ABU isolates are all confirmed ABU strains, i.e., they were isolated from the urine of asymptomatic women with bacteriuria, defined as two consecutive voided urine specimens with isolation of the same bacterial strain in counts of >105 CFU/ml. The cystitis and pyelonephritis strains have been isolated in monocultures from the urine of patients diagnosed with acute cystitis and acute pyelonephritis, respectively, all at Hvidovre Hospital (Denmark). The urosepsis isolates were cultured from the blood of patients suffering from UTI-derived sepsis at the Princess Alexandra Hospital (Brisbane, Australia). All strains were grown in modified LB medium (39) prior to genomic-DNA isolation and phenotypic characterization. Three well-characterized strains commonly reported in the literature, namely, CFT073 (pyelonephritis), 83972 (ABU), and Nissle 1917 (commensal/probiotic; Mutaflor, Ardeypharm GmbH), were included as reference strains for comparative purposes.
Table 1.
Phenotypic and genotypic characteristics of strains
| Strain | Source | Phylo- genetic group | Reference | fima | YAf | papa | papGUMN026a | papG83972a | papGIAI39a | HAb | Hemolysisc | Motility (mm)d | tcpCa | PAI-serU | sfa-foc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MG1655 | Commensal | A | BEAICDFGH+ | + | − | − | − | − | − | − | 47 | − | − | − | |
| B2A | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | + | 80 | + | + | C+ |
| B3 | Urosepsis | B2 | This study | BDFGH+ | − | IBAHCDJKEFG+ | + | + | − | − | + | 80 | − | − | CBADEFGH+ |
| B6 | Urosepsis | B2 | This study | BEAICDFGH+ | + | − | − | − | − | − | − | 76 | − | + | |
| B8 | Urosepsis | B2 | This study | BEAICDFGH+ | + | − | − | − | − | − | + | 80 | + | + | CBADEFGH+ |
| B18 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBCDJKEFG+ | − | + | − | − | + | 0 | + | + | C+ |
| B20 | Urosepsis | B2 | This study | BEACDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 32 | − | − | |
| B26 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 80 | + | + | CBADEFGH+ |
| B30 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 77 | − | − | S+ |
| B31 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBHCDJKEFG+ | − | + | − | + | + | 0 | + | + | |
| B34 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 31 | − | − | CBDEFSH+ |
| B37 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | + | − | − | + | 0 | + | + | CBADEFGH+ |
| B39 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBACDJKEFG+ | − | + | − | − | + | 0 | + | + | C+ |
| B40 | Urosepsis | B2 | This study | BEAICDFGH+ | + | IBCDJKEFG+ | − | + | − | − | + | 0 | + | + | CBADEFSH+ |
| B41 | Urosepsis | B2 | This study | BEACDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 36 | − | − | |
| CFT073 | Urosepsis/pyelonephritis | B2 | 30 | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | + | 19 | + | + | CBADEFGH+ |
| VR150 | Pyelonephritis | B2 | 14 | BEAICDFGH+ | + | F+ | − | − | − | − | + | 78 | + | + | CBADEFGH+ |
| VR151 | Pyelonephritis | B2 | 14 | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | + | − | + | + | 10 | + | + | CBADEFGH+ |
| VR152 | Pyelonephritis | B1 | 14 | BEAICDFGH+ | + | A+ | − | − | − | − | − | 80 | − | − | |
| VR153 | Pyelonephritis | B2 | 14 | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 10 | − | − | |
| VR154 | Pyelonephritis | D | 14 | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 65 | − | − | |
| VR155 | Pyelonephritis | D | 14 | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | + | − | 54 | − | − | |
| VR156 | Pyelonephritis | B2 | 14 | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | + | − | + | + | 80 | − | − | CBADEFGH+ |
| VR157 | Pyelonephritis | B2 | 14 | BEACDFGH+ | + | IBCDJKEFG+ | + | − | − | + | + | 75 | + | + | C+ |
| VR145 | Cystitis | B2 | 40 | BEAICDFGH+ | + | − | − | − | − | − | + | 80 | − | − | CBADEFGH+ |
| VR146 | Cystitis | B2 | 40 | BEAICDFGH+ | + | I+ | − | − | − | − | + | 69 | − | − | CBDEFH+ |
| VR147 | Cystitis | D | This study | BEAICDFGH+ | + | IBAHCDJKEFG+ | + | − | − | − | − | 38 | − | − | |
| VR148 | Cystitis | A | This study | BEAICDFGH+ | − | − | − | − | − | − | − | 80 | − | − | |
| VR149 | Cystitis | D | This study | BEAICDFGH+ | + | − | − | − | − | − | − | 80 | − | − | |
| VR158 | Cystitis | B2 | This study | BEAICDFGH+ | + | − | − | − | − | − | − | 68 | − | + | |
| VR159 | Cystitis | B2 | 40 | BEAICDFGH+ | + | IBA+ | − | − | − | − | + | 0 | − | − | CBDEFH+ |
| VR160 | Cystitis | D | This study | BEAICDFGH+ | − | IBAHCDJKEFG+ | + | − | − | + | − | 58 | − | − | |
| VR161 | Cystitis | B1 | This study | BEAICDFGH+ | − | − | − | − | − | − | − | 80 | − | − | |
| 83972 | ABU | B2 | 1, 27 | BDFGH+ | − | IBAHCDJKEFG+ | − | + | − | − | − | 0 | − | − | CBADEFGH+ |
| VR11 | ABU | B2 | 7 | BEAICDFGH+ | + | − | − | − | − | − | + | 80 | − | − | CBADEFGH+ |
| VR12 | ABU | B1 | 7 | BEAICDFGH+ | + | − | − | − | − | − | − | 80 | − | − | |
| VR89 | ABU | D | 33 | BEAICDFGH+ | − | EFG++ | − | − | + | − | − | 0 | − | − | |
| VR90 | ABU | D | 33 | BEAICDFGH+ | + | IBAG++ | − | − | + | − | − | 0 | − | − | C+ |
| VR91 | ABU | B1 | 33 | DH+ | − | A+ | − | − | − | − | − | 70 | − | − | |
| VR92 | ABU | B1 | 33 | BEAICDFGH+ | + | − | − | − | − | − | − | 0 | − | − | |
| VR94 | ABU | B2 | 33 | BEAICDFGH+ | + | I+ | − | − | − | − | − | 0 | − | − | CBADEFSH+ |
| VR95 | ABU | A | 33 | BEAICDFGH+ | − | −e | − | − | − | − | − | 8 | − | − | |
| VR96 | ABU | B1 | 33 | H+ | − | − | − | − | − | − | − | 62 | − | − | |
| VR135 | ABU | B2 | 14 | BEAICDFGH+ | + | I+ | − | − | − | − | − | 57 | − | − | CBADEFSH+ |
| VR136 | ABU | B2 | 14 | BEACDFGH+ | + | − | − | − | − | − | + | 24 | + | + | CADEFGSH+ |
| VR137 | ABU | D | 14 | BEAICDFGH+ | + | IBAHCDJKFG+ | + | − | + | + | − | 0 | − | − | C+ |
Operon genes presented, based on CGH data. +, present; −, absent.
HA, hemagglutination. The same results were obtained with and without mannose. +, present; −, absent.
All nonhemolytic strains lacked the hlyABCD genes, according to CGH data, with the exception of strain 83972, which carries the genes but is unable to produce functional hemolysin due to sequence variations. +, present; −, absent.
Average motility on urine plates (four replicates); 80 indicates maximum motility (i.e., the plates were completely covered after 16 h).
Most likely carries some other variant of papG (33).
YA, yeast agglutination. +, present; −, absent.
Microarray design and sample preparation.
The detailed design of the custom microarray has been described elsewhere (40); the 31 E. coli genomes used for designing the microarray included one urosepsis/pyelonephritis (CFT073), one pyelonephritis (strain 536), four cystitis (F11, IAI39, UMN026, and UTI89), and two ABU (VR50 and 83972) strains, as well as other pathogenic E. coli strains (such as avian pathogenic E. coli [APEC] and enterohemorrhagic E. coli [EHEC]) and a number of K-12 isolates. Of the 31 strains, 6, 8, 11, and 3 belonged to the phylogenetic groups A, B1, B2, and D, respectively. The microarray consisted of 134,285 probe sets (of 50- to 75-mers) representing 16,098 E. coli target genes. The genomic DNA was isolated using the Illustra bacterial genomic Prep Spin Kit (GE Healthcare; 28-9042-58), and the samples were diluted to the recommended concentration. Sample preparation was then carried out using the NimbleGen Arrays User's Guide for CGH analysis. All isolates, except the reference strains CFT073 and 83972, were run as single samples.
CGH data analysis.
Data analysis was performed in R (statistical software), using the “oligo” package for analysis of oligonucleotide arrays at the probe level (Bioconductor) (9). The RMA algorithm was used to perform background subtraction, normalization, and expression calculation (with output on the log2 scale). BLAST atlases were created using the GeneWiz whole-genome visualization tool (31). Hierarchical agglomerative clustering was performed in R, using the “hclust” clustering method, a Euclidean distance measure, and the complete genomic profile of each strain (log values for all genes represented by four or more probes). For interpretation of the CGH data, the following cutoff values (log) were selected for presence/absence calls of the individual probes: log values of 6 to 8 were considered negative, 8 to 10 borderline/unknown, and 10 to 12 positive. The cutoff values were selected based on comparison of the CGH data obtained for CFT073 (run in triplicate) and the known genome sequence of the strain.
PCR analysis.
The microarray data were verified by PCR amplification using primers targeting genes located upstream and downstream of pathogenicity island (PAI)-CFT073-serU.
Phylogenetic group determination.
For identification of phylogenetic group associations, the rapid triplex PCR method was employed, using primers targeting two genes (chuA and yjaA) and one anonymous DNA fragment (TspE4.C) (4).
Statistical analysis.
Statistical analysis was performed mainly by use of Fisher's exact test. For multigroup comparisons, a 4-group Fisher's exact test was performed. For upper versus lower UTI comparisons, the prevalence data obtained for the urosepsis and pyelonephritis isolates were compared with the data obtained for the cystitis and ABU isolates; P values of < 0.05 were considered significant.
Yeast agglutination and hemagglutination tests.
The presence of type 1 fimbriae was assayed by the ability of the bacteria to agglutinate yeast (Saccharomyces cerevisiae) cells on glass slides. Ten microliters of fresh overnight cultures was mixed with 10 μl 5% yeast cells. The experiment was repeated after the cells had been washed and resuspended in LB containing 50 mM methyl-α-d-mannopyranoside.
The capacity of bacteria to express P fimbriae was assayed by hemagglutination with human type A red blood cells (RBCs). The RBCs were washed twice with phosphate-buffered saline (PBS), and 10 μl of 5% RBCs was mixed with a single bacterial colony (freshly grown on LB plates) suspended in PBS on glass slides. Any strain showing positive hemagglutination was tested again after 30 min of incubation with 1% d-mannose to rule out type 1 fimbriae and to further support the notion that agglutination was likely mediated by P fimbriae.
Motility on urine plates.
One microliter of overnight culture was stabbed into urine plates (pooled human urine) containing 0.3% agar. The distance of migration (the diameter of the ring around the inoculation site) was measured after 16 h of incubation at 37°C. The assay was performed in duplicate and repeated twice in different batches of urine.
Hemolytic activity on blood agar plates.
Isolated colonies were spot inoculated, and the production of hemolysin was detected by determining a zone of lysis under each colony on tryptone soy agar plates with sheep blood (Oxoid Deutschland GmbH) after overnight incubation of the tested strains.
Microarray data accession numbers.
The supporting microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) with the accession numbers E-MEXP-3089 and E-MTAB-212.
RESULTS
Genomic diversity among urinary tract isolates.
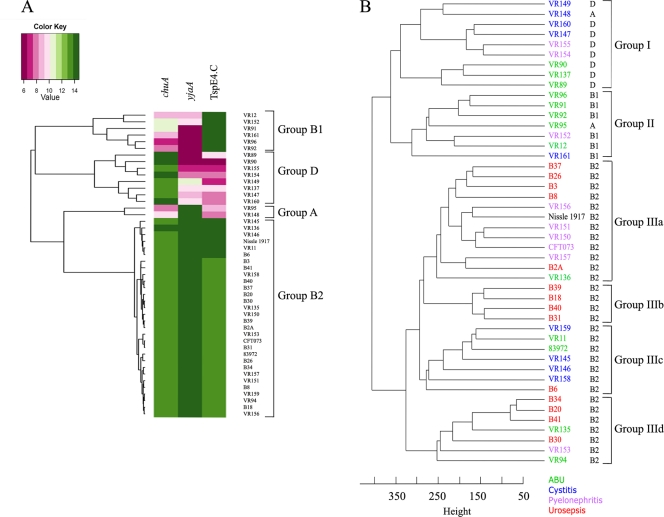
Forty-three E. coli strains were studied (12 ABU, 9 cystitis, 8 pyelonephritis, and 14 urosepsis strains). For comparison we also included three reference strains: CFT073, a pyelonephritis strain; 83972, an ABU strain; and the closely related probiotic strain Nissle 1917. The phylogenetic distribution of these strains within each disease category was as follows: ABU, 38% B2, 31% B1, 23% D, and 8% A; cystitis, 45% B2, 11% B1, 33% D, and 11% A; pyelonephritis, 67% B2, 11% B1, and 22% D; and urosepsis, 100% B2. CGH analysis of the 46 E. coli strains revealed that they clustered into three major groups (Fig. 1). Group I and II strains were predominantly from phylogenetic groups D and B1, respectively. In contrast, all group III strains belonged to phylogenetic group B2. The group III strains could be further subdivided into four subgroups; notably, groups IIIb and IIId contained a high proportion of urosepsis strains (100% and 71%, respectively).
Fig. 1.
Phylogenetic relationships between UTI isolates. (A) Phylogenetic group association based on the three group identifiers, chuA, yjaA, and TspE4.C. The CGH data obtained three phylogenetic identifiers correlated with the PCR-based analysis, giving a perfect distribution into phylogenetic groups. (B) Hierarchical clustering of the strains based on the whole-genome CGH profiles. Clustering was performed in R.
Prevalence of genomic islands.
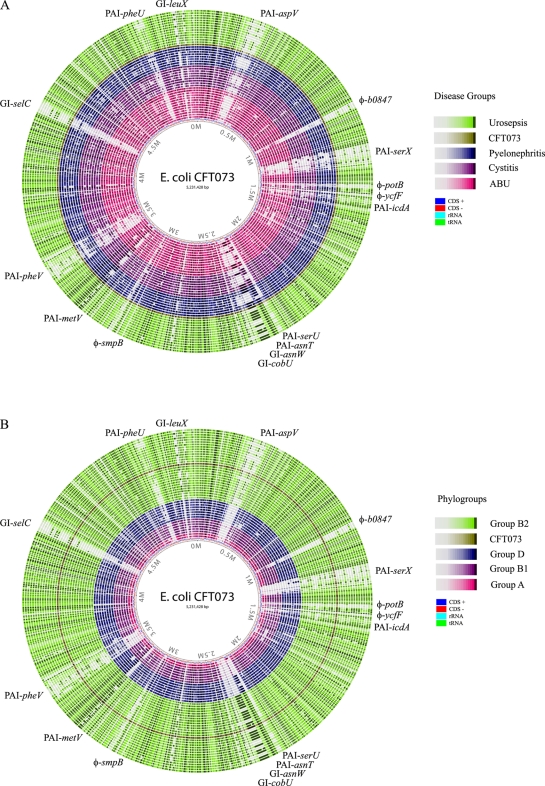
CGH analysis provides a powerful tool for studying the prevalence of genes associated with genomic islands (GIs). The CFT073 genome was included in the design of our microarray and thus acted as an excellent reference for comparison of the UTI isolates. The GIs of the highly virulent pyelonephritis strain CFT073 are very well characterized (28, 29). In order to study the prevalence of CFT073-associated GIs among the UTI isolates, the genomic profiles of the isolates were compared with CFT073 with respect to CFT073-associated probes. A permissive cutoff value for the presence of an island (≥2/3 of the genes in an island predicted to be present) was selected to allow for some cross-hybridization between generic island elements, e.g., transposases and integrases (summarized in Tables 2 and 3). Furthermore, the genomic profiles were analyzed with the GeneWiz BLAST atlas program, which was used to compare the inferred genomic profile of each UTI isolate to the genomic sequence of CFT073 (Fig. 2). The UTI isolates were compared with respect to disease category, as well as phylogenetic group association.
Table 2.
Presence of GIs and phage regions of CFT073 in different UTI isolates
| Island name | Common name | CDSs in island | Virulence/fitness gene(s) within islands | No. of CDSs represented on chip | % of isolates carrying island in disease categorya: |
Pc |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Urosepsis (15) | Pyelonephritis (8) | Cystitis (9) | ABU (13) | All | Upper vs. lower | |||||
| PAI-CFT073-aspV | PAI-IIICFT073 | c0253-c0368 | fpbABCD, cdiA, picU, tosCBDA, vat | 83 | 40 (6) | 44 (4) | 44 (4) | 23 (3) | 0.602 | 0.542 |
| φ-CFT073-b0847 | c0932-c0979 | 42 | 13 (2) | 0 (0) | 0 (0) | 31 (4) | 0.137 | 0.414 | ||
| PAI-CFT073-serX | c1165-c1293 | mchBCDEF, sfa/foc, iroBCDEN, flu | 102 | 33 (5) | 33 (3) | 22 (2) | 23 (3) | 0.835 | 0.514 | |
| φ-CFT073-potB | c1400-c1475 | 51 | 47 (7) | 56 (5) | 33 (3) | 38 (5) | 0.670 | 0.373 | ||
| φ-CFT073-ycfD | c1481-c1507 | 14 | 7 (1) | 0 (0) | 11 (1) | 0 (0) | 0.803 | 1.000 | ||
| PAI-CFT073-icdA | c1518-c1601 | sitABCD | 74 | 80 (12) | 56 (5) | 44 (4) | 31 (4) | 0.060 | 0.017 | |
| PAI-CFT073-serU | c2392-c2416 | tcpC | 19 | 60 (9) | 33 (3) | 0 (0) | 8 (1) | 0.002 | 0.001 | |
| PAI-CFT073-asnT | HPICFT073 | c2418-c2436 | fyuA | 15 | 100 (15) | 78 (7) | 67 (6) | 69 (9) | 0.046 | 0.022 |
| GI-CFT073-asnW | c2449-c2475 | pks | 26 | 67 (10) | 44 (4) | 33 (3) | 38 (5) | 0.363 | 0.139 | |
| GI-CFT073-cobU | c2482-c2528 | hma, fpbABCD_2 | 37 | 93 (14) | 44 (4) | 56 (5) | 38 (5) | 0.011 | 0.033 | |
| φ-CFT073-smpB | c3143-c3206 | 49 | 20 (3) | 11 (1) | 44 (4) | 31 (4) | 0.475 | 0.189 | ||
| PAI-CFT073-metV | c3385-c3410 | hcp, clpB, c3405 to c3409 | 25 | 100 (15) | 56 (5) | 33 (3) | 38 (5) | 0.000 | 0.001 | |
| PAI-CFT073-pheV | PAI-ICFT073 | c3556-c3698 | sisA, hlyA, pap, iha, sat, iutA, iucABCD, flu, kpsTM | 124 | 33 (5) | 56 (5) | 33 (3) | 15 (2) | 0.182 | 0.208 |
| GI-CFT073-selC | c4491-c4581 | sisB | 70 | 33 (5) | 11 (1) | 0 (0) | 0 (0) | 0.030 | 0.022 | |
| PAI-CFT073-pheU | PAI-IICFT073 | c5143-c5216 | pap_2 | 61 | 53 (8) | 22 (2) | 0 (0) | 8 (1) | 0.008 | 0.004 |
| GI-CFT073-leuXb | c5371-c5386 | 15 | 33 (5) | 22 (2) | 22 (2) | 15 (2) | 0.746 | 0.491 | ||
Percentage of isolates predicted to carry the islands (isolates which filtered more than 2/3 of the island CDSs present). The actual number of isolates predicted to carry the island is in parentheses.
The boundary of GI-CFT073-leuX was redefined as c5371-c5386 based on comparisons with E. coli MG1655 and UPEC 536.
P values below 0.05 are indicated in bold.
Table 3.
Presence of prototypical GIs and phage regions of CFT073 in different UTI E. coli isolates based on phylogenetic group
| Island name | % of isolates carrying island in phylogenetic groupa: |
P |
% of isolates carrying island among B2 isolates in disease categorya: |
Pc |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B2 (29) | D (8) | B1 (6) | A (2) | All | B2 vs. others | Urosepsis (15) | Pyelonephritis (5) | Cystitis (4) | ABU (5) | Commensalsb (2) | All | Upper vs. lower | |
| PAI-CFT073-aspV | 59 (17) | 0 (0) | 0 (0) | 0 (0) | 0.001 | 0.000 | 40 (6) | 80 (4) | 100 (4) | 60 (3) | 0 (0) | 0.134 | 0.234 |
| φ-CFT073-b0847 | 7 (2) | 13 (1) | 50 (3) | 0 (0) | 0.050 | 0.166 | 13 (2) | 0 (0) | 0 (0) | 0 (0) | 50 (1) | 1.000 | 1.000 |
| PAI-CFT073-serX | 45 (13) | 0 (0) | 0 (0) | 0 (0) | 0.013 | 0.001 | 33 (5) | 60 (3) | 50 (2) | 60 (3) | 0 (0) | 0.568 | 0.688 |
| φ-CFT073-potB | 41 (12) | 88 (7) | 17 (1) | 0 (0) | 0.018 | 0.755 | 47 (7) | 60 (3) | 0 (0) | 40 (2) | 0 (0) | 0.395 | 0.234 |
| φ-CFT073-ycfD | 3 (1) | 0 (0) | 17 (1) | 0 (0) | 0.356 | 1.000 | 7 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1.000 | 1.000 |
| PAI-CFT073-icdA | 76 (22) | 38 (3) | 0 (0) | 0 (0) | 0.000 | 0.000 | 80 (12) | 80 (4) | 50 (2) | 80 (4) | 50 (1) | 0.748 | 0.642 |
| PAI-CFT073-serU | 45 (13) | 0 (0) | 0 (0) | 0 (0) | 0.013 | 0.001 | 60 (9) | 60 (3) | 0 (0) | 20 (1) | 0 (0) | 0.109 | 0.020 |
| PAI-CFT073-asnT | 100 (29) | 88 (7) | 0 (0) | 50 (1) | 0.000 | 0.000 | 100 (15) | 100 (5) | 100 (4) | 100 (5) | 100 (2) | 1.000 | 1.000 |
| GI-CFT073-asnW | 76 (22) | 0 (0) | 0 (0) | 0 (0) | 0.000 | 0.000 | 67 (10) | 80 (4) | 75 (3) | 100 (5) | 0 (0) | 0.617 | 0.382 |
| GI-CFT073-cobU | 93 (27) | 0 (0) | 0 (0) | 50 (1) | 0.000 | 0.000 | 93 (14) | 80 (4) | 100 (4) | 100 (5) | 50 (1) | 0.741 | 1.000 |
| φ-CFT073-smpB | 14 (4) | 63 (5) | 50 (3) | 0 (0) | 0.013 | 0.014 | 20 (3) | 0 (0) | 0 (0) | 20 (1) | 0 (0) | 0.889 | 1.000 |
| PAI-CFT073-metV | 97 (28) | 0 (0) | 0 (0) | 0 (0) | 0.000 | 0.000 | 100 (15) | 100 (5) | 75 (3) | 100 (5) | 50 (1) | 0.138 | 0.310 |
| PAI-CFT073-pheV | 34 (10) | 63 (5) | 0 (0) | 0 (0) | 0.080 | 1.000 | 33 (5) | 60 (3) | 25 (1) | 20 (1) | 0 (0) | 0.640 | 0.431 |
| GI-CFT073-selC | 21 (6) | 0 (0) | 0 (0) | 0 (0) | 0.499 | 0.075 | 33 (5) | 20 (1) | 0 (0) | 0 (0) | 50 (1) | 0.423 | 0.137 |
| PAI-CFT073-pheU | 34 (10) | 13 (1) | 0 (0) | 0 (0) | 0.293 | 0.067 | 53 (8) | 40 (2) | 0 (0) | 0 (0) | 0 (0) | 0.069 | 0.011 |
| GI-CFT073-leuX | 38 (11) | 0 (0) | 0 (0) | 0 (0) | 0.053 | 0.004 | 33 (5) | 40 (2) | 50 (2) | 40 (2) | 0 (0) | 0.942 | 0.694 |
Percentage of isolates predicted to carry the islands (isolates which filtered more than 2/3 of the island CDSs present). Values in parentheses are the actual number of isolates predicted to carry the island.
Probe sequences were used in BLAST analysis against the two sequenced B2 strains SE15 and ED1a.
P values below 0.05 are indicated in bold.
Fig. 2.
Atlases generated using BLAST to compare the inferred genomic sequences of the 45 UTI isolates to CFT073, based on their disease categories (A) and phylogenetic groups (B). The strains were as follows: (A) B2A, B3, B6, B8, B18, B20, B26, B30, B31, B34, B37, B39, B40, B41, CFT073, VR150, VR151, VR152, VR153, VR154, VR155, VR156, VR157, VR145, VR146, VR147, VR148, VR149, VR158, VR159, VR160, VR161, VR1, VR11, VR12, VR89, VR90, VR91, VR92, VR94, VR95, VR96, VR135, VR136, and VR137; and (B) B2A, B3, B6, B8, B18, B20, B26, B30, B31, B34, B37, B39, B40, B41, CFT073, VR150, VR151, VR153, VR156, VR157, VR145, VR146, VR158, VR159, VR1, VR11, VR94, VR135, VR136, VR154, VR155, VR147, VR149, VR160, VR89, VR90, VR137, VR152, VR161, VR12, VR91, VR92, VR96, VR148, and VR95.
Several of the GIs, including PAI-CFT073-icdA, PAI-CFT073-serU, PAI-CFT073-asnT, GI-CFT073-cobU, PAI-CFT073-metV, PAI-CFT073-selC, and PAI-CFT073-pheU (Table 2 and Fig. 2), were found to be significantly more associated with the urosepsis and pyelonephritis isolates than with the cystitis and ABU isolates, suggesting that these islands are associated with highly virulent UPEC strains. However, the majority of these islands were also significantly linked to the phylogenetic group origin (Table 3). Separate analysis of the B2 isolates revealed a significantly greater prevalence of PAI-CFT073-serU and PAI-CFT073-pheU among the urosepsis and pyelonephritis isolates than among the cystitis and ABU isolates (Table 3). One island, viz., PAI-CFT073-asnT, or the high-pathogenicity island (HPI), was found in all B2 isolates. Arguably, this suggests that the island was acquired early in the evolution of the lineage (Table 3). Most sequenced B2 strains, including the two commensal isolates SE15 and ED1a, also carry this island. However, the HPI is not restricted to group B2; it was found, in particular, among the group D isolates and in one of the two group A isolates. Another island, PAI-CFT073-metV, was also present in all but one of the group B2 isolates (cystitis isolate VR158). PAI-CFT073-metV has previously been shown to contribute to upper urinary tract infection (in a mouse model), an activity shown to be associated with the last six genes in the island (28). In fact, although VR158 was predicted to lack the majority of the island (resulting in an overall absence call), the strain contained five of these genes (c3405 to c3409; c3410 was not on the microarray). The B2 commensal strain SE15 also carries this island, and the other B2 commensal strain, ED1a, carries the last six genes. Several islands were found exclusively among the B2 UTI strains (as compared to the sequenced non-UTI strains [see Fig. S3 in the supplemental material]), i.e., PAI-CFT073-aspV, PAI-CFT073-serX, PAI-CFT073-serU, GI-CFT073-asnW, GI-CFT073-selC, and GI-CFT073-leuX. Two PAIs, PAI-CFT073-asnT and PAI-CFT073-pheV, were particularly prevalent among the group D strains (Table 3). The group A strains had very few islands; only two islands were predicted to be present, i.e., PAI-CFT073-asnT in ABU strain VR95 and GI-CFT073-cobU in cystitis strain VR148. Although these strains may carry other non-CFT073-like islands, it is possible that these islands contribute to persistence in the urinary tract.
Although the majority of the PAIs (63%) were more commonly associated with the urosepsis and pyelonephritis isolates than with the cystitis and ABU strains, a considerable fraction of the cystitis and ABU isolates contained multiple PAIs, and none of these strains were completely devoid of genes from these PAIs (Table 2). The average number of GIs per strain decreased according to the severity of the infection caused by the strain; urosepsis, pyelonephritis, cystitis, and ABU strains carried on average 8.1, 6.4, 4.4, and 4.1 GIs per strain, respectively. However, there was no significant difference between the different disease categories within the same phylogenetic group; in the B2 group, which contained strains from all four disease categories, the urosepsis, pyelonephritis, cystitis, and ABU isolates carried on average 8.1, 8.6, 6.3, and 7.4 islands, respectively. On the other hand, the two commensal strains in the B2 group contained only 3.5 GIs per strain (Table 3). The average number of islands present per strain from each phylogenetic group was 7.8, 3.6, 1.3, and 1.0 for groups B2, D, B1, and A, respectively. Altogether, although the number of GIs is significantly higher in a UTI strain than in a commensal strain, our results suggest that the number of GIs present in a UTI strain is linked to the phylogenetic group association rather than to the disease category.
Distribution of genes encoding virulence and fitness factors.
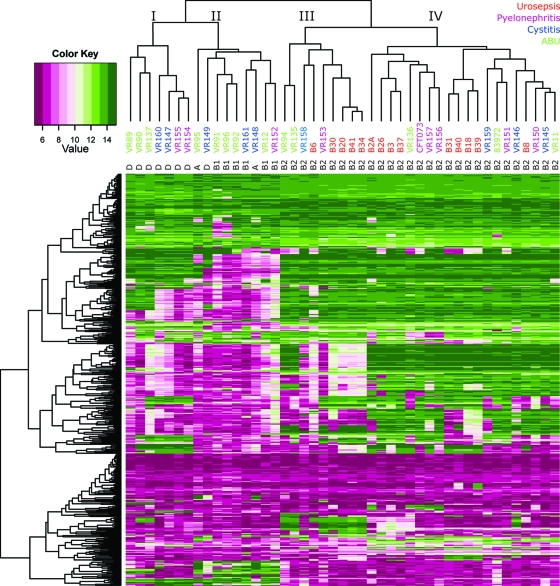
We then constructed a heat map based on 509 genes that encode factors that have been shown to be involved in the virulence and/or fitness of UPEC and ExPEC strains, as well as putative virulence/fitness factors (Fig. 3). The CGH data obtained for these factors clustered the B2 strains separately, while another subgroup contained only D strains (group I); the remaining subgroup contained all B1 and A strains, as well as one D strain (group II) (Fig. 3). The isolates belonging to group A, B1, and D possessed fewer virulence/fitness genes than the B2 strains, in particular, the subgroup with B1 and A strains (group II). This observation fits with the notion that a high proportion of B2 isolates are more virulent, simply because they possess a high proportion of virulence/fitness factors. For the majority of the virulence/fitness genes, the ABU and cystitis isolates belonging to B2 showed virulence gene profiles very similar to those of the virulent isolates. The B2 isolates could be split into two subgroups, groups III and IV (Fig. 3), where group III contains fewer virulence/fitness factors than group IV. Interestingly, both B2 groups III and IV contain strains from all four disease categories and approximately the same proportion of urosepsis and ABU isolates, i.e., 56% and 45% urosepsis and 22% and 11% ABU strains in groups III and IV, respectively. This suggests that the number of virulence/fitness factors per se cannot determine the pathogenic potential of a UTI strain.
Fig. 3.
Heat map of the 45 UTI isolates constructed based on the CGH signals obtained for 509 established and putative virulence and fitness factors of ExPEC strains.
Since there was a clear correlation between the numbers of PAIs and virulence/fitness factors and the phylogenetic group origin, we treated each phylogenetic group separately when screening for virulence/fitness factors specific to each disease category. Due to the low number of isolates in groups B1 and A, no statistically significant results were obtained for these groups. Among the B2 isolates, 33 probes showed significantly different prevalences among the four disease categories or among upper versus lower UTIs (see Table S1 in the supplemental material). A third of these probes were pap operon genes representing all pap genes encoding P fimbriae with the exception of papA, encoding the major subunit. P fimbriae have previously been shown to be important for UTI colonization, and the cognate receptor is abundant in the kidneys (reviewed in reference 21). This correlated well with phenotypic data, where only one cystitis and none of the ABU isolates agglutinated red blood cells while the majority of the pyelonephritis and urosepsis isolates did (Table 1). Four probes specific for phosphoglycerate transport genes (pgt) were also identified; these were particularly predominant in the urosepsis isolates. Interestingly, in CFT073, the pgt operon is located on PAI-CFT073-pheU, which also carries one of the two pap operons present in CFT073. Whether the pgt genes confer any virulence properties is unknown. Other probes differentially distributed and associated with upper UTI included two probes specific for the antigen 43 (Ag43) gene, which encodes an autotransporter protein that contributes to long-term colonization of the urinary tract (38). Six probes were significantly associated with cystitis and ABU isolates; they included genetic variants of fimA and kpsU, encoding the major subunit of type 1 fimbriae and a capsular protein, respectively. (Due to genetic divergence between the genomes for these genes, representatives of each genetic variant were included on the CGH microarray.) This suggests that antigenic and perhaps functional variations between important surface structures, such as type 1 fimbriae and capsule, may underlie the differences observed between the isolates. The ibeA gene, encoding a protein associated with invasion by newborn meningitis-causing E. coli (15) and identified as a virulence factor in avian-pathogenic E. coli (10), was also positively associated with lower rather than upper UTI (see Table S1 in the supplemental material).
Among the group D isolates, 34 probes were identified that displayed a significantly different distribution among the three disease groups (no urosepsis strains belonged to group D). Five of these 34 probes were more strongly associated with pyelonephritis and cystitis isolates than with ABU isolates (three probes specific for the ybg fimbrial operon and two probes specific for genes that encode hypothetical proteins). On the other hand, 29 of the 34 probes were more strongly associated with ABU isolates than with pyelonephritis or cystitis isolates; these included several fimbrial genes: the ycbRSTU, yadKMN, and yfcQRSUV genes (see Table S1 in the supplemental material). Interestingly, the ycb and yad fimbrial operons have been implicated in biofilm formation, while the yfc operon genes have been shown to confer adherence to T24 bladder epithelial cells (24).
UTI-specific and phylogenetic group-specific genes.
The issue of UTI-specific genetic markers has received considerable attention over the years, and several attempts to identify UPEC-specific genes have been made. Using a bioinformatics approach, Chen et al. identified 29 genes that were subjected to positive selection among UPEC strains (3). Furthermore, using a CGH approach, Lloyd et al. identified 131 genes that were present in all UPEC strains but not in any of the fecal or commensal strains examined (29). Since both of these studies focused entirely on virulent UTI isolates, it was not clear if any of these genes were restricted to pathogens or prevalent among UTI isolates in general. We assessed our CGH data to test for a possible correlation between the UTI categories and the prevalence of these UPEC-specific genes.
Based on the 29 genes identified by Chen et al., the isolates grouped almost entirely according to their phylogenetic groups, with little or no correlation with their disease categories; all B2 strains and D strains clustered in one group each, and the third group contained all B1 and A strains (see Fig. S1 in the supplemental material). A similar trend, although not as pronounced, was seen when examining the prevalence of the 131 genes identified by Lloyd et al.; while the B2 isolates grouped separately, the remaining phylogenetic groups were less segregated. Here, three group D ABU strains clustered with the B2 isolates instead of with the other D, A, and B1 isolates. No clear delineation between disease categories was apparent (see Fig. S2 in the supplemental material). The 131 genes were particularly prevalent among the B2 strains; this was also clearly reflected in the clustering of our 45 isolates (see Fig. S2 in the supplemental material). No genes that were uniformly present in the 45 UTI isolates examined in this study were absent in all six non-ExPEC sequenced strains. The same was true when we focused only on the isolates that caused symptomatic UTI. This supports the notion that uropathogenesis and urinary tract infectivity are multifactorial traits.
Fitness factors.
By virtue of their avirulent nature and their ability to colonize the urinary tract and grow in urine, ABU strains must possess fitness factors rather than virulence factors. To search for fitness factor-encoding genes, we compared the inferred genomic contents of ABU isolates from phylogenetic groups A and B1 with six sequenced nonpathogenic non-UTI E. coli commensal strains, including the prototypic K-12 strain MG1655. No genes shared among the ABU isolates of the two groups were absent in the six strains used for comparison. However, each individual isolate was found to carry numerous genes that were not present in the commensal isolates, clearly signifying that the ABU strains are genetically diverse. Genes present in some ABU isolates included several iron-scavenging genes, as well as the pco genes involved in copper resistance. We also screened for genes shared by all ABU isolates within phylogenetic group B1. This analysis identified six genes, which were all related to propanediol utilization (pdu); the exact role of these genes in E. coli is unknown, but in Salmonella enterica serovar Typhimurium, the pdu genes have been implicated in fitness during in vivo infection (5).
There was only one ABU strain that belonged to phylogenetic group A, i.e., VR95. This strain was compared directly to the two commensal group A strains MG1655 and HS. In total, 91 probes were specific for VR95. Among these probes were genes associated with HPI. The HPI genes are expressed during in vivo growth of the ABU isolate 83972 (32). This suggests that the HPI is a fitness island rather than a pathogenicity island per se, a notion that is supported by the presence of this island in the two B2 commensal isolates (see Fig. S3 in the supplemental material). Interestingly, genes of the HPI are induced during growth in urine in another ABU group A isolate, viz., VR50, which shows high resemblance to commensal isolates (12). In summary, all ABU strains examined contained between one and nine of the CFT073 islands or phage regions (on average, 4.1 per strain) (Table 2), suggesting that fitness is acquired by compilation of different genes and not by acquiring any specific set of genes.
Phenotypic characterization.
Expression of functional fimbriae has been shown to be important for colonization of the different compartments of the urinary tract. In particular, three UPEC class fimbriae have been identified; type 1 fimbriae, encoded by the fim genes, which bind specifically to mannose targets found on the surfaces of uroepithelial bladder cells (41); P fimbriae, encoded by the pap genes, which bind to the α-d-galactopyranosyl-(1-4)-β-d-galactopyranoside receptor epitope in the globoseries of glycolipids of the upper urinary tract (18, 26); and F1C fimbriae, encoded by the foc genes, which bind to galactosylceramide targets present on epithelial cells in the bladder and kidneys, as well as globotriaosylceramide, present exclusively in the kidneys (2, 20). The different binding patterns of these three fimbriae correlated well with the presence of fimbrial gene clusters among the disease categories. The fim genes were present in the majority of strains in all four disease categories; 93%, 100%, 100%, and 77% of the urosepsis, pyelonephritis, cystitis, and ABU strains carried the fim genes, respectively. While all of the urosepsis and pyelonephritis strains carrying the fim genes expressed functional type 1 fimbriae as assessed by yeast cell agglutination, three and two of the cystitis and ABU strains, respectively, contained the fim genes but failed to express the phenotype (Table 1). All four phylogenetic groups contained strains not expressing type 1 fimbriae. Furthermore, three ABU strains and one urosepsis isolate were predicted to carry eroded fim clusters. They included two B2 isolates, i.e., the ABU strain 83972 and the urosepsis strain B3, which were both predicted to lack the same fim genes, namely, fimEAIC. This 4.5-kb deletion has been verified by sequencing of the fim cluster of 83972 (23).
The pap genes were present in 86%, 78%, 22%, and 15% of the urosepsis, pyelonephritis, cystitis, and ABU strains, respectively. While all the pyelonephritis isolates carrying the pap genes agglutinated human RBCs, two of the urosepsis isolates failed to express the phenotype. Of the two cystitis isolates predicted to carry all pap genes (both from group D), only one expressed the phenotype. Two ABU strains, one group B2 (strain 83972) and one group D (VR137) isolate, contained the pap genes; only VR137 agglutinated RBCs. Strain 83972 is known to carry point mutations in the papG gene rendering it incapable of producing functional P fimbriae (23). Overall, only group B2 and D strains expressed the phenotype; none of the A strains and only one B1 strain contained the pap genes.
The foc genes, encoding F1C fimbriae, were present in 33 to 44% of all four disease groups. All 18 strains that contained the foc genes were B2 isolates (Table 1), which corresponds to 62% of the B2 isolates.
Motility and hemolysis of RBCs are two other phenotypes often associated with UPEC. All of the pyelonephritis isolates and 89% of the cystitis isolates were motile; in contrast only 64% of the urosepsis and 46% of the ABU isolates were motile (Table 1). The ability to cause hemolysis of RBCs was variable and decreased according to disease severity: 57%, 56%, 33%, and 15% of the urosepsis, pyelonephritis, cystitis, and ABU isolates, respectively (Table 1). All hemolysis-positive strains were B2 isolates.
In summary, the ABU isolates contained the lowest proportion of strains that showed functional type 1 and P fimbriae, motility on human urine plates, and hemolytic activity. The highest proportion of strains expressing functional type 1 and P fimbriae and displaying motility in urine were found among the pyelonephritis strains, while hemolytic activity was seen at a similar prevalence among sepsis and pyelonephritis isolates. Hemolysis was clearly associated with isolates of phylogenetic group B2.
DISCUSSION
The E. coli genome is undergoing constant evolution and reflects the interplay between environmental selection pressure and gene content. Gene acquisition and gene loss are the key factors in genome evolution and are involved in the formation of new strains. It is important to note that the urinary tract is normally sterile and introduction of bacteria is “accidental.” Infectious strains are generally believed to originate from the fecal flora. The environmental conditions in the intestines and the urinary tract are quite different; arguably, they impose different selection pressures on the bacteria. Thus, the evolution of UPEC is not driven by a requirement to cause disease but rather by a trade-off between environmental requirements in a particular niche and the genetic composition of the bacterial strain. E. coli strains acquire new genetic information via horizontal gene transfer, often in the form of plasmids or GIs. A subgroup of GIs is termed PAIs because, when identified, they were found to harbor virulence-associated genes (11). Likewise, loss of genetic information can occur via loss of plasmids or GIs, but also by smaller genetic lesions, like point mutations. A classic example is the ABU strain 83972, which is a deconstructed pathogen with an ancestor that closely resembled the bona fide pyelonephritis strain CFT073 (22, 40).
In this study, we used a multigenome CGH microarray for a comparative genomics study of 45 E. coli strains isolated from four different types of urinary tract infections. The major advantage of this approach was that a 31 E. coli strain large pangenome was represented on the microarray, significantly improving the application range of the array. The microarray included probes representing all four UTI disease categories employed in this study, i.e., the genomes of one urosepsis/pyelonephritis, one pyelonephritis, four cystitis, and two ABU strains were included in the microarray design (40). This allowed us to estimate the genomic contents of the UTI strains and the genetic variations among the strains. An inherent limitation of the microarray is that novel genes, or genes that are too heterologous, cannot be detected. It should also be noted that our analysis did not take plasmid-borne genes into account. Therefore, the possibility that plasmid-specific virulence or fitness genes could have increased the discriminating power of the data cannot be excluded.
From our CGH analysis, it was apparent that the single most important determinant for the UTI isolates was their phylogenetic group origin. Generally, the data suggested that the B2 strains represent a highly virulent lineage of isolates that are genotypically distinct from the other phylogenetic groups. Although horizontally acquired genetic elements in general did not disrupt the clonal structure of the species, the group A strains did not cluster separately from the B1 and D groups.
No clear differences between symptomatic and ABU isolates were apparent. We did, however, find a significant correlation between disease severity and certain PAIs. Thus, PAI-CFT073-serU and PAI-CFT073-pheU were found to be significantly more associated with the urosepsis and pyelonephritis isolates than with the cystitis and ABU isolates independent of phylogenetic group origin. One island, PAI-CFT073-metV, was found exclusively in B2 isolates. This island has previously been reported to contribute to upper UTI due to at least six genes on the island (28). The average number of PAIs was significantly higher among the group B2 isolates (7.8 PAIs compared with 1.0 to 3.6 PAIs for groups A, B1, and D), while no correlation between the number of PAIs and disease severity was apparent within group B2 (6.3 to 8.6 PAIs for all four disease categories). However, the commensal B2 isolates carried an average number of PAIs that was significantly lower (3.5 PAIs) than for strains in any of the disease categories, indicating that specific genes are required for colonization of the urinary tract but that the severity of disease might be impossible to predict by estimating the number of virulence/fitness genes carried by the strain.
A picture of the genetic profiles of different classes of UTI strains is slowly emerging. In this study, the most virulent strains (which belonged to phylogenetic group B2) generally had the most virulence factors. Despite this, no specific constellation of virulence factors was found in the most virulent strains. Rather, an assortment of factors was associated with high virulence. This assortment of genes included those encoding adhesins, iron uptake systems, toxins, capsule, and flagella. A similar picture emerged regarding fitness factors. Arguably, ABU strains that are nonpathogenic yet grow well in urine and colonize the urinary tract should have urinary tract-related fitness factors compared with commensal gut isolates. We found that the genes residing on the HPI genomic island could be associated with fitness genes rather than virulence. Furthermore, genes involved in iron acquisition seem to be obligatory for urinary tract fitness. However, as in the case of virulence factors, it appeared that urinary tract fitness is acquired through an assortment of genes rather than through a specific gene set. Previous studies have suggested that many ABU strains have lost critical virulence determinants or show reduced expression of such genes (13, 32). We note that this study did not involve a detailed phenotypic analysis of the strains or take into account the immune status of the infected patient. Individual patient parameters, including immune status, could, for example, contribute to ABU. Taken together, our study supports the notion that urovirulence and fitness factors can be a mixture of factors taken from a rich menu of genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carsten Friis for excellent support for bioinformatics analysis. We thank Birthe Jul Bondo for expert technical assistance.
This work was supported by Lundbeckfonden (grant no. R17-A1603). M.A.S. is supported by an ARC Future Fellowship and a grant from the National Health and Medical Research Council of Australia (569676).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Andersson P., et al. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59:2915–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bäckhed F., et al. 2002. Identification of target tissue glycosphingolipid receptors for uropathogenic, F1C-fimbriated Escherichia coli and its role in mucosal inflammation. J. Biol. Chem. 277:18198–18205 [DOI] [PubMed] [Google Scholar]
- 3. Chen S. L., et al. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. U. S. A. 103:5977–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clermont O., Bonacorsi S., Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conner C. P., Heithoff D. M., Julio S. M., Sinsheimer R. L., Mahan M. J. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. U. S. A. 95:4641–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dobrindt U., Chowdary M., Krumbholz G., Hacker J. 2010. Genome dynamics and its impact on evolution of Escherichia coli. Med. Microbiol. Immunol. 199:145–154 [DOI] [PubMed] [Google Scholar]
- 7. Ferrières L., Hancock V., Klemm P. 2007. Biofilm exclusion of uropathogenic bacteria by selected asymptomatic bacteriuria Escherichia coli strains. Microbiology 153:1711–1719 [DOI] [PubMed] [Google Scholar]
- 8. Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S–13S [DOI] [PubMed] [Google Scholar]
- 9. Gentleman R. C., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Germon P., et al. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179–1186 [DOI] [PubMed] [Google Scholar]
- 11. Hacker J., Kaper J. B. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641–679 [DOI] [PubMed] [Google Scholar]
- 12. Hancock V., Ferrières L., Klemm P. 2008. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology 154:167–175 [DOI] [PubMed] [Google Scholar]
- 13. Hancock V., Klemm P. 2007. Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect. Immun. 75:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hancock V., Nielsen E. M., Krag L., Engberg J., Klemm P. 2009. Comparative analysis of antibiotic resistance and phylogenetic group patterns in human and porcine urinary tract infectious Escherichia coli. APMIS 117:786–790 [DOI] [PubMed] [Google Scholar]
- 15. Huang S., Wan Z., Chen Y., Jong A. Y., Kim K. S. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 183:1071–1078 [DOI] [PubMed] [Google Scholar]
- 16. Johansen T., et al. 2006. Hospital acquired urinary tract infections in urology departments: pathogens, susceptibility and use of antibiotics: data from the PEP and PEAP-studies. Int. J. Antimicrob. Agents 28:S91–S107 [DOI] [PubMed] [Google Scholar]
- 17. Johnson J. R., Russo T. A. 2002. Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. J. Infect. Dis. 186:859–864 [DOI] [PubMed] [Google Scholar]
- 18. Kallenius G., et al. 1981. Structure of carbohydrate part of receptor on human uroepithelial cells for pyelonephritogenic Escherichia coli. Lancet ii:604–606 [DOI] [PubMed] [Google Scholar]
- 19. Kaper J. B., Nataro J. P., Mobley H. L. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 20. Khan A. S., et al. 2000. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect. Immun. 68:3541–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klemm P., Hancock V., Schembri M. A. 2010. Fimbrial adhesins from extraintestinal Escherichia coli. Environ. Microbiol. Rep. 2:628–640 [DOI] [PubMed] [Google Scholar]
- 22. Klemm P., Hancock V., Schembri M. A. 2007. Mellowing out: adaptation to commensalism by Escherichia coli asymptomatic bacteriuria strain 83972. Infect. Immun. 75:3688–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klemm P., Roos V., Ulett G. C., Svanborg C., Schembri M. A. 2006. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74:781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Korea C.-G., Badouraly R., Prevost M.-C., Ghigo J.-M., Beloin C. 2010. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 12:1957–1977 [DOI] [PubMed] [Google Scholar]
- 25. Larsen B., Monif G. R. 2001. Understanding the bacterial flora of the female genital tract. Clin. Infect. Dis. 32:e69–e77 [DOI] [PubMed] [Google Scholar]
- 26. Leffler H., Svanborg-Eden C. 1981. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect. Immun. 34:920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindberg U., et al. 1975. Asymptomatic bacteriuria in schoolgirls. II. Differences in Escherichia coli causing asymptomatic bacteriuria. Acta Paediatr. Scand. 64:432–436 [DOI] [PubMed] [Google Scholar]
- 28. Lloyd A. L., Henderson T. A., Vigil P. D., Mobley H. L. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 191:3469–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lloyd A. L., Rasko D. A., Mobley H. L. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 189:3532–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mobley H. L. T., et al. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pedersen A. G., Jensen L. J., Brunak S., Stærfeldt H.-H., Ussery D. W. 2000. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 299:907–930 [DOI] [PubMed] [Google Scholar]
- 32. Roos V., Klemm P. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect. Immun. 74:3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roos V., Nielsen E. M., Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol. Lett. 262:22–30 [DOI] [PubMed] [Google Scholar]
- 34. Schappert S. M. 1999. Ambulatory care visits to physician offices, hospital outpatient departments, and emergency departments: United States, 1997. Vital Health Stat. 13:1–39 [PubMed] [Google Scholar]
- 35. Smith J. L., Fratamico P. M., Gunther N. W. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4:134–163 [DOI] [PubMed] [Google Scholar]
- 36. Stamm W. E. 1991. Catheter-associated urinary tract infections: epidemiology, pathogenesis, and prevention. Am. J. Med. 91:65S–71S [DOI] [PubMed] [Google Scholar]
- 37. Stamm W. E., Martin S. M., Bennett J. V. 1977. Epidemiology of nosocomial infection due to Gram-negative bacilli: aspects relevant to development and use of vaccines. J. Infect. Dis. 136:S151–S160 [DOI] [PubMed] [Google Scholar]
- 38. Ulett G. C., et al. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 75:3233–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vejborg R. M., Bernbom N., Gram L., Klemm P. 2008. Anti-adhesive properties of fish tropomyosins. J. Appl. Microbiol. 105:141–150 [DOI] [PubMed] [Google Scholar]
- 40. Vejborg R. M., Friis C., Hancock V., Schembri M. A., Klemm P. 2010. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol. Genet. Genomics 283:469–484 [DOI] [PubMed] [Google Scholar]
- 41. Wu X. R., Sun T. T., Medina J. J. 1996. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl. Acad. Sci. U. S. A. 93:9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.