Abstract
Glycerol dibiphytanyl glycerol tetraether (GDGT)-based intact membrane lipids are increasingly being used as complements to conventional molecular methods in ecological studies of ammonia-oxidizing archaea (AOA) in the marine environment. However, the few studies that have been done on the detailed lipid structures synthesized by AOA in (enrichment) culture are based on species enriched from nonmarine environments, i.e., a hot spring, an aquarium filter, and a sponge. Here we have analyzed core and intact polar lipid (IPL)-GDGTs synthesized by three newly available AOA enriched directly from marine sediments taken from the San Francisco Bay estuary (“Candidatus Nitrosoarchaeum limnia”), and coastal marine sediments from Svalbard, Norway, and South Korea. Like previously screened AOA, the sedimentary AOA all synthesize crenarchaeol (a GDGT containing a cyclohexane moiety and four cyclopentane moieties) as a major core GDGT, thereby supporting the hypothesis that crenarchaeol is a biomarker lipid for AOA. The IPL headgroups synthesized by sedimentary AOA comprised mainly monohexose, dihexose, phosphohexose, and hexose-phosphohexose moieties. The hexose-phosphohexose headgroup bound to crenarchaeol was common to all enrichments and, in fact, the only IPL common to every AOA enrichment analyzed to date. This apparent specificity, in combination with its inferred lability, suggests that it may be the most suitable biomarker lipid to trace living AOA. GDGTs bound to headgroups with a mass of 180 Da of unknown structure appear to be specific to the marine group I.1a AOA: they were synthesized by all three sedimentary AOA and “Candidatus Nitrosopumilus maritimus”; however, they were absent in the group I.1b AOA “Candidatus Nitrososphaera gargensis.”
INTRODUCTION
The discovery of ammonia-oxidizing archaea (AOA) and their apparent importance in the biogeochemical cycling of nitrogen (8, 19, 50) has encouraged the ongoing study of their ecology and efforts to enrich and cultivate individual species. AOA perform the aerobic oxidation of ammonia to nitrite—the first and rate-limiting step in the process of nitrification—and are abundant in many natural environments, often outnumbering their ammonia-oxidizing bacterial counterparts (based on quantitative comparisons of 16S rRNA and amoA genes and on in situ hybridization techniques) (21, 26, 50). To date, cultivation efforts have resulted in the enrichment and characterization of three AOA, representing three phylogenetic lineages within the group I AOA: “Candidatus Nitrosopumilus maritimus SCM1” (19), “Candidatus Nitrosocaldus yellowstonii” (6), and “Candidatus Nitrososphaera gargensis” (13). In addition, “Candidatus Cenarchaeum symbiosum,” an organism belonging to the group I AOA living in symbiosis with the marine sponge Axinella mexicana, has not been cultivated but has been well characterized (10, 11, 31).
It has been recently shown that all enriched AOA synthesize the membrane lipid crenarchaeol (6, 29, 33, 41)—a unique glycerol dibiphytanyl glycerol tetraether (GDGT) including four cyclopentane moieties and a cyclohexane moiety (Fig. 1)—which has not been encountered in any cultivated (hyper)thermophilic Crenarchaeota or Euryarchaeota until now (41). These include the three cultivated species mentioned above and “Ca. Cenarchaeum symbiosum” (41), in addition to uncharacterized AOA enriched from the North Sea and the Indian Ocean (32, 49). A close association between crenarchaeol and putative AOA has been previously noted in diverse environments (e.g., 5, 21, 28), and this, coupled with its occurrence in cultivated representatives, suggests that it may in fact be specific to AOA (6, 29).
Fig. 1.
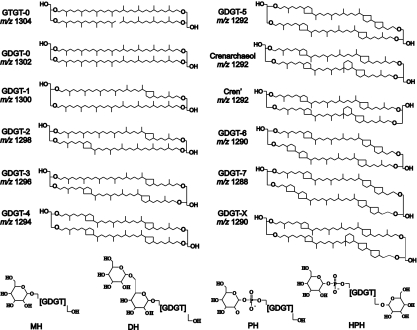
Structures of diglycerol di(tri)alkyl tetraethers GD(T)GTs commonly synthesized by Archaea and molecular weights shown as m/z values. GDGT-X was recently identified as a major core GDGT in the soil AOA “Ca. Nitrososphaera gargensis”; however, it has not been observed in other cultivated Archaea or environmental GDGT distributions thus far. Below, IPLs identified in this work (and also in “Ca. Nitrosopumilus maritimus” and “Ca. Nitrososphaera gargensis”); polar headgroups are shown in full structure: monohexose (MH), dihexose (DH), phosphohexose (PH), and hexose + phosphohexose (HPH). It should be noted that the exact positions of the polar headgroups are unknown.
Aside from crenarchaeol, AOA also synthesize GDGTs with 0 to 3 cyclopentane moieties (Fig. 1). Generally, GDGT-0 and crenarchaeol predominate, with lower relative abundances of GDGT-1 to GDGT-3, and similar GDGT distributions have been recovered from marine suspended particulate matter and sediments (reference 35 and references therein; 34). The occurrence of GDGT-4 has also been noted in “Ca. Nitrosopumilus maritimus,” “Ca. Nitrososphaera gargensis” and “Ca. Nitrosocaldus yellowstonii,” but it is typically present in small relative amounts (6, 29, 33). GDGT analysis has been used to provide DNA-independent information on the structure and function of archaeal communities in numerous marine environments, and it has proved useful in substantiating or complementing other molecular evidence (1, 5). Incorporation of 13C-labeled bicarbonate into the GDGTs of a marine crenarchaeote enriched from the North Sea provided the first direct evidence for an autotrophic lifestyle of planktonic mesophilic AOA (48), and determination of the radiocarbon content of GDGTs in the marine water column suggested that autotrophy accounts for approximately 83% of the carbon assimilated by mesopelagic AOA, the rest coming from heterotrophic consumption of organic carbon (17).
Compared to core GDGTs (as described above), their intact polar lipid (IPL) counterparts [i.e., GDGTs bound to a polar headgroup(s)] may give more specific information on putative microbial community structure. IPLs are thought to derive from living (or recently living) organisms, as the covalently bound polar headgroups are relatively rapidly lost upon cell senescence (12, 46). GDGT-based IPLs have been used previously as tracers of AOA in marine sediments and water columns (1, 38), to determine the viability of microbes where conventional molecular approaches may show a relative bias (23), and in quantitatively distinguishing between living and dead cell material (16, 28). The additional structural information obtained from polar headgroup analysis of IPLs, as opposed to core lipid analysis only, may also be potentially useful for inferring the presence of certain phylogenetic lineages. Overall, IPLs give more relevant information regarding the local viable organisms than analysis of core lipids, which often largely represent fossilized organic matter.
Of the four AOA cultures that have been profiled for their GDGT composition, only “Ca. Nitrosopumilus maritimus,” a group I.1a AOA isolated from an aquarium, and “Ca. Nitrososphaera gargensis,” a group I.1b AOA enriched from a hot spring, have been profiled for their GDGT-based IPLs (29, 33). “Ca. Nitrososphaera gargensis” has an unusual GDGT profile and is phylogenetically more closely related to soil AOA than marine AOA. Although “Ca. Nitrosopumilus maritimus” is closely related to sequences of marine AOA, it was enriched from sea aquarium gravel, not a natural marine environment. Further development and progress in the application of IPL analysis to marine AOA ecology necessitates the study of GDGTs and their associated IPLs of AOA actually enriched from marine environments similar to those commonly studied.
Here we have analyzed three new cultures of group I.1a AOA enriched from marine sediments off the coast of Svalbard, Norway (“AR”), and South Korea (“SJ”) (25), as well as sediments from the San Francisco Bay estuary (“Ca. Nitrosoarchaeum limnia”) (2), for their IPL and core GDGTs. Although group I AOA were first identified in the marine water column (7, 9), a large abundance and diversity of sedimentary AOA have been demonstrated (8). Comparisons among our findings and with those of previously studied AOA contribute additional information on the diversity of marine AOA GDGTs and IPLs and give insight into lipid biomarkers specific to this phylogenetic lineage of Archaea that recently was proposed on the basis of whole-genome analyses to represent a new phylum, the Thaumarchaeota (4, 42).
MATERIALS AND METHODS
Cultivation of archaeal ammonia oxidizers.
Enrichment cultures of “Ca. Nitrosoarchaeum limnia SFB1” were grown as previously described (2). Briefly, surface sediments (0 to 5 cm) collected in the northern part of the San Francisco Bay estuary (site SU001S; 38°5′55.75″N, 122° 2′47.40″W) were inoculated into a modified low-salinity version of synthetic crenarchaeota medium (19) as described by Mosier and Francis (24). At the time of sampling, nutrient concentrations in the bottom water just above the sediments were 2 μM ammonia, 14 μM nitrate, and 0.9 μM nitrite. Salinity was 7.9 practical salinity units (psu), temperature was 21.6°C, and the sediment C:N ratio was 15.8 (data courtesy of the San Francisco Bay Regional Monitoring Program). Molecular and microscopy-based methods showed that the enrichment was dominated (>80%) by one archaeal species (2). For lipid analysis, 585 ml of culture was filtered over one 0.22-μm-pore-size Duopore filter and 1,710 ml of culture was filtered over two 0.7-μm-pore-size GF/F filters (diameter, 47 mm).
Marine sediments (ca. 100 g of sediment from a depth of 0 to 1 cm) collected from Donghae (128°35′E, 38°20′N; depth, 650 m) and Svalbard (Arctic region, 016°28′E, 78°21′N; depth, 78 m) were mixed in the field with 1 liter of local seawater in sterilized glass bottles by stirring with a sterile spatula. Approximately 10 ml of each sediment slurry was transferred to a sterile conical tube and transported back to the laboratory at 4°C. Cultures were grown aerobically in natural seawater medium, which contains autoclaved coastal seawater from Donghae (South Korea) supplemented with ammonium chloride (1 mM), sodium thiosulfate (0.1 mM), sodium bicarbonate (2.5 mM), potassium phosphate (0.1 mM), trace element mixture (1×) (47), and vitamin solution (1×) (47). The pH was adjusted to 8.2 using 1 N NaOH or HCl. The cultures from Donghae, South Korea (SJ), and the Arctic sea of Svalbard (AR) were incubated at a static condition with daily intermittent inverting instead of continuous shaking. After oxidation of ammonia (typically after 2 weeks), 5% of the total culture volume was routinely transferred to new seawater medium at 25°C in dark conditions. After batch culture, the pH of this medium was not significantly changed (8.0 to 8.2). The purity of the AOA in the SJ and AR cultures has been described previously (25), and we checked the purity for each batch culture using denaturing gradient gel electrophoresis (DGGE) of an archaeal 16S rRNA gene sequence. The archaeal cells (ca. 2 × 1010 cells) used for lipid work were harvested from 0.2-liter cultures by centrifugation at 8,000 rpm (5,200 × g) for 1 h.
Intact polar lipid analysis.
IPLs of “Ca. Nitrosoarchaeum limnia” were extracted separately from the 0.2-μm Duopore and 0.7-μm GF/F filters. Whole-cell pellets of AR and SJ biomass were extracted as such. IPLs were extracted using a modified Bligh and Dyer technique (3): a known volume of single-phase solvent mixture of methanol (MeOH)-dichloromethane (DCM)-phosphate buffer (2:1:0.8, vol/vol/vol) was added to the sample in a centrifuge tube, and the tube was placed in an ultrasonic bath for 10 min. The extract and residue were separated by centrifuging at 2,500 rpm for 5 min, and the solvent mixture was collected in a separate flask (three times). DCM and phosphate buffer were added to the single-phase extract to give a new MeOH-DCM-phosphate buffer ratio of 1:1:0.9 (vol/vol/vol) and to induce phase separation. The extract was centrifuged at 2,500 rpm for 5 min. The DCM phase was collected in a round-bottom flask, and the MeOH:phosphate buffer phase was washed two additional times with DCM. The combined DCM phases were reduced under rotary vacuum and evaporated to dryness under a stream of N2.
Residual biomass of “Ca. Nitrosoarchaeum limnia” left on both Duopore and GF/F filters was reextracted, substituting 5% trichloroacetic acid (TCA) for the phosphate buffer as described by Sturt et al. (43), and analyzed separately to determine the effectiveness and efficiency of the regular Bligh and Dyer extraction. After Bligh and Dyer extraction of AR and SJ biomass, there was not enough residual biomass to perform a TCA extraction.
High-performance liquid chromatography (HPLC)-electrospray ionization (ESI)/mass spectrometry (MS).
IPL-GDGTs were analyzed according to conditions described previously (33) as modified from the method of Sturt et al. (43). For the analysis an Agilent (Palo Alto, CA) 1100 series LC equipped with a thermostat auto-injector was coupled to a Thermo TSQ Quantum EM triple quadrupole mass spectrometer equipped with an Ion Max source with an ESI probe. Detection was achieved using positive-ion ESI/MS by scanning mass range m/z 1,000 to 2,000.
Core lipid analysis.
Acid hydrolysis was performed on aliquots of Bligh and Dyer extracts obtained from filtered “Ca. Nitrosoarchaeum limnia” biomass and on whole cells of SJ and AR, to cleave polar headgroups and release core GDGTs. Biomass was refluxed in 2 ml of 5% HCl in MeOH for 3 h. The cooled solution was adjusted to pH 5 with 2 N KOH-MeOH (1:1, vol/vol). Bidistilled water was added to H2O-MeOH to a final ratio of 1:1 (vol/vol), and this mixture was washed three times with DCM. The DCM fractions were collected and dried over Na2SO4. The extract was dissolved in hexane-propanol (99:1, vol/vol) and filtered over a 0.4-μm-pore-size PTFE filter before analysis by high-performance liquid chromatography atmospheric pressure chemical ionization mass spectrometry (HPLC-APCI/MS).
HPLC-APCI/MS.
Core GDGTs were analyzed using a procedure modified from those of Hopmans et al. (15) and Schouten et al. (36). GDGTs were detected with full-scan analysis from m/z 900 to 1,400 and were quantified using single-ion monitoring (SIM) of glycerol trialkyl glycerol tetraethers (GTGTs) with m/z 1,304 and GDGTs with m/z 1,302, 1,300, 1,298, 1,296, 1,294, 1,292, 1,290, and 1,288. The relative abundance was determined by integration of the extracted peak areas of their [M+H]+. Because crenarchaeol and GDGT-4 coelute and naturally occurring isotopes of crenarchaeol contribute to the GDGT-4 signal, the integrated peak area was corrected by subtracting a response equal to 45% of the crenarchaeol peak from the 1294 peak, according to theoretical isotope distributions described by Hopmans et al. (15).
RESULTS
Core GDGT distributions.
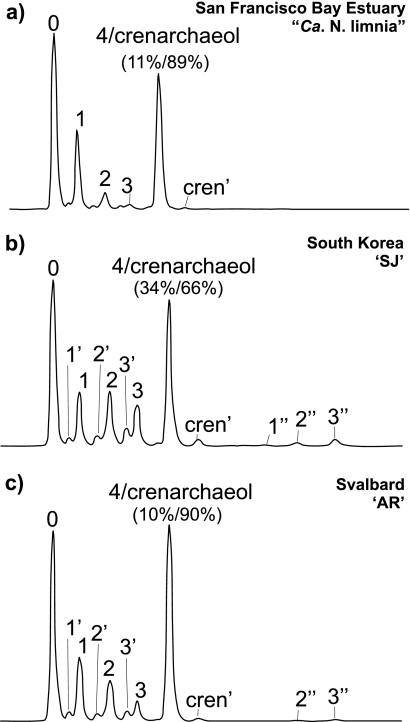
HPLC-APCI-MS analysis of acid-hydrolyzed enrichment culture material revealed the presence of GDGT-0 to GDGT-4, crenarchaeol, and the regioisomer of crenarchaeol in all three cultures (Fig. 2). Although SIM parameters were set to detect GDGT-6, -7, and -X (Fig. 1), none of these GDGTs were detected. Trace amounts of the tribiphytanyl glycerol tetraether, GTGT-0 (Fig. 1), were detected in enrichment cultures AR and “Ca. Nitrosoarchaeum limnia,” whereas it was not detected in the SJ enrichment (Table 1). GDGTs of all AOA enrichments were dominated by GDGT-0 and crenarchaeol, but the relative amounts of the other GDGTs were quite different. The South Korean enrichment (SJ) showed a large contribution of GDGT-4 (34% of the peak area representing coeluting GDGT-4 and crenarchaeol), while the enrichments from San Francisco Bay and coastal Svalbard sediments (AR) contained much less (10 to 11% of total peak area) (Fig. 2 and Table 1). “Ca. Nitrosoarchaeum limnia” also showed a distinctly higher relative abundance of GDGT-1 (Fig. 2a) than SJ and AR, which showed a somewhat similar relative contribution of GDGT-1 to -3 (Fig. 2b and c). In addition, the SJ and AR enrichments contained notable contributions of early-eluting isomers of GDGT-1 to -3 (i.e., 1′, 2′, and 3′), which were present, although in lower relative abundance, in “Ca. Nitrosoarchaeum limnia” (Table 1). Interestingly, relatively low but notable amounts of GDGTs with molecular masses similar to those of GDGT-1 to -3, but with much later elution times, were present in SJ and AR (labeled as 1″, 2″, and 3″ in Fig. 2b and c), suggesting the presence of novel GDGTs and/or isomers which have not previously been reported.
Fig. 2.
Base peak chromatogram of core GDGT profiles of San Francisco Bay enrichment “Ca. Nitrosoarchaeum limnia” (a), coastal South Korea enrichment SJ (b), and coastal Svalbard, Norway, enrichment AR (c). Numbers over peaks correspond to GDGT structures shown in Fig. 1. Numbers with ′ or ″ represent unknown structures with the same m/z as the corresponding GDGT. Percentages listed below peak 4/crenarchaeol indicate the respective contribution of each GDGT to the area of that peak, as corrected according to the method of Hopmans et al. (15).
Table 1.
Fractional abundance of core GDGTs (and GTGT-0) synthesized by the sedimentary AOA analyzed in this work
| Enrichment | GTGT-0 | GDGT fractional abundancea |
TEX86b | Tempc (°C) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1′ | 1″ | 2 | 2′ | 2″ | 3 | 3′ | 3″ | 4 | 4′ | cren | cren′ | ||||
| “Ca. Nitrosoarchaeum limnia” | 0.001 | 0.35 | 0.17 | 0.01 | ND | 0.05 | 0.005 | ND | 0.01 | 0.006 | ND | 0.04 | 0.005 | 0.34 | 0.004 | 0.27 | −0.6 |
| SJ | 0.00 | 0.26 | 0.08 | 0.01 | 0.004 | 0.11 | 0.02 | 0.01 | 0.08 | 0.03 | 0.03 | 0.12 | ND | 0.24 | 0.01 | 0.70 | 28.0 |
| AR | 0.001 | 0.31 | 0.10 | 0.01 | 0.002 | 0.09 | 0.01 | 0.003 | 0.04 | 0.02 | 0.008 | 0.04 | ND | 0.35 | 0.006 | 0.56 | 21.4 |
GDGTs with ′ or ″ indicate isomers identified in the respective chromatograms (exact structures unknown); numbers correspond to the structures shown in Fig. 1. cren, crenarchaeol. ND, not detected.
TEX86 values were calculated according to the method of Schouten et al. (35).
Temperature was calculated according to the method of Kim et al. (18) using the TEX86H calibration for SJ and AR and the TEX86L calibration for “Ca. Nitrosoarchaeum limnia.”
IPL-GDGT distributions.
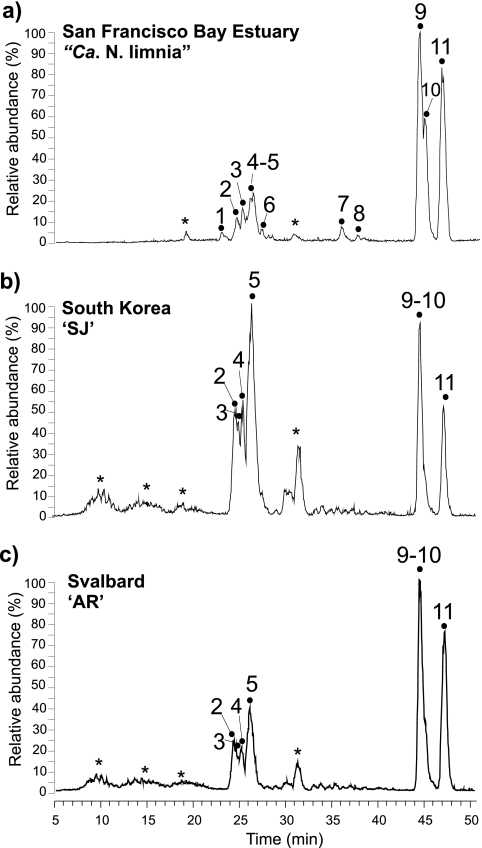
HPLC-ESI/MS analysis of the Bligh and Dyer extracts of the three enrichment cultures revealed the presence of two major clusters of peaks and a number of minor peaks (Fig. 3). GDGT-based IPLs were tentatively identified via mass spectral analysis and showed that all three enriched AOA synthesized similar IPLs containing both glyco and phospho headgroups, although the relative abundances of these IPL classes differed somewhat between cultures. Identification of the major core GDGTs associated with the different IPLs was based on molecular weight and diagnostic fragments of core GDGTs in the mass spectra. This has been previously shown to reveal the same distributions as GDGTs analyzed after acid hydrolysis of IPL fractions prepared from HPLC (33).
Fig. 3.
Base peak chromatogram of GDGT-based IPL profiles (m/z 1,000 to 2,000) of San Francisco Bay enrichment “Ca. Nitrosoarchaeum limnia” (a), coastal South Korea enrichment SJ (b), and coastal Svalbard, Norway enrichment “AR” (c). Peak numbers correspond to IPL structures described in Table 2. *, nonidentified peak.
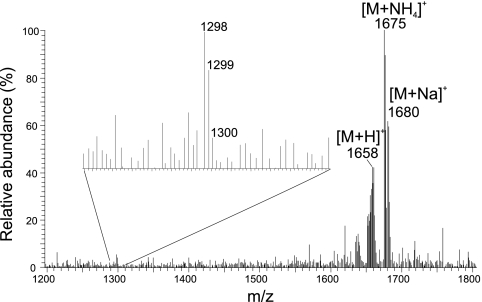
All of the peaks in the first cluster of polar lipids (represented by peaks 1 to 6 in Fig. 3) were identified as GDGT-based glycolipids (GL). Peaks 1 to 4 represent dihexose (DH) GDGTs, with each peak associated with a different major core lipid (Table 2). The main ions in the mass spectra of each IPL corresponded to the ammoniated (NH4+), [M+18]+, and sodiated (Na), [M+23]+, adducts of the IPL, and minor ions represented the core GDGTs. Mass spectra of peak 5 were consistent with a monohexose (MH) IPL with an additional headgroup of 180 Da (MH+180), with GDGT-1 to -3 as the major core lipids, in all three cultures. This IPL, containing the 180-Da headgroup, has been identified previously in “Ca. Nitrosopumilus maritimus SCM1” and has also been recovered from marine sediments (22, 33); however, the precise structure of this headgroup remains unknown. In SJ, this IPL was present in much higher relative abundances than in “Ca. Nitrosoarchaeum limnia” and AR, dominating the GL cluster of the base peak chromatogram. In general, the GL cluster in SJ represented a much higher proportion of the GDGT-based IPLs than the other two AOA. Peak 6 was very small and coeluted with the tail of peak 5 observed in “Ca. Nitrosoarchaeum limnia,” and it was not identified in SJ or AR. The dominant ions were consistent with an IPL containing either two of the unknown 180-Da headgroups or an unknown headgroup of 360 Da coupled to a GDGT-2 core lipid (Fig. 4). In this case, the [M+H]+ ion was quite apparent (m/z 1,658), along with the [M+NH4]+ and [M+Na]+ ions. GDGT-2 appeared to be the dominant core GDGT, as revealed by the m/z 1,298 in the mass spectrum. To the best of our knowledge, this particular IPL has not been previously reported.
Table 2.
Main mass spectra ions, polar headgroups, and major core GDGTs associated with each IPL peak (Fig. 3)
| Fig. 3 peak | Main ions (m/z) | IPL headgroupa | Main core GDGTs |
|---|---|---|---|
| 1 | 1302–1304, 1643–1645, 1648–1650 | DH | GTGT-0, GDGT-0 |
| 2 | 1298–1300, 1639–1641, 1644–1646 | DH | GDGT-2 |
| 3 | 1294–1296, 1635–1637, 1640–1642 | DH | GDGT-3 |
| 4 | 1294–1296, 1635–1637, 1640–1642 | DH | GDGT-4 |
| 5 | 1296–1300, 1638–1642, 1655–1659, 1660–1664 | MH+180 | GDGT-1 to -3 |
| 6 | 1298, 1658, 1675, 1680 | 180 (n = 2) | GDGT 2 |
| 7 | 1534, 1544, 1561, 1566 | PH | GDGT-0 |
| 8 | 1534, 1551, 1556 | PH | Crenarchaeol |
| 9 | 1544, 1706, 1723, 1728 | HPH | GDGT-0 |
| 10 | 1542, 1704, 1721, 1726 | HPH | GDGT-1 |
| 11 | 1534, 1696, 1713, 1718 | HPH | Crenarchaeol |
DH, dihexose; MH, monohexose; PH, phosphohexose; HPH, hexose-phosphohexose; 180, an additional moiety of 180 Da.
Fig. 4.
Mass spectrum of peak 6 in the base peak chromatogram of the HPLC/MS analysis of the extract of “Ca. Nitrosoarchaeum limnia,” showing ions consistent with an IPL containing either two 180-Da headgroups or an unknown headgroup of 360 Da in combination with GDGT-2 as its core lipid.
Peaks 7 and 8 indicated IPLs with a phosphohexose (PH) headgroup coupled to GDGT-0 and crenarchaeol, respectively. These peaks were unique to “Ca. Nitrosoarchaeum limnia,” present in apparently low relative amounts in comparison to the other IPLs. Peaks 9 to 11 are striking in their dominance in the IPL profiles of all three AOA cultures, representing IPLs with a hexose headgroup in addition to a phosphohexose headgroup (HPH) coupled to GDGT-0, GDGT-1, and crenarchaeol, respectively. The distribution of these three HPH-IPLs was nearly identical for the three cultures, despite variable distributions of the GLs. Furthermore, the distributions of the HPH IPLs share similarities with the total core GDGT distributions (Fig. 2), which were dominated by GDGT-0 and crenarchaeol, with smaller amounts of GDGT-1. Indeed, in general it appears that GDGT-0 and crenarchaeol are more closely associated with the HPH headgroups, rather than with the DH and 180 headgroups, which were more commonly associated with the minor GDGTs 2 to 4. A similar trend was noticed in “Ca. Nitrosopumilus maritimus SCM1” (33).
Comparison of filter pore size and BDE and TCA extraction on IPLs of “Ca. Nitrosoarchaeum limnia” culture.
Archaeal lipids in the natural environment are typically studied by filtering water through GF/F filters (5, 40). To test the effects of filter material and pore size on the collection and extraction of IPLs, suspended “Ca. Nitrosoarchaeum limnia” culture was filtered onto both Duopore (DUO) and GF/F filters, with nominal pore sizes of 0.22 μm and 0.7 μm, respectively. The results from equivalent amounts of injected lipid extract from the four separate extracts are summarized in Table 3. The sequential BDE and TCA extracts of the DUO filter and the BDE extract of the GF/F-filter showed almost identical IPL profiles, while only two IPL-GDGT peaks were detected in the TCA extract of the GF/F-filtered cells, possibly because of the small amounts recovered (Table 3). The overall extraction yield (BDE + TCA) from GF/Fs (based on summed IPL response per ml culture; Table 3) was approximately eight times lower than that of the DUO filters, indicating that significantly more cells per ml of culture filtered were captured on the DUO filters. In addition, TCA extraction of the DUO filters yielded much more additional GDGTs (41% of total IPL-GDGTs) than the TCA-extracted GF/F filter (6% of total IPL-GDGTs).
Table 3.
Yields from sequential Bligh and Dyer (BDE) and trichloroacetic acid (TCA) extractions of “Ca. Nitrosoarchaeum limnia” culture filtered over Duopore and GF/F filtersa
| GDGT IPL peak | 0.22-μm Duopore yield |
0.7-μm GF/F yield |
||
|---|---|---|---|---|
| BDE | TCA | BDE | TCA | |
| Distribution (% summed peak areas 1–11) | ||||
| 1–4 | 9.5 | 9.7 | 9.4 | 0.0 |
| 5 | 11 | 10 | 11 | 0.0 |
| 6 | 1.1 | 1.0 | 1.2 | 0.0 |
| 7 | 1.7 | 1.7 | 1.9 | 0.0 |
| 8 | 0.6 | 0.3 | ND | 0.0 |
| 9–10 | 47 | 44 | 42 | 39 |
| 11 | 29 | 33 | 35 | 61 |
| Yield (% total BDE + TCA signal) | ||||
| 1–4 | 58 | 42 | 100 | 0 |
| 5 | 60 | 40 | 100 | 0 |
| 6 | 62 | 38 | 100 | 0 |
| 7 | 59 | 41 | 100 | 0 |
| 8 | 72 | 28 | ND | ND |
| 9–10 | 61 | 39 | 93 | 7 |
| 11 | 56 | 44 | 89 | 11 |
| Total yield (%) | 59 | 41 | 94 | 6 |
| Total response/ml culture (×105)b | 8.4 | 5.9 | 1.6 | 0.14 |
ND, not detected.
Sum of integrated peak areas.
DISCUSSION
Core GDGTs.
Research over the past few years has highlighted the omnipresence of crenarchaeol in the marine environment, as simultaneous studies have called attention to the ubiquity and potential importance of AOA in these environments using molecular ecological approaches. While a few empirical studies have pointed to a link between AOA and the occurrence of crenarchaeol in marine water columns, marine sponges, and soils (5, 21, 41, 49), crenarchaeol synthesis by an AOA was only unambiguously confirmed by its discovery in “Ca. Nitrosopumilus maritimus SCM1” (33), an AOA isolated from tropical aquarium gravel (19). Subsequent confirmation of crenarchaeol synthesis by two other AOA, “Ca. Nitrosocaldus yellowstonii” (6) and “Ca. Nitrososphaera gargensis” (29), led to the proposal that crenarchaeol is synthesized exclusively by AOA and is therefore a specific biomarker for this crenarchaeal lineage (6, 29). The weight of this hypothesis, however, has relied mainly on analysis of the aforementioned AOA, which were enriched from nonmarine sources (Table 4).
Table 4.
Overview of AOA analyzed for GDGTsb
| Enrichment/isolate | Location | Source | Temp (°C) |
NH4+ (μM) | pH | Reference | |
|---|---|---|---|---|---|---|---|
| In situ | Culture | ||||||
| “Ca. Nitrosoarchaeum limnia” | San Francisco Bay estuary | Estuarine sediment | 21.6 | 22 | 500 | 7.0–7.2 | 2; this work |
| SJ′ | Donghae, South Korea | Coastal marine sediment | 0.5 | 25 | 100 | 8.2 | 25; this work |
| AR′ | Svalbard, Norway | Coastal marine sediment | −0.5 | 25 | 100 | 8.2 | 25; this work |
| “Ca. Nitrosopumilus maritimus SCM1” | Tropical aquarium | Gravel | 21–23 | 28 | 500 | 7.0–7.2 | 19, 33 |
| “Ca. Nitrososphaera gargensis” | Siberia Garga hot spring | Microbial mat | <60 | 46 | 500 | 7.8 | 13, 29 |
| “Ca. Nitrosocaldus yellowstonii”a | Yellowstone Heart Lake hot spring | Sediment | 70–80 | 72 | 95 | 8.3 | 6 |
| “Ca. Cenarchaeum symbiosum”a | Santa Barbara, CA | Marine sponge A. mexicana | 8–18 | 10 | n.s. | n.s. | 31, 41 |
Analyzed for core lipids, not for IPLs.
n.s., not specified.
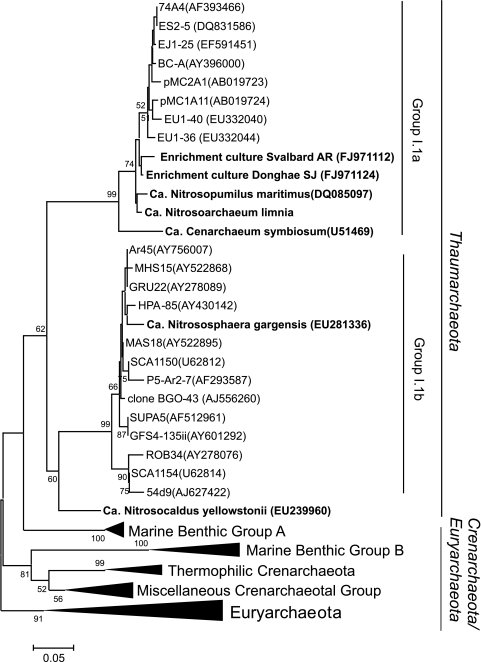
A significant amount of work on AOA has centered on the marine realm where AOA are ubiquitously abundant; however, GDGT data derived from lipid analysis of AOA enriched directly from marine suspended particulate matter and sediments have been lacking until now. Confirmation of crenarchaeol and crenarchaeol regioisomer synthesis by “Ca. Nitrosoarchaeum limnia,” SJ, and AR (enriched directly from marine sediments) further substantiates the possible specificity of crenarchaeol to AOA and provides the strongest evidence thus far that crenarchaeol recovered from marine environments derives from AOA living there. The phylogeny based on the 16S rRNA gene of all of the AOA analyzed for GDGTs to date is shown in Fig. 5: crenarchaeol synthesis in three thaumarchaeotal lineages indicates that it is widely distributed. Crenarchaeol synthesis by multiple representatives from the group I.1a Thaumarchaeota further implies the likelihood that crenarchaeol synthesis is widely occurring throughout this group, regardless of species and/or habitat (e.g., planktonic or sedimentary). All of the Thaumarchaeota enrichments thus far are AOA, implying the possibility that crenarchaeol synthesis may actually be exclusive to archaea involved in ammonia oxidation. Verification of this would require lipid analysis of enriched, nonammonia-oxidizing representatives of the Thaumarchaeota and other mesophilic Crenarchaeota; however, no such cultures exist at this time.
Fig. 5.
Summarized archaeal phylogeny based on the 16S rRNA gene showing detailed groupings with previously cultivated representatives (bold type) in addition to those analyzed in this work. For phylogenetic analysis, the gene sequences of related taxa were obtained from the GenBank database, and multiple alignments were performed using the Clustal X program (44). The phylogenetic trees were constructed by using the MEGA 3 computer software program (20).
Based on the recently proposed new classification of AOA into a new phylum, the Thaumarchaeota (4, 42), it was also suggested that crenarchaeol should be renamed thaumarchaeol (42). As discussed above, crenarchaeol biosynthesis seems indeed to fall within the thaumarchaeotal phylum (Fig. 4), which would argue for the renaming of crenarchaeol. On the other hand, nicknames of chemical components should not be changed immediately following each phylogenetic reclassification. For example, archaeol is still called archaeol even though only one of the three archaeal phyla (i.e., the Euryarchaeota) contains archaeol as a major membrane lipid. Therefore, we suggest that the name crenarchaeol be retained and that this name be reevaluated when more information on archaeal phylogeny and membrane lipid biosynthesis becomes available.
In addition to crenarchaeol, all AOA that have been screened for core GDGTs also synthesize GDGT-0 to GDGT-4 and, with the exception of “Ca. Nitrososphaera gargensis,” produce GDGT-0 in high relative abundances often comparable to that of crenarchaeol. GDGT-1 to -4 are usually produced in smaller amounts than GDGT-0 and crenarchaeol; their comparative abundances vary significantly between cultures. Similarly, our data yielded different distributions of GDGT-1 to -4 for all three enrichment cultures (Fig. 3 and Table 1). The high abundance of GDGT-4, particularly in SJ (34% of the combined crenarchaeol + GDGT-4 peak; Fig. 2b) is intriguing, as it has not been observed in such high abundances in any other AOA. GDGT-4 is often the most abundant GDGT in (hyper) thermophilic Archaea, presumably because its structure (containing four cyclopentane moieties [Fig. 1]) promotes dense packing of lipids which helps maintain the integrity of the cell membrane at high temperatures where it would otherwise be compromised. Of the three AOA studied here, AR and SJ were enriched from among the lowest in situ temperatures (−0.5 to 0.5°C), and in comparison with abundances of other AOA isolated and cultivated at much higher temperatures (i.e., “Ca. Nitrososphaera gargensis and “Ca. Nitrosocaldus yellowstonii”; Table 4), the relative abundance of GDGT-4 in especially SJ is still anomalously high. The occurrence of such relatively large amounts of GDGT-4 in an archaeon enriched from such a low temperature is unusual and difficult to explain at present.
Precisely why GDGT profiles of thaumarchaeotal enrichments differ from each other is still largely unknown and may have to do with a number of physical conditions. Temperature, specifically, has been shown to exhibit control over GDGT composition in both (hyper)thermophilic Crenarchaeota and Euryarchaeota (45) and mesophilic Thaumarchaeota (32, 49). An empirical correlation between marine sea surface temperature (SST) and sedimentary GDGT distribution, comprised of GDGT-1 to -3 and the crenarchaeol regioisomer, has led to the development of the TEX86 temperature proxy (35), which is based on the premise that planktonic marine Thaumarchaeota adapt their GDGT distribution according to temperature and this signal is recorded in fossil GDGTs that reach the sediments. The cultures of “Ca. Nitrosoarchaeum limnia,” SJ, and AR represent relevant AOA upon which to test whether sedimentary AOA adapt their membranes in a similar way.
Despite similar cultivation temperatures (22 to 25°C), the TEX86 values of “Ca. Nitrosoarchaeum limnia,” SJ, and AR were quite different (Table 1). The TEX86-derived temperatures for SJ and AR (28°C and 21°C [Table 1]) are around the cultivation temperatures (25°C). These sedimentary AOA do follow the marine TEX86 calibration recently presented by Kim et al. (18), consistent with findings reported for “Ca. Nitrosopumilus maritimus” and “Ca. Nitrososphaera gargensis,” a group I.1b thaumarchaeote enriched from a hot spring (29). In contrast, the TEX86-derived temperature of “Ca. Nitrosoarchaeum limnia” is ca. 0°C, far below the cultivation and in situ temperatures of 22°C and 21.6°C, respectively. The reason for this is presently unclear but may have to do with the fact that it was cultivated at a lower pH than SJ and AR (Table 4). Shimada et al. (39) reported that Thermoplasma acidophilum cultivated in the pH range 1.2 to 3.0 decreased the number of cyclopentane moieties in the membrane GDGTs at lower pH, the same response as is observed here for the sedimentary AOA. Cultivation at a lower salinity (i.e., “Ca. Nitrosoarchaeum limnia” was grown at ∼8‰ salinity) is an unlikely explanation for the low TEX86 value of “Ca. Nitrosoarchaeum limnia,” since Wuchter et al. (49) already demonstrated for a marine thaumarchaeotal enrichment that salinity has no effect on TEX86. This is further corroborated by the identical marine and lacustrine TEX86-temperature calibrations (30).
Lipids with the same molecular weights as GDGT-1 to -3 but with slightly earlier elution times (usually leading the main GDGT peak) have been recovered from the environment (e.g., see reference 28) and appear similar to those observed in the sedimentary AOA enrichments (Fig. 2b and c, GDGTs 1′, 2′, and 3′). More commonly reported is a lipid similar to GDGT-4 (27, 41); however, GDGT-4′ was not detected in SJ and AR and was present in trace amounts in “Ca. Nitrosoarchaeum limnia” (Table 1). Unambiguous identification of these compounds has yet to be performed; however, tandem MS (MS-MS) data show that in terms of mass spectra they are identical to the later-eluting GDGTs and therefore represent structural isomers (E. Hopmans, J. Weijers, and S. Schouten, unpublished data). In some soils and hot spring sediments, these isomers can be as abundant as the known GDGTs (28); however, they are not abundant in marine suspended particulate matter and surface sediments. Their abundance in the AOA studied here, compared to that in other AOA, points to sedimentary AOA as primary producers of these unknown lipids in marine environments; however, their sources in terrestrial environments remain unknown. In addition, some other late-eluting lipids were present in the SJ and AR cultures (Fig. 2b and c, GDGTs 1″, 2″, and 3″). At the moment, however, their structures remain enigmatic. A late-eluting lipid related to crenarchaeol with an additional cyclopentane moiety (m/z 1,290, GDGT X; Fig. 1) was recently identified in the moderate thermophile “Ca. Nitrososphaera gargensis,” which falls into the soil group I.1b AOA (29). We did not identify this GDGT in either of the AOA enrichments analyzed in this study.
IPL-GDGTs.
IPL headgroups synthesized in notable amounts in all three cultures include DH, MH+180, and HPH moieties, suggesting that they are important in the cell membranes of marine AOA. Trace MH and only small apparent amounts of PH headgroup moieties were detected in “Ca. Nitrosoarchaeum limnia,” which were completely absent in SJ and AR, signifying that these headgroups may not be widely synthesized by marine AOA, despite apparently larger relative abundances of these IPLs in “Ca. Nitrosopumilus maritimus” (33). Comparison of the IPL composition of “Ca. Nitrosoarchaeum limnia,” SJ, and AR AOA with those identified previously for “Ca. Nitrosopumilus maritimus” and “Ca. Nitrososphaera gargensis” showed that they all make IPL-GDGTs with DH and HPH headgroups (Table 3). Typically, duplicates of batch cultures of microorganisms may show some subtle variations in the relative abundance of lipids, but such differences will not affect this conclusion. Further comparison of the GDGTs associated with each headgroup indicated that HPH bound to crenarchaeol represents the only IPL common to all five species, consequently rendering HPH-crenarchaeol the best biomarker lipid for the detection of living AOA. In addition to its uniqueness to AOA, PLs in general are superior to GLs as indicators of living organisms due to their lower stability outside the cell membrane (i.e., upon cell senescence) (12, 37).
GDGTs associated with particular headgroups seem to occur in a way similar to those in “Ca. Nitrosopumilus maritimus”: GDGT-0, GDGT-1, and crenarchaeol are largely associated with PH and HPH headgroups, while minor GDGTs 2 to 4 are usually associated with DH and MH+180 moieties (Table 2). Thus, synthesis of IPL-GDGTs by sedimentary AOA is not necessarily different from that of pelagic AOA.
The implications drawn from differences between glycosidic and phosphate-based IPLs raise additional interest in other structurally enigmatic headgroup moieties. Common among the AOA is a MH+180 Dalton headgroup (Table 2), which was originally identified in marine sediments (1, 43) prior to confirmation of its biosynthesis by “Ca. Nitrosopumilus maritimus” (33). With the exception of “Ca. Nitrososphaera gargensis,” all four other AOA synthesize IPL-GDGTs containing this headgroup, signifying that it may be a commonly synthesized moiety among group I.1a AOA. IPL analysis of additional AOA from other group I lineages, however, is needed to confirm this. Nevertheless, MH+180 bound to crenarchaeol has recently been detected in water column suspended particulate matter (29a). Interestingly, “Ca. Nitrosoarchaeum limnia” synthesizes what we have tentatively identified as a GDGT-based IPL with a double 180-Da moiety (Fig. 3, Table 3), which has not been previously reported as far as we are aware.
Effects of filter type and TCA extraction on IPLs of “Ca. Nitrosoarchaeum limnia.”
In contrast to sediments, which are typically freeze-dried and extracted as such, GDGTs present in the marine water column are often recovered by filtration of SPM over GF/F filters; however, the reported size of marine Thaumarchaeota (e.g., 0.5 by 0.15 μm for “Ca. Nitrosopumilus maritimus” [19]) is often smaller than the GF/F nominal pore size (0.7 μm) typically used for particulate matter filtration intended for lipid analysis. The low level of recovery of suspended culture of “Ca. Nitrosoarchaeum limnia” on GF/Fs compared to that on the DUO filters (which have 0.7-μm and 0.22-μm pore sizes, respectively) (Table 3) suggests that when planktonic Thaumarchaeota are not particle associated (as they often are in more turbid waters, e.g., the North Sea [50]), a large proportion of cells present will not be recovered on those filters. However, since the effective pore size will decrease with an increasing load on the filter, field experiments filtering turbid waters should result in much higher levels of recovery than we observed here (14). While in our study, nominal pore size had no major observable effect on the IPL distribution (which was not surprising, as the culture was uni-archaeal), it could result in quantitative biases in GDGT/IPL distributions recovered from the environment, where differences could exist in the cell size and/or shape and the propensity of local Thaumarchaeota to associate with particles.
Sequential extraction of the DUO filters with TCA resulted in a substantial additional recovery of IPLs (i.e., 41% of the total recovered IPLs), with the same distribution as those extracted using Bligh and Dyer solvents, in contrast to results with the GF/F filters, where TCA extraction did not yield substantial additional amounts. Previously, it was noted that sequential extraction of whole-cell biomass of “Ca. Nitrososphaera gargensis” with TCA did not result in a significant increase in IPL yields (29), thereby suggesting that IPL profiles of SJ and AR are representative and that BDE alone is sufficient for a qualitative assessment of IPLs in culture material, particulate matter, and sediments. However, for quantitative analysis, filtration of water over filters with smaller pore size and additional extraction with TCA may need to be performed.
Conclusions.
We report the core and IPL-GDGTs synthesized by three sedimentary marine group I.1a AOA, enriched from sediments derived from the San Francisco Bay estuary (“Ca. Nitrosoarchaeum limnia”), the South Korean coast (SJ) and Svalbard, Norway (AR). These cultures represent the first AOA enriched directly from the marine environment. They all synthesize crenarchaeol and minor amounts of the crenarchaeol regioisomer, confirming crenarchaeol as a widespread biomarker for AOA in the marine environment both in the water column and in surface sediments. Major GDGT-based IPLs synthesized by “Ca. Nitrosoarchaeum limnia,” SJ, and AR contained HPH headgroups attached primarily to crenarchaeol and GDGT-0, in addition to a variety of sugar-based headgroups bound mainly to minor GDGTs 2 to 4. The occurrence of HPH-crenarchaeol in all AOA analyzed for IPLs thus far points to its utility as a general biomarker for viable AOA. Systematic differences in GDGT-headgroup associations may have implications for our understanding of environmental GDGT distributions, particularly when trying to dissociate “dead” from “living” archaeal lipid signals using core and GL-GDGT distributions, respectively.
ACKNOWLEDGMENTS
Irene Rijpstra is thanked for analytical assistance.
The Darwin Center for Biogeosciences partially funded this project.
Footnotes
Published ahead of print on 25 March 2011.
Publication number DW-2011-1005 of the Darwin Center for Biogeosciences.
REFERENCES
- 1. Biddle J. F., et al. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U. S. A. 103:3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blainey P. C., Mosier A. C., Potanina A., Francis C. A., Quake S. R. 2011. Genome of low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLos ONE 6:e16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 4. Brochier-Armanet C., Boussau B., Gribaldo S., Forterre P. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 5. Coolen M. J. L., et al. 2007. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ. Microbiol. 9:1001–1016 [DOI] [PubMed] [Google Scholar]
- 6. de la Torre J. R., Walker C. B., Ingalls A. E., Könneke M., Stahl D. A. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810–818 [DOI] [PubMed] [Google Scholar]
- 7. DeLong E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuhrman J. A., McCallum K., Davis A. A. 1992. Novel major archaebacterial group from marine plankton. Nature 356:148–149 [DOI] [PubMed] [Google Scholar]
- 10. Hallam S. J., et al. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:520–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hallam S. J., et al. 2006. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U. S. A. 103:18296–18301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harvey H. R., Fallon R. D., Patton J. S. 1986. The effect of organic matter and oxygen on the degradation of bacterial membrane lipids in marine sediments. Geochim. Cosmochim. Acta 50:795–804 [Google Scholar]
- 13. Hatzenpichler R., et al. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herfort L., Schouten S., Boon J. P., Sinninghe Damsté J. S. 2006. Application of the TEX86 temperature proxy to the southern North Sea. Org. Geochem. 37:1715–1726 [Google Scholar]
- 15. Hopmans E. C., Schouten S., Pancost R. D., van der Meer M. T. J., Sinninghe Damsté J. S. 2000. Analysis of intact tetraether lipids in archaeal cell material and sediments by high performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 14:585–589 [DOI] [PubMed] [Google Scholar]
- 16. Huguet C., Martens-Habbena W., Urakawa H., Stahl D. A., Ingalls A. E. 2010. Comparison of extraction methods for quantitative analysis of core and intact polar glycerol dialkyl glycerol tetraethers (GDGTs) in environmental samples. Limnol. Oceanogr. Methods 8:127–145 [Google Scholar]
- 17. Ingalls A. E., et al. 2006. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. U. S. A. 103:6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim J. H., et al. 2010. New indices and calibrations derived from the distribution of crenarchaeal isoprenoid tetraether lipids: implications for past sea surface temperature reconstructions. Geochim. Cosmochim. Acta 74:4639–4654 [Google Scholar]
- 19. Könneke M. E., de la Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 20. Kumar S., Tamura K., Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 21. Leininger S., et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809 [DOI] [PubMed] [Google Scholar]
- 22. Lipp J. S., Hinrichs K. U. 2009. Structural diversity and fate of intact polar lipids in marine sediments. Geochim. Cosmochim. Acta 73:6816–6833 [Google Scholar]
- 23. Lipp J. S., Morono Y., Inagaki F., Hinrichs K. U. 2008. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454:991–994 [DOI] [PubMed] [Google Scholar]
- 24. Mosier A. C., Francis C. A. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002–3016 [DOI] [PubMed] [Google Scholar]
- 25. Park B.-J., et al. 2010. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 76:7575–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park S.-J., Park B.-J., Rhee S.-K. 2008. Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12:605–615 [DOI] [PubMed] [Google Scholar]
- 27. Pearson A., et al. 2008. Factors controlling the distribution of archaeal tetraethers in terrestrial hot springs. Appl. Environ. Microbiol. 74:3523–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pitcher A., Schouten S., Sinninghe Damsté J. S. 2009. In situ production of crenarchaeol in two California hot springs. Appl. Environ. Microbiol. 75:4443–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pitcher A., et al. 2010. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic group I.1b Archaeon. ISME J. 4:542–552 [DOI] [PubMed] [Google Scholar]
- 29a. Pitcher A., et al. Niche segregation of ammonia-oxidizing Archaea and anammox bacteria in the Arabian Sea oxygen minimum zone as determined by a combined intact polar lipid and gene-based approach. ISME J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Powers L., et al. 2010. Applicability and calibration of the TEX86 paleothermometer in lakes. Org. Geochem. 41:404–413 [Google Scholar]
- 31. Preston C. M., Wu K. Y., Molinski T. F., DeLong E. F. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. U. S. A. 93:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schouten S., Forster A., Panoto F. E., Sinninghe Damste J. S. 2007. Towards calibration of the TEX86 palaeothermometer for tropical sea surface temperatures in ancient greenhouse worlds. Org. Geochem. 38:1537–1546 [Google Scholar]
- 33. Schouten S., et al. 2008. Intact membrane lipids of “Candidatus Nitrosopumilus maritimus,” a cultivated representative of the cosmopolitan mesophilic group I crenarchaeota. Appl. Environ. Microbiol. 74:2433–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schouten S., Hopmans E. C., Pancost R. D., Sinninghe Damsté J. S. 2000. Widespread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc. Natl. Acad. Sci. U. S. A. 97:14421–14426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schouten S., Hopmans E. C., Schefuß E., Sinninghe Damsté J. S. 2002. Distributional variations in marine crenarchaeotal membrane lipids: a new tool for reconstructing ancient sea water temperatures? Earth Planet. Sci. Lett. 204:265–274 [Google Scholar]
- 36. Schouten S., Huguet C., Hopmans E. C., Kienhuis M. V. M., Sinninghe Damsté J. S. 2007. Analytical methodology for TEX86 paleothermometry by high-performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal. Chem. 79:2940–2944 [DOI] [PubMed] [Google Scholar]
- 37. Schouten S., Middelburg J. J., Hopmans E. C., Sinninghe Damsté J. S. 2010. Fossilization and degradation of intact polar lipids in deep subsurface sediments: a theoretical approach. Geochim. Cosmochim. Acta 74:3806–3814 [Google Scholar]
- 38. Schubotz F., Wakeham S. G., Lipp J. S., Fredricks H. F., Hinrichs K. U. 2009. Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ. Microbiol. 11:2720–2734 [DOI] [PubMed] [Google Scholar]
- 39. Shimada H., Nemoto N., Shida Y., Oshima T., Yamagishi A. 2008. Effects of pH and temperature on the composition of polar lipids in Thermoplasma acidophilum HO-62. J. Bacteriol. 190:5404–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sinninghe Damsté J. S., et al. 2002. Distribution of membrane lipids of planktonic Crenarchaeota in the Arabian Sea. Appl. Environ. Microbiol. 68:2997–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinninghe Damsté J. S., Schouten S., Hopmans E. C., van Duin A. C. T., Geenevasen J. A. J. 2002. Crenarchaeol: the characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota. J. Lipid Res. 43:1641–1651 [DOI] [PubMed] [Google Scholar]
- 42. Spang A., et al. 2010. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 18:331–340 [DOI] [PubMed] [Google Scholar]
- 43. Sturt H. F., Summons R. E., Smith K., Elvert M., Hinrichs K.-U. 2004. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18:617–628 [DOI] [PubMed] [Google Scholar]
- 44. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uda I., Sugai A., Itoh Y. H., Itoh T. 2001. Variation in molecular species of polar lipids from Thermoplasma acidophilum depends on growth temperature. Lipids 36:103–105 [DOI] [PubMed] [Google Scholar]
- 46. White D. C., Davis W. M., Nickels J. S., King J. D., Bobbie R. J. 1979. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40:51–62 [DOI] [PubMed] [Google Scholar]
- 47. Widdel F., Bak F. 1992. Gram-negative mesophilic sulfate reducing bacteria, p. 3352–3378In Balows A., Truper H. G., Dworkin M., Harder W., Schleifer K. H. (ed.), The prokaryotes, 2nd ed. Springer, New York, NY [Google Scholar]
- 48. Wuchter C., Schouten S., Boschker H. T. S., Sinninghe Damsté J. S. 2003. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol. Lett. 219:203–207 [DOI] [PubMed] [Google Scholar]
- 49. Wuchter C., Schouten S., Coolen M. J. L., Sinninghe Damsté J. S. 2004. Temperature-dependent variation in the distribution of tetraether membrane lipids of marine Crenarchaeota: implications for TEX86 paleothermometry. Paleoceanography 19:PA4028, doi: 10.1029/2004PA001041 [Google Scholar]
- 50. Wuchter C., et al. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]