Abstract
Stx bacteriophages in 68 samples of beef and salad were quantified by real-time quantitative PCR (qPCR). Stx phages from the samples were propagated in Escherichia coli C600, E. coli O157:H7, and Shigella strains and further quantified. Fifty percent of the samples carried infectious Stx phages that were isolated from plaques generated by lysis.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) causes severe human diseases (22). The most probable natural reservoir of STEC is cattle. Contamination can occur at different levels of the food chain (5, 13). Transmission of STEC is due mainly to consumption of fecally contaminated food or water (4, 5, 22).
Shiga toxin (Stx) is a cardinal virulence factor of STEC. Shiga toxin genes (stx1, stx2, or variants) are carried by lambdoid phages (12, 21). The induction and regulation of Stx phages are involved directly in Stx production and in the pathogenicity of the strains (7, 29). Free Stx phages occur in different environments (9, 15, 19, 20, 23), and they are more persistent than bacteria (15, 19). Stx phages are involved in the horizontal transduction of stx in vitro (24) and in vivo (6, 27), which has also been demonstrated in food matrices (14). Transduction of stx leads to the emergence of new Stx-producing bacteria, and therefore, the occurrence of Stx phages in the environment increases the risk of the emergence of new STEC strains. In order to guarantee food quality, it is necessary to consider the potential risk arising from the possible presence of infectious Stx phages in food. In this study, we detected and quantified Stx phages in commercial beef and salad samples by quantitative PCR (qPCR) and evaluated their infectivity.
Bacteriophage 933W was used as positive control (21). E. coli O157:H7 strain ATCC 43888, E. coli C600, and Shigella sonnei strain 866 (18) were used as stx-negative host strains for the propagation of Stx phages. Aerobic colony counts were performed on Trypticase soy agar (TSA) at 37°C; and E. coli counts were performed on Chromocult coliform agar (Merck, Darmstadt, Germany). Somatic coliphages were determined according to the ISO standard method (1), based on the double-agar-layer method for the analysis of phages in a sample, using E. coli strain WG5 as a host. The presence of stx2 gene-carrying bacteria among coliform bacteria grown on Chromocult agar was determined by colony blotting using a specific probe to detect the stx2 gene (11). Enumeration of Stx phages on agar plates was performed by the double-agar-layer method and plaque blot hybridization using the stx2 gene probe as previously described (20).
Food samples were purchased from local retailers located in the city of Barcelona (Spain). The supermarkets and grocery stores were selected at random, and each sample was obtained from a different retailer to give a more general overview. In most cases, the stores had different suppliers. Thirty-six samples of ground beef (containing only beef) were freshly minced on request at local supermarkets. The 32 salad samples were from a commercially prepared mixture of salads, mostly containing frisée lettuce, corn salad, and radicchio (red chicory) from different suppliers. The salad mixtures presented some variations in salad composition, depending on the supermarket and the suppliers. Twenty-five grams of minced beef or salad was diluted 1:4 (wt/vol) in one-fourth-strength Ringer's solution. The mixture was placed in stomacher bags with filters (Afora, Barcelona, Spain) and homogenized for 2 min (masticator; IUL Instruments GmbH, Königswinter, Germany). Fifty milliliters of the homogenate was used to purify bacteriophages and for the measurement of microbiological parameters.
Bacteriophages were purified from the mixtures by filtration through low-protein-binding 0.22-μm-pore-size membranes (Millex-GP; Millipore, Bedford, MA) and concentrated as described previously (16). Bacteriophage suspension was treated with DNase (100 units·ml−1) and incubated at 37°C for 1 h. An aliquot of the sample at this stage was amplified by qPCR to confirm that free DNA containing stx had been removed and that only phage DNA from phage particles was amplified.
To assess the presence of infectious Stx phages in the food samples, 5 ml of a culture (optical density at 600 nm [OD600] = 0.5) of strains E. coli O157:H7, E. coli C600, and S. sonnei 866 was added to 30 ml of LB broth, and 15 ml of the bacteriophages was purified from the samples. Cultures were incubated at 37°C for 18 h. Bacteriophages were purified from the supernatant of the enrichment cultures as described above.
DNA was isolated from the phages by proteinase K digestion and phenol extraction (16). The presence of stx2 in phage DNA was qualitatively determined by conventional PCR with primers UP378/LP378 (19) for a first-round PCR and internal primers for a second-round nested PCR (19), and amplified fragments were sequenced (20). This set of primers amplified all stx2 variants except stx2f. For sequencing purposes, primers S2Aup/lp and GK4 (18), amplifying the A and B subunits of the stx2 operon (18), were used.
The number of stx copies in phage DNA was quantified by a custom TaqMan qPCR (15, 16). The qPCR is located within the 169-bp amplicon generated with the internal primers described above (19). Since Stx phages are known to carry only one stx copy, the stx gene copy (GC) values can be extrapolated to be the number of Stx phages in each sample. For quantification by qPCR, standards were prepared (15), and the number of stx copies in the samples was calculated by reference to this standard. The GC values presented are the average value from the three replicates performed for each sample. To screen for PCR inhibition, dilutions of the standard used for quantification were spiked with DNA extracted from the food samples, and the empirical difference was compared to the number of copies of stx in the standards (15). Inhibition of the PCR by sample DNA was not detected. Evaluation of sensitivity and efficiency of the methods was reported previously (15, 16). The detection limit of the technique obtained with the standard indicated that our reaction detected 5.29 stx gene copies, which corresponds to a threshold cycle (CT) of 32.
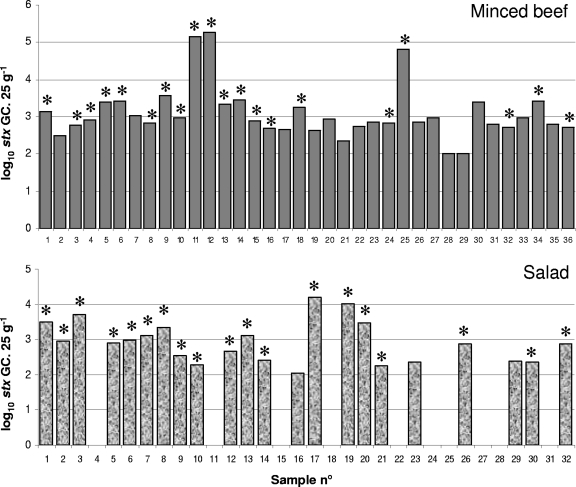
Shiga toxin phages were detected by qPCR in all the beef samples and in the 68.7% of salad samples (Fig. 1). Conventional PCR, even when using nested PCR, showed fewer positive results, with 55.6% of beef samples and 59.4% of salad samples testing positive. The range of CT values obtained from the samples was from 24 to 30 for beef samples and from 26 to 32 in salad samples. Samples with higher CT values than these were not considered positive, since they exceeded the detection limit.
Fig. 1.
Qualitative occurrence of Stx phages determined by conventional PCR and quantitative evaluation of Stx phages (number of log10 GCs·25 g−1) evaluated by qPCR in minced beef and salad samples.
The differences between qPCR and conventional PCR could be due to the differences in the methods used and the fact that the amplicon used in qPCR was 65 bp (15, 16), while the amplicon used in nested PCR was 169 bp (3). Therefore, qPCR was more sensitive than conventional PCR, at least for this particular assay. qPCR provides speed, robustness, reproducibility, and a lower contamination risk than nested PCR, in addition to the information yielded by the quantification of stx.
The sequence of the stx2 gene (A and B subunits) carried by the phages isolated from food was determined by amplification and sequencing. Phage DNA from some of the samples with greater densities of stx2 phages was selected for these studies. The sizes of the amplicons sequenced and used for comparison against previously described sequences varied depending on the sample, and as observed in Table 1, some samples did not allow amplification of long amplimers with the primers used. Nevertheless, the samples showed the presence of stx2 and stx2 variants (mostly stx2c and stx2d) (Table 1). In some cases, the sequences of amplified fragments showed 100% homology with several variant sequences available in GenBank, so it was impossible to determine the type of variant carried by the phage DNA of these samples.
Table 1.
Comparison of the stx2 fragment sequence amplified from phage DNA isolated from salad and ground beef with previously described sequences
| Sample no. | Fragment size (bp)a | stx variant(s) | Sequence homologyc | GenBank accession no. |
|---|---|---|---|---|
| Salad | ||||
| 1 | 732 | stx2, stx2c, stx2db | Escherichia coli O157:H7 00F078 | GU983682.2 |
| Stx2-converting phage 1717 | FJ188381.1 | |||
| Escherichia coli strain N5578 | GQ429168.1 | |||
| Escherichia coli strain TS06/08 | FM998848.1 | |||
| 8 | 692 | stx2, stx2c, stx2d | Escherichia coli O157:H7 00F078 | GU983682.2 |
| Stx2-converting phage 1717 | FJ188381.1 | |||
| Escherichia coli strain N5578 | GQ429168.1 | |||
| Escherichia coli strain TS06/08 | FM998848.1 | |||
| 13 | 1,211 | stx2c | Stx2-converting phage 1717 | GU983682.2 |
| Escherichia coli O157:H7 00F078 | FJ188381.1 | |||
| 17 | 725 | stx2, stx2c, stx2d | Bacteriophage A312 | AY633461.1 |
| Stx2-converting phage 1717 | FJ188381.1 | |||
| Escherichia coli O157:H7 | AB071845.1 | |||
| 19 | 521 | stx2 variant not defined | Escherichia coli OX3:H21 | L11078.1 |
| Minced beef | ||||
| 1 | 756 | stx2, stx2c, stx2d | Escherichia coli O157:H7 00F078 | GU983682.2 |
| Escherichia coli TS09/08 | FM998850.1 | |||
| Escherichia coli strain N5578 | GQ429168.1 | |||
| 5 | 1,024 | stx2 | Bacteriophage 933W | AF125520.1 |
| 6 | 1,023 | stx2 | Bacteriophage 933W | AF125520.1 |
| 11 | 498 | stx2, stx2c, stx2d | Stx2-converting phage 1717 | FJ188381.1 |
| Escherichia coli isolate P1332 | EU816442.1 | |||
| Bacteriophage A312 | AY633461.1 | |||
| 12 | 699 | stx2, stx2c, stx2d | Bacteriophage A312 | AY633461.1 |
| Stx2-converting phage 1717 | FJ188381.1 | |||
| Escherichia coli O157:H7 stx2vhdA | AB071845.1 | |||
| 14 | 740 | stx2 | Bacteriophage 933W | AF125520.1 |
| 18 | 733 | stx2 | Bacteriophage 933W | AF125520.1 |
| 25 | 705 | stx2, stx2c, stx2d, stx2g | Escherichia coli clone pEHEC400 | AY443052.1 |
| Escherichia coli strain 5021/96 | AJ567994.1 | |||
| Escherichia coli O28ab:H28 | AY095209.1 | |||
| Escherichia coli isolate EC173b serotype O174:H21 | AF500190.1 |
Size of the fragment sequenced and used for BLAST analysis.
The stx2 sequence of the fragment matches with those of all three variants.
Presented those sequences showing 100% homology with the sequence of the fragment and listed in the first position after BLAST analysis.
Despite the abundance of Stx phages in these samples, they showed acceptable microbiological levels (Table 2) for human consumption by following European and U.S. regulations (2, 28). European regulation determines aerobic colony counts between 5 × 105 and 5 × 106·g−1, E. coli counts between 50 and 500 CFU·g−1 in minced beef, and values of E. coli between 100 to 1,000 CFU·g−1 in precut vegetables (ready to eat) as acceptable. Most of these samples were also within the current limits imposed by U.S. regulation (standard plate counts of 1.0 × 106 CFU·g−1 and E. coli counts of 100 CFU·g−1 in boneless beef) (28). For salads, the U.S. Food and Drug Administration (FDA) (www.fda.gov) requires E. coli counts of <100 most probable number (MPN)·g−1. There are no reports that these samples were associated with any kind of food-borne disease.
Table 2.
Microbiological parameters and Stx phage density in minced beef and salad samplesb
| Sample | No. of samples | Microbiological parameters (no. of CFU or PFU·25 g−1) |
Stx phage density (no. of stx GCs·25 g−1)a |
||||
|---|---|---|---|---|---|---|---|
| Aerobic colonies | E. coli bacteria | Somatic coliphages | Average | Max | Min | ||
| Beef | 36 | 8.75 × 106 (6.02 × 105) | 7.38 × 103 (1.05 × 103) | 5.00 × 102 (2.00 × 102) | 1.27 × 104 (4.00 × 104) | 1.78 × 105 | 1.01 × 102 |
| Salad | 32 | 1.57 × 106 (2.52 × 105) | 4.25 × 103 (4.16 × 103) | < 0.1 | 2.3 × 103 (3.86 × 103) | 1.57 × 104 | 1.82 × 102 |
Max and min, samples showing the maximum and minimum values for Stx phages, respectively.
Values shown in parentheses are standard deviations.
Additionally, stx2 gene-carrying bacteria were not detected in the samples. Somatic coliphages, proposed as viral fecal indicators (1), were included in this study to evaluate the levels of the viral fecal load in the samples. Somatic coliphages showed very low values, indicating that samples were not polluted with viruses of fecal origin. Since the values of somatic coliphages do not correlate with the values of Stx phages (data not shown), we conclude that Stx phages do not always have a fecal origin. This lack of correlation between somatic coliphages and Stx phages was observed previously in environmental samples (15).
However, the general abundance of Stx phages in the samples analyzed does not determine whether these phages are infectious. To assess the infectivity of Stx phages, three bacterial host strains were selected according to their suitability for Stx phage propagation (19, 20). We compared the densities of Stx phages from the original sample with the values of Stx phages obtained from the same volume but after propagating them in the bacterial enrichment culture.
Minced beef and salad samples were selected at random from the total pool of positive samples. We incubated the samples in an enrichment culture and compared the number of Stx phages (stx GCs) with that in the original sample (Table 3). The number of Stx phages was considered to have been increased by enrichment when they were significantly different (P < 0.05) from those counted before the enrichment, as calculated by paired t test.
Table 3.
Increase in the densities of Stx phages in some of the samples evaluated before and after their propagation in host strains
| Host strain | Parameter | Value for sample group |
|||||
|---|---|---|---|---|---|---|---|
| Minced beef (25 samples) |
Salad (18 samples) |
||||||
| Geometric mean | Max | Min | Geometric mean | Max | Min | ||
| E. coli C600 | No. of GCs before enrichment | 5.00 × 102 | 1.78 × 105 | 1.00 × 102 | 1.08 × 102 | 7.53 × 102 | 1.82 × 102 |
| No. of GCs after enrichment | 5.74 × 103 | 2.42 × 105 | 7.90 × 102 | 1.68 × 103 | 8.43 × 103 | 1.92 × 102 | |
| GC increasea | 4.35 × 103 | 2.34 × 105 | 3.79 × 102 | 1.52 × 103 | 7.48 × 103 | 2.81 × 103 | |
| % positiveb | 56 | 39 | |||||
| E. coli O157:H7 (stx negative) | No. of GCs before enrichment | 3.73 × 102 | 7.70 × 103 | 1.43 × 102 | 2.38 × 102 | 2.98 × 103 | 1.82 × 102 |
| No. of GCs after enrichment | 3.83 × 103 | 1.11 × 104 | 3.98 × 102 | 6.51 × 103 | 1.09 × 104 | 3.17 × 103 | |
| GC increasea | 2.40 × 103 | 1.09 × 104 | 2.89 × 102 | 6.03 × 103 | 1.05 × 103 | 2.97 × 103 | |
| % positiveb | 56 | 61 | |||||
| S. sonnei 866 | No. of GCs before enrichment | 3.72 × 102 | 3.44 × 103 | 1.03 × 102 | 2.58 × 102 | 7.53 × 102 | 2.00 × 102 |
| No. of GCs after enrichment | 9.32 × 103 | 1.48 × 107 | 3.37 × 102 | 4.58 × 103 | 9.79 × 103 | 2.71 × 103 | |
| GC increasea | 6.93 × 103a | 1.48 × 107 | 1.97 × 102 | 3.73 × 103 | 9.74 × 103 | 1.00 × 103 | |
| % positiveb | 52 | 65 | |||||
Shows the geometrical mean of the increase observed in those samples that have shown phage propagation.
Percentage of samples in which a significant (P < 0.05) increase in the number of Stx phages was observed after propagation in the host strain.
In minced beef samples, 52 to 56% showed an increase in the densities of Stx phages after their propagation with the host strains. In salad samples, 39 to 65% showed a significant (P < 0.05) increase in the number of Stx phages after propagation (Table 3). The increase indicates that, in many samples, the Stx phages were infective, although the level of propagation varied depending on the samples and the host strain. Furthermore, the presence of infectious Stx phages in 16 meat samples and 12 salad samples was enumerated by the double-agar-layer method using the three host strains. The presence of Stx phages was tested by plaque blot hybridization with the stx2 probe. We detected only positive signals when using S. sonnei as the host strain but not when using the other two hosts. The Stx-positive plaques were excised from the agar's top layer and suspended in 50 μl of double-distilled water, and the phage DNA was extracted (20). The presence of the stx2 gene in the phage DNA isolated from the plaques was further confirmed by PCR.
All together, we visualized and confirmed 40 positive Stx plaques of lysis. Of these, 8 plaques were isolated from 7 different salad samples, and 32 plaques were isolated from 21 different meat samples. Stx plaques were also observed in 20% of the supernatants of the corresponding enrichment cultures. We attempted to further propagate these phages, again using S. sonnei in a new culture, and to evaluate the propagation by qPCR. Although some increase in the number of GCs was detected after the culture enrichment, the difference in this case was not significant (P > 0.05), and we cannot confirm that the phages from the positive plaques of lysis were more infective. Another possibility would be that phages, instead of replicating in the host strains causing lysis, remained integrated in the bacterial genome and generated lysogens. However, after several attempts, no lysogens were isolated from the lytic spot area on the agar plate.
To our knowledge, there is no information about the burst size of Stx phages, which limits our ability to predict the increase in a replication cycle. The lack of replication of the Stx phages isolated from the plaques of lysis and the difficulty in visualizing the plaques hampered the burst-size studies. Some samples increased by only 102 stx GCs. We conclude that in these samples, only a few infectious particles were present, which induced few replication cycles. Otherwise, we would expect higher stx GC values. This might be because the Stx phage progeny generated after the first replication cycle was not further infective or because only a few particles from the progeny were infective. Although there are no experimental data to support it, a plausible hypothesis would be abortive infection in the host strain (10). In abortive infection, the host cell interrupts phage development, resulting in the release of few or no progeny particles and the death of the infected cells. This lack of replication was also observed in the infectious Stx phages detected by plaque blotting and isolated from the plaques of lysis. In other samples, however, more Stx phages were detected after propagation (up to 107 GCs·25 g−1). In these samples, infectious particles might have been originally present, or alternatively, the phage progeny generated might not have been the result of an abortive infection and the new phages might have been able to propagate effectively in the next replication cycles. Additionally, differences between the Stx phages detected must be considered, given that the term Stx phages refers to a wide, heterogeneous family (18, 26) comprising different kinds of phages with the sole common trait of carrying the stx gene.
The ratio of the number of infectious Stx phages to that of Stx phages detected by qPCR was estimated in previous studies (16) to lie between 1/10 and 1/1,000, depending on the Stx phage and the host strain used. If we apply this estimation to food samples, with densities of Stx phages exceeding 102 GCs/ml (Fig. 1), some of these phages can be expected to be infectious.
A more complete morphological and genetic characterization of Stx phages was impossible, since attempts to propagate them were largely unsuccessful. This difficulty in isolating and propagating Stx phages has been reported previously (8, 20), and it supports the use of culture-independent methods for their detection (15, 17, 23). In addition, no stx lysogens were detected when we tested all the supernatants of the enrichment cultures using methods previously described (18, 25). This was not surprising, since the generation of lysogens occurs with a low frequency in many cases and the number of free Stx phages in the samples did not guarantee the generation of lysogens (20, 25).
The origin of the Stx phages found in food is unknown. However, high densities of Stx phages have been found in human and animal environments (15, 19), pointing to an environmental origin. Many treatments and protocols applied in food treatment aim to eliminate pathogenic bacteria effectively, but they fail to inactivate Stx phages, if present, or the possible effect on induction and release of prophages from their lysogenic strains. The significance of this study is that infectious free Stx phages are present in commercial food samples, and to our knowledge, this feature has not been considered previously. The presence of Stx phages in food can cause stx transduction to the bacterial flora present in the matrices, as we previously demonstrated (14), generating new STEC strains in the samples, perhaps during storage. In addition, ingestion of free Stx phages present in food can also lead to conversion of commensal gut bacteria, since stx transduction has previously been reported in vivo (6, 27) and because stx transduction in vivo seems to be more effective than that in vitro (27). As far as we know, there are no reports indicating that stx transduction in food could be a relevant concern, probably due to the low frequency of phage and bacteria encounters in a given environment. However, it is possible under appropriate conditions and should not be ruled out.
Acknowledgments
This study was supported by the Generalitat de Catalunya (grant 2009SGR1043), by the Spanish Ministry of Education and Science (grant AGL2009-07576), and by the Xarxa de Referència en Biotecnologia (XRB). L. Imamovic is the recipient of a grant from the Spanish Ministry of Education and Science (grant FPI 20060054361).
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Anonymous. 2000. ISO 10705-2: water quality. Detection and enumeration of bacteriophages–part 2: enumeration of somatic coliphages. International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 2. Anonymous. 2005. Commission regulation (EC) no. 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Off. J. Eur. Communities 338:1–25 [Google Scholar]
- 3. Bastien P., Procop G. W., Reischl U. 2008. Quantitative real-time PCR is not more sensitive than “conventional” PCR. J. Clin. Microbiol. 46:1897–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brabban A. D., Hite E., Callaway T. R. 2005. Evolution of foodborne pathogens via temperate bacteriophage-mediated gene transfer. Foodborne Pathog. Dis. 2:287–303 [DOI] [PubMed] [Google Scholar]
- 5. Caprioli A., Morabito S., Brugère H., Oswald E. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289–311 [DOI] [PubMed] [Google Scholar]
- 6. Cornick N. A., Helgerson A. F., Mai V., Ritchie J. M., Acheson D. W. 2006. In vivo transduction of an Stx-encoding phage in ruminants. Appl. Environ. Microbiol. 72:5086–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Sablet T., et al. 2008. Differential expression of stx2 variants in Shiga toxin-producing Escherichia coli belonging to seropathotypes A and C. Microbiology 154:176–186 [DOI] [PubMed] [Google Scholar]
- 8. Döpfer D., et al. 2010. Pathogenic potential and horizontal gene transfer in ovine gastrointestinal Escherichia coli. J. Appl. Microbiol. 108:1552–1562 [DOI] [PubMed] [Google Scholar]
- 9. Dumke R., Schroter-Bobsin U., Jacobs E., Roske I. 2006. Detection of phages carrying the Shiga toxin 1 and 2 genes in waste water and river water samples. Lett. Appl. Microbiol. 42:48–53 [DOI] [PubMed] [Google Scholar]
- 10. Emond E., Molineau S. 2007. Bacteriophages and food fermentations. In McGrathe S., Van Sinderen D. (ed.), Bacteriophages, genetics and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 11. García-Aljaro C., Muniesa M., Jofre J., Blanch A. R. 2004. Prevalence of the stx2 gene in coliform populations from aquatic environments. Appl. Environ. Microbiol. 70:3535–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herold S., Karch H., Schmidt H. 2004. Shiga toxin-encoding bacteriophages-genomes in motion. Int. J. Med. Microbiol. 294:115–121 [DOI] [PubMed] [Google Scholar]
- 13. Hussein H. S., Bollinger L. M. 2005. Prevalence of Shiga toxin-producing Escherichia coli in beef cattle. J. Food Prot. 68:2224–2241 [DOI] [PubMed] [Google Scholar]
- 14. Imamovic L., Jofre J., Schmidt H., Serra-Moreno R., Muniesa M. 2009. Phage-mediated Shiga toxin 2 gene transfer in food and water. Appl. Environ. Microbiol. 75:1764–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imamovic L., Ballesté E., Jofre J., Muniesa M. 2010. Quantification of Shiga toxin-converting bacteriophages in wastewater and in fecal samples by real-time quantitative PCR. Appl. Environ. Microbiol. 76:5693–5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imamovic, Serra-Moreno L. R., Jofre J., Muniesa M. 2010. Quantification of Shiga toxin 2-encoding bacteriophages, by real-time PCR and correlation with phage infectivity. J. Appl. Microbiol. 108:1105–1114 [DOI] [PubMed] [Google Scholar]
- 17. McDonald J. E., Smith D. L., Fogg P. C., McCarthy A. J., Allison H. E. 2010. High-throughput method for rapid induction of prophages from lysogens and its application in the study of Shiga toxin-encoding Escherichia coli strains. Appl. Environ. Microbiol. 76:2360–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muniesa M., et al. 2004. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 150:2959–2971 [DOI] [PubMed] [Google Scholar]
- 19. Muniesa M., Jofre J. 1998. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl. Environ. Microbiol. 64:2443–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muniesa M., Serra-Moreno R., Jofre J. 2004. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx genes appeared conserved. Environ. Microbiol. 6:716–725 [DOI] [PubMed] [Google Scholar]
- 21. O'Brien A. D., et al. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694–696 [DOI] [PubMed] [Google Scholar]
- 22. Paton J. C., Paton A. W. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rooks D. J., et al. 2010. Development and validation of a qPCR-based method for quantifying Shiga toxin-encoding and other lambdoid bacteriophages. Environ. Microbiol. 12:1194–1204 [DOI] [PubMed] [Google Scholar]
- 24. Schmidt H., Bielaszewska M., Karch H. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage Φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Serra-Moreno R., Jofre J., Muniesa M. 2007. Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J. Bacteriol. 189:6645–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith D. L., et al. 2007. Multilocus characterization scheme for Shiga toxin-encoding bacteriophages. Appl. Environ. Microbiol. 73:8032–8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toth I., et al. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69:7242–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. U.S. Department of Agriculture (USDA) 2009. Technical requirements schedule for fresh-chilled boneless beef for further processing. Livestock and seed program. Agricultural Marketing Service, Washington, DC [Google Scholar]
- 29. Wagner P. L., Acheson D. W., Waldor M. K. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]