Abstract
In spite of technical advances that have provided increases in orders of magnitude in sequencing coverage, microbial ecologists still grapple with how to interpret the genetic diversity represented by the 16S rRNA gene. Two widely used approaches put sequences into bins based on either their similarity to reference sequences (i.e., phylotyping) or their similarity to other sequences in the community (i.e., operational taxonomic units [OTUs]). In the present study, we investigate three issues related to the interpretation and implementation of OTU-based methods. First, we confirm the conventional wisdom that it is impossible to create an accurate distance-based threshold for defining taxonomic levels and instead advocate for a consensus-based method of classifying OTUs. Second, using a taxonomic-independent approach, we show that the average neighbor clustering algorithm produces more robust OTUs than other hierarchical and heuristic clustering algorithms. Third, we demonstrate several steps to reduce the computational burden of forming OTUs without sacrificing the robustness of the OTU assignment. Finally, by blending these solutions, we propose a new heuristic that has a minimal effect on the robustness of OTUs and significantly reduces the necessary time and memory requirements. The ability to quickly and accurately assign sequences to OTUs and then obtain taxonomic information for those OTUs will greatly improve OTU-based analyses and overcome many of the challenges encountered with phylotype-based methods.
INTRODUCTION
The application of ecological theory developed for macroscopic organisms to microorganisms is challenged by difficulties in defining the appropriate levels of spatial, temporal, and taxonomic scales. Ascertaining an appropriate taxonomic scale is particularly troubling because of the inability to systematically define various taxonomic levels across the Bacteria when relatively few bacterial taxa have ever been cultured. Often, taxonomic outlines reflect biases within the field and battles between taxonomic “lumpers” and “splitters” (3, 25). Considering the now widespread use of next-generation sequencing technology that allows investigators to interrogate bacterial populations previously inaccessible due to their rarity, the challenge of placing 16S rRNA gene sequences from uncultured bacteria into a bacterial taxonomy is even more acute. Two general approaches have been widely pursued for binning sequences into microbial populations. The first method relies upon reference taxonomic outlines to classify sequences to taxonomic bins (i.e., phylotypes) (10, 16, 24). The second method allows the data to “speak for themselves” by assigning sequences to operational taxonomic units (OTUs) based on the similarity of sequences within a data set to each other (20, 21, 23).
Many microbiologists prefer phylotype-based methods because they enable an investigator to place a label onto a sequence indicating its relationship to previously cultured and characterized microbes. Although an appealing approach, there are myriad examples of organisms that belong to the same species that have different phenotypes and organisms with the same phenotype belonging to different taxonomic lineages. For example, the assignment of a 16S rRNA gene sequence to the genus Pseudomonas could indicate the presence of either a beneficial or pathogenic bacterium in a sample. Furthermore, because most taxonomy outlines are based on what is known of already cultured organisms, members of candidate phyla (e.g., TM7) or difficult-to-culture phyla (e.g., Acidobacteria) lack a well-defined taxonomy that extends to the genus or species level. Finally, building upon the inability to develop a coherent definition for a bacterial species, it is impossible to consistently define bacterial genera, families, orders, classes, or phyla. The result being that there are at least three widely used curated taxonomy outlines that contain significant conflicts with each other (4). Despite these limitations, phylotype-based methods are computationally efficient, lend themselves well to parallelization, and provide a stable classification. Numerous studies have shown that diverse approaches to classification are robust (10, 16, 24). Although one may debate the merits of what the classification means, there is little debate over the quality of the classifications.
OTU-based methods overcome a number of limitations associated with phylotypes. Namely, because a taxonomy outline is not used, one is not restricted to the bins described by the outline. Thus, it becomes possible to assign all sequences to bins on the same basis, regardless of whether the sequence is represented by references within a taxonomy outline or whether there is conflict in how different outlines classify the most similar reference sequences. Because the methods used are cluster based and not classification based, whether two sequences are found in the same OTU depends on the other sequences in the data set. OTU-based methods also assume that bacterial 16S rRNA genes evolve at the same rate regardless of their taxonomic affiliation, whereas taxonomists would debate differential rates of evolution to split or lump a taxonomy, OTU-based methods remain agnostic and do not take such considerations into account (14). This can be seen either as a strength or a weakness, depending on one's perspective. Interpretation of OTUs is complicated by the lack of a consistent method for converting between the thresholds used to define OTUs and taxonomic levels. For example, the operational definition of a species, 3% dissimilarity, is often cited but controversial (6, 8, 11, 22). Perhaps the most significant limitation to using OTU-based methods is that the clustering algorithms are computationally intensive, relatively slow, and can require significant amounts of memory (20, 21, 23).
A more general problem faced by OTU-based methods is the choice of what method to use to cluster sequences into OTUs. The nearest (i.e., single-linkage), furthest (i.e., complete linkage), weighted neighbor, and average neighbor (i.e., unweighted-pair group method using average linkages [UPGMA]) hierarchical clustering algorithms are commonly used in various disciplines to assign individuals to bins (14). Within the field of microbial ecology, the furthest neighbor algortihm was originally suggested because it gave the most conservative estimate of how much additional sampling was required to complete a census of a community (20). Others have recently noticed that the furthest neighbor algorithm is sensitive to sequencing artifacts and suggested using the average neighbor algortihm based on empirical observations from sequencing a collection of reference 16S rRNA gene fragments (9). Improvements have been made to these algorithms that focus on reducing the memory and processing time requirements. Most notable among these is the use of sparse matrices that only represent the unique sequences in a data set as input (21) and an online algorithm that requires a small memory footprint (23). Due to the computationally intensive nature of these approaches, others have developed and employed heuristics to assign 16S rRNA gene sequences to OTUs (5, 15, 23). Unfortunately, none of these clustering methods have been vetted by assessing the quality of the sequence assignments using 16S rRNA gene sequences, and so researchers select an approach based on speed, ease of use, and personal experience.
This study explores and proposes solutions to current challenges experienced in applying OTU-based methods. First, we explore whether it is feasible to derive distance-based cutoffs that would permit one to translate between OTU- and phylotype-based analyses. Supporting the conventional wisdom, we assert that it is not possible to define distance-based delineations for different taxonomic levels and instead propose a method for labeling an OTU with a taxonomic label. Second, faced with the challenge of determining the most robust method of assigning sequences to OTUs, we apply a taxonomy-independent metric to demonstrate that the average neighbor clustering algorithm outperforms other deterministic and heuristic approaches. Third, after implementing numerous algorithmic modifications to improve the speed and memory requirements of these clustering algorithms without sacrificing accuracy, we describe a novel heuristic that overcomes these issues with minimal effect on clustering accuracy. Throughout this study, we sought to blend the independent and interacting contributions of OTU- and phylotype-based methods to improve the analysis of 16S rRNA gene sequences.
MATERIALS AND METHODS
Data set.
We analyzed a collection of 14,956 unique, full-length, high-quality, well-aligned 16S rRNA gene sequences (18). To analyze regions that are tractable using the popular 454 FLX Titanium sequencing technology, we extracted the V13 and V35 regions from the full-length sequences based on their alignment coordinates. For V13 sequences, Escherichia coli positions 28 through 514 were considered, and for the V35 sequences, positions 357 and 906 were considered. These positions were based on the sites where commonly used PCR primers anneal (13). Full-length sequences spanned E. coli positions 28 through 1491. There were 13,217 unique V13 sequences and 12,387 unique V35 sequences. Ribosomal Database Project (RDP) classifications were determined by classifying sequences with the Bayesian classifier.
Bayesian classifier.
We implemented the naïve Bayesian classifier proposed by Wang and colleagues (24). Whereas the original implementation was written in the Java programming language, our version was written in C++. Our implementation allows users to classify their sequences by using any reference database and taxonomy. Furthermore, the version available within mothur can utilize multiple processors for parallel processing. Classification of test sequences by using the RDP training set yielded similar results to those provided by using the original Java version. We used the RDP-supplied training set, which was released on 20 March 2010 (http://sourceforge.net/projects/rdp-classifier/). The RDP classification scheme provides a traditional Linnaean hierarchy that is more easily standardized than the greengenes (4)- or SILVA (17)-based taxonomies; therefore, we decided to use the RDP-based outline for the remainder of our analysis. The RDP training set contains 8,127 bacterial sequences distributed among 35 phyla, 72 classes, 107 orders, 288 families, and 1,585 genera. Following the suggestions described by the RDP (http://rdp.cme.msu.edu), we used the last taxonomic level for a sequence that had a pseudo-bootstrap value of at least 80%. We used 1,000 pseudo-bootstrap replications, which would result in a standard error of 1.3% for pseudo-bootstrap values of 80.0%.
Hierarchical clustering.
We tested several permutations of the traditional hierarchical clustering approach, which used pairwise distance matrices as input. Distance matrices were calculated by assuming that consecutive insertions or deletions represented one mutation event (18). We calculated OTUs for distance thresholds ranging between 0.00 and 0.10 with increments of 0.01; all distance thresholds represented a hard cutoff with no rounding. We evaluated four hierarchical clustering algorithms (14). The furthest neighbor algorithm (i.e., complete linkage) requires that the distance between every sequence within an OTU be within the specified threshold. The nearest neighbor algorithm (i.e., single linkage) requires that all sequences within the specified threshold of any other sequence belong to the same OTU. The weighted neighbor algorithm (i.e., weighted arithmetic average clustering) gives equal weights to the distances between OTUs when they are joined to form a new OTU. In contrast, the average neighbor algorithm (i.e., unweighted arithmetic average clustering) weighs the OTUs to be joined by the number of sequences within each OTU. For each algorithm, ties between equal distances were broken by randomly selecting a distance to use for the next clustering step.
The four clustering algorithms were implemented as part of four general clustering approaches. First, we used a traditional approach that processes a PHYLIP-formatted distance matrix using an approach previously used in DOTUR (20). Second, we used an approach that makes use of a sparse matrix format that has previously been described (21). Third, we implemented an “on-the-fly” approach that is used in ESPRIT (23). We expanded this approach from the original method described for the furthest neighbor algorithm to utilize all four clustering algorithms. Finally, we developed a method that splits a distance matrix into nonoverlapping submatrices, which could then be processed in series or parallel. As none of these methods employ heuristics, we confirmed that the same clustering algorithm (e.g., average neighbor) gave the same output regardless of the approach (i.e., traditional, sparse, on-the-fly, or matrix split).
Heuristics.
We implemented four nonhierarchical clustering algorithms that utilize heuristics and have previously been used to assign 16S rRNA gene sequences to OTUs. First, we assigned sequences to OTUs using CD-HIT-EST version 4.3 with the default settings (15). Second, we assigned sequences to OTUs using BlastClust version 2.2.16 and the default settings, with the exception that we forced the program to utilize 8 processors (http://www.ncbi.nlm.nih.gov/IEB/ToolBox/C_DOC/lxr/source/doc/blast/blastclust.html). Third, we assigned sequences to OTUs using the 32-bit release of UClust 3.0.617 (5). We evaluated UClust's default settings as well as the predefined exact and optimal settings and found that the exact and optimal settings generated similar OTUs that were more robust than the default settings. Because the optimal setting was the fastest, we utilized that setting for this study. Finally, we assigned sequences to OTUs using the LINUX version of ESPRIT that was updated on 20 July 2009 (23). ESPRIT's default settings were used, with the exception that we used a kmer distance threshold of 0.40 and did not perform the ESPRIT sequence preprocessing steps. In addition to the modifications we made to the default parameters for each of these programs, we adjusted the clustering threshold to generate OTU assignments for distance thresholds between 0.00 and 0.10 incremented by 0.01.
Assessment of clustering quality.
To overcome the challenge of assessing clustering quality by using an objective standard, we implemented methods used in machine learning control theory. For a set of sequences assigned to OTUs at a specific distance threshold clustered by one of the clustering algorithms, we counted the number of sequence pairs that could be considered true positives (TPs), true negatives (TNs), false positives (FPs), and false negatives (FNs). A pair of sequences was considered a true positive (TP) if the distance between the sequences was smaller than the distance threshold and they belonged to the same OTU; a false positive (FP) was a pair of sequences that belonged to the same OTU but had a pairwise distance larger than the threshold. A pair of sequences was considered as a true negative (TN) if their pairwise distance was larger than the threshold and they did not belong to the same OTU; a false negative (FN) was a pair of sequences that belonged to different OTUs, but had a pairwise distance smaller than the threshold. There are numerous methods to weigh these four values. To evenly balance the terms, we utilized the Matthew's correlation coefficient (MCC):
The MCC coefficient can vary between −1 and +1 and represents the correlation between the observed and predicted values of the clustering scheme (2).
Computation.
All analyses were performed using mothur v.1.14 and custom Perl scripts on a cluster of compute nodes, where each node contained dual quad core 2.26-GHz Intel Nehalem central processing units (CPUs) with access to 48 GB of random-access memory (RAM). The reported times for analyses represent the minimum “wall time” for three executions of the same setting and should only be interpreted for assessment of relative performance.
RESULTS
What are appropriate distance thresholds for an OTU-based analysis?
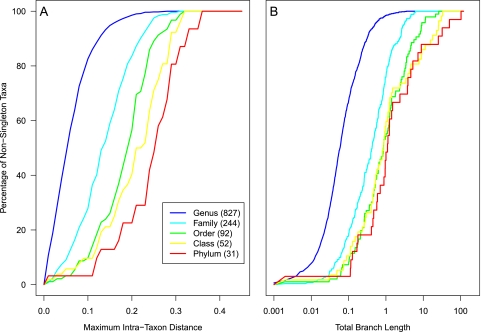
We found that the genetic distance between the most disparate full-length 16S rRNA gene sequences within a named taxonomic group represented a continuum for each level in the hierarchy (Fig. 1). Furthermore, the distances within a taxonomic group are not evenly distributed within the group. Genera such as Bacillus (n = 360 sequences, maximum distance [max] = 0.14, mean distance = 0.07, standard deviation of distances [SD]= 0.02), Bacteroides (n = 50, max = 0.17, mean = 0.09, SD = 0.03), Clostridium (n = 99, max = 0.15, mean = 0.08, SD = 0.02), and Pseudomonas (n = 514, max = 0.10, mean = 0.04, SD = 0.02) were very broad. In contrast, genera such as Bradyrhizobium (n = 76, max = 0.06, mean = 0.02, SD = 0.01), Cetobacterium (n = 86, max = 0.02, mean = 0.01, SD = 0.003), Pseudoalteromonas (n = 101, max = 0.07, mean = 0.02, SD = 0.01), and Staphylococcus (n = 43, max = 0.05, mean = 0.03, SD = 0.01) were much tighter. The 663 genera in which the maximum intragenus distance was greater than 0.094 represented more than 50% of the sequences. Figure 1A also indicates that there is considerable overlap in the maximum intrataxon distances between taxonomic levels. For example, there are groups at every level in the taxonomic outline where the maximum intragroup distance is less than 0.15. Similarly, we observed that the variation in phylogenetic diversity represented at each taxonomic level also represented a continuum (Fig. 1B). When applied to the V13 and V35 regions, we observed the same general trends that were observed for the full-length sequences (see Fig. S1 and S2 in the supplemental material). As would be expected based on earlier work (18), the intrataxon distances obtained with V13 sequences were generally larger and the intrataxon distances obtained with the V35 sequences were generally smaller than those calculated with the full-length sequences (see Fig. S1 and S2 in the supplemental material). Based on these results, it was clearly impossible to select single thresholds to operationally define a grouping and give it a position in a Linnaean taxonomy. Alternatively stated, there is no consistent relationship between the phenotypically derived bacterial taxonomy and genetic diversity for full-length 16S rRNA gene sequences.
Fig. 1.
Cumulative fraction of taxa that had a specified maximum intrataxon distance (A) and total branch length (B) for each taxonomic level when full-length 16S rRNA gene sequences were analyzed. At each taxonomic level, sequences that did not affiliate with a known lineage (i.e., incertae sedis) were excluded. The numbers in parentheses next to the name of each taxonomic level indicate the number of taxa within that level that we observed. (See Fig. S1 and S2 in the supplemental material for the same analysis using the V13 and V35 sequences, respectively.)
A method for assigning a taxonomic label to an OTU.
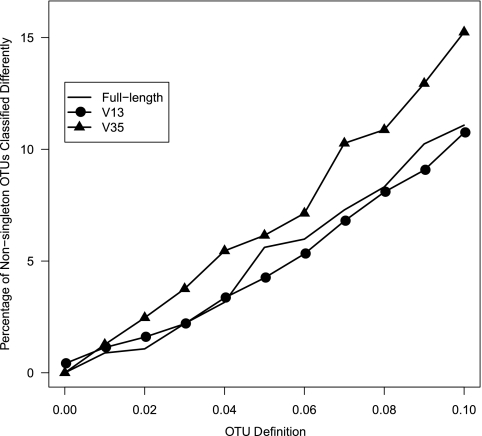
Because of the difficulty in relating distance-based thresholds to taxonomic levels, we tested two methods for classifying OTUs. One algorithm for applying a taxonomic label to an OTU involves identifying a sequence within an OTU that has the smallest distance from all of the other sequences in the OTU and then classifying that sequence. Such a sequence is called a representative sequence for the OTU. Yet, if an OTU represents sequences from multiple taxa, classification of a representative sequence could result in a false taxonomic labeling of the OTU. An alternative algorithm, which we propose here, is to classify every sequence in an OTU and then to identify the majority consensus taxonomy of the sequences within the OTU. This consensus-based algorithm can be modified by increasing the level of consensus required to assign a taxonomic label to an OTU. To evaluate the merits of each approach, we applied the two algorithms to OTUs identified for the full-length, V13, and V35 sequences using the average neighbor algorithm. We found that as the OTU cutoff increases, the fraction of differently classified OTUs increased (Fig. 2). Had a stricter consensus definition been applied, the discrepancy between the two algorithms would have been greater.
Fig. 2.
Fraction of OTUs calculated for a 0.03-cutoff level that were represented by more than one sequence and had different classifications when we classified the OTU using a representative sequence from the OTU or by determining the majority consensus taxonomy for the full-length, V13, and V35 16S rRNA gene sequence data sets.
As described above, each taxonomic level contains considerable range in the maximum intrataxonomic group genetic diversity (Fig. 1). For example, the maximum intragenus genetic distance for most genera is less than 0.15. This would suggest that using OTU thresholds below 0.15 would yield multiple OTUs that could be assigned to the same genus. We found that for full-length sequences, there were 4.4 OTUs per genus at the 0.03 distance threshold, 3.5 at the 0.05 threshold, and 2.7 at the 0.10 threshold. Similarly for V13 sequences, there were averages of 5.1, 4.1, and 3.1 OTUs per genus at the 0.03, 0.05, and 0.10 thresholds. Averages of 4.0, 3.2, and 2.7 OTUs per genus were observed at the same thresholds for V35 sequences. Although these do not represent species, strictly speaking, OTUs defined at these thresholds do provide a more refined definition of subgenus populations.
Because there were no clear thresholds to define taxonomic levels, we also expected some OTUs to represent sequences from multiple lineages. When assigning full-length sequences to OTUs using the average neighbor algorithm, there were 3,566 OTUs defined at the 0.03 threshold with only one sequence (i.e., singletons) and 1,906 nonsingleton OTUs. Among the nonsingleton OTUs, there were 140 OTUs whose sequences' taxonomies did not all agree. Within this set of discordant OTUs, 24 OTUs had sequences that classified to the same taxonomic depth, 98 had sequences that varied in their taxonomic depth by one level, 12 had sequences that varied by two levels, 4 had sequences that varied by three levels, and one each had sequences that varied by four or five levels. Similar percentages of OTUs with these levels of variation in taxonomic depth were also observed for the V13 and V35 sequences. The high concordance of taxonomies represented within OTUs confirms the assertion that OTUs can provide a more refined analysis than is possible by the phylotype-based approach.
What is the best method to assign sequences to OTUs?
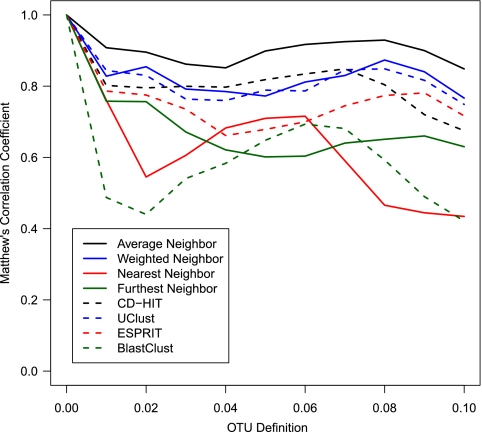
One significant challenge in assigning sequences to OTUs is identifying an algorithm that balances the inclusion of sequences into an OTU that are within a specified genetic distance while excluding those that are greater than that distance. To assess the quality of the clusters, we utilized the Matthew's correlation coefficient. Among the four classic hierarchical clustering algorithms we tested, the average neighbor algorithm (i.e., UPGMA) was considerably better than the weighted, nearest, and furthest neighbor algorithms, regardless of the region and distance threshold that we tested (Fig. 3; see Fig. S3 and S4 in the supplemental material). The nearest and furthest neighbor algorithms produced OTUs that yielded similar MCC values; however, their clustering was worse than was observed for the weighted neighbor OTUs. The average neighbor algorithm also performed better than four published heuristics (i.e., CD-HIT, UClust, ESPRIT, and BlastClust) that have been used to cluster 16S rRNA gene sequences into OTUs. In general, the MCC values for CD-HIT and UClust were comparable to each other, followed by ESPRIT and BlastClust. The outputs from CD-HIT and UClust were comparable to that observed with the weighted neighbor algorithm, while ESPRIT and BlastClust were comparable to that observed with the furthest and nearest neighbor algorithms. Next, we used the average neighbor algorithm to assign V13 and V35 sequences to OTUs and calculated the MCC values based on the pairwise distances calculated with the full-length sequences. For the V13 sequences, the MCC values were 0.67, 0.70, and 0.70 for distance thresholds of 0.03, 0.05, and 0.10. For the V35 sequences, the MCC values were 0.65, 0.81, and 0.71. These results are aligned with a previous analysis that showed a generally poor correlation between the pairwise distance calculated by using full-length and V13 and V35 sequences (18).
Fig. 3.
Variation in the Matthew's correlation coefficient calculated for OTUs identified by using eight classification algorithms at genetic distances varying between 0.00 and 0.10 for full-length 16S rRNA gene sequences. (See Fig. S3 and S4 in the supplemental material for the same analysis using the V13 and V35 sequences, respectively.)
Improving OTU assignment algorithms without heuristics.
We utilized several observations to accelerate the speed of the clustering process and to reduce the RAM requirements for storing the distance matrix without changing the clustering observed when using the traditional algorithm. First, we noted that as sequencing coverage increases, so does the probability that duplicate sequences will be observed. Therefore, it is possible to only assign unique sequences to OTUs and then map onto them the identity of the duplicate sequences. Although we initially screened our full-length sequences to only include unique sequences, when we analyzed the V13 and V35 sequences we observed a considerable speedup (Table 1). Second, we noted that researchers are typically only interested in analyzing OTUs clustered below a specified distance threshold. Considering distances above the threshold are not necessary to calculate OTUs, the distance matrix can be represented in a more efficient “sparse” format, effectively reducing the computational complexity and memory requirements. Again, for all regions, this observation resulted in an accelerated speedup in the time required to assign sequences to OTUs (Table 1). Third, we recognized that it is possible to split a sparse distance matrix into sections that do not overlap with each other. Each section of a sparse matrix could be clustered into OTUs separately and the results combined. The advantage of this approach is that the clustering could be done in parallel or series with the benefits of reducing the effective number of sequences (N) and speeding up the clustering rate, minimizing memory usage, and potentially using the multiple processors found on many computers to further accelerate the OTU assignment step. Splitting a distance matrix yielded mixed results for speed and showed little difference when using 1 or 8 processors. This was because of the considerable time required to split the matrix and merge the outputs of the separately clustered OTUs and because the collection of submatrices still contained large matrices that required lengthy processing times. Finally, we borrowed an observation from Sun et al. (23) that sequences could be assigned to OTUs without reading in the entire sparse distance matrix at once by using an “on-the-fly” clustering algorithm. The resulting times were comparable for the furthest and nearest neighbor algorithms and required a negligible amount of RAM; however, the time required for the weighted and average neighbor algorithms was excessive and although storage of the actual distance matrix was not necessary, it was necessary to store an expansive mapping matrix that required large amounts of RAM for storage (Table 1). As the degree of connectedness among sequences varies between data sets, the modifications outlined above to the classical algorithm may vary in their performance.
Table 1.
Comparison of times required to cluster sequences into OTUs for distance cutoffs ranging between 0.00 and 0.10 for various clustering algorithms and input data formats when applied to full-length, V13, and V35 16S rRNA gene sequencesa
| Algorithm | Approachb | Wall time (min) for sequence: |
||
|---|---|---|---|---|
| Full length | V13 | V35 | ||
| Average neighbor | Traditional | 61.63 | 59.22 | 65.77 |
| Unique | 61.63 | 42.68 | 38.17 | |
| Sparse | 27.25 | 8.12 | 30.58 | |
| Split-8 | 24.82 | 11.43 | 30.90 | |
| On-the-fly | 6,085.97 | 2,848.80 | 6,035.52 | |
| Weighted neighbor | Traditional | 63.87 | 59.63 | 63.67 |
| Unique | 63.87 | 43.17 | 38.28 | |
| Sparse | 20.30 | 7.75 | 24.28 | |
| Split-8 | 24.70 | 11.50 | 28.73 | |
| On-the-fly | 7,597.98 | 3,396.17 | 7,852.87 | |
| Furthest neighbor | Traditional | 61.27 | 56.50 | 62.85 |
| Unique | 61.27 | 43.23 | 39.00 | |
| Sparse | 0.53 | 0.15 | 0.25 | |
| Split-8 | 2.80 | 1.32 | 1.92 | |
| Online | 3.28 | 1.33 | 2.57 | |
| Nearest neighbor | Traditional | 65.30 | 61.90 | 66.72 |
| Unique | 65.30 | 45.38 | 39.83 | |
| Sparse | 0.53 | 0.15 | 0.25 | |
| Split-8 | 2.80 | 1.35 | 1.92 | |
| On-the-fly | 3.25 | 1.28 | 2.50 | |
| CD-HIT | UniqSeq | 88.13 | 15.90 | 10.00 |
| UClust | UniqSeq | 11.85 | 2.98 | 2.63 |
| ESPRIT | UniqSeq | 6,361.85 | 228.45 | 390.70 |
| BlastClust | UniqSeq | 919.52 | 165.67 | 187.47 |
| Phylotype | UniqSeq | 46.38 | 10.38 | 12.08 |
Although the V13 and V35 16S rRNA gene sequences are comparable in length, the V35 16S rRNA gene sequences took longer to cluster because there were more pairwise distances among sequences in that region that were smaller than 0.10 than were found in the other data sets. All times represent the “wall time” in minutes required for each analysis using the computer system described in Materials and Methods.
The “traditional” approach represented all 14,956 sequences according to a PHYLIP-formatted lower-triangular distance matrix. The “unique” approach only used the sequences that were identical to each other over their full length according to a PHYLIP-formatted lower-triangular-distance matrix. The “sparse” approach only used the sequences that were not identical to each other over their full length according to a sparse matrix format. The “split-8” approach split the sparse data format into mutually exclusive submatrices and clustered the submatricies in parallel by using 8 processors. The “on-the-fly” data format used the sparse data format but processed the distance matrix without reading the entire matrix into memory. The “UniqSeq” approach represented the data by only using unique, unaligned, FASTA-formatted sequences.
Leveraging observations to create a better heuristic.
Above, we demonstrated that heuristic algorithms do not generate OTUs that are as well formed as the average neighbor algorithm (Fig. 3; see Fig. S3 and S4 in the supplemental material); however, the average neighbor algorithm can be considerably slower and perhaps require computational resources that are beyond the means of many investigators (Table 1). Based on earlier results in this study, we made two important observations that could enable us to create an accurate and fast clustering heuristic: (i) lineages within a taxonomic level (e.g., genus) are more different from each other than the thresholds commonly used for OTU-based analyses (i.e., less than 0.10), and (ii) a distance matrix can be split and the submatrices processed in series or in parallel. Our simple heuristic involves either splitting a collection of aligned sequences or a distance matrix based on the taxonomic assignment of each sequence, clustering the sequences, and then merging the OTU assignments. For example, the 14,956 full-length sequences could be assigned to 33 phyla plus one pool of sequences that could not be assigned to a phylum. Then the sequence affiliated within each phylum could be used to calculate pairwise distances to each other and clustered into OTUs. Finally, the resulting OTU lists from each phylum would be merged.
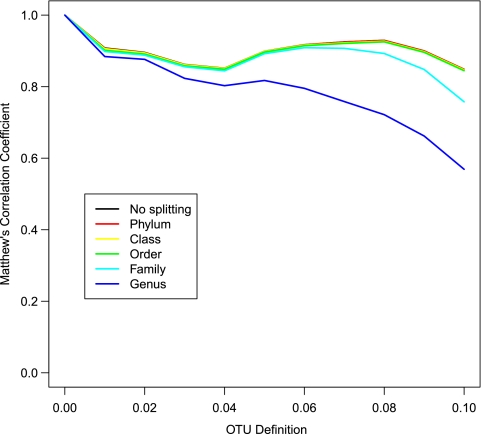
We evaluated this new heuristic by splitting either the aligned sequences or the sparse distance matrix at the five taxonomic levels between phylum and genus. The MCC values were the same regardless of whether we split the sequences or the distance matrix. The MCC values were indistinguishable from the classical average neighbor algorithm when splitting down to the level of order and clustering to the 0.10 threshold (Fig. 4; see Fig. S5 and S6 in the supplemental material). Using 8 processors and splitting aligned sequences, we were able to obtain OTUs that were as robust as the average neighbor using the full-length, V13, and V35 sequences, respectively (Table 2). Even if only one processor was used, the new heuristic was still considerably faster and produced more robust OTU assignments than any of the heuristics.
Fig. 4.
Comparison of the Matthew's correlation coefficients for OTUs calculated from a threshold of 0.00 to 0.10 when using the phylotype-OTU heuristic for full-length 16S rRNA gene sequences. For each region, cutoff, and taxonomic level used to split the sequences, the correlation coefficients overlapped with each other, except for the family and genus taxonomic levels. (See Fig. S5 and S6 in the supplemental material for the same analysis using the V13 and V35 sequences, respectively.)
Table 2.
Time required to use the phylotype-OTU heuristic when splitting sequences or distance matrices at various taxonomic depths for full-length, V13, and V35 16S rRNA gene sequences
| Region | Input | No. of processors | Wall time (min) for calculation of distance to levela: |
||||
|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | |||
| Full length | Distances | 1 | 26.82 | 9.20 | 3.43 | 1.40 | 1.25 |
| 8 | 20.38 | 7.38 | 3.47 | 1.35 | 1.22 | ||
| Sequences | 1 | 50.43 | 16.48 | 5.97 | 1.72 | 1.13 | |
| 8 | 17.38 | 4.50 | 1.77 | 0.73 | 0.80 | ||
| V13 | Distances | 1 | 11.77 | 5.30 | 2.23 | 0.95 | 0.85 |
| 8 | 8.93 | 3.88 | 2.07 | 0.87 | 0.75 | ||
| Sequences | 1 | 15.82 | 5.73 | 2.23 | 0.70 | 0.52 | |
| 8 | 6.83 | 1.75 | 0.95 | 0.42 | 0.57 | ||
| V35 | Distances | 1 | 24.72 | 5.37 | 2.52 | 1.43 | 1.20 |
| 8 | 19.65 | 4.78 | 2.25 | 1.28 | 1.12 | ||
| Sequences | 1 | 24.43 | 5.30 | 1.80 | 0.60 | 0.52 | |
| 8 | 13.35 | 2.20 | 0.80 | 0.43 | 0.57 | ||
All times represent the wall time in minutes required for each analysis using the computer system described in Materials and Methods. Using 8 processors, the full-length, V13, and V35 unique sequences sets required 15.07, 4.50, and 2.67 min, respectively, to calculate the sparse distance matrix.
DISCUSSION
The field of microbial ecology has benefited from a growth in the number of tools available to analyze the growing number of 16S rRNA gene sequences. As we have shown in this study, both OTU- and phylotype-based methods have unique challenges that affect one's ability to implement the method and interpret the results. The results presented in this study enable researchers to better interpret and overcome these challenges. There are a several extensions of this research that deserve further consideration.
First, the primary limitation of the phylotype-based approach is that taxonomic outlines are not well suited for the analysis of novel sequences from previously unidentified lineages. Even among the reference sequences used by the RDP to train the Bayesian classifier, 6.6% of the sequences do not have a genus-level name. Among the full-length sequences we analyzed, 19.1% of the sequences did not have a genus-level name. A related problem is that the classifier is only capable of classifying to the extent that it is provided for by the reference taxonomies. For instance, it would be impossible to assign sequences to the level of species because the taxonomies end at the level of genus. Researchers can partially overcome this by adding sequences to their reference database representing the groups they are interested in and by extending their taxonomy to the species level. Alternatively, a researcher could classify their sequences to the deepest possible taxonomic level and then to use an OTU-based approach to subdivide those groups further (e.g., the genus data in Fig. 4, Fig. S5 and S6 in the supplemental material, and Table 2). The challenges of this approach include the inability to compare results of research groups that use different taxonomies and the lingering difficulty with mapping distance thresholds to distinguish between groups within the same taxonomic level.
Second, others have observed that OTU-based methods are more sensitive to sequencing errors than phylotype-based methods (9, 12). Based on the results we have presented, phylotype-based methods are less sensitive to such errors because they operate at a much broader level than OTU-based methods. This leads to an overall muting of the effects of sequencing errors. Interestingly, when others have resequenced mock communities and used broad OTU definitions, the effects of sequencing errors are minimized (9, 12). Thus, it is not that phylotype-based methods are not sensitive to sequencing errors, rather that the commonly used genus-level cutoff represents such a broad distance that it masks the amount of sequencing error.
Third, the dependence upon heuristics to overcome technical limitations of methods used to assign sequences to OTUs has resulted in a sacrifice of accuracy. Instead we have developed a new heuristic that makes use of elements commonly used in a typical sequence analysis pipeline to minimize computational overhead. First, a common feature of the previous heuristics was that they are implemented without a multiple sequence alignment. Such an alignment is necessary for identifying chimeric sequences (1, 7). Furthermore, NAST-based aligners have been parallelized and are capable of aligning 18 full-length sequences per second (19). Second, sequence classification is routinely used to describe the taxonomic structure of a community by using robust classifiers (e.g., the Bayesian classifier [24]) that can be parallelized and are capable of classifying 0.7 full-length sequence per second per processor with 1,000 bootstrapping iterations. Although the algorithmic improvements made to the classical average-neighbor algorithm are to be preferred to any heuristic, we have demonstrated that the phylotype-OTU approach is the best heuristic for when sequencing capacities overwhelm computational resources.
As we have discussed previously, the genetic distances calculated for 16S rRNA gene fragments are mediocre surrogates for distances between full-length sequences (18). That it is impossible to relate genetic distances to taxonomic data underscores the observation that pyrotag data are a marker for a marker (i.e., the 16S rRNA gene) of taxonomic diversity within a community. Instead we have demonstrated how researchers can perform a robust analysis using OTU-based methods and link the resulting OTUs to taxonomic data to leverage the wealth of phenotypic data related to those lineages. This merger of taxonomy-independent and -dependent methods will significantly enhance future experiments analyzing communities by using 16S rRNA gene sequence data.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anders Andersson for helpful conversations that helped us to formulate our method of assessing the quality of OTU assignments.
This project was funded by the School of Medicine at the University of Michigan and a grant from the National Science Foundation (award 0743432).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baldi P., Brunak S., Chauvin Y., Andersen C. A., Nielsen H. 2000. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics 16:412–424 [DOI] [PubMed] [Google Scholar]
- 3. Cohan F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457–487 [DOI] [PubMed] [Google Scholar]
- 4. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 6. Goris J., et al. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57:81–91 [DOI] [PubMed] [Google Scholar]
- 7. Huber T., Faulkner G., Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 8. Hugenholtz P., Goebel B. M., Pace N. R. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huse S., Welch D. M., Morrison H., Sogin M. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huse S. M., et al. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Konstantinidis K. T., Ramette A., Tiedje J. M. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1929–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kunin V., Engelbrektson A., Ochman H., Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12:118–123 [DOI] [PubMed] [Google Scholar]
- 13. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY [Google Scholar]
- 14. Legendre P., Legendre L. 1998. Numerical ecology. Elsevier, New York, NY [Google Scholar]
- 15. Li W., Godzik A. 2006. CD-HIT: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed] [Google Scholar]
- 16. Liu Z., DeSantis T. Z., Andersen G. L., Knight R. 2008. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schloss P. D. 2010. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput. Biol. 6:e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schloss P. D. 2009. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One 4:e8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schloss P. D., Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stackebrandt E., Goebel B. M. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 23. Sun Y., et al. 2009. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 37:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ward D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271–277 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.