Abstract
Orientia tsutsugamushi is the etiological agent of scrub typhus, a mite-borne, febrile illness that occurs in the Asia-Pacific region. We conducted strain characterization of O. tsutsugamushi isolates from chiggers obtained from rodents based the nucleotide sequence of the 56-kDa outer membrane protein gene. With the use of PCR, a total of 68 DNA sequences of 56-kDa antigen genes were amplified. Phylogenetic analysis revealed that there were at least six definable clusters among the 68 isolates: 37% Karp-related strains (25/68), 27% TA763 strains (18/68), 12% JG-related strains (8/68), 19% Kato-related strains (13/68), 4% divergent strains (3/68), and 1% representing a Gilliam prototype strain (1/68). Overall, the O. tsutsugamushi genotypes exhibited a high degree of diversity, similar to that seen in strains from the rest of the areas where scrub typhus is endemic. Moreover, the 56-kDa protein sequence similarity between O. tsutsugamushi isolates from mites and those from human patients (H. Y. Lu et al., Am. J. Trop. Med. Hyg. 83:658-663, 2010) were striking, thus highlighting potential risk factors for this emerging zoonotic disease.
INTRODUCTION
Orientia tsutsugamushi is an obligate intracellular, Gram-negative bacterium that causes scrub typhus, an acute febrile illness characterized by a typical primary lesion (eschar), lymphadenopathy, and nonspecific symptoms such as chills, headache, and malaise (10, 24). The disease is endemic in Asian countries, including Japan, South Korea, China, Thailand, and Taiwan (6, 13). An estimated 1 million cases occur annually (22). However, the precise incidence of this disease is unknown, as the available diagnostic facilities in most of the region of endemicity are limited (32). In nature, the bacterial agent is maintained within a life cycle involving wild mammals and a mite vector, primarily of the Leptotrombidium genus (14). Humans are accidentally infected by the bites of a larval mite, known as a chigger, during feeding (30).
The 56-kDa type-specific antigen (TSA) gene encodes a dominant membrane protein, which accounts for 10% to 15% of the total protein of O. tsutsugamushi (17). The deduced size of the 56-kDa protein varies from 516 to 541 amino acids, according to gene sequences deposited in the GenBank (11). It is highly immunogenic, and the sera of most patients with scrub typhus are reactive. Serological analyses show that the antigenicity of the 56-kDa protein is diverse and includes variants such as the representative Karp-, Gilliam-, and Kato-type strains and other isolates (2, 8, 18). The immune responses to the 56-kDa protein provide protection against Scrub typhus, suggesting that it is a potential candidate for vaccine development (3, 16, 33). No commercially licensed vaccine for scrub typhus is currently available.
In the present study, we cloned and sequenced the 56-kDa TSA gene of O. tsutsugamushi isolated from the chigger mite obtained from a rodent in order to understand the genetic types as well as the relationships with other known strains. Our results demonstrated that the genotypes of O. tsutsugamushi isolates appeared highly diverse and widely and geographically distributed in Taiwan.
MATERIALS AND METHODS
Study site and arthropod collection.
All the arthropods used in this study were collected from rodents captured by randomly setting live traps in the fields of Taiwan's mainland and off shore islands, such as Penghu, Kinmen, and Lan-Yu. The rodent species found in this study were diverse, including Bandicota indica, Rattus norvegicus, Rattus losea, Rattus flavipectus, Rattus mindanensis, and Suncus murinus; the predominance of the rat species depended on the geographic location. R. losea was commonly found in all study sites except Lanyu. The dominant rodent species consist of R. flavipectus in Kinmen and R. mindanensis in Lanyu.
The mites were collected from the captured rodents by brushing the animal's fur thoroughly using a metal comb over the plastic bag. The collected mites were placed in an Eppendorf tube and delivered to the laboratory.
DNA extraction and isolation of bacteria.
The mites carried in each individual rodent were grouped and were processed for DNA extraction in accordance with the procedures described previously (28). Briefly, the mites (10 to 50 mites) collected from individual rodents were pooled, their surfaces were disinfected in a solution of 70% methanol containing 0.2% iodine triturated in a 0.2- to 1-ml sucrose-phosphate-glutamate buffer, and were then subjected to DNA extraction and cell culture. The DNA was extracted using a QIAamp DNA minikit (Qiagen, Hilden, Germany) and then stored at −70°C for later use. O. tsutsugamushi was isolated from the mite homogenates by inoculation into L929 cells by use of the Shell-vial method (12), and infected monolayer cells were examined for O. tsutsugamushi using an indirect fluorescence assay (IFA).
PCR amplification.
Nested PCR was performed to detect O. tsutsugamushi DNA in the DNA extract. The primer sequences were designed based on the DNA sequence from the Karp strain (GenBank accession no. AY956315) coding for the 56-kDa TSA gene, as described previously (5, 7). The two pairs of primers used were as follows: pair 1, comprising F6 (5′-GTTGGAGGAATGAATTACTGG-3′) (nucleotides 406 to 425) and R6 (5′-AGCGCTAGGTTTATTAGCAT-3′) (nucleotides 1059 to 1040), and pair 2, comprising F7 (5′-AGGATTAGAGTGTGGTCCTT-3′) (nucleotides 369 to 388) and R7 (5′-ACAGATGCACTATTGGCAA-3′) (nucleotides 1175 to 1156). Approximately 4% of the first PCR products were used as a template DNA for the second PCR; purified Karp strain DNA and distilled water were used instead of the specimen DNA as positive and negative controls, respectively. PCR targeting DNA was visualized with a UV transilluminator after agarose gel electrophoresis and staining with ethidium bromide (see Fig. S1 in the supplemental material).
PCR amplification of the complete 56-kDa TSA gene was performed using primers TF1 (5′-AGAATGAAAAAAATTATGTTAATTGC-3′) and TR1 (5′-AAACTAGAAGTTATAGCGYACAC-3′). The PCR program was started with denaturation at 94°C for 3 min, followed by 35 cycles consisting of 40 s at 94°C, 30 s at 55°C, and 1 min 40 s at 72°C, with a final extension step of 10 min at 72°C. The PCR was conducted in a PTC-200 peltier thermal cycler (MJ Research, Reno, NV). The PCR products were purified using a PCR purification kit (Qiagen, Hilden, Germany) and cloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). PCR products were sequenced by using an ABI 3730 XL DNA analyzer at Genomics BioSci & Tech, Inc., Taipei, Taiwan. Strands were sequenced in the forward and reverse directions using primers T7P (5′-TAA TAC GAC TCA CTA TAG G-3′) and T3P (5′-GCAATTAACCCTCACTAAAGGG-3′), corresponding to the priming sites on TA cloning vector.
Phylogenetic analysis.
Multiple sequence alignment and analysis of the genetic relationships between isolates and reference strains were performed using Bionumerous version 4.6 and the CLUSTAL_X program (26), and the resulting dendrogram was constructed with the software program MEGA4 (25), using the neighbor-joining method and bootstrap test on 1,000 replicates. The complete nucleotide sequences of the 56-kDa gene for the O. tsutsugamushi strains isolated from different regions and the references for each type of strain are listed in Table 1. A percentage nucleotide identity matrix was constructed using the DNAStar MEGALIGN 6.1 program (DNAStar, Inc., Madison, WI).
Table 1.
Description of Orientia tsutsugamushi isolates and reference strains examineda
| Isolate | Mo/yr | Source | Location | Country | No. of bases | GenBank accession no. | Strain | Source or reference |
|---|---|---|---|---|---|---|---|---|
| HL01 | 01/2008 | Chiggers | Hualien | Taiwan | 1,566 | GU120139 | TA763 | This study |
| HL02-1 | 04/2008 | Chiggers | Hualien | Taiwan | 1,605 | GU120140 | Karp related | This study |
| HL02-2 | 04/2008 | Chiggers | Hualien | Taiwan | 1,566 | GU120141 | TA763 | This study |
| HL03-1 | 01/2008 | Chiggers | Hualien | Taiwan | 1,566 | GU120142 | TA763 | This study |
| HL03-2 | 01/2008 | Chiggers | Hualien | Taiwan | 1,596 | GU120143 | Kato related | This study |
| HL04 | 01/2008 | Chiggers | Hualien | Taiwan | 1,605 | GU120144 | Karp related | This study |
| HL05 | 01/2008 | Chiggers | Hualien | Taiwan | 1,605 | GU120145 | Karp related | This study |
| KM01 | 2001 | Chiggers | Kinmen | Taiwan | 1,611 | GU120146 | Karp related | This study |
| KM02 | 2002 | Chiggers | Kinmen | Taiwan | 1,575 | GU120147 | Gilliam | This study |
| KM03 | 2003 | Chiggers | Kinmen | Taiwan | 1,590 | GU120148 | Kato related | This study |
| KM04 | 01/2008 | Chiggers | Kinmen | Taiwan | 1,587 | GU120149 | TA763 | This study |
| KM05 | 03/2008 | Chiggers | Kinmen | Taiwan | 1,611 | GU120150 | Karp related | This study |
| KM06 | 03/2008 | Chiggers | Kinmen | Taiwan | 1,575 | GU120151 | Kato related | This study |
| KM07 | 03/2008 | Chiggers | Kinmen | Taiwan | 1,608 | GU120152 | Karp related | This study |
| KM08 | 03/2008 | Chiggers | Kinmen | Taiwan | 1,605 | GU120153 | TA763 | This study |
| KM09 | 03/2008 | Chiggers | Kinmen | Taiwan | 1,608 | GU120154 | Karp related | This study |
| KM10-1 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,605 | GU446588 | TA763 | This study |
| KM10-2 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,605 | GU446589 | TA763 | This study |
| KM11-1 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,611 | GU446590 | Karp related | This study |
| KM11-2 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,608 | GU446591 | Karp related | This study |
| KM12 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,605 | GU446592 | Karp related | This study |
| KM13 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,608 | GU446593 | Karp related | This study |
| KM14 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,572 | GU446594 | JG related | This study |
| KM15-1 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,593 | GU446595 | Divergent | This study |
| KM15-2 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,593 | GU446596 | Divergent | This study |
| KM16-1 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,632 | GU446597 | Divergent | This study |
| KM16-2 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,596 | GU446598 | JG related | This study |
| KM17-1 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,611 | GU446599 | Karp related | This study |
| KM17-2 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,611 | GU446600 | Karp related | This study |
| KM18 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,611 | GU446601 | Karp related | This study |
| KM19-1 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,611 | GU446602 | Karp related | This study |
| KM19-2 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,584 | GU446603 | TA763 | This study |
| KM20 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,605 | GU446604 | TA763 | This study |
| KM21-1 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,575 | GU446605 | Kato related | This study |
| KM21-2 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,560 | GU446606 | TA763 | This study |
| KM21-3 | 10/2009 | Chiggers | Kinmen | Taiwan | 1,605 | GU446607 | TA763 | This study |
| MZ01-1 | 07/2007 | Chiggers | Matsu | Taiwan | 1,602 | GU120155 | Karp related | This study |
| MZ01-2 | 07/2007 | Chiggers | Matsu | Taiwan | 1,602 | GU120156 | Karp related | This study |
| MZ02 | 07/2007 | Chiggers | Matsu | Taiwan | 1,572 | GU120157 | JG related | This study |
| OI01 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,572 | GU446608 | Kato related | This study |
| OI010 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,566 | GU446620 | TA763 | This study |
| OI011 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,566 | GU446621 | TA763 | This study |
| OI02 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,572 | GU446609 | JG related | This study |
| OI03-1 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,566 | GU446610 | Kato related | This study |
| OI03-2 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,566 | GU446611 | Kato related | This study |
| OI04 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,578 | GU446612 | Kato related | This study |
| OI05-1 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,605 | GU446613 | TA763 | This study |
| OI05-2 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,611 | GU446614 | Karp related | This study |
| OI06-1 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,581 | GU446615 | Kato related | This study |
| OI06-2 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,602 | GU446616 | Karp related | This study |
| OI07 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,602 | GU446617 | Karp related | This study |
| OI08 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,608 | GU446618 | Karp related | This study |
| OI09 | 12/2009 | Chiggers | Lan-Yu | Taiwan | 1,575 | GU446619 | Kato related | This study |
| PH01 | 07/2007 | Chiggers | Penghu | Taiwan | 1,608 | GU120158 | Karp related | This study |
| PH02 | 05/2008 | Chiggers | Penghu | Taiwan | 1,608 | GU120159 | Karp related | This study |
| PH03 | 05/2008 | Chiggers | Penghu | Taiwan | 1,605 | GU120160 | TA763 | This study |
| PH04 | 05/2008 | Chiggers | Penghu | Taiwan | 1,608 | GU120161 | Karp related | This study |
| PH05 | 05/2008 | Chiggers | Penghu | Taiwan | 1,605 | GU120162 | TA763 | This study |
| TT01-1 | 07/2008 | Chiggers | Taitung | Taiwan | 1,575 | GU120163 | Kato related | This study |
| TT01-2 | 07/2008 | Chiggers | Taitung | Taiwan | 1,572 | GU120164 | JG related | This study |
| TT02-1 | 10/2008 | Chiggers | Taitung | Taiwan | 1,572 | GU120165 | JG related | This study |
| TT02-2 | 10/2008 | Chiggers | Taitung | Taiwan | 1,572 | GU120166 | JG related | This study |
| TT03-1 | 12/2008 | Chiggers | Taitung | Taiwan | 1,593 | GU120168 | Divergent | This study |
| TT03-2 | 12/2008 | Chiggers | Taitung | Taiwan | 1,572 | GU120169 | Kato related | This study |
| TT04 | 12/2008 | Chiggers | Taitung | Taiwan | 1,566 | GU120170 | Kato related | This study |
| TT05 | 12/2008 | Chiggers | Taitung | Taiwan | 1,572 | GU120171 | JG related | This study |
| TT06-1 | 12/2008 | Chiggers | Taitung | Taiwan | 1,605 | GU120172 | TA763 | This study |
| TT06-6 | 12/2008 | Chiggers | Taitung | Taiwan | 1,605 | GU120173 | TA763 | This study |
| Hualien-1 | Hualien | Taiwan | 1,842 | AY243357 | JG related | 11 | ||
| Hualien-3 | Hualien | Taiwan | 1,875 | AY636101 | Kato related | 11 | ||
| Hualien-4 | Hualien | Taiwan | 1,742 | AY714315 | Kato related | 11 | ||
| Hualien-7 | Hualien | Taiwan | 1,857 | AY834393 | JG related | 11 | ||
| Hualien-8 | Hualien | Taiwan | 1,863 | DQ323174 | JG related | 11 | ||
| Hualien-11 | Hualien | Taiwan | 1,889 | DQ323175 | TA763 | 11 | ||
| Hualien-12 | Hualien | Taiwan | 1,890 | DQ323176 | Karp related | 11 | ||
| Hualien-13 | Hualien | Taiwan | 1,856 | DQ789360 | JG related | 11 | ||
| Hualien-14 | Hualien | Taiwan | 1,851 | DQ852664 | TA763 | 11 | ||
| Taitung-1 | Taitung | Taiwan | 1,889 | AF516948 | Karp related | 11 | ||
| Taitung-2 | Taitung | Taiwan | 1,857 | AY335819 | JG related | 11 | ||
| Taitung-4 | Taitung | Taiwan | 1,889 | AY787232 | TA763 | 11 | ||
| Taitung-5 | Taitung | Taiwan | 1,863 | AY834392 | JG related | 11 | ||
| CDC Gilliam | Taiwan | 1,860 | DQ485289 | Gilliam | 11 | |||
| CDC Karp | New Guinea | 1,884 | AY956315 | Karp | 11 | |||
| CDC Kato | Japan | 1,875 | AY836148 | Kato | 11 | |||
| TA 763 | 1963 | Rodent | Thailand | 1,581 | U80636 | TA763 | 11 | |
| Kawasaki | Japan | 1,569 | M63383 | JG related | ||||
| UT176 | 07/2004 | Human | Udorn Thani | Thailand | 1,602 | EF213081 | Karp related | 4 |
| UT302 | 08/2004 | Human | Udorn Thani | Thailand | 1,587 | EF213078 | TA763 | 4 |
| UT316 | 10/2004 | Human | Udorn Thani | Thailand | 1,611 | EF213078 | Karp related | 4 |
| UT329 | 07/2005 | Human | Udorn Thani | Thailand | 1,596 | EF213099 | JG related | 4 |
| UT336 | 07/2005 | Human | Udorn Thani | Thailand | 1,599 | EF213089 | Karp related | 4 |
| TW-1 | 05/2006 | Human | Kinmen | Taiwan | 1,608 | GQ332742 | Karp related | 15 |
| TW-2 | 10/2006 | Human | Taoyuan County | Taiwan | 1,605 | GQ332743 | Karp related | 15 |
| TW-3 | 07/2006 | Human | Taipei County | Taiwan | 1,605 | GQ332744 | Karp related | 15 |
| TW-6 | 09/2006 | Human | Kaohsiung City | Taiwan | 1,608 | GQ332747 | Karp related | 15 |
| TW-7 | 06/2006 | Human | Kaohsiung City | Taiwan | 1,608 | GQ332748 | Karp related | 15 |
| TW-8 | 11/2007 | Human | Changhua County | Taiwan | 1,692 | GQ332749 | Karp related | 15 |
| TW-11 | 07/2007 | Human | Nantou County | Taiwan | 1,584 | GQ332752 | TA763 | 15 |
| TW-12 | 05/2007 | Human | Taitung County | Taiwan | 1,593 | GQ332753 | Divergent | 15 |
| TW-13 | 11/2007 | Human | Nantou County | Taiwan | 1,557 | GQ332754 | JG related | 15 |
| TW-14 | 11/2007 | Human | Taitung County | Taiwan | 1,551 | GQ332755 | JG related | 15 |
| TW-15 | 12/2007 | Human | Pingtung County | Taiwan | 1,569 | GQ332756 | JG related | 15 |
| TW-16 | 07/2007 | Human | Kaohsiung City | Taiwan | 1,572 | GQ332757 | JG related | 15 |
| TW-17 | 07/2007 | Human | Taipei City | Taiwan | 1,596 | GQ332758 | JG related | 15 |
| TW-18 | 06/2007 | Human | Kaohsiung City | Taiwan | 1,596 | GQ332759 | JG related | 15 |
| TW-20 | 05/2006 | Human | Hsinchu County | Taiwan | 1,572 | GQ332761 | Kato related | 15 |
| TW-21 | 07/2006 | Human | Kinmen | Taiwan | 1,590 | GQ332762 | Kato related | 15 |
| TW-22 | 06/2006 | Human | Kaohsiung City | Taiwan | 1,575 | GQ332763 | Kato related | 15 |
“Gilliam” refers to the Gilliam prototype; “JG related” refers to the Gilliam variant.
Nucleotide sequence accession numbers.
All the DNA sequences used in this study were deposited in the GenBank database under the accession numbers indicated in Table 1.
RESULTS
Prevalence of O. tsutsugamushi in chigger mites from rodents.
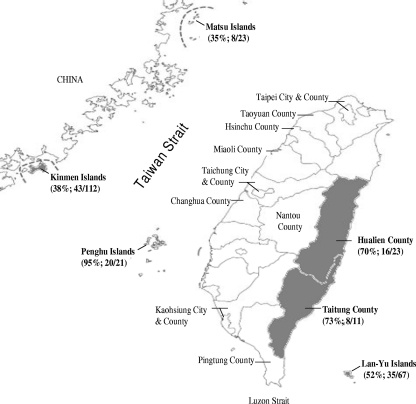
Figure 1 shows the Taiwan map indicating the geographic areas of O. tsutsugamushi strains isolated from chigger mites collected from rodents as well as the strains isolated from a human scrub typhus patient (15) that were used in this study. The numbers of mites carried by each individual rodent, i.e., the mite indexes, differed drastically, from a few to several hundreds. Mite pools collected from 130 out of 257 (51%) rodents were found to be positive for the O. tsutsugamushi 56-kDa gene fragment based on nested PCR. The prevalence of O. tsutsugamushi in rodents among the study sites ranged from 35 to 95%. A relatively high prevalence rate in Penghu was observed in the present study. Among the nest PCR-positive specimens, 49/95 (52%) mite homogenates that had been isolated from 19 R. losea organisms, 13 R. mindanensis organisms, 13 R. flavipectus organisms, 3 Suncus murinus organisms, and 1 Bandicota indica organism were successfully cultured for bacterial pathogens as examined by IFA using O. tsutsugamushi-specific antibody. A total 68 complete 56-kDa TSA gene sequences were amplified from these samples by PCR.
Fig. 1.
Map of the geographic areas in Taiwan showing O. tsutsugamushi strains isolated in this study and from a human scrub typhus patient. For each study site, the number of captured rodents and the prevalence of O. tsutsugamushi DNA fragment in mite-hosting rodents are indicated.
O. tsutsugamushi characteristics.
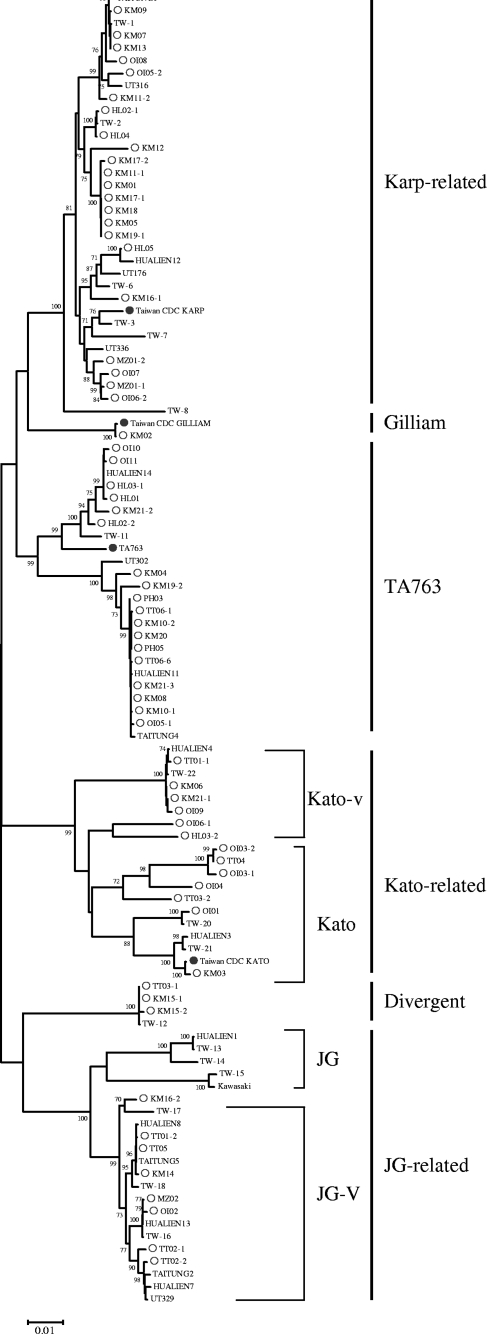
The results of analysis of the 56-kDa TSA gene sequences of the O. tsutsugamushi isolates used in this study are shown in Tables 1 and 2. These O. tsutsugamushi strains are related to either the Karp- or Gillian-type strains or Kato and the Thai-type strain TA763 (Fig. 2). The majority of isolates (38%; 25/68) were related to the Karp-type reference strain (percent nucleotide identity range [PNIR] for Karp strain, 94.4% to 96.8%). Only one isolate was related to the Gilliam prototype strain (PNIR for Gilliam strain, 99.8%). The amino acid sequence identities ranged from 73.2% to 100% among the 68 isolates.
Table 2.
Comparison of the amino acid homologies of the entire 56-kDa TSA gene between the Orientia tsutsugamushi strains isolated from Taiwan and the reference strainsa
| Isolate | % Homology to indicated O. tsutsugamushi reference strain |
|||||||
|---|---|---|---|---|---|---|---|---|
| KARP | KATO | Gilliam | UT316 | TW-22 | TA763 | UT329 | TW-12 | |
| HL01 | 80.9 | 75.9 | 81.3 | 89.7 | 78.5 | 90.6 | 76.2 | 76.9 |
| HL02-1 | 93.0 | 76.9 | 84.4 | 96.3 | 76.0 | 84.0 | 78.2 | 76.3 |
| HL02-2 | 82.3 | 77.5 | 82.6 | 90.1 | 78.7 | 91.9 | 77.6 | 78.3 |
| HL03-1 | 81.3 | 76.3 | 81.7 | 89.9 | 78.9 | 91.0 | 76.6 | 77.3 |
| HL03-2 | 77.2 | 89.9 | 78.2 | 85.1 | 81.0 | 76.6 | 78.0 | 76.9 |
| HL04 | 93.0 | 76.9 | 84.4 | 96.5 | 76.0 | 84.2 | 78.2 | 76.3 |
| HL05 | 93.0 | 76.9 | 84.4 | 96.5 | 76.0 | 84.2 | 78.2 | 76.3 |
| KM01 | 93.2 | 77.8 | 85.7 | 96.3 | 76.3 | 83.4 | 79.2 | 76.3 |
| KM02 | 84.1 | 77.0 | 99.6 | 89.6 | 75.1 | 82.8 | 87.8 | 77.5 |
| KM03 | 75.5 | 99.6 | 76.6 | 84.4 | 79.3 | 75.0 | 75.8 | 77.4 |
| KM04 | 79.6 | 75.2 | 80.3 | 87.8 | 77.2 | 81.7 | 74.5 | 76.1 |
| KM05 | 93.2 | 77.8 | 85.7 | 96.2 | 76.3 | 83.4 | 79.2 | 76.3 |
| KM06 | 76.1 | 79.5 | 74.9 | 85.7 | 99.4 | 77.2 | 73.9 | 73.5 |
| KM07 | 92.2 | 75.4 | 82.9 | 98.4 | 75.2 | 85.2 | 77.5 | 75.0 |
| KM08 | 80.0 | 74.9 | 81.5 | 88.6 | 75.9 | 82.2 | 73.9 | 76.1 |
| KM09 | 92.2 | 75.4 | 82.9 | 98.4 | 75.2 | 85.2 | 77.5 | 75.0 |
| KM10-1 | 80.0 | 74.9 | 81.5 | 88.5 | 75.9 | 82.2 | 73.9 | 76.1 |
| KM10-2 | 80.0 | 74.9 | 81.5 | 88.5 | 75.9 | 82.2 | 73.9 | 76.1 |
| KM11-1 | 93.2 | 77.8 | 85.7 | 96.3 | 76.3 | 83.4 | 79.2 | 76.3 |
| KM11-2 | 92.4 | 76.6 | 83.7 | 98.4 | 75.9 | 84.5 | 78.5 | 76.1 |
| KM12 | 90.6 | 74.2 | 83.8 | 94.8 | 73.4 | 81.3 | 75.7 | 73.3 |
| KM13 | 92.2 | 75.4 | 82.9 | 98.4 | 75.2 | 85.2 | 77.5 | 75.0 |
| KM14 | 76.8 | 77.4 | 85.3 | 85.8 | 73.5 | 75.8 | 96.7 | 76.3 |
| KM15-1 | 73.4 | 77.6 | 77.5 | 86.0 | 73.5 | 75.2 | 75.3 | 99.6 |
| KM15-2 | 73.0 | 77.2 | 76.9 | 85.9 | 72.9 | 74.9 | 74.7 | 99.2 |
| KM16-1 | 91.0 | 74.6 | 84.0 | 95.0 | 72.7 | 83.1 | 77.1 | 74.0 |
| KM16-2 | 77.4 | 75.8 | 86.2 | 85.7 | 72.7 | 76.4 | 95.7 | 76.3 |
| KM17-1 | 93.2 | 77.8 | 85.7 | 96.2 | 76.3 | 83.4 | 79.2 | 76.3 |
| KM17-2 | 92.8 | 77.4 | 85.3 | 96.2 | 75.9 | 83.0 | 78.8 | 76.0 |
| KM18 | 93.2 | 77.8 | 85.7 | 96.2 | 76.3 | 83.4 | 79.2 | 76.3 |
| KM19-1 | 93.2 | 77.8 | 85.7 | 96.2 | 76.3 | 83.4 | 79.2 | 76.3 |
| KM19-2 | 78.0 | 74.0 | 79.5 | 87.2 | 75.6 | 80.9 | 73.4 | 75.6 |
| KM20 | 80.0 | 74.9 | 81.5 | 88.5 | 75.9 | 82.2 | 73.9 | 76.1 |
| KM21-1 | 76.1 | 79.5 | 74.9 | 85.6 | 99.4 | 77.2 | 73.9 | 73.5 |
| KM21-2 | 80.7 | 75.5 | 82.2 | 89.6 | 78.7 | 89.8 | 76.9 | 75.9 |
| KM21-3 | 80.0 | 74.9 | 81.5 | 88.5 | 75.9 | 82.2 | 73.9 | 76.1 |
| MZ01-1 | 94.5 | 75.5 | 82.6 | 95.5 | 74.6 | 83.2 | 76.1 | 76.3 |
| MZ01-2 | 94.5 | 75.7 | 84.5 | 95.3 | 74.4 | 83.2 | 77.2 | 75.2 |
| MZ02 | 76.0 | 76.0 | 86.6 | 85.1 | 72.5 | 75.2 | 97.3 | 74.5 |
| OI01 | 75.9 | 89.3 | 78.5 | 84.5 | 79.7 | 74.5 | 76.1 | 77.6 |
| OI02 | 76.0 | 76.0 | 86.2 | 85.2 | 72.5 | 75.2 | 96.9 | 74.1 |
| OI03-1 | 74.2 | 82.0 | 73.9 | 84.6 | 78.4 | 71.7 | 72.3 | 72.8 |
| OI03-2 | 73.6 | 82.0 | 73.5 | 84.8 | 78.0 | 71.7 | 72.3 | 72.8 |
| OI04 | 78.1 | 83.8 | 76.5 | 83.5 | 80.4 | 75.3 | 75.1 | 73.5 |
| OI05-1 | 79.8 | 74.7 | 81.3 | 88.5 | 75.7 | 81.8 | 73.8 | 75.9 |
| OI05-2 | 89.5 | 74.9 | 82.1 | 98.5 | 74.9 | 83.0 | 76.2 | 75.6 |
| OI06-1 | 76.8 | 83.3 | 75.4 | 84.8 | 83.6 | 77.9 | 75.5 | 74.5 |
| OI06-2 | 94.2 | 75.3 | 82.3 | 95.4 | 74.4 | 83.2 | 75.9 | 75.8 |
| OI07 | 94.4 | 75.5 | 82.6 | 95.3 | 74.6 | 83.0 | 76.1 | 76.1 |
| OI08 | 91.4 | 74.5 | 82.1 | 97.8 | 74.5 | 84.7 | 76.2 | 75.0 |
| OI09 | 75.5 | 79.0 | 74.3 | 85.3 | 99.0 | 76.6 | 73.3 | 72.9 |
| OI10 | 80.7 | 75.5 | 80.9 | 89.7 | 78.1 | 90.4 | 75.8 | 76.6 |
| OI11 | 80.9 | 75.9 | 81.3 | 89.7 | 79.1 | 90.6 | 76.4 | 76.9 |
| PH01 | 91.6 | 74.9 | 82.3 | 98.2 | 74.7 | 84.7 | 76.9 | 74.4 |
| PH02 | 91.8 | 75.0 | 82.5 | 98.3 | 74.9 | 84.9 | 77.1 | 74.6 |
| PH03 | 80.2 | 74.7 | 81.6 | 88.7 | 76.1 | 82.0 | 74.1 | 75.9 |
| PH04 | 91.8 | 75.0 | 82.5 | 98.3 | 74.9 | 84.9 | 77.1 | 74.6 |
| PH05 | 80.0 | 74.9 | 81.5 | 88.5 | 75.9 | 82.2 | 73.9 | 76.1 |
| TT01-1 | 75.9 | 79.3 | 74.7 | 85.5 | 99.4 | 77.0 | 73.5 | 73.3 |
| TT01-2 | 76.8 | 77.4 | 85.3 | 85.8 | 73.5 | 75.8 | 96.6 | 76.3 |
| TT02-1 | 77.9 | 77.2 | 86.6 | 85.4 | 74.3 | 76.8 | 98.7 | 75.9 |
| TT02-2 | 77.2 | 76.6 | 87.2 | 85.2 | 74.1 | 76.0 | 99.4 | 75.1 |
| TT03-1 | 73.6 | 77.6 | 77.5 | 86.0 | 73.5 | 75.4 | 75.3 | 99.8 |
| TT03-2 | 78.1 | 79.3 | 78.7 | 86.7 | 77.6 | 76.8 | 76.1 | 82.7 |
| TT04 | 74.2 | 82.4 | 73.9 | 84.8 | 78.4 | 72.1 | 72.7 | 73.2 |
| TT05 | 77.0 | 77.6 | 85.5 | 86.0 | 73.7 | 76.0 | 96.9 | 76.4 |
| TT06-1 | 80.0 | 74.9 | 81.5 | 88.5 | 75.9 | 82.2 | 73.9 | 76.1 |
| TT06-6 | 80.0 | 74.9 | 81.5 | 88.5 | 75.9 | 82.2 | 73.9 | 75.9 |
The homologies between the 56-kDa TSA protein sequences of the reference strains and the strains within the same genotype group are indicated in bold.
Fig. 2.
Phylogenetic tree of O. tsutsugamushi based on the nucleotide sequences of the 56-kDa cell surface antigen gene. The subset of the phylogenetic tree is made up of isolates (strains from this study are indicated by open circles) associated with clades with sequence divergence from the reference strains (filled circles), such as Karp, Gilliam-related strains, and Kato, and strains isolated from humans (no mark). Isolates are identified by their GenBank accession numbers. Phylogenetic analyses were conducted using MEGA4. The evolutionary history was inferred using the neighbor-joining method and a bootstrap test on 1,000 replicates.
O. tsutsugamushi distribution by geographical region.
The majority of O. tsutsugamushi isolates (29/68) studied were from Kinman Island, which represented a high proportion of Karp-related strains (44%; 13/29), eight TA763 strains (28%; 8/29), two Gilliam-related strains (7%; 2/29), one single Gilliam isolate, and two divergent strains. In contrast, 14 isolates (6 [43%; 6/14] Kato-related strains, 4 [29%; 4/14] Karp-related strains, 3 [21%; 3/14] TA763-related strains, and 1 JG-related strain) were found on Lan-Yu, a small island in the Pacific Ocean southeast of Taiwan. Ten isolates (four [40%; 4/10] JG-, three [30%; 3/10] Kato-, and two TA763-related strains and 1 divergent strain) were from Taitung County. Seven isolates were from Hualien (three [43%; 3/7] TA763 strains, two Karp-related strains, and one Kato-related strain). Five isolates (three Karp and two TA763 strains) were from the Penghu islands. Three isolates (two Karp-related strains and one JG-related strain) were from the Matsu islands. The genotypes of O. tsutsugamushi isolates from Taiwan varied widely, with the main Karp-type strains widely distributed in different geographic regions.
Taiwanese O. tsutsugamushi isolates and international strains.
The phylogenetic relationships among Taiwan O. tsutsugamushi isolates and strains from other countries were investigated using MEGA4. Figure 2 shows that the phylogenetic tree based on the 56-kDa TSA gene sequences is divided into six definable groups. The analysis showed that the Taiwan isolates within the group of Karp-related strains were more similar to Thailand strain UT316 than to Karp-CDC (New Guinea). Consistently, a large number of isolates belonged to the TA763 clade, a strain type commonly present in Thailand but not in Japan and Korea.
It is noteworthy that the 56-kDa TSA gene sequences detected in this study were closely related to those found in a previous study of blood samples from Taiwan scrub typhus patients (15), suggesting the pathogenic potential of O. tsutsugamushi strains. The results are shown in Table 2, indicating that among Kato-related strains, KM-06, KM21-1 OI-09, and TT01-1 were 99% or more identical to the TW-22 strain. Three isolates, KM15-1, KM15-2, and TT03-1, clustered with the divergent group and had 99.2 to 99.8% identity to the TW-12 strain.
DISCUSSION
We demonstrated a high level of genetic diversity for O. tsutsugamushi strains isolated in Taiwan. According to DNA sequencing of the 56-kDa type-specific antigen gene among the 68 O. tsutsugamushi isolates, all can be grouped into six known genotypes, those representing Karp (37%), TA763 (27%), Kato (19%), JG-related strains (12%), and two minority groups, comprising divergent strains (4%) and one Gilliam-type strain (1%).
Molecular diagnosis of scrub typhus is performed by PCR amplification of target genes from O. tsutsugamushi, including a 60-kDa heat shock protein (GroEL) (19, 20, 23) and 47-kDa (9) and 56-kDa outer membrane protein genes (1, 4). Based on the restriction fragment length polymorphism (RFLP) pattern of the 56-kDa gene, Qiang et al. (21) performed a preliminary study of genetic typing of O. tsutsugamushi isolates from mites found on wild rodents. These researchers suggested that various types of O. tsutsugamushi were indigenous in Taiwan. Our results support this early observation and show that Karp- and TA763-related strains represent two predominant strain types among Taiwan isolates. The TA763 strain, which originated from rodents, is prevalent in Thailand but not Korea and Japan. In addition, the protein sequences of strains KM15-2, KM15-1, and TT03-1 were less than 90% identical to the known sequences, indicating that these variants could be new and region-specific genotypes. Various genotypes are found in different geographic regions in Taiwan. Nonetheless, we found no significant association between strain type and locality, most likely due to limitation of the sample size.
Interestingly, the 56-kDa protein sequences of three Karp-related strains (KM09, KM07, and KM13) were identical, as was the case for four TA763-related strains (KM01, KM17-1, KM17-3, KM8, and KM19-1). This suggests that these types of strains might be predominant in Kinmen. These similarities were very unlikely to have resulted from cross contamination during PCR amplification, because mite specimens were separately prepared under tightly controlled conditions in a biosafety level 3 cabinet.
The clinical significance of the strains among the isolates is largely unknown. The 56-kDa-protein sequence comparison showed that some strains, such as TT02-1 and TT02-2, were 98.7% or more identical to the human isolate UT329 from Thailand, indicating that these strains might play an epidemiological role for this infectious disease. Similarly, the 56-kDa-TSA-gene sequences of other strains in this study were shown to be highly similar to that isolated from the scrub typhus patient in Taiwan. This finding thus is not only suggesting the pathogenic potential of these O. tsutsugamushi isolates but also emphasizing the risk of acquiring scrub typhus due to mite bites.
The rodent species were found to be predominant but geographically specific, widely distributed in offshore islands, especially R. mindanensis, which was unique to Lanyu. As no characterization was carried out, the specific mite species were not known. Many mite species, including Leptotrombidium scutellare, Walchia chinensis, Leptotrombidium yui, and Odontacarus majesticus, have been identified to be the vector of scrub typhus in Taiwan, but Leptotrombidium deliense is the most significant in terms of prevalence (29). However, significant association between bacterial strain and mite species was not found. The average number of mites/rodent (the mite index) was found to be positively related to O. tsutsugamushi antibody titers in the rodent sera (data not shown). The mite-hosting rodents coinfested with ticks were observed frequently, ranging from 22.2% in Hualien to 42.1% in Kinmen. Therefore, coinfection of O. tsutsugamushi and other important tick-borne pathogens in rodents may have occurred. Recent study demonstrates that the detection of spotted fever group rickettsiae and Ehrlichia chaffeensis infection in rodent ticks likely supports this assumption (28, 31). To avoid underestimating the mix-infected bacterial strains within the pooled mite specimens, more than one single amplicon was elected and sequenced during PCR cloning.
In this case, rodents were caught by random sampling via live traps in distinct regions. Thus, ecological factors such as terrain features and seasonal variations (27) could have had a great influence on the rodent capture rate and the numbers of subsequent mite specimens collected, and that could potentially skew the results observed.
This large-scale surveillance of O. tsutsugamushi DNA sequences isolated from chigger mites provides an appreciable database for diagnosis for scrub typhus. The result also provides insight into the distribution of this zoonotic pathogen in Taiwan.
In conclusion, this study adds the knowledge of a high degree of diversity of O. tsutsugamushi genotypes and strains in wild rodents. The most common type is related to the Karp-type reference strain that is widely distributed in different geographic regions of Taiwan. The isolation from chiggers of O. tsutsugamushi strains closely related in genotype to the strains isolated from scrub typhus patients should warrant further investigation of the pathogenicity of these bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank the rodent-related infectious disease surveillance team of the Institute of Preventive Medicine for the collection of field samples.
This research was funded by the Ministry of National Defense of the Republic of China.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Blacksell S., et al. 2008. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol. Med. Microbiol. 52:335–342 [DOI] [PubMed] [Google Scholar]
- 2. Cao M., et al. 2007. Preparation of recombinant antigen of O. tsutsugamushi Ptan strain and development of rapid diagnostic reagent for scrub typhus. Am. J. Trop. Med. Hyg. 76:553–558 [PubMed] [Google Scholar]
- 3. Chattopadhyay S., et al. 2005. Scrub typhus vaccine candidate Kp r56 induces humoral and cellular immune responses in cynomolgus monkeys. Infect. Immun. 73:5039–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eremeeva M. E., Madan A., Shaw C. D., Tang K., Dasch G. A. 2005. New perspectives on rickettsial evolution from new genome sequences of rickettsia, particularly R. Canadensis and Orientia tsutsugamushi. Ann. N. Y. Acad. Sci. 1063:47–63 [DOI] [PubMed] [Google Scholar]
- 5. Furuya Y., Yoshida Y., Katayama T., Yamamoto S., Kawamura A., Jr 1993. Serotype-specific amplification of Rickettsia tsutsugamushi DNA by nested polymerase chain reaction. J. Clin. Microbiol. 31:1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gale J., et al. 1974. Scrub typhus in eastern Taiwan, 1970. Am. J. Trop. Med. Hyg. 23:679–684 [DOI] [PubMed] [Google Scholar]
- 7. Horinouchi H., et al. 1996. Genotypic identification of Rickettsia tsutsugamushi by restriction fragment length polymorphism analysis of DNA amplified by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 54:647–651 [DOI] [PubMed] [Google Scholar]
- 8. Jang W. J., Huh M. S., Park K. H., Choi M. S., Kim I. S. 2003. Evaluation of an immunoglobulin M capture enzyme-linked immunosorbent assay for diagnosis of Orientia tsutsugamushi infection. Clin. Diagn. Lab. Immunol. 10:394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang J., et al. 2004. Development of quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am. J. Trop. Med. Hyg. 70:351–356 [PubMed] [Google Scholar]
- 10. Kelly D. J., Richards A. L., Temenak J., Strickman D., Dasch G. A. 2002. The past and present threat of rickettsial diseases to military medicine and international public health. Clin. Infect. Dis. 34(Suppl. 4):S145–S169 [DOI] [PubMed] [Google Scholar]
- 11. Kelly D. J., Fuerst P. A., Ching W. M., Richards A. L. 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 48:S203–S230 [DOI] [PubMed] [Google Scholar]
- 12. Kelly P. J., Raoult D., Mason P. R. 1991. Isolation of spotted fever group rickettsia from triturated ticks using a modification of the centrifugation-shell vial technique. Trans. R. Soc. Trop. Med. Hyg. 85:397–398 [DOI] [PubMed] [Google Scholar]
- 13. Lee Y. S., Wang P. H., Tseng S. J., Ko C. F., Teng H. J. 2006. Epidemiology of scrub typhus in eastern Taiwan, 2000–2004. Jpn. J. Infect. Dis. 59:235–238 [PubMed] [Google Scholar]
- 14. Lerdthusnee K., et al. 2002. Efficiency of Leptotrombidium chiggers at transmitting Orientia tsutsugamushi to laboratory mice. J. Med. Entomol. 39:521–525 [DOI] [PubMed] [Google Scholar]
- 15. Lu H. Y., et al. 2010. Phylogenetic analysis of 56-kDa type-specific antigen gene of Orientia tsusugamushi isolates in Taiwan. Am. J. Trop. Med. Hyg. 83:658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ni Y. S., et al. 2005. Protection against scrub typhus by plasmid vaccine encoding the 56-KD outer membrane protein gene. Am. J. Trop. Med. Hyg. 73:936–941 [PubMed] [Google Scholar]
- 17. Ohashi N., Nashimoto H., Ikeda H., Tamura A. 1990. Cloning and sequencing of the gene (tsg56) encoding a type-specific antigen from Rickettsia tsutsugamushi. Gene 91:119–122 [DOI] [PubMed] [Google Scholar]
- 18. Ohashi N., Nashimoto H., Ikeda H., Tamura A. 1992. Diversity of immunodominant 56-kDa Type-specific antigen of Rickettsia tsutsugamushi. J. Biol. Chem. 267:12728–12735 [PubMed] [Google Scholar]
- 19. Paris D. H., Blacksell S. D., Newton P. N., Nicholas P. J. 2008. Simple, rapid and sensitive detection of Orientia tsutsugamushi by loop-isothermal DNA amplification. Trans. R. Soc. Trop. Med. Hyg. 58:1360–1368 [DOI] [PubMed] [Google Scholar]
- 20. Paris D. H., Aukkanit N., Jenjaroen K., Blacksell S. D., Day N. P. 2009. A highly sensitive quantitative real-time PCR assay based on the groEL gene of contemporary Thai strains of Orientia tsutsugamushi. Clin. Microbiol. Infect. 15:488–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiang Y., et al. 2003. Phylogenetic characterization of Orientia tsutsugamushi isolated in Taiwan according to the sequence homologies of 56-kDa type-specific antigen genes. Microbiol. Immunol. 47:577–583 [DOI] [PubMed] [Google Scholar]
- 22. Silpapojakul K. 1997. Scrub typhus in the Western Pacific region. Ann. Acad. Med. Singapore 26:794–800 [PubMed] [Google Scholar]
- 23. Stover C. K., Marana D. P., Dasch G. A., Oaks E. V. 1990. Molecular cloning and sequence analysis of the Sta 58 major antigen gene of Rickettsia tsutsugamushi: sequence homology and antigen comparison of Sta58 to the 60-kilodalton family of stress proteins. Infect. Immun. 58:1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamura A., Ohashi N., Urakami H., Miyamura S. 1995. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int. J. Syst. Bacteriol. 45:589–591 [DOI] [PubMed] [Google Scholar]
- 25. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 26. Thompson J. D., Gibson T., Plewniak F., Jeanmougin F., Higgins G. D. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Traub R., Wisseman W. C. 1968. Ecological considerations in scrub typhus. Bull. World Health Organ. 39:219–230 [PMC free article] [PubMed] [Google Scholar]
- 28. Tsui P. Y., et al. 2007. Molecular detection and characterization of spotted fever group rickettsiae in Taiwan. Am. J. Trop. Med. Hyg. 77:883–890 [PubMed] [Google Scholar]
- 29. Wang H. C., Chung C. L., Lin T. H., Wang C. H., Wu W. J. 2004. Studies on the vectors and pathogens of scrub typhus on murine-like animals in Kinmen County, Taiwan. Formosan Entomol. 24:257–272 (In Chinese.) [Google Scholar]
- 30. Watt G., Parola P. 2003. Scrub typhus and tropical rickettsioses. Curr. Opin. Infect. Dis. 16:429–436 [DOI] [PubMed] [Google Scholar]
- 31. Weng M. H., et al. 2010. Ehrlichia chaffeensis infection in rodent ticks-Kinmen. Taiwan Epidemol. Bull. 26:170–176 [Google Scholar]
- 32. World Health Organization 2010. Frequently asked questions—scrub typhus. World Health Organization, Regional Office for South-East Asia, New Delhi, India [Google Scholar]
- 33. Yu Y., Wen B., Niu D. S., Chen M., Qiu L. 2005. Induction of protective immunity against scrub typhus with a 56-kDa recombinant antigen of fused with a 47 kilodalton antigen of Orientia tsutsugamushi Karp. Am. J. Trop. Med. Hyg. 72:458–464 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.