Abstract
During fermentation of sugars, a number of bacterial species are able to switch from mixed acid production to acetoin and 2,3-butanediol production in order to avoid lethal acidification of their environment, although the regulation of this switch is only poorly understood. In this study, we report the identification of the budAB structural operon, involved in acetoin production in Serratia plymuthica RVH1, and its activation by a LysR-type regulator encoded by budR, immediately upstream of this operon. In addition, the regulation of budR transcription was elucidated and found to be subject to negative control by BudR itself and to positive control by external stimuli such as N-(3-oxohexanoyl)-l-homoserine lactone (OHHL) quorum sensing signaling molecules and acetate. Interestingly, however, we observed that induction of budR transcription by OHHL or acetate did not require BudR, indicating the involvement of additional regulatory factors in relaying these environmental signals to the budR promoter.
INTRODUCTION
A wide variety of strains are able to switch to acetoin and 2,3-butanediol production during fermentative growth to counteract the lethal acidification that typically accompanies mixed acid production (3, 17, 21). Butanediol is produced by first converting two molecules of pyruvate into α-acetolactate by α-acetolactate synthase, after which α-acetolactate is processed to acetoin by α-acetolactate decarboxylase. Finally, acetoin can be converted reversibly to 2,3-butanediol through the action of acetoin reductase (25). Often, the structural genes for 2,3-butanediol production are organized into an operon that is controlled by a neighboring and divergently transcribed LysR homologue (6, 12, 15). Nevertheless, the regulatory aspects, and especially the integration of environmental cues into the control of this pathway, remain largely unexplored.
In this article, we report on the identification and regulation of the acetoin operon in Serratia plymuthica RVH1 and on its activation at the molecular level by signals of acidification and quorum sensing (QS).
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and S. plymuthica RVH1 strains were cultured at 30°C in Luria-Bertani (LB) broth or agar (1.5% agar) or in M9 minimal medium (14). Where necessary, LB medium was acidified using 1 M HCl or buffered with 100 mM phosphate buffer at pH 7.0. As described previously (24), growth was monitored using a Bioscreen C microbiology reader (Thermo Life Sciences, Brussels, Belgium), while acetoin production and medium acidification were determined by the Voges-Proskauer and methyl red assays, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant features | Reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| MG1655 | F−rph-1 | 5 |
| S17-1 λpir | recAthiprohsd(r− m+) RP4:2-Tc:Mu:Km Tn7 λpir Tpr Smr | 16 |
| S. plymuthica strains | ||
| RVH1 | Wild type; natural isolate | 20, 23 |
| RVH1 splI | splI::aacC1 Gmr | 22 |
| RVH1 budB | budB::EZ-Tn5 Kmr | This study |
| RVH1 budR | budR::cat Cmr | This study |
| RVH1 budA::lacZ | RVH1 with chromosomal lacZ reporter fusion to budAB promoter; Kmr | This study |
| RVH1 splI budA::lacZ | RVH1 splI with chromosomal lacZ reporter fusion to budAB promoter; Gmr Kmr | This study |
| RVH1 budR budA::lacZ | RVH1 budR with chromosomal lacZ reporter fusion to budAB promoter; Cmr Kmr | This study |
| Plasmids | ||
| pTRC99a | Cloning vector carrying IPTG-inducible trc promoter (Ptrc); Apr | 2 |
| pTRC99a-Ptrc-budAB | pTRC99a carrying the budAB operon fused downstream of Ptrc | This study |
| pFPV25 | Cloning vector carrying promoterless gfp; Apr | 19 |
| pFPV25-PbudR-gfp | pFPV25 carrying the budR promoter fused upstream of gfp | This study |
| pUC18 | Cloning vector; Apr | 26 |
| pUC18-budA::lacZ-kan | pUC18 carrying the budAB promoter fused to lacZ | This study |
| pSF100 | Pir-dependent replicon; Apr Kmr | 13 |
| pSF100-budR::cat | pSF100 carrying budR::cat | This study |
| pSF100-budA::lacZ-kan | pSF100 carrying the lacZ reporter cloned into the budA locus | This study |
The following concentrations of sugar and/or antibiotics (Applichem, Darmstadt, Germany) were used when appropriate: glucose, 0.5%; ampicillin, 100 μg/ml; carbenicillin, 300 μg/ml; kanamycin, 50 μg/ml; and gentamicin, 20 μg/ml. The synthetic N-acyl-l-homoserine lactone (AHL) used in this study, N-(3-oxohexanoyl)-l-homoserine lactone (OHHL) (5 μM solution in water), was purchased from Sigma (Bornem, Belgium).
Identification and sequencing of the RVH1 bud locus.
A random knockout library of S. plymuthica RVH1 was constructed using an EZ-Tn5 KAN-2 Tnp transposome kit as described by the manufacturer (Epicentre, BiozymTC BV, Landgraaf, Netherlands). Circa 4,000 independent clones were purified, grown in LB-glucose, and screened for acetoin production by use of the Voges-Proskauer test and for acidification by use of the methyl red assay. One mutant that exhibited a yellow coloration in the former and a red coloration in the latter test, indicative of a loss of acetoin production and increased acidification of the growth medium, was selected. Genomic DNA of this clone was isolated using a Qiagen 100/G genomic tip according to the protocol supplied by the manufacturer. Subsequently, the insertion site was determined by sequencing (VIB Sequencing Facility, Antwerp, Belgium) the flanking site of the transposon, using KAN-2 RP-1 from an EZ-Tn5 KAN-2 Tnp transposome kit. Comparison of the obtained sequence with the GenBank nucleotide sequence database by use of BLASTX (1) revealed that the transposon was located in a gene homologous to an α-acetolactate synthase gene. In addition, part of the neighboring α-acetolactate decarboxylase gene sequence was also obtained.
To fully obtain the RVH1 sequences of these two genes and the neighboring lysR homologue (whose presence was suspected through comparison with the sequence of Serratia proteamaculans 568 [18]), primers budR_FW and budB_Rev were designed based on the nucleotide sequence of S. proteamaculans 568 and were combined with S. plymuthica RVH1-based primers budA_Rev and budA_FW, respectively. The resulting PCR products were sequenced using these four primers and the additional primers budR_gap, bud_intergen, and budB_final in order to close the gaps. The three genes were named budR, budA, and budB, among which budA and budB form an operon separated by 382 bp from budR, which is divergently transcribed (GenBank accession number HQ602654). All primers used in this study are summarized in Table 2.
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| KAN-2 RP-1 | GCAATGTAACATCAGAGATTTTGAG |
| budR_FW | TCACTCCCAGCTCGGGGGCT |
| budB_Rev | TTAAATCATCTGGCTGAA |
| budA_Rev | CGTGGTCGAGCTGATAATCC |
| budA_FW | AACCGACCTTTGCCTTTCTC |
| budR_gap | CGCCCGCTATGTTGAAC |
| bud_intergen | GTTCAACATAGCGGGCG |
| budB_final | GATCTCCAACGGCCAGCAG |
| budA_Fus | CCCCTTAGAAACCCTGCACCAAATG |
| pUC47 | CGCCAGGGTTTTCCCAGTCACGAC |
| pUC24bis | GAGTCAGTGAGCGAGGAAGGC |
| budAB_start | ATGAACGAAAAGCAGGGTGG |
| budR_Fus | CCCCTTAGGCGTCATTCATCTTTGC |
| Up-bud-XhoI | CCCCCTCGAGCATTACGTACTCCTCCACCAG |
| Down-bud-XhoI | CCGGCTCGAGAAGCAGGGTTGTTCCTGTGCG |
| pKD3/4-P1-FW | GTGTAGGCTGGAGCTGCTTC |
| pKD3/4-P2-Rev | CATATGAATATCCTCCTTAG |
| lacZ-FW_XhoI | TCGGCTCGAGACCATGATTACGGATTCACT |
| lacZ-RV_XhoI | TCCGCTCGAGTTTTTGACACCAGACCAACT |
| Up-bud-SalI | GGGGGTCGACGGGGCCTACGGACGGTAAGAG |
| Down-bud-SalI | GCGCGTCGACCGCAGAACAGGTTCGGTGAAG |
Construction of budR mutant.
To construct a budR-deficient mutant, primers budR_FW and budA_Fus were used to amplify the budR locus, after which this PCR product was ligated with pUC18 digested with SmaI (26). A unique MunI restriction site in the center of the budR gene was used to digest the resulting construct. After the ends were blunted, a chloramphenicol resistance cassette obtained from λ NK1324 (7) by BamHI cleavage and blunting was cloned in this location. PCR amplification of the resulting budR::cat allele by use of primers pUC47 and pUC24bis, followed by ligation with an EcoRV-digested pSF100 suicide plasmid (13), resulted in plasmid pSF100-budR::cat. E. coli S17-1 λpir (16) carrying this plasmid was conjugated with S. plymuthica RVH1, and exconjugants were selected on M9 minimal medium containing chloramphenicol. Colonies in which double homologous recombination had occurred were selected by loss of ampicillin or carbenicillin resistance. All primers used in this study are summarized in Table 2.
Construction of a chromosomal budAB promoter reporter fusion to lacZ.
A budAB promoter reporter fusion to lacZ was constructed by PCR amplification of the intergenic region between budR and budA together with the first ca. 400 bp of budA, using the up-bud-SalI and down-bud-SalI primers, after which the obtained PCR product was digested with SalI and ligated with a SalI-digested pUC18 plasmid (26). PCR amplification of this construct was performed using the primers up-bud-XhoI and down-bud-XhoI, after which the resulting fragment was ligated to a phosphorylated kanamycin resistance gene obtained through PCR amplification of a pKD4 plasmid (4) by use of the pKD3/4-P1-Fw and pKD3/4-P2-Rev primers. From the resulting plasmid, an amplicon was prepared with primers down-bud-XhoI and pKD3/4-P1-Fw and was ligated to a phosphorylated amplicon of the lacZ gene that was obtained through PCR amplification of genomic DNA of E. coli with primers lacZ-Fw and lacZ-Rev. The resulting plasmid had lacZ and the kanamycin resistance gene inserted at the 5′ end of the budA gene. After restriction analysis to confirm the correct orientation of the lacZ gene with regard to the budAB promoter, the plasmid was designated pUC18-budA::lacZ-Km. Next, the entire fragment, containing the budA promoter fusion coupled to lacZ and the kanamycin resistance gene, was amplified by a PCR using the up-bud-SalI and down-bud-SalI primers and was ligated to a SalI-digested pSF100 plasmid (13) from which the kanamycin resistance gene was previously removed by SalI digestion. Using E. coli S17-1 λpir (16), the resulting pSF100-budA::lacZ-Km plasmid was conjugated to the S. plymuthica RVH1 wild type and its splI and budR derivatives. Only transconjugants resistant to kanamycin and sensitive to carbenicillin were retained, as in these clones double homologous recombination via the ca. 300- to 400-bp flanking regions mediated genomic insertion of the lacZ reporter and the kanamycin resistance marker into the budA gene (referred to as budA::lacZ). Proper genomic insertion of the budA::lacZ fusion was further confirmed by PCR analysis and by loss of acetoin production in RVH1.
Construction of pTRC99a-Ptrc-budAB and pFPV25-PbudR-gfp.
To construct pTRC99a-Ptrc-budAB, a plasmid containing an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible budAB operon, the budAB genes were PCR amplified using the budAB_start and budB_Rev primers. Next, this product was ligated to a SmaI-digested pTRC99a plasmid (2). Sequence analysis confirmed complete and correct insertion of the budAB operon. Heterologous expression of the budAB genes from this plasmid in E. coli MG1655 (5), which does not naturally produce acetoin, imposed acetoin production, as confirmed by the Voges-Proskauer assay.
A budR promoter reporter fusion to gfp (i.e., PbudR-gfp) was constructed by PCR amplification of the intergenic region between budR and budA, using the budA_Fus and budR_Fus primers, after which the obtained PCR product was ligated into a SmaI-digested pFPV25 plasmid (19) which contained a promoterless gfp gene. Sequence analysis confirmed correct insertion of the fragment. All primers used in this study are summarized in Table 2.
Measurement of fluorescence and β-galactosidase.
Fluorescence was measured by either a flow cytometer (Guava Easycyte Plus; Guava Technologies, Millipore, Brussels, Belgium) or a microplate reader (Fluoroskan Ascent FL apparatus; Thermo Life Sciences, Brussels, Belgium). For flow cytometry, cells were harvested and diluted in 10 mM phosphate buffer (pH 7.0) to obtain a concentration of 500 to 2,500 cells/μl. Subsequently, the mean green fluorescent protein (GFP) content of ca. 5,000 cells was determined using a Guava Easycyte Plus flow cytometer, using excitation and emission filters of 488 nm and 525 nm, respectively. For the microplate reader, 300 μl of a cell suspension was placed in a well of a 96-well microplate, and GFP fluorescence was recorded at 520 nm, using an excitation wavelength of 480 nm. Finally, the production of β-galactosidase was measured using the chromogenic substrate ortho-nitrophenyl-β-d-galactosidase (ONPG) as described by Miller (11).
RESULTS AND DISCUSSION
Both quorum sensing and low pH regulate acetoin production in S. plymuthica RVH1.
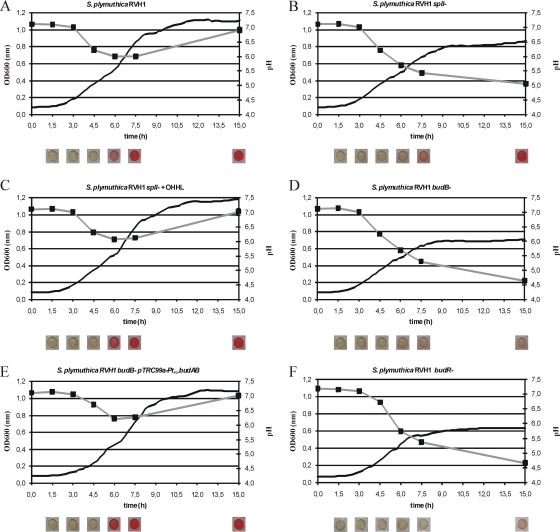
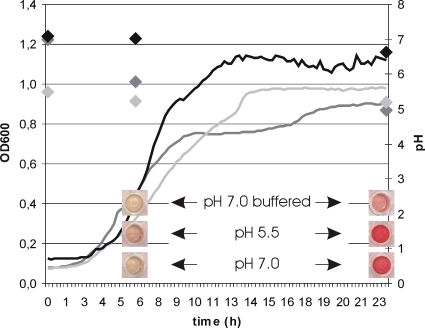
Following up on our previous discovery that an AHL-based QS system activates acetoin and 2,3-butanediol production in Serratia plymuthica RVH1 (24) (Fig. 1 A to C), we observed that a mutant deficient in the production of QS signaling molecules (RVH1 splI) still exhibited residual, though delayed, acetoin (i.e., precursor of 2,3-butanediol) production (Fig. 1B), indicating that QS does not constitute the only regulation of acetoin production in RVH1. Since the RVH splI mutant strongly acidified the medium, and since low pH was previously observed to affect acetoin production in Bacillus subtilis and Klebsiella terrigena (10, 12), we compared the effects of acidic and neutral environments on acetoin production in the RVH1 splI mutant (Fig. 2). As such, we could clearly see that for cultures starting growth under acidic conditions (pH 5.5), acetoin became detectable earlier and rose to higher levels than those for cultures starting at a pH of 7.0 (buffered or unbuffered). Moreover, despite the more or less constant pH of ca. 5.5, the cell density even exceeded that for the same strain starting growth at a nonbuffered pH of 7.0. Finally, while clearly attenuated, the residual acetoin production of RVH1 splI grown in LB-glucose buffered at pH 7.0 most likely stemmed from the slight drop in pH experienced by the culture as the buffering capacity was exceeded.
Fig. 1.
Growth curves (black lines), pH profiles (gray lines; black boxes represent measurement points), and acetoin production (images below each graph) at indicated time points for different S. plymuthica RVH1 mutants. Representative results of three independent experiments are shown.
Fig. 2.
Growth curves for an RVH1 splI mutant in LB containing 0.5% glucose with an initial pH of 7.0 (dark gray), 5.5 (light gray), or 7.0 buffered with 100 mM phosphate buffer (black). Measurements of pH (diamonds in corresponding colors) and acetoin production (images below each graph) are shown at the indicated time points. Representative results of three independent experiments are shown.
Together, these results demonstrate that acetoin production in S. plymuthica RVH1 is responsive to both pH and QS, a feature that was previously also established for Vibrio cholerae (8). In the latter, however, QS regulation is mechanistically different from that in S. plymuthica, since it consists of an AI-2 and CAI-1 signal cascade instead of an AHL system. This indicates that this dual regulation arose more than once during evolution, which advocates the apparent benefit of both cues providing input in the regulation of 2,3-butanediol production. Most likely, QS allows cells to anticipate and circumvent the potentially lethal acidification of a progressively densifying culture engaged in mixed acid fermentation by switching in a timely manner to 2,3-butanediol production.
Identification of the bud locus in S. plymuthica RVH1.
In order to further dissect the regulation of acetoin production at the genetic level, we searched for the corresponding structural genes by screening a random transposon knockout library of RVH1 for mutants unable to produce acetoin and counteract acidification during the fermentation of glucose. As such, a clone was obtained whose transposon insertion site was mapped to a gene homologous to an α-acetolactate synthase gene (designated budB), which allowed the subsequent identification of a neighboring LysR gene homologue (designated budR) and an α-acetolactate decarboxylase gene (designated budA). More specifically, the budA and budB genes form an operon separated by 382 bp from the upstream and divergently transcribed budR gene. The predicted gene products show a high degree of homology with the corresponding proteins of S. proteamaculans 568: 74% for BudR (LysR), 92% for BudA, and 92% for BudB.
Whereas growth of the RVH1 wild type on glucose was characterized by an initial pH decrease (from 7.1 to 6.0) followed by an increase (from 6.0 to 7.0) (Fig. 1A), the RVH1 budB mutant showed a continuous pH decrease (from 7.1 to 4.5) and concomitantly displayed hampered growth (Fig. 1D). Furthermore, while acetoin was already clearly detectable in the wild-type strain after 6 h of growth, its production was completely abolished in the budB mutant, which indicates that the isolated bud locus is absolutely required for acetoin production in RVH1. Finally, it should also be noted that acidification, loss of acetoin production, and impaired growth of the budB mutant could be reversed completely by constitutive expression of the budAB genes from a plasmid (Fig. 1E).
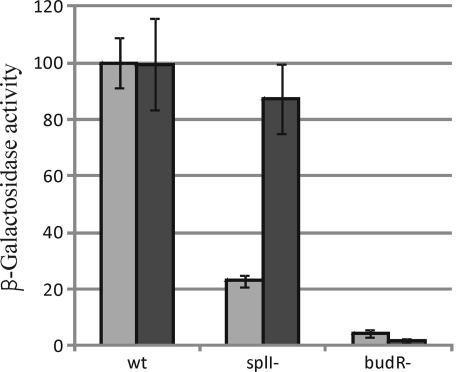
Interestingly, a de novo-constructed RVH1 budR mutant growing on glucose was phenotypically indistinguishable from a budB mutant and likewise failed to produce any acetoin (Fig. 1F). To confirm this at the genetic level, a chromosomal budAB-lacZ promoter fusion was constructed and validated to be responsive to quorum sensing (Fig. 3). Moreover, knocking out budR decreased expression levels of a chromosomal budAB-lacZ promoter fusion to circa 4% of those observed in the RVH1 wild type (Fig. 3). Together, these results indicate that BudR is an essential activator of the budAB operon, which is in agreement with BudR functionality in Bacillus subtilis, Klebsiella terrigena, and Vibrio cholerae (8, 10, 12).
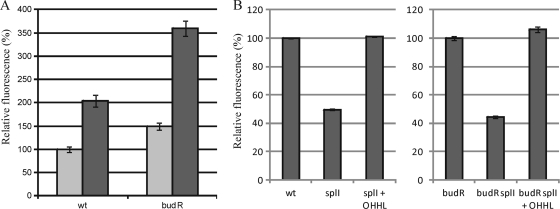
Fig. 3.
Expression of a chromosomal budA::lacZ promoter fusion in wild-type RVH1 (wt), RVH1 splI, and RVH1 budR grown in LB without (light gray) or with (dark gray) added OHHL. Stationary-phase cultures were diluted 1/10 and exposed for 4 h to the indicated conditions. β-Galactosidase activity was calculated in Miller units and expressed relative to the activity in wild-type RVH1 without OHHL, which was arbitrarily set at 100%.
Regulation of budR transcription in S. plymuthica RVH1.
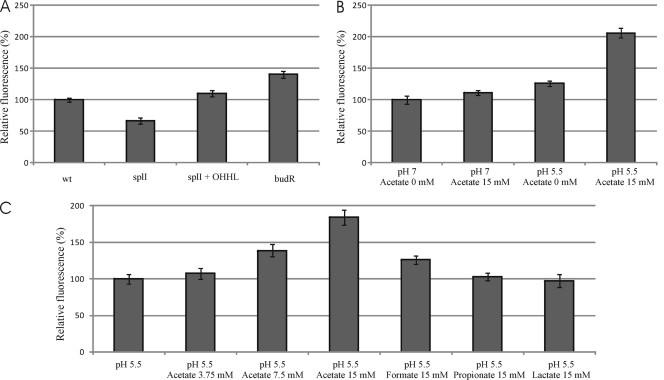
The abolition of acetoin production and budAB expression in the budR knockout strain also implied that the regulation of budAB by both QS and pH must be mediated by BudR. Therefore, we decided to construct a plasmid-borne budR promoter fusion to the gfp gene (i.e., PbudR-gfp) to examine the regulation of budR transcription more closely. As such, we observed that PbudR activity in an RVH1 splI mutant was considerably attenuated compared to that in the wild-type strain but could be rescued by the addition of OHHL signaling molecules (Fig. 4 A), clearly demonstrating that QS control of 2,3-butanediol production is genetically integrated at the PbudR promoter.
Fig. 4.
Relative expression of plasmid-borne PbudR-gfp in wild-type RVH1, RVH1 splI, RVH1 splI complemented with OHHL, and RVH1 budR grown in LB at pH 7.0 (A); in wild-type RVH1 grown in LB medium at pH 5.5 or 7.0 in the presence or absence of 15 mM acetate (B); and in RVH1 splI grown in LB medium at pH 5.5 in the presence of 0, 3.75, 7.5, or 15 mM acetate or in the presence of 15 mM formate, propionate, or lactate (C). Stationary-phase cultures were diluted 1/100 and exposed for 4 h to the indicated conditions before GFP expression was measured. In each graph, GFP expression of the control strain (i.e., first bar) was arbitrarily set at 100%. Average values and standard deviations for three independent experiments are shown.
In subsequently examining the effect of low pH on budR transcription, no significant difference in PbudR activity could be observed between wild-type RVH1 exposed to LB at pH 5.5 or pH 7.0 (Fig. 4B), indicating that it was not the effect of low pH itself that was conveyed to the budR promoter. However, when acetate was examined as a possible trigger, it emerged that PbudR activity could indeed be increased considerably in the presence of 15 mM acetate. However, this induction occurred only at a low pH (pH 5.5) and completely disappeared at neutral pH, suggesting that budR transcription is responsive to acetate only once its protonated and neutral form can enter the cell. Finally, while a clear dose-response relationship between acetate concentration and budR transcription could be established (Fig. 4C), limited or no induction could be obtained with formate, propionate, or lactate at pH 5.5 (Fig. 4C), although it should be noted that the dissociation constants of these molecules differ.
Interestingly, when the PbudR-gfp reporter was transformed into an RVH1 budR mutant, we observed higher PbudR activities than those in the corresponding wild-type strain (Fig. 4A), suggesting a feedback mechanism in which BudR represses its own transcription. This is in contrast to its role as an activator of the budAB operon but in agreement with previous observations in Klebsiella terrigena (10). In general, most lysR homologues seem to interfere with their own expression (6, 15), since they typically bind the promoter regions of the genes they regulate and thus obstruct access to their own neighboring and divergently oriented promoters.
Surprisingly, however, we observed that in an RVH1 budR mutant, budR transcription was still similarly responsive to acetate (at pH 5.5) and OHHL as that in the wild-type parent (Fig. 5 A and B), demonstrating that BudR itself is not required to convey these signals to the budR promoter. Accordingly, additional regulatory factors likely play an important role in directing budR transcription. This contrasts with currently forwarded hypotheses in which intracellular acetate (or one of its downstream metabolites) is assumed to associate with the BudR protein in order to activate it (8, 10, 12). Nevertheless, although many LysR homologues indeed appear to depend on inducer ligands (6, 9), direct binding of acetate to BudR has never been shown experimentally.
Fig. 5.
(A) Relative expression of plasmid-borne PbudR-gfp in wild-type RVH1 and RVH1 budR in LB medium at pH 7.0 (light gray) or pH 5.5 in the presence of 15 mM acetate (dark gray). Stationary-phase cultures were diluted 1/100 and exposed for 4 h to the indicated conditions before GFP expression was measured. (B) Relative expression of plasmid-borne PbudR-gfp in wild-type RVH1, RVH1 splI, and RVH1 splI complemented with OHHL (left) and in RVH1 budR, RVH1 splI budR, and RVH1 splI budR complemented with OHHL (right). Stationary-phase cultures were grown under the indicated conditions before GFP expression was measured. In each graph, GFP expression of the control strain (i.e., first bar) was arbitrarily set at 100%. Average values and standard deviations for three independent experiments are shown.
Conclusions.
In this study, we report on the identification of structural (budAB) and regulatory (budR) genes responsible for acetoin production in S. plymuthica RVH1. Moreover, we demonstrate that dual regulation of acetoin production by QS and intracellular acetate seems to be integrated in the expression of the budR gene, encoding the activator of the budAB operon. However, while BudR was shown to be an autorepressor, activation of budR transcription by QS and acetate proceeded independently of BudR.
ACKNOWLEDGMENTS
P.M. was funded by a Ph.D. grant of the Agency for Innovation by Science and Technology (IWT-Vlaanderen). R.V.H. worked on this project as a research assistant from the Research Foundation Flanders (FWO-Vlaanderen) and as a postdoctoral fellow from the K.U. Leuven Research Fund. A.A. acknowledges support from the Research Foundation Flanders (FWO-Vlaanderen; grant KAN2008 1.5.258.08).
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Altschul S., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amann E., Ochs B., Abel K. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315 [DOI] [PubMed] [Google Scholar]
- 3. Celińska E., Grajek W. 2009. Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnol. Adv. 27:715–725 [DOI] [PubMed] [Google Scholar]
- 4. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guyer M., Reed R., Steitz J., Low K. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135–140 [DOI] [PubMed] [Google Scholar]
- 6. Henikoff S., Haughn G., Calvo J., Wallace J. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. U. S. A. 85:6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kleckner N., Gottesman J. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139–180 [DOI] [PubMed] [Google Scholar]
- 8. Kovacikova G., Lin W., Skorupski K. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57:420–433 [DOI] [PubMed] [Google Scholar]
- 9. Maddocks S., Oyston P. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623 [DOI] [PubMed] [Google Scholar]
- 10. Mayer D., Schlensog V., Böck A. 1995. Identification of the transcriptional activator controlling the butanediol fermentation pathway in Klebsiella terrigena. J. Bacteriol. 177:5261–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 12. Renna M., Najimudin N., Winik L., Zahler S. 1993. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rubirés X., et al. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 15. Schell M. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597–626 [DOI] [PubMed] [Google Scholar]
- 16. Simon R., Priefer U., Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 17. Syu M. 2001. Biological production of 2,3-butanediol. Appl. Microbiol. Biotechnol. 55:10–18 [DOI] [PubMed] [Google Scholar]
- 18. Taghavi S., et al. 2009. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75:748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valdivia R., Falkow S. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367–378 [DOI] [PubMed] [Google Scholar]
- 20. Van Houdt R., Aertsen A., Jansen A., Quintana A. L., Michiels C. W. 2004. Biofilm formation and cell-to-cell signaling in Gram-negative bacteria isolated from a food processing environment. J. Appl. Microbiol. 96:177–184 [DOI] [PubMed] [Google Scholar]
- 21. Van Houdt R., Aertsen A., Michiels C. W. 2007. Quorum-sensing-dependent switch to butanediol fermentation prevents lethal medium acidification in Aeromonas hydrophila AH-1N. Res. Microbiol. 158:379–385 [DOI] [PubMed] [Google Scholar]
- 22. Van Houdt R., et al. 2007. Characterization of a luxI/luxR-type quorum sensing system and N-acyl-homoserine lactone-dependent regulation of exo-enzyme and antibacterial component production in Serratia plymuthica RVH. Res. Microbiol. 158:150–158 [DOI] [PubMed] [Google Scholar]
- 23. Van Houdt R., Moons P., Jansen A., Vanoirbeek K., Michiels C. W. 2005. Genotypic and phenotypic characterisation of a biofilm-forming Serratia plymuthica isolate from a raw vegetable processing line. FEMS Microbiol. Lett. 246:265–272 [DOI] [PubMed] [Google Scholar]
- 24. Van Houdt R., Moons P., Hueso Buj M., Michiels C. W. 2006. N-Acyl-l-homoserine lactone quorum sensing controls butanediol fermentation in Serratia plymuthica RVH1 and Serratia marcescens MG1. J. Bacteriol. 188:4570–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao Z., Xu P. 2007. Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33:127–140 [DOI] [PubMed] [Google Scholar]
- 26. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]