Abstract
Staphylococcus aureus is a major bovine mastitis pathogen. Although the reported antimicrobial resistance was generally low, the emergence of new genetic clusters in bovine mastitis requires examination of the link between antimicrobial resistance and genotypes. Here, amplified fragment length polymorphism (AFLP) profiles and standard antimicrobial resistance profiles were determined in order to characterize a total of 343 S. aureus cow mastitis isolates from two geographically close regions of Switzerland and France. AFLP profiles revealed similar population compositions in the two regions, with 4 major clusters (C8, C20, C97, and C151), but the proportions of isolates in each cluster significantly diverged between the two countries (P = 9.2 × 10−9). Antimicrobial resistance was overall low (<5% resistance to all therapeutically relevant molecules), with the exception of penicillin resistance, which was detected in 26% of the isolates. Penicillin resistance proportions differed between clusters, with only 1 to 2% of resistance associated with C20 and C151 and up to 70% associated with bovine C97. The prevalence of C20 and C8 was unexpectedly high and requires further investigation into the mechanism of adaptation to the bovine host. The strong association of penicillin resistance with few clusters highlights the fact that the knowledge of local epidemiology is essential for rational choices of antimicrobial treatment in the absence of susceptibility testing. Taken together, these observations argue in favor of more routine scrutiny of antimicrobial resistance and antibiotic-resistant clones in cattle and the farm environment.
INTRODUCTION
Staphylococcus aureus is a colonizer and opportunistic pathogen of humans and numerous animal species. In cattle, it is responsible for about one-third of the cases of clinical and subclinical mastitis (2, 3), a disease causing major economic loss in the dairy industry worldwide (26). It is also known to be contagious, spreading in a farm from cow to cow during the milking process by contaminated machines, equipments, or milkers' hands. The presence of such pathogens in food-producing animals or in raw milk raises the question of the potential transmissibility of specific resistant clones or antimicrobial resistance determinants, as well as the possible routes of transmission from the animal to humans or vice versa (5, 7, 10).
Over the past decade, several molecular studies have described the population structure and diversity of S. aureus causing bovine mastitis (9, 12–14, 29). In general, little overlap was found between the compositions of S. aureus populations of human and animal origin. However, the various genotyping methods that are used, such as pulsed-field gel electrophoresis (PFGE), multilocus sequence type (MLST), amplified fragment length polymorphism (AFLP), and spa typing, assess different characteristics of the bacterial genome, which may result in different strain clustering and genealogies that are difficult to compare (9, 14). Likewise, it was initially proposed that a relatively small number of broadly distributed S. aureus genotypes were responsible for the majority of cases of bovine mastitis (10, 28). More recently, however, the increasing number of studies from diverse locations found a number of only locally prevalent clusters in addition to globally present genotypes (13, 18).
The antimicrobial resistance profiles are part of the epidemiological knowledge that constitutes key information for decisions on therapeutic strategies. While low incidences of resistance among S. aureus from bovine mastitis were generally reported (1, 2, 16), multiresistant phenotypes and methicillin-resistant S. aureus (MRSA) have increasingly been described recently (6, 31). However, apart from these sporadic reports on MRSA, which were of sequence type (ST) 398, data on the link between the genetic background and antimicrobial resistance in cattle are scarce.
The present study attempted to better understand the ecology of bovine mastitis S. aureus isolates in two neighboring Swiss and French regions, along the Rhône-Alpes valley. A collection of 343 epidemiologically distinct S. aureus isolates from suspected mastitis was sampled without biases related to disease severity or treatment failure. The collection of isolates recovered from both geographical areas was analyzed for its population structure and antibiotic resistance phenotypes, and links between genotypes and antibiotic resistance phenotypes were determined.
MATERIALS AND METHODS
Sample collection.
A total of 343 S. aureus isolates originated from 142 Swiss and 138 French farms, respectively. Isolates from milk samples received by diagnostic veterinary laboratories where the cases of suspected subclinical and clinical bovine mastitis are investigated were collected for this study. The collection area encompassed the Rhône-Alpes region (from western Switzerland to eastern France). The two regions are separated by a distance of ca. 150 kilometers. The Swiss isolates were collected between June 2007 and July 2009 by the Institut Galli Valério in Lausanne (O. Sakwinska, M. Giddey, M. Moreillon, D. Morisset, A. Waldvogel, and P. Moreillon, submitted for publication), and the French ones between January 2007 and October 2009 by the Food Safety Agency in Lyon. One isolate of a distinct AFLP type per farm was selected for the present study: 193 Swiss and 150 French isolates were included. All isolates originated from milk samples and were identified according to standard methods as described elsewhere (15).
Antimicrobial susceptibility testing.
Bacteria were grown 18 to 24 h on a nonselective medium and tested for their antimicrobial susceptibility using the disk diffusion method on Mueller-Hinton agar (Bio-Rad, Marnes la Coquette, France) as recommended by the Antibiogram Committee of the French Society of Microbiology (CA-SFM) (4). The antimicrobial agents tested (penicillin, cefoxitin, kanamycin, gentamicin, erythromycin, spiramycin, lincomycin, pristinamycin, vancomycin, teicoplanin, fusidic acid, trimethoprim-sulfamethoxazole, rifampin, linezolid, and enrofloxacin) were chosen according to their medical or veterinary interest. Results were classified as susceptible, intermediate, or resistant, according to the approved clinical breakpoints. S. aureus ATCC 25923 was used as a quality control. PCR screening for an internal fragment of the mecA gene was systematically performed using the primers mecA_Sa_fw_865 (5′-AAA AAG CTC CAA CAT GAA GA-3′) and mecA_Sa_rv_1211 (5′-GTT GAA CCT GGT GAA GTT GT-3′).
Genotyping.
AFLP genotyping was performed as previously described (23), except that only one set of selective primers was used (Csp6I with a G extension and MboI with a G extension). In brief, approximately 100 to 150 ng of chromosomal DNA was digested with the restriction enzymes MboI and Csp61 (Fermentas). Oligonucleotide linkers for both enzymes were ligated to the digestion mixture, and a nonselective round of amplification was performed, followed by selective amplification with the above-mentioned primers. The repeat region of the spa gene was amplified according to previously described methods (8, 27), and spa types were assigned with an online spa database (http://www.spaserver.ridom.de/).
Data analysis.
Electropherograms of AFLP patterns were analyzed with GeneMapper software (Applied Biosystems) as previously described (23). Each AFLP marker was scored as either 0 or 1. Clustering was performed with Bayesian phylogeny using MrBayes (21). Swiss strains have been assigned previously to AFLP clusters equivalent to MLST clonal complexes (Sakwinska et al., submitted). Clusters were numbered according to corresponding MLST clonal complexes (http://eburst.mlst.net). Statistics were performed using chi-square (χ2) tests, with Yates' continuity correction when needed, using the free R, version 11.1, software (19).
RESULTS
Population structure.
According to AFLP analysis, the 343 isolates were grouped into 19 clusters, among which 17 could be readily identified (Table 1). One spa-type t3750 and three t3576 isolates could not be assigned cluster numbers since they did not cluster with an isolate of a known MLST. Two isolates (one French and one Swiss) were nontypeable by AFLP (abnormal chromatograms and a very high run-to-run variability) but could be grouped with the C8 and C97 clusters based on their spa types (t2953 and t524, respectively).
Table 1.
Diversity of spa types among the different clusters
| Cluster | spa type | spa repeats | No. of Swiss isolates | No. of French isolates |
|---|---|---|---|---|
| 1 | t127 | 07-23-21-16-34-33-13 | 3 | 0 |
| 5 | t002 | 26-23-17-34-17-20-17-12-17-16 | 0 | 1 |
| t311 | 26-23-17-34-20-17-12-17-16 | 0 | 1 | |
| 7 | t091 | 07-23-21-17-34-12-23-02-12-23 | 2 | 0 |
| 8 | t008 | 11-19-12-21-17-34-24-34-22-25 | 0 | 3 |
| t024 | 11-12-21-17-34-24-34-22-25 | 6 | 3 | |
| t711 | 04-21-17-34-24-34-22-25 | 11 | 0 | |
| t2953 | 11-12-21-17-34-24-34-22-25-25 | 27 | 13 | |
| t3802 | 04-21-17-34-24-34-22-25-25 | 0 | 2 | |
| t5268 | 11-21-17-34-24-34-22-25-25 | 7 | 0 | |
| t5270 | 11-12-21-17-34-24-34-22-25-25-25 | 3 | 0 | |
| t5271 | 11-17-34-24-34-22-25-25 | 6 | 0 | |
| t5694 | 11-12-17-34-24-34-22-25-25 | 7 | 0 | |
| t6281 | 04-21-17-34-24-34-22-25-25-75 | 2 | 0 | |
| Unknown | 11-17-34-24-34-22-25-25-75 | 0 | 1 | |
| 15 | t084 | 07-23-12-34-34-12-12-23-02-12-23 | 2 | 0 |
| 20 | t164 | 07-06-17-21-34-34-22-34 | 15 | 53 |
| t195 | 07-06-17-21-34-34 | 0 | 1 | |
| t881 | 07-06-17-34-34-22-34 | 0 | 1 | |
| t1544 | 07-22-34 | 1 | 2 | |
| t1987 | 07-06-17-21-34 | 0 | 1 | |
| t2094 | 26-06-17-21-34-34-22-34 | 10 | 2 | |
| t3277 | 07-17-21-34-34-22-34 | 0 | 1 | |
| t5993 | 15-34-34-22-34 | 0 | 1 | |
| Unknown | 07-06-17-21-34-34-02-34 | 0 | 1 | |
| 25 | t078 | 04-21-12-41-20-17-12-12-17 | 0 | 1 |
| 45 | t015 | 08-16-02-16-34-13-17-34-16-34 | 1 | 0 |
| 72 | t148 | 07-23-12-21-12-17-20-17-12-12-17 | 0 | 1 |
| 97 | t267 | 07-23-12-21-17-34-34-34-33-34 | 0 | 3 |
| t524 | 04-17 | 19 | 17 | |
| t528 | 4 | 0 | 3 | |
| t3992 | 26-23-12-21-17-34-34-33-34 | 0 | 1 | |
| t5695 | 07-23-12-21-17-34-34-34-34-34 | 2 | 0 | |
| t6280 | 26-23-12-21-17-34-34-34-34-34 | 1 | 0 | |
| Unknown | 07-23-12-21-17-34-34-34-34-34-34 | 0 | 2 | |
| 101 | t1312 | 04-20-12-17-17 | 1 | 0 |
| 121 | t645 | 14-44-13-12-17-23-18-17 | 0 | 1 |
| t1773 | 04-82-17-25-17 | 0 | 1 | |
| 151 | t529 | 04-34 | 60 | 25 |
| 188 | t189 | 07-23-12-21-17-34 | 1 | 0 |
| 398 MRSA | t011 | 08-16-02-25-34-24-25 | 3 | 0 |
| 398 MSSA | t011 | 08-16-02-25-34-24-25 | 0 | 1 |
| t034 | 08-16-02-25-02-25-34-24-25 | 0 | 1 | |
| Unknown | 08-16-02-25-02-25-34-24-25-25 | 0 | 1 | |
| 479 | t453 | 04-20-17 | 2 | 0 |
| New1 | t3750 | 04-20-25-23-24-17 | 0 | 1 |
| New2 | t3576 | 04-31-17-25-17 | 0 | 3 |
Figure 1 shows that the majority of the Swiss (91.7%) and the French isolates (90.7%) were concentrated in only four different clusters, including C8, C20, C97, and C151. However, the proportion of the isolates in each of these clusters was significantly different between the two countries (P = 9.2 × 10−9). Cluster 8, which has only recently been described for bovine mastitis in Switzerland (Sakwinska et al., submitted), was less frequent in France (14.7% versus 35.8% in Switzerland, P = 2.1 × 10−5). Cluster 151 was also significantly less frequent in France than in Switzerland (16.7% versus 31.1%, P = 0.003). Conversely, cluster 20 was more prevalent in France (42% versus 13.5%, P = 2.1 × 10−5), whereas cluster 97 was equally frequent in both countries (17.3% versus 11.4%, P = 0.16). Minor clusters represented strains that are usually found in humans (clusters 5 or 45) or in swine (cluster 398) (Table 1).
Fig. 1.
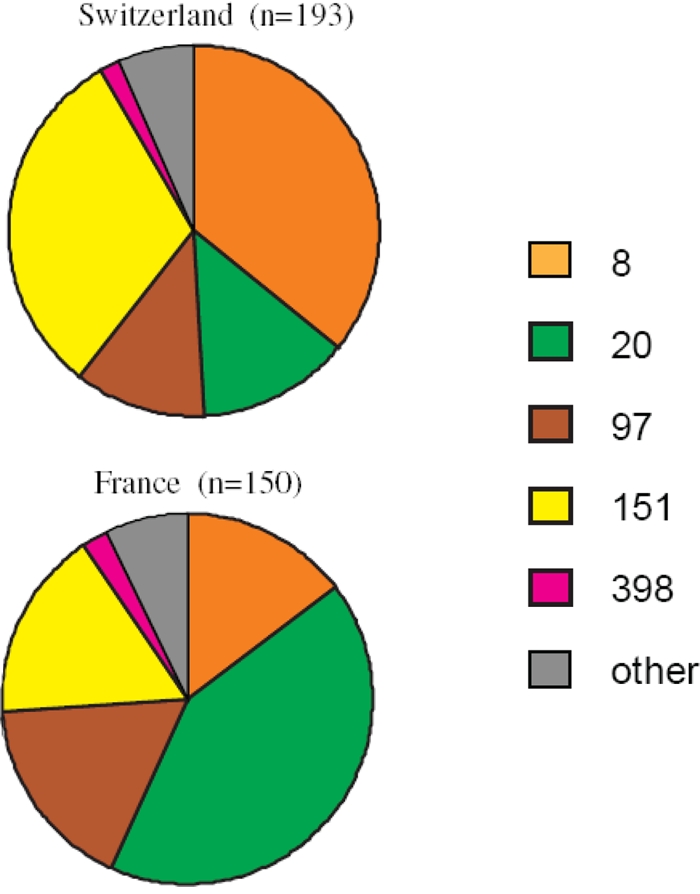
Genotype composition, according to clusters, of S. aureus isolated from bovine mastitis.
spa typing revealed finer-scale differences in the population compositions between the two studied regions (Table 1). A total of 49 different spa types were identified, but only 7/49 (14.3%) were found in both countries, and these were exclusively distributed in the four major types, C8, C20, C97, and C151. This indicates that these major clusters encompassed clones that were shared by both countries, whereas the strain diversity was due to minor clusters.
Yet this clone sharing was not absolute. For example, C8 encompassed 11 spa types, of which only t2953 and t024 were found among both Swiss and French isolates. In particular, t2953 was present in 27/69 (39%) of the Swiss strains and 13/21 (62%) of the French strains, making it a prevalent bilateral clone. In contrast, t024 had a low prevalence (<10%) in both areas. Likewise, C20 encompassed 8 spa types, of which only t164 and t2094 were found in both countries. Indeed, t164 was found in 15/26 (57%) of Swiss isolates and 53/82 (85%) of French isolates, whereas t2094 was found in 10/26 (38%) of Swiss isolates but only 2/62 (0.3%) of French isolates. Eventually, C97 segregated in 6 spa types, of which only t524 was shared, and was present in 19/22 or 86% of Swiss isolates and 17/24 or 71% of French isolates, thus representing a major strain in both places. On the other hand, C151 clustered in only a single spa type (t529) present in both areas, making it a uniquely shared strain.
Thus, in addition to diversity, substantial shared clonality existed between both areas. Indeed, taking these together, 137/193 (71%) Swiss isolates had identical counterparts (in terms of cluster and spa types) in French isolates, and 113/150 (75%) French isolates had identical counterparts in Swiss isolates.
Antimicrobial resistance.
Resistance to non-beta-lactam molecules, such as tetracyclines, macrolides, and aminoglycosides, was quasi-anecdotic (Table 2). Only penicillin resistance was consistently detected in 88/343 (25.7%) total tested isolates and in proportions that did not significantly differ between Switzerland (43/193, 22%; including 3 MRSA) and France (45/150, 30%) (P = 0.1). However, proportions of penicillin-resistant isolates differed greatly and significantly among the clusters (P = 10−6) (Table 3). Indeed, only 2/89 (2.2%) C20 and 1/85 (1.2%) C151 isolates were penicillin resistant. In contrast, 33/91 (36.3%) C8 and 34/48 (70.8%) C97 isolates were penicillin resistant. Moreover, in both C8 and C97, all the resistant strains belonged to the unique spa type shared between Swiss and French isolates, i.e., t2953 for C8 and t524 for C97 (Table 2). Thus, 20/40 (50%) isolates of spa-type t2953 (C8) and 29/36 (80%) isolates of spa-type t524 (C97) were penicillin resistant.
Table 2.
Antimicrobial resistance frequencies of Swiss and French S. aureus isolates
| Antimicrobial agents | Switzerland (n = 193) |
France (n = 150) |
||
|---|---|---|---|---|
| No. of isolates | % | No. of isolates | % | |
| Penicillin | 43 | 22.3 | 45 | 30.0 |
| Cefoxitin | 3 | 1.6 | 0 | 0.0 |
| Kanamycin | 2 | 1.0 | 0 | 0.0 |
| Gentamicin | 2 | 1.0 | 0 | 0.0 |
| Erythromycin | 1 | 0.5 | 4 | 2.7 |
| Lincomycin | 1 | 0.5 | 1 | 0.7 |
| Spiramycin | 1 | 0.5 | 1 | 0.7 |
| Pristinamycin | 0 | 0.0 | 0 | 0.0 |
| Tetracycline | 6 | 3.1 | 8 | 5.3 |
| Vancomycin | 0 | 0.0 | 0 | 0.0 |
| Teicoplanin | 0 | 0.0 | 0 | 0.0 |
| Fusidic acid | 0 | 0.0 | 1 | 0.7 |
| Bactrim | 0 | 0.0 | 0 | 0.0 |
| Rifampin | 0 | 0.0 | 0 | 0.0 |
| Linezolid | 0 | 0.0 | 0 | 0.0 |
| Enrofloxacin | 0 | 0.0 | 0 | 0.0 |
Table 3.
Distribution of penicillin resistance among different clusters
| Cluster | No. of isolates | Penicillin-resistant strains |
|
|---|---|---|---|
| No. | % | ||
| 8 | 91 | 33 | 36.3 |
| 20 | 89 | 2 | 2.2 |
| 97 | 48 | 34 | 70.8 |
| 151 | 85 | 1 | 1.2 |
| Others | 30 | 19 | 60.0 |
| Total | 343 | 88 | 25.7 |
Another cluster with increased antimicrobial resistance was C398. Three Swiss C398 isolates were MRSA (24), while isolates from France were all methicillin-sensitive S. aureus (MSSA) (Table 4). All six isolates were resistant to more than one antimicrobial agent, including beta-lactams and tetracycline. One MRSA isolate displayed additional resistance to kanamycin and gentamicin, and another carried resistance to macrolides and lincosamides.
Table 4.
Cluster 398: resistance and spa types
| Country | Genotype | spa type | Associated resistance(s)a |
|---|---|---|---|
| France | MSSA | t011 | Tet |
| MSSA | t034 | Tet | |
| MSSA | Unknown | Tet | |
| Switzerland | MRSA | t011 | Tet |
| MRSA | t011 | Tet, Kan, Gen | |
| MRSA | t011 | Tet, Ery, Lin, Sp |
Tet, tetracycline; Kan, kanamycin; Gen, gentamicin; Ery, erythromycin; Lin, lincomycin; Sp, spiramycin.
DISCUSSION
We characterized the AFLP types and antimicrobial resistance phenotypes of 343 distinct S. aureus isolates from bovine mastitis of two Swiss and French regions along the Rhône valley. Despite the diversity of spa types, 90% of all isolates belonged to only 4 major clusters, i.e., C8, C20, C97, and C151. The presence of C97 and C151 was congruent with the findings of recent publications, which reported them as the most predominant clusters associated with bovine mastitis in a number of countries worldwide (9, 16, 28). C20 was described for human carriage (22) and infection (33), as well as for bovine mastitis in southern Germany and Japan (10, 16). C8 is typically a human genotype which gave rise to prominent MRSA (20), but it has only sporadically been reported for animals, including horses in Germany (MRSA ST254) (32) and bovine mastitis in the Netherlands (ST8, 8% of isolates) (30). In Switzerland, where C8 was predominant in bovine mastitis, it was not detected in farmers' colonization, suggesting that it is indeed fully capable of transmission among cattle (Sakwinska et al., submitted).
The overall population compositions of S. aureus from the two geographic regions were very similar regarding the detected clones. However, the proportions of these clones significantly diverged, which might reflect the evolution and dissemination of clones due to both therapeutic practices and animal movements. The remarkable predominance of C20 in France cannot be easily linked to the presence of this cluster in human colonization; indeed, a recent study did not detect this or similar genotypes in human colonization in Western Europe, including France (22). Moreover, spa typing revealed that although the 4 major clusters were shared, 3 of them (i.e., clusters 8, 20, and 97) were composed of several subtypes that were different between the two places. This might indicate that clones that were originally identical at both locations are now evolving independently.
From the antimicrobial resistance standpoint, proportions and specific drug resistances were low (<5% for most drugs) and similar in both countries. The only exception was resistance to first-line-treatment penicillin G, which reached 23.3% and 30% in Switzerland and in France, respectively. Although this is still far from the ca. 80% level of penicillin-resistant S. aureus in humans (17), it is noteworthy that penicillin resistance in mastitis isolates was highly clonal. Indeed, up to 80% of resistant isolates concentrated in two particular spa types of cluster 8 (i.e., t2953) and cluster 97 (t524). This finding is far from trivial in terms of therapeutic implications and also not restricted to this geographical region (30). Indeed, S. aureus responsible for bovine mastitis is known to be frequently contagious in a given herd, and thus, several animals in the same farm have shared the same clone (28). As a result, penicillin treatment failing in one animal infected with a penicillin-resistant clone is likely to fail in all the other infected animals from the same farm. In contrast, it might not fail in neighboring farms, where cows are colonized with penicillin-susceptible clones. Therefore, since antimicrobial susceptibility tests are not systematically recommended for bovine mastitis in a number of European countries (11), the circulation of several clones presenting diverging resistance patterns might generate a variety of therapeutic attitudes, as already reported (25). If penicillin resistance continues to increase, routine antimicrobial susceptibility tests may become a necessity. In parallel, well-designed studies and analyses are necessary for a more profound understanding of the local epidemiology, in order to promote coordinated therapeutic practices.
Only 3 MRSA ST398 isolates were detected in Switzerland, but the likely transmission source was identified in only one case (Sakwinska et al., submitted). In parallel, gentamicin-kanamycin resistance arose twice in Switzerland (never in France), whereas erythromycin resistance was detected four times in France (and once in a Swiss ST398 isolate). Although the origin of these rare cases remains unknown, they interestingly correlated with therapeutic usage, since aminoglycosides are prescribed as second-line antimicrobial agents in Switzerland and macrolides in France. Other resistances can be considered negligible, since they are only sporadically encountered.
In conclusion, there are four major bovine mastitis clusters of S. aureus present in various proportions in two contiguous regions of Switzerland and France. The predominance of C97 and C151 was expected, as these clusters are known to be cow adapted. On the other hand, the presence of C20 and C8, their mechanisms of adaptation to animals, and their transmission require further studies. In addition, one salient observation of this study is the low prevalence of antimicrobial resistance in the two Swiss and French strain collections. This is of good promise at a time when agriculture is denounced as a major culprit for promoting antimicrobial resistance selection (5). However, the results also highlight that resistance to first-line penicillin is concentrated in two predominant clones which, if they disseminate successfully, might render this molecule inactive. Surveillance of such clones is important and argues in favor of more routine drug resistance testing to give accurate treatment options to veterinarians. This should be associated with more research on molecular types for better knowledge of both local and global epidemiologies of S. aureus clones.
ACKNOWLEDGMENTS
This work was supported by grant 32003B-113854 to P.M. from the Swiss National Science Foundation and by the French Food Safety Agency (Anses).
We thank Marlyse Giddey and Martine Moreillon for technical support and sample collection. We also thank all the French department laboratories of the Resapath network that participated in this study. We are particularly grateful to Emilie Gay, who kindly performed the statistical analyses.
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Bengtsson B., et al. 2009. Antimicrobial susceptibility of udder pathogens from cases of acute clinical mastitis in dairy cows. Vet. Microbiol. 136:142–149 [DOI] [PubMed] [Google Scholar]
- 2. Botrel M. A., et al. 2010. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhone-Alpes, France. Foodborne Pathog. Dis. 7:479–487 [DOI] [PubMed] [Google Scholar]
- 3. Bradley A. J., Leach K. A., Breen J. E., Green L. E., Green M. J. 2007. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet. Rec. 160:253–258 [DOI] [PubMed] [Google Scholar]
- 4. CA-SFM 2010. Recommandations 2009 du Comité de l'Antibiogramme de la Société Française de Microbiologie, p. 1–49 Comité de l'Antibiogramme de la Société Française de Microbiologie, Paris, France [Google Scholar]
- 5. Cassone M., Giordano A. 2009. Resistance genes traveling the microbial internet: down the drain, up the food chain? Expert Rev. Anti Infect. Ther. 7:637–639 [DOI] [PubMed] [Google Scholar]
- 6. Fessler A., et al. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619–625 [DOI] [PubMed] [Google Scholar]
- 7. Haenni M., et al. 2011. Staphylococcal bovine mastitis in France: enterotoxins, resistance and the human Geraldine methicillin-resistant Staphylococcus aureus clone. J. Antimicrob. Chemother. 66:216–218 [DOI] [PubMed] [Google Scholar]
- 8. Harmsen D., et al. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hasman H., et al. 2010. spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet. Microbiol. 141:326–331 [DOI] [PubMed] [Google Scholar]
- 10. Hata E., et al. 2010. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J. Clin. Microbiol. 48:2130–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoe F. G., Ruegg P. L. 2005. Relationship between antimicrobial susceptibility of clinical mastitis pathogens and treatment outcome in cows. J. Am. Vet. Med. Assoc. 227:1461–1468 [DOI] [PubMed] [Google Scholar]
- 12. Ikawaty R., et al. 2009. Characterization of Dutch Staphylococcus aureus from bovine mastitis using a Multiple Locus Variable Number Tandem Repeat Analysis. Vet. Microbiol. 136:277–284 [DOI] [PubMed] [Google Scholar]
- 13. Jørgensen H. J., Mork T., Caugant D. A., Kearns A., Rorvik L. M. 2005. Genetic variation among Staphylococcus aureus strains from Norwegian bulk milk. Appl. Environ. Microbiol. 71:8352–8361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kapur V., Sischo W. M., Greer R. S., Whittam T. S., Musser J. M. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maes N., Magdalena J., Rottiers S., De Gheldre Y., Struelens M. J. 2002. Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures. J. Clin. Microbiol. 40:1514–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monecke S., Kuhnert P., Hotzel H., Slickers P., Ehricht R. 2007. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet. Microbiol. 125:128–140 [DOI] [PubMed] [Google Scholar]
- 17. Nimmo G. R., et al. 2003. Antimicrobial resistance in Staphylococcus aureus in Australian teaching hospitals, 1989–1999. Microb. Drug Resist. 9:155–160 [DOI] [PubMed] [Google Scholar]
- 18. Rabello R. F., et al. 2007. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 56:1505–1511 [DOI] [PubMed] [Google Scholar]
- 19. R Development Core Team 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 20. Robinson D. A., Enright M. C. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 22. Ruimy R., et al. 2009. Comparisons between geographically diverse samples of carried Staphylococcus aureus. J. Bacteriol. 191:5577–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sakwinska O., et al. 2009. Genetic diversity and ecological success of Staphylococcus aureus strains colonizing humans. Appl. Environ. Microbiol. 75:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakwinska O., Waldvogel A., Giddey M., Moreillon M., Moreillon P. 2008. Farmers frequently acquire Staphylococcus aureus from cow mastitis, abstr. C2-1828, p. 445 Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC), American Society for Microbiology, Washington, DC [Google Scholar]
- 25. Sawant A. A., Sordillo L. M., Jayarao B. M. 2005. A survey on antibiotic usage in dairy herds in Pennsylvania. J. Dairy Sci. 88:2991–2999 [DOI] [PubMed] [Google Scholar]
- 26. Seegers H., Fourichon C., Beaudeau F. 2003. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 34:475–491 [DOI] [PubMed] [Google Scholar]
- 27. Shopsin B., et al. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith E. M., et al. 2005. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol. 43:4737–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smyth D. S., et al. 2009. Molecular genetic typing reveals further insights into the diversity of animal-associated Staphylococcus aureus. J. Med. Microbiol. 58:1343–1353 [DOI] [PubMed] [Google Scholar]
- 30. van den Borne B. H., et al. 2010. Host adaptation of bovine Staphylococcus aureus seems associated with bacteriological cure after lactational antimicrobial treatment. J. Dairy Sci. 93:2550–2558 [DOI] [PubMed] [Google Scholar]
- 31. Vanderhaeghen W., et al. 2010. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet. Microbiol. 144:166–171 [DOI] [PubMed] [Google Scholar]
- 32. Walther B., et al. 2009. Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 47:704–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yao D., et al. 2010. Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections (SSTIs). BMC Infect. Dis. 10:133. [DOI] [PMC free article] [PubMed] [Google Scholar]


