Abstract
We applied molecular, microscopic, and culture techniques to characterize the microbial communities in snow and air at remote sites in the Canadian High Arctic (Ward Hunt Island, Ellesmere Island, and Cornwallis Island, latitudes 74 to 83oN). Members of the Bacteria and Eukarya were prevalent in the snow, and their small subunit (SSU) rRNA gene signatures indicated strong local aerial transport within the region over the preceding 8 months of winter snowpack accumulation. Many of the operational taxonomic units (OTUs) were similar to previously reported SSU rRNA gene sequences from the Arctic Ocean, suggesting the importance of local aerial transport processes for marine microbiota. More than 47% of the cyanobacterial OTUs in the snow have been previously found in microbial mats in the region, indicating that this group was also substantially derived from local sources. Viable cyanobacteria isolated from the snow indicated free exchange between the snow and adjacent mat communities. Other sequences were most similar to those found outside the Canadian Arctic but were from snow, lake and sea ice, glaciers and permafrost, alpine regions, Antarctica, and other regions of the Arctic, supporting the concept of global distribution of microbial ecotypes throughout the cold biosphere.
INTRODUCTION
As a result of their microscopic size, resistance to environmental extremes, and large populations, free-living microbes have high dispersal rates that favor cosmopolitanism (19). According to a longstanding conjecture (6), the same microbial species will occupy similar environments throughout the biosphere because there are no effective barriers to their planetary dispersal. Several studies based on morphological and small subunit (SSU) rRNA gene sequence analyses have suggested that microbial genera are globally distributed (17, 29, 50, 55). However, other recent studies that have examined specific groups in detail indicate geographical patterning of microbial communities (36, 44, 66). Although the differing levels of taxonomic resolution from different markers have clouded the interpretation of global distribution patterns, SSU rRNA gene sequences are the most commonly available and are useful in comparing geographic and environmental distribution patterns (29, 35).
Microbial transport occurs via flowing rivers, percolating groundwater, thermohaline and surface oceanic circulation, convection currents and wind in the atmosphere, animal migration, and anthropogenic activities. Aerial transport has long been viewed as a major transport route enabling microbes to colonize remote habitats. Spore formers, such as Gram-positive bacteria and fungi, in particular, should be able to survive long-range transport. In remote sites under background atmospheric conditions, bacterial and fungal cell concentrations can reach 104 and 103 cells/m3, respectively (5). The presence of microbes in the atmosphere varies temporally, with higher concentrations during the summer months, and spatially, with higher concentrations in urban areas than in rural areas. Viable microorganisms have been found in the stratosphere, at altitudes as high as 77 km (24, 65). The relative contribution of bacteria compared to those of fungi and pollen grains increases with altitude, likely due to their smaller cell size and consequent lower sinking velocity (5). This suggests that bacteria are better dispersed than eukaryotes in the air since they remain for longer periods of time in the atmosphere. In addition to implications for dispersal, microbes in the atmosphere are potentially involved in cloud chemistry due to ice nucleation activity of some species and their potential to grow on simple compounds present in cloud droplets (9, 15, 52).
The cold biosphere, defined as the ensemble of habitats over planet Earth that experience prolonged cold and freezing, acts as a severe ecological filter for all immigrant and resident organisms. These ecosystems are mostly microbial and are dominated by cold-adapted (psychrophilic) and cold-tolerant (psychrotolerant) microbes. Surveys of glaciers, snow, lake ice, sea ice, and atmospheric clouds have revealed the recurrent presence of a number of bacterial phyla: Bacteriodetes (previously referred to as the Cytophaga-Flavobacteria-Bacteroides or CFB cluster), Actinobacteria, Firmicutes, and Proteobacteria, including representatives from the Alpha-, Beta-, and Gammaproteobacteria classes (4). The cryosphere bacteria must contend with a severe combination of environmental stresses that include low nutrient concentrations, high solar UV radiation, freeze/thaw cycles, and limited liquid water. The bacterial activity reported in supercooled cloud droplets (52) suggests that cold-dwelling microbes are better adapted to atmospheric transport than other microbes, and the cryosphere therefore provides an attractive environment for evaluating microbial dispersal and biogeography. However, environmental filters take time to select adapted communities, and in the poor growth environment of the snow, more local transport processes would favor organisms already abundant in the local environment and would mask the signal from global cryosphere microbes. There are three major biomes in the coastal Arctic that could contribute to local inocula in the snow: (i) cyanobacterial mats characteristic of Arctic freshwater environments, (ii) marine microbes from the adjacent sea, and (iii) halotolerant sea ice flora of the Arctic Ocean. Given the distinct phylogenetic signatures of most freshwater and marine microorganisms (23, 33, 34), marine microbes in snow would indicate local transport.
The objectives of this research were to investigate the diversity and likely source environments of microbes in the snow and air in the High Arctic Canada. We sampled two polar desert sites, including at the remote, northern limit of the North American Arctic, and compared our SSU rRNA gene results with those from temperate and cold biosphere sites elsewhere, including Antarctica.
MATERIALS AND METHODS
Study sites.
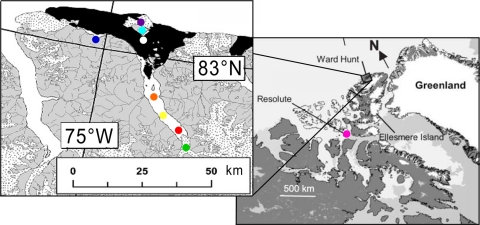
Samples were collected from two locations in the Canadian High Arctic: the northern coast of Ellesmere Island in the vicinity of Ward Hunt Island (83°05′N, 74°09′W) and 1,000 km to the south, at Char Lake (CL) (74°42′N, 94°53′W) in the Resolute Bay region of Cornwallis Island (Fig. 1). Ellesmere Island sampling was over a 50-km transect extending inland from Ward Hunt Island (near sea level) to the Disraeli Glacier (DG) (850 m above sea level). Ward Hunt Island is near the northern terrestrial limit of Canada (Cape Aldrich on Ellesmere Island, at 83°07′00"N, 69°35′00"W; 61 km east of Ward Hunt Island) and is surrounded by the Ward Hunt Ice Shelf (WHIS). Microbial mats of the WHIS, which subsist under the snow during winter and then grow in summer in elongated meltwater ponds, have been investigated previously (8, 29, 42). WHIS lies at the seaward end of Disraeli Fjord (DF), which is 5 km wide and extends 30 km inland (30). Disraeli Glacier is one of three glaciers that extend as floating ice tongues into the head of Disraeli Fjord. Snow was also sampled on the ice cover of Lake A, a meromictic lake on the northern coast of Ellesmere Island, 15 km west of Disraeli Fjord (for further information, see reference 40) and of Ward Hunt Lake (details in reference 7). Snow samples from Char Lake were collected from the frozen lake surface at two sites.
Fig. 1.
Locations of the sampling sites in High Arctic Canada. On the northern coast of Ellesmere Island, Quttinirpaaq Lagoon (QL, purple dot), Ward Hunt Island (light blue dot), including Ward Hunt Lake (WHL) and the air sampling site (WHI), Ward Hunt Ice Shelf (WHIS, white dot), Lake A (LA, dark blue dot), Disraeli Fjord site C (DFC, orange dot), site B (DFB, yellow dot) and site A (DFA, red dot), Disraeli Glacier (DG, green dot) are shown. The WHIS (black area) surrounds Ward Hunt Island. On Cornwallis Island, Char Lake (CL, pink dot) is in the vicinity of Resolute Bay. This figure was modified from the work of Mueller et al. (39) and Van Hove et al. (59).
Meteorological data.
Automated meteorological stations were situated on the north shore of Ward Hunt Island (83°05′33"N, 74°07′48"W) and on the eastern shore of Lake A, Ellesmere Island (83°00′08"N, 75°23′22"W) and provided continuous year-round data (for details, see reference 64). Snow depth was measured with a Sonic SR50 sensor (Campbell Scientific Canada Corp.). Hourly averages of wind direction and speed measurements were measured with an R.M. Young Co. wind sensor (model 05103) at a height of 10 m (Ward Hunt Island) or 3 m (Lake A).
Snow sampling.
Snow samples for the molecular analysis were collected in May and June 2008 from the surface of Lake A (LA) at three sites, Quttinirpaaq Lagoon (QL), Ward Hunt Lake (WHL), Disraeli Fjord (DF) at three sites, Char Lake (CL) at two sites, Disraeli Glacier (DG), and on Ward Hunt Ice Shelf (WHIS) (Fig. 1). Each site was sampled in duplicate, with samplings spaced 50 m apart. There was no discoloration of the snow indicating snow algae or sediment patches at any of the sites, and sampling was done at random over the visually homogeneous surface. The operator wore sterile gloves and used an ethanol-flame-sterilized shovel to transfer snow from throughout the snow profile (up to 88 cm deep) into ethanol-flame-sterilized polyethylene boxes, and the samples were then allowed to thaw over 1 to 2 days. Sixty liters of packed snow per sample resulted in 12 to 30 liters of snowmelt. Melting-snow samples were frequently mixed to maintain a homogeneous water temperature of ≤4°C. Twelve liters of snowmelt sample were filtered through a 0.2-μm-pore-size Sterivex filter unit (small fraction; Millipore) after prefiltration through a 47-mm-diameter, 3-μm-pore-size polycarbonate filter (large fraction). The filters were then stored in lysis buffer (50 mM tris, 40 mM EDTA, and 750 mM sucrose) at −20°C until further analysis at Université Laval. Autoclave-sterilized Milli-Q water was treated in tandem with the snow samples as a negative-control field blank.
For microscopic analysis of the snow, samples were collected on top of the ice cover of the Lake A, Ward Hunt Lake, and Char Lake sites in July 2009 (with a snow depth ranging between 3 and 10 cm). Ten sites per lake were chosen in order to assess the horizontal patchiness. The snow was collected into wide-mouth 4-liter Nalgene bottles with a small, ethanol-flame-sterilized shovel. After melting in darkness, conductivity of the meltwater was measured with a Water Analyser probe (Oakton). A 50-ml subsample of snowmelt was fixed in glutaraldehyde (final concentration, 1%) for 1 h at 4°C. The samples were then passed through 0.2-μm black 25-mm-diameter polycarbonate filters (Poretics) and stained with 4′,6-diamidino-2-phenylindole (DAPI) (5 μg/ml, final concentration; Invitrogen Inc.) for 15 to 20 min (47). Filters were mounted on slides with nonfluorescent mounting oil (Immersol 518 M). The slides were stored in the dark at 4°C and subsequently transferred to a −20°C freezer until enumeration by epifluorescence microscopy. Counts were made at magnification ×1,000 using an Olympus BX51 fluorescence microscope.
Air sampling.
Arctic air samples were taken at Ward Hunt Island (WHI) in July 2009 using a Burkard multivial cyclone air sampler (Burkard Manufacturing Co., Rickmansworth, United Kingdom) installed 1 m above bare ground and 100 m from the automated meteorological station. The operator wore sterile gloves, a clean hat, a mask, and a laboratory coat to avoid contamination. Prior to the sampling, the sampler was disinfected with 70% ethanol and purged for 24 h. During the sampling period, airborne cells were collected in tubes under a 16.5-liters/min airflow for 50 h. After sampling, lysis buffer was added to the tubes, which were stored at −20°C for subsequent DNA analysis.
SSU rRNA gene clone libraries.
Nucleic acids were extracted following a standard salt protocol (1). Briefly, the microbial cells in lysis buffer were digested with lyzozyme (1 mg/ml, final concentration), proteinase K (0.2 mg/ml), and sodium dodecyl sulfate (0.01%). DNA was separated from the other organic phases by centrifugation in a supersaturated NaCl solution (3 M). The DNA was then precipitated with 70% ethanol and dissolved in 1× TE buffer (1 mM Tris-HCl and 0.1 mM EDTA). The DNA concentration in the extracts was quantified with a fluorescence technique using PicoGreen and the Turner BioSystems TBS-380 fluorometer, following the manufacturer's recommendations. Sites were selected to build clone libraries according to the DNA concentration and the localization. The 16S rRNA gene was amplified by PCR using the universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) and the Bacteria-specific primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (20). Since cyanobacteria are poorly recovered using these primers, they were specifically targeted using the forward primer 27F1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and the cyanobacterium-specific primer 809R (5′-GCTTCGGCACGGCTCGGGTCGATA-3′) (28). The 18S rRNA gene was amplified with the eukaryote-specific primers NSF 4/18 (5′-CTGGTTGATYCTGCCAGT-3′) and EukB (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) (37). For bacteria, the initial denaturation step at 94°C for 3 min was followed by 35 cycles of DNA denaturation at 94°C for 30 s, primer annealing at 55°C for 30 s, and strand elongation at 72°C for 1 min, with a final extension at 72°C for 5 min. For the Disraeli Fjord C clone library, additional PCR products that had been amplified following subjection to an annealing temperature of 60°C were pooled. For Disraeli Fjord B, two clone libraries were made, one with an annealing temperature of 55°C and another with an annealing temperature of 60°C. For cyanobacteria, the initial denaturation step at 94°C for 4 min was followed by 35 cycles of DNA denaturation at 94°C for 20 s, primer annealing at 55°C for 30 s, strand elongation at 72°C for 1 min, and a final extension at 72°C for 7 min. For eukaryotes, the initial denaturation step at 94°C for 3 min was followed by 30 cycles of DNA denaturation at 94°C for 45 s, primer annealing at 55°C for 1 min, strand elongation at 72°C for 3 min, and a final extension at 72°C for 10 min. PCR products amplified from several DNA concentrations (ranging from 13 to 750 pg/μl in the PCR mix for the snow samples and from 1 to 4 pg/μl for the air sample) were pooled, cleaned with the Qiagen purification kit, and cloned using the TA cloning kit (Invitrogen, California) or the Strataclone PCR cloning kit (Stratagene, California) following the manufacturers' directions. Positive clones were screened for restriction fragment length polymorphisms (RFLP) with HaeIII (Gibco BRL, Maryland) for the universal bacterial and eukaryotic sequences and with AluI and HpaII (Fermentas, New Hampshire) for the cyanobacterial sequences. Clones with the same RFLP pattern were considered to be members of the same phylotype. Multiple clones per RFLP pattern were sequenced to confirm that each pattern represents only one operational taxonomic unit (OTU). The 16S rRNA gene was sequenced in both directions using primers for the vector M13 sites, resulting in nearly full-length 16S rRNA sequences. Cyanobacterial and eukaryotic sequences were sequenced using the promoter T7 in the M13 vector and the internal primer 528F (16), respectively.
Isolation of cyanobacteria.
Snow samples collected May 2008 were used to test for the presence of viable cyanobacteria using a method modified from that of Vézina and Vincent (61). Snow from the Ward Hunt Lake, Lake A, and Char Lake sites was melted as described above, and between 4 and 8 liters was filtered onto 47-mm-diameter, 0.2-μm-pore-size polycarbonate filters. Isolates of cyanobacteria were obtained by incubating these filters in the liquid culture medium BG-11 (49) under a range of conditions (with and without nitrate, with and without preincubation in liquid medium in the field) to maximize the diversity of isolates. Cycloheximide (40 mg/liter, final concentration) was added to inhibit eukaryotic growth.
Ward Hunt Lake and Lake A cultures initially begun in the field were exposed to natural 24-h daylight at temperatures ranging between 3 and 10°C for 2 weeks. For the second part of the field season, the cultures were maintained at ∼16°C under natural light at Resolute Bay. Once back at Université Laval, the cultures were transferred to a constant-temperature growth cabinet and maintained at 10°C with continuous irradiance under 50-μmol photons m−2 s−1 cool white fluorescent light.
Genomic DNA was extracted from unialgal cultures. Approximately 100 mg of cells were resuspended in 300 μl XS buffer (1% potassium-ethyl-xanthogenate, 800 mM ammonium acetate, 20 mM EDTA, 1% sodium dodecyl sulfate, 100 mM Tris-HCl [pH 7.4]) (57). The mixture was vortex mixed and incubated for 2 h. The extracts were cooled on ice for 10 min, and cell debris was removed by centrifugation at 12,000 rpm for 10 min. The supernatant was collected, and DNA was precipitated overnight by addition of 1 volume of isopropanol and 1/10 volume of 4 M ammonium acetate at −20°C. The precipitated DNA was pelleted by centrifugation at 12,000 rpm for 10 min and washed with 70% ethanol. The extracted DNA was then resuspended in 100 μl of sterile water. PCR amplification was done as for the clone library analysis, and sequencing was done using the 809R internal primer (28).
Diversity and community composition.
The SSU rRNA gene sequences from the snow and air samples were compared to those in the NCBI GenBank database using BLASTn (3) to identify closest matches and their source environments. Sequences were checked using the Chimera check software program at Ribosomal Data Project II (Michigan State University; http://wdcm.nig.ac.jp/RDP/cgis/chimera.cgi?su=SSU), and suspected chimeras were excluded from further analysis. The remaining sequences were manually trimmed and aligned using the MUSCLE software program (www.ebi.ac.uk/Tools/muscle/index.html).
OTUs were defined as having ≥97%, ≥98%, or ≥99% similarity for the bacterial (including cyanobacteria), eukaryotic, and chloroplastic sequences, respectively, and were determined using the Mothur software program, v.1.7.2 (http://www.mothur.org/wiki) (54). Thresholds for eukaryotic and chloroplastic sequences were set higher, because 18S rRNA and chloroplastic 16S rRNA genes are more conserved than the bacterial 16S rRNA gene, where the threshold is 97% based on DNA-DNA hybridization to differentiate species (51). This is based on empirical comparisons with classical taxonomy and 18S rRNA gene similarities (38). The same program was used to calculate the Shannon index and Chao1 richness. Equal numbers of sequences were randomly selected per site for intersite diversity and richness comparison. Similarity between communities was assessed by Bray-Curtis cluster analyses using the software program PAST v.1.90 (25).
Nucleotide sequence accession numbers.
The nucleotide sequences data reported in the present study were submitted to GenBank under the accession numbers HQ230103 to HQ230240 and HQ529495 to HQ529499.
RESULTS
Snowpack characteristics.
Snow sampling for molecular analyses in the Ward Hunt Island region was carried out in late spring, at the time of maximum snow accumulation and immediately prior to the rapid snowpack loss by melting (Fig. 2). At all sites, the snow was sampled to the depth of hard ice or firn, which ranged from 10 cm on Char Lake to 85 cm on Disraeli Glacier. Wavelike ridges of hard snow (termed “kimugiyuk” or “sastrugi” and normally formed perpendicular to the direction of the wind) were observed at all sites except Disraeli Fjord. On Ward Hunt Lake and Quttinirpaaq Lagoon, the snow cover was dense and uniform and had a polystyrene-like texture, with large crystals of snow on the surface. On Lake A, the snowpack had a dense layer at the surface (5 to 10 cm thick), a loose intermediate layer with powdery to granular snow, and another dense layer at the bottom. At Disraeli Fjord A and B, the snow was wet and heavy and contained cylindrical ice crystals at the surface and granular crystals at the base of the snow pits. At Disraeli Fjord C and Disraeli Glacier, the snowpack was multilayered. The snow on Char Lake showed the onset of melt: it was wet and cohesive, and liquid water was discernible at the ice-snow interface. In July 2009, conductivity of the melted snow ranged from 11.1 to 60.5 μS/cm for Char Lake snow, 1.5 to 16.2 μS/cm for Lake A snow, and 3.4 to 9.9 μS/cm for Ward Hunt Lake snow.
Fig. 2.
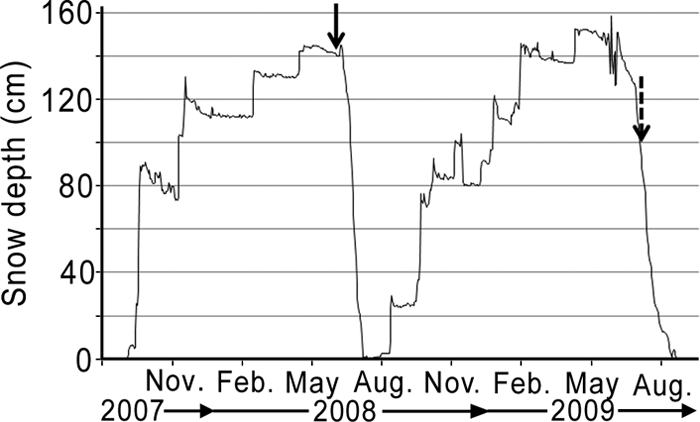
Depth of snow over the ground on the northern shore of Ward Hunt Island between August 2007 and August 2009. Arrows indicate the moment of snow sampling for the molecular analyses (plain arrow) and the microscopic analyses, as well as the air sampling (dotted arrow).
Wind conditions.
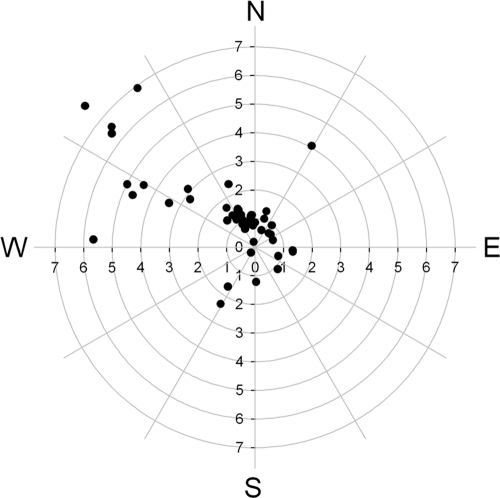
For the snow accumulation period prior to sampling (September 2007 to June 2008), the wind at Lake A came predominantly from the northeast quarter (35% of the hourly mean samples). The wind came from the southwest, northwest, and southeast quarters 29%, 17%, and 19% of the time, respectively. There was a similar pattern for the same period in 2008 and 2009, with a predominance of wind from the northeast quarter (45% of the time). There was no clear trend in wind direction on Ward Hunt Island. In the 2007-2008 snow accumulation period, wind came from the northwest, southwest, southeast, and northeast quarters 32%, 28%, 28%, and 12% of the time. For the same period over 2008 and 2009, wind came from the southwest, northwest, southeast, and northeast quarters 31%, 29%, 27%, and 13% of the time. During the air sampling, the meteorological station recorded winds from predominantly the north: 67% from the northwest quarter and 18% of the time from the northeast quarter, with the strongest winds coming from the northwest (Fig. 3).
Fig. 3.
Direction and speed (m/s) of winds during the air sampling from 7 to 9 July 2009. Dots indicate the hourly averages of wind speed (values on the axes) and direction that the wind came from.
Microbial DNA and cells.
DNA quantification and prokaryote cell concentrations estimated from the DAPI-stained cells indicated that the microbiota were nonhomogeneously distributed in the snowpack in both sampling years. In June 2008, relative extracted DNA concentrations varied from undetectable to 47 pg/ml (all values are for the melted snow water) (Fig. 4). Median concentrations were 2.2 and 2.9 pg/ml for the small and large fractions, respectively. These DNA concentrations varied by up to 23-fold between sites distant by tens of meters and by up to 31-fold between sites that were tens of kilometers distant. Prokaryote cell concentrations in 2009 samples were also heterogeneous (Fig. 5). Consistent with the DNA results from the previous year, cell concentrations were low for the Lake A and Char Lake samples. The medians between the lakes differed by a factor of 1.2. Overall, the counts ranged from 0.02 × 103 to 2.07 × 103 prokaryotic cells/ml, with substantial variation at both small and large length scales. Higher but similarly variable concentrations were recorded in WHL snow, with cell counts ranging from 1.52 × 103 to 53.2 × 103 cells/ml. Sites a few meters apart varied by up to a factor of 5, while those distant by tens of meters varied by up to 2 orders of magnitude.
Fig. 4.
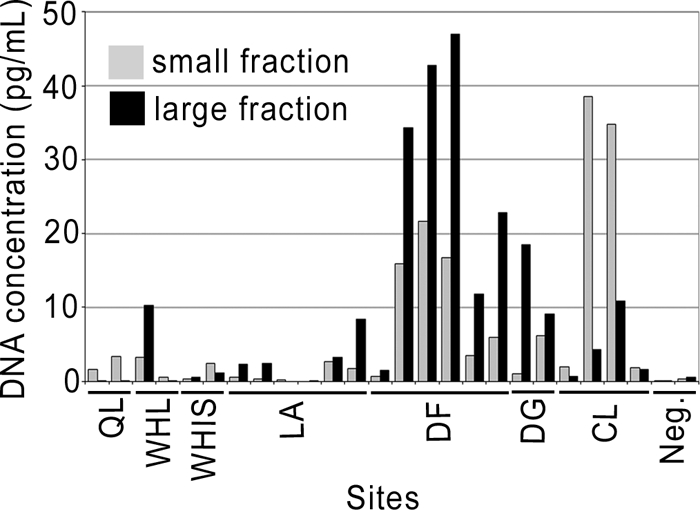
Extracted DNA concentrations in the small (0.2 to 3 μm) and large (>3 μm) fractions from High Arctic snow samples collected in May and June 2008 (QL, Quttinirpaaq Lagoon; WHL, Ward Hunt Lake; WHIS, Ward Hunt Ice Shelf; LA, Lake A; DF, Disraeli Fjord; DG, Disraeli Glacier; CL, Char Lake; Neg., field blanks). The sites were sampled in duplicate at a distance of tens of meters apart.
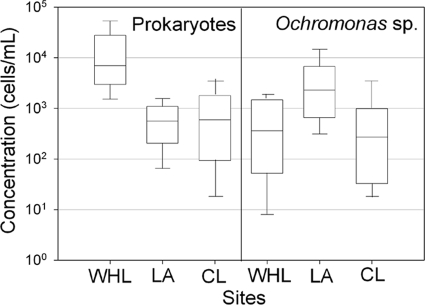
Fig. 5.
Concentration of prokaryotic and Ochromonas cells (note the log scale) in the snow cover of Ward Hunt Lake (WHL), Lake A (LA), and Char Lake (CL) in July 2009, determined by DAPI slide counts.
Ochromonas-like microbial eukaryotes were also identified in the DAPI slides from the three sites, and as with the prokaryotes, the cells were patchy. The median cell concentrations varied by a factor of 8.5 among sites (Fig. 5) and ranged from 0.01 × 103 to 1.73 × 103 cells/ml in Ward Hunt Lake snow, from 0.28 × 103 to 8.50 × 103 cells/ml in Lake A snow, and from 0.02 × 103 to 2.11 × 103 cells/ml in Char Lake snow.
Bacteria in the snow.
We recovered 285 16S rRNA gene clones from the bacterial libraries constructed from five of the sampling sites. No PCR amplification was obtained using DNA extracts from the field blanks, and these results therefore appear to be free of contamination. The bacterial phylotypes were distributed into 18 OTUs at the >97% similarity level (see Table S1 in the supplemental material). Proteobacteria was the dominant phylum among all OTUs, in particular the order Burkholderiales in the class Betaproteobacteria, which represented 64% of bacterial clone sequences. Other OTUs belonged to the phyla Bacteriodetes and Cyanobacteria and the Deinococcus group. All but one of the OTUs matched sequences that had been previously recovered from cold environments such as glaciers, alpine lakes, snow, and cold soils. The exception was an OTU (represented by the clone DFCb18 in Table S1) that had no >97% match in GenBank.
In many sites, OTUs belonging to genera with known psychrophilic taxa were detected: Polaromonas, Aquaspirillum, Octadecabacter, and Glaciecola. However, there were differences in clonal abundance and diversity among the sites (Table 1; see also Table S1 in the supplemental material). The lowest diversity was found in the Lake A and Char Lake A and B snow samples. Even though these sampling sites were a thousand kilometers apart, their microbial snow communities were highly similar as shown by the Bray-Curtis cluster analysis (Fig. 6). Aquaspirillum arcticum was the dominant taxon, representing 96% of the bacterial clone sequences in the Lake A snow sample, 89% for Char Lake A, and the only bacterial OTU for Char Lake B. Another Aquaspirillum was found in the Disraeli Fjord B snow sample but had no close match to cultivated strains in GenBank.
Table 1.
Bacterial and eukaryotic diversity indices for samples at each site
| Site | Shannon indexa |
Chao1 richness |
||
|---|---|---|---|---|
| Bacteria | Eukarya | Bacteria | Eukarya | |
| Lake A | 0.18 ± 0.19 | 0.18 ± 0.27 | 2 | 2 |
| Disraeli Fjord site C | 1.44 ± 0.22 | 1.77 ± 0.36 | 8 | 23 |
| Disraeli Fjord site B | 1.86 ± 0.22 | NA | 10.5 | NA |
| Disraeli Fjord site A | NAb | 0.89 ± 0.25 | NA | 6 |
| Disraeli Glacier | NA | 0.93 ± 0.25 | NA | 4 |
| Char Lake site A | 0.36 ± 0.25 | NA | 4 | NA |
| Char Lake site B | 0 ± 0 | 0 ± 0 | 1 | 2 |
The values after “±” specify the lower and upper bounds of a 95% confidence interval.
NA, no clone library was made for these samples.
Fig. 6.
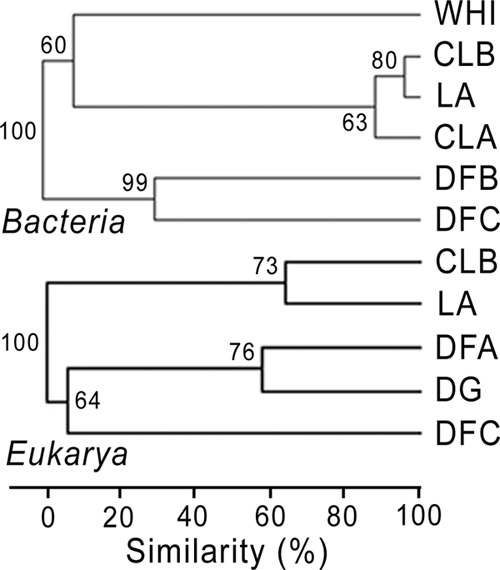
Bray-Curtis cluster analysis comparing the bacterial (top) and eukaryotic (bottom) communities detected in the snow in May and June 2008 at Char Lake sites A and B (CLA and CLB), Lake A (LA), Disraeli Fjord sites A, B, and C (DFA, DFB, and DFC), and Disraeli Glacier (DG) and in the air of Ward Hunt Island (WHI) in July 2009. The comparison takes into account the relative abundance of each OTU in the clone libraries and results from 1,000 replicates (values indicated at the nodes).
Higher diversity indices for bacteria were recorded in the Disraeli Fjord B and C samples (Table 1). These snow communities were dominated by sequences closest to those of marine groups that require sea salts for growth. The Disraeli Fjord B bacterial community was dominated by Glaciecola pallidula (24%) and Polaribacter irgensii (26%), while the Disraeli Fjord C clone library was dominated by Glaciecola psychrophila (35%) and by Colwellia piezophila (28%).
The bacterial rarefaction curves (see Fig. S1 in the supplemental material), with the exception of the DFB bacterial library (DFBb), reached an asymptote, indicating that there was a reasonable sampling of the species richness. The Chao1 nonparametric diversity estimator confirmed this finding by showing that 100% of the predicted number of OTUs was recovered except for DFBb, for which the number of OTUs recovered corresponded to 86% of the predicted asymptote.
In addition to the bacterial 16S rRNA gene clones, 195 sequences were from the chloroplast 16S rRNA gene (see Table S2 in the supplemental material). Several of these showed matches to the algal phyla Cryptophyta and Bacillariophyta; however, most of the chloroplast sequences could not be matched to any particular group currently in GenBank.
Cyanobacteria were detected in cultures of all the different snow samples tested. Among the cultures, we isolated viable cyanobacteria belonging to the orders Oscillatoriales, Nostocales, and Chroococcales. The most common taxa in the cultures were filamentous oscillatorians. We also constructed a clone library from one of the Ward Hunt Lake snow samples using the same primers that have been used to determine the cyanobacterial community composition in the benthic microbial mats of Ward Hunt Lake and in meltpools on the adjacent ice shelves (29). We detected a total of 17 cyanobacterial OTUs (see Table S3 in the supplemental material), and 8 of these had >97% identity with those from microbial mats in Ward Hunt Lake, Markham and Ward Hunt ice shelves, the inflow of Lake A, and Antoniades Pond, a small water body in the catchment of Lake A. Five snow sequences representing five different OTUs had their closest match in GenBank to the Jungblut et al. (29) mat sequences sampled in 2007, with up to 100% identity. The clone library for the large fraction also indicated the presence of the Gloeobacterales in the snow. Many sequences in the clone libraries did not match the 16S rRNA gene sequences in GenBank at the >97% level, most of them having only a 92% similarity to sequences of their closest cultivated neighbors.
Eukaryotes in the snow.
Eukaryotic clone libraries were constructed for snow from five of the sites, and we obtained 333 clones. The 18S rRNA gene sequences indicated the presence of ciliates, Chrysophyceae, Pelagophyceae, Bacillariophyta, Dinophyceae, Cercozoa, Basidiomycota, Cryptophyta, Streptophyta, and Chlorophyceae (see Table S4 in the supplemental material). Twelve of the 23 OTUs (52%) were previously recorded in samples from cold environments, such as cold marine waters, sea ice, mountain stream sediment, and snow. Eight OTUs (35%) had no matches with >98% similarity to other sequences in GenBank. The eukaryotic clones mostly belonged to phototrophic taxa, notably Ochromonas, Pelagomonas, Ancylonema, Chloromonas, and Chlamydomonas. Species known to form resting stages were also present, such as Polarella glacialis, Chloromonas sp., Chlamydomonas sp., and Ochromonas sp.
The Shannon diversity index indicated the same high variability as for the bacterial communities, ranging from extremely low diversity in the Lake A and Char Lake snow samples to higher values in the fjord vicinity (Table 1). The low-diversity communities of Lake A and Char Lake snow were dominated by the same eukaryotic OTU, which was 99.8% similar to Ochromonas sp. CCMP1899, isolated from sea ice in Antarctica; this taxon accounted for 96% and 64% of the clones, respectively. This dominance is consistent with the abundance of the Ochromonas cell type observed by fluorescence microscopy 1 year later (see “Microbial DNA and cells”).
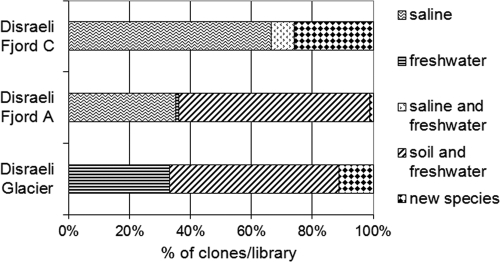
Similar to the bacterial results, for the site nearest to the sea on Disraeli Fjord (DFC), a large proportion of its community was represented by marine taxa (7/16 OTUs or 38/88 clones) (Fig. 7). Conversely, the communities of the innermost sites (DFA and DG) had sequences with closest matches to a number of terrestrial representatives (notably the genera Leucosporidium, Ancylonema, and Chloromonas), although the marine signal was still detectable in the Disraeli Fjord A sample.
Fig. 7.
Source environment of eukaryotic clones according to sampling site and compilation of the isolation sources of all the sequences in GenBank having >98% similarity with a specific clone representing one OTU. Clone abundances were determined from the RFLP pattern repeats. The numbers of clones were 88 for Disraeli Fjord C, 91 for Disraeli Fjord A, and 27 for Disraeli Glacier.
DFAe and DFCe were the only clone libraries for which the rarefaction curves did not reach an asymptote (see Fig. S1 in the supplemental material). The Chao1 values were 83% for DFAe and 70% for DFCe but 100% for all of the other samples, indicating good coverage.
Bacteria in the atmosphere.
For the Ward Hunt air sampling, field blanks and PCR-negative controls all showed undetectable amounts of DNA, implying an absence of contamination. The air sample extract contained 16 ng DNA/ml, corresponding to a total of 0.64 ng of DNA in ∼49 m3 of sampled air (13 pg/m3). We retrieved 71 16S rRNA gene clones from the air sample library. These clustered into 14 OTUs (see Table S1 in the supplemental material). The taxa belonged to the major phyla Bacteriodetes, Acidobacteria, Firmicutes, and Proteobacteria with representatives from the Alpha-, Beta-, and Gammaproteobacteria classes. Eleven of the OTUs were previously found in cold environments, including two OTUs previously isolated only from the sea. These included Roseobacter and uncultured Cytophagales. Two other OTUs belonged to the genera Lactobacillus and Staphylococcus, and the final OTU had no match >97% similar in GenBank but showed 95.4% similarity to an Acidobacteria isolate. Four OTUs detected in snow the preceding year were also found in this air sample. They were related to Aquaspirillum arcticum, Janthinobacterium, Rhodoferax, and Pseudomonas syringae. One Polaromonas sequence was also found in a chimera, revealing its presence in the air, but its sequence was excluded from the analysis. As in the snow bacterial clone libraries, chloroplast 16S rRNA gene sequences were detected with eight sequences that clustered into the algal class Prasinophyceae and an unknown phylum (see Table S2).
DISCUSSION
Microbial cells and DNA were detected in the snow at all locations, and microbial DNA was also collected in the 24-h air sampling on Ward Hunt Island. These observations imply that microbiota are widely dispersed via wind and precipitation across the High Arctic and are a common feature of the snowpack environment. The clone library analysis revealed a relatively diverse ensemble of taxa with a total of 25 bacterial OTUs in the snow and air and 23 eukaryotic OTUs in the snow. No previous molecular studies have been conducted on the snow and aerial microbiology at these remote, far northern sites, and there are few data of this type from the polar regions in general. The annual snow cover melts at these sites each summer, and the microbial content of the snow is therefore the accumulation of aerial transport by wind and precipitation, and possibly growth, over the preceding months.
Spatial distribution of cells.
Extracted DNA and prokaryotic cell concentrations were at or below the lower end of the range of published values for snow, which vary from 200 cells/ml in South Pole snow (14) to 7.2 × 105 cells/ml in the snow cover of Zadong Glacier on the Tibetan Plateau (32). The low microbial concentrations in High Arctic snow may reflect the remote geographic location of the sampling sites relative to those of temperate snow packs that receive much greater microbial loading from surrounding ecosystems and the persistent low-temperature regime of the high-latitude environment that inhibits in situ growth rates. Postdepositional processes can have a strong influence on microbial abundance in snow (67). DNA quantification and the DAPI cell counts showed that the spatial distribution of microbes was nonuniform across the snow cover. This high degree of patchiness is consistent with the distribution of sediments on the Ward Hunt Ice Shelf near our Ellesmere Island sampling sites (39). In 2001, sediment cover varied from 12% of the western surface of the ice shelf to 14% in the middle and 2% on the eastern sector. This trend was also reported for other Ellesmere Island ice shelves and is reflected in the patchy spatial distribution of microbial mats that grow in these environments. On Markham Ice Shelf, the sediment content of the 20-mm-thick mats was substantial and constituted 75 to 91% of the mat dry weight (63). Eolian processes are likely to contribute to this patchy distribution of sediments and microbes. Although our snow samples contained fine sediment, which was visible after melting of the snow, because of logistic constraints this was not quantified. Wind transport has also been shown to be an important vector of microbes in temperate locations (22). There is also likely to be a positive feedback, with nutrients released from the sediments enhancing the local growth of associated, wind-blown microbes and heat absorption by the sediments causing meltwater production for microbial growth (see below). These microbial hot spots of colonization and growth may also act as a local inoculum for broader dissemination across the snowpack.
Spatial distribution of taxa.
Samples grouped into subclusters according to the potential source environments for their communities (Fig. 6). In the vicinity of the fjord, most sequences matched to obligate marine taxa. In the northernmost site (DFC), both bacterial and eukaryotic communities were rich in marine species. The Disraeli Fjord site B community was also made up of marine bacteria, such as Glaciecola spp. and Loktanella sp. The clustering of Disraeli Fjord site A and the Disraeli Glacier site, located southward, is likely due to the presence of characteristic terrestrial species (Leucosporidium sp. and Chloromonas sp.). Polarella sp., a sea ice dinoflagellate (38), was the only taxon found in the snow of both site A and site C, with a relative abundance of at least 20% in each library. These observations suggest that the fjord valley may channel the wind through to these sites. The wavelike ridges observed at the surface of the glacier oriented in the direction of the valley also support this idea.
Interestingly, the communities in Lake A and Char Lake snow were highly similar. The two lakes have similar landscapes but vary greatly in area (Char Lake, 0.53 km2 [53]; Lake A, 5 km2 [59]) and lie 1,000 km apart.
The low level of similarity between the bacterial communities of the air and the snow samples could reflect the different time spans of sampling, since most of the snow microbiota would have accumulated over weeks to months prior to being sampled, while the air sampling provided only a snapshot of the dispersed microbiota. However, some taxa (Aquaspirillum arcticum, Rhodoferax sp., and Polaromonas sp.) were detected in both the air and snow, suggesting that these bacteria are especially abundant and commonly dispersed.
Local sources.
Despite harsh environmental conditions in the Canadian High Arctic, this region supports several ecosystem types in the vicinity of our sampling sites (see reference 64 and references therein), and these may provide local sources of inocula for the snow microbial assemblages. In particular, microbial mats form biomass-rich communities on ice shelves, in wetlands, and at the bottom of lakes and ponds. These mats consist of diverse microbial assemblages consolidated by intertwined trichomes of cyanobacteria, and they include bacteria, archaebacteria, eukaryotic microbes, viruses, and metazoans such as rotifers, tardigrades, nematodes, and the free-living tubellarian platyhelminths, all contained within a cohesive matrix of exopolymeric substances (63). Metagenomic analysis has shown that these mats are functionally as well as taxonomically diverse and that they are genetically dominated by the Proteobacteria and Cyanobacteria (60).
We detected cyanobacteria in the snow by molecular analysis and by cultivation, and many of these OTUs had a close match to sequences previously determined for microbial mats in this High Arctic region. The bacterial communities of Ward Hunt and Markham ice shelf microbial mats were examined by clone library analysis of their 16S rRNA genes in 2005 and 2006 (8). Again, some of those taxa were highly similar to our High Arctic sequences: four snow OTUs were 97.2% to 100% similar to the mat sequences. In particular, the genera Polaromonas, Rhodoferax, and Aquaspirillum were similar in both snow and ice shelf mats. One sequence of Brevundimonas detected in the air was 98.9% similar to that of a mat isolate. These results suggest that Arctic mat microbes are dispersed in the atmosphere and across the snow cover. This dispersion aids new mat development when the bacteria or cyanobacteria colonize snow or ice regions with favorable conditions. As the mats grow, this would result in locally reduced albedo and more rapid meltwater generation, in turn favoring mat growth and the attrition of snow and ice. The role of cyanobacterial mats as accelerators of ice melt was noted by Nordenskiöld on the Greenland Ice Cap (31) and by Mueller and Vincent (41) for the Ellesmere ice shelves. The present results extend these observations by showing that these microbial mat-forming agents are widely dispersed in Arctic snow.
Three of the bacterial OTUs collected by the air sampler on WHI were affiliated with taxa often associated with humans: Stenotrophomonas maltophilia, Lactobacillus delbrueckii, and Staphylococcus hominis (see Table S1 in the supplemental material). However, although S. maltophilia is commonly isolated from clinical specimens, it is also widely distributed in natural environments, especially in the soil and plant rhizosphere (27), and can utilize simple organic compounds, such as acetate, succinate, and propionate, as its sole carbon source (56). In contrast, Lactobacillus and Staphylococcus spp. are more likely to come from anthropogenic activities. For example, S. hominis is a member of the airborne bacterial population within the confined Antarctic Base Concordia (58) and L. delbrueckii is used in the manufacturing of dairy products. Our air sampler was located 500 m from the camp on Ward Hunt Island, which has a 50-year history of intermittent summer occupation by military personnel, scientists, and explorers, and it is likely that human-associated microbes have been widely dispersed across the site. Pearce et al. (45) described bacterial populations collected over a 2-week period in the atmosphere of the Halley V research station in Antarctica, with 35% of sequences appearing to be related to human-associated bacteria. During summer, Halley Station receives up to 70 people, and human-associated microbiota are a likely component of the aerobiology, masking to some extent the natural signal of this otherwise pristine environment.
Although it is possible that the communities resulted from long-range dispersal of cosmopolitan taxa followed by environmental selection for growth of cold-adapted genotypes in the snow, the most parsimonious explanation is that the biota in the snow was predominantly from the local sources that they resembled. The transient nature of the snowpack in which the microbial load is renewed each year leads to only restricted opportunities for growth. The air samples provided more direct evidence of local sources. Growth in the air sampler is unlikely, and conditions are very different from those in either the sea or terrestrial mats and lakes. Air was constantly pumped through the tube over the whole sampling period, likely leading to cell desiccation. Microbes detected in this sample were predominantly cold taxa, many similar to those found in the snow the year before (Aquaspirillum sp., Rhodoferax sp., and Polaromonas sp.), including some obligate marine microbes.
Regional sources.
Some of the snow samples contained a relatively high abundance of bacteria characteristic of cold oceanic waters, notably the genera Glaciecola, Colwellia, Loktanella, and Polaribacter. This unexpected observation, given the low conductivity of the snow, suggests an input of bacteria originating from marine aerosols, and it provides evidence of dispersal of marine microbiota via the atmosphere within the High Arctic. Snow samples also contained eukaryotic microbes that have been isolated exclusively from the marine environment in the past. Furthermore, the air sample contained chloroplast DNA of Micromonas, a major component of the Arctic Ocean phytoplankton (36), and sequences of obligate marine bacteria, providing evidence of the active aerial transport of marine microbiota. All of the sampling sites were at or near the coast, and it is likely that the Arctic Ocean and sea ice served as an inoculum. Sea ice with extremely high concentrations of cells, especially diatoms, within brine channels is often more productive per cubic meter than the underlying pelagic zone (12). In the present study, the Disraeli Fjord C clone library contained a large proportion of marine taxa, with the eukaryotic community dominated by a marine diatom. Many sequences corresponding to the chloroplast 16S rRNA gene of marine diatoms were also found in the bacterial clone libraries of Disraeli Fjord B and C (see Table S2 in the supplemental amterial). Brinkmeyer et al. (11) found Octadecabacter sp. in both Antarctic and Arctic sea ice, but it was more abundant in the latter. In our Disraeli Fjord B snow sample, one OTU (represented by DFBb6 in Table S1) accounted for 13% of the clones bearing bacterial sequence in the library and was identical (1,390/1,390 bp) with those reported by Brinkmeyer et al. (11). Although some salty microenvironments in the snow may exist in microfilms around snow and sediment granules and could support marine taxa to some extent, the overall detection of marine microbes in the Arctic atmosphere strongly suggests a local input from the Arctic Ocean, especially since these taxa were also detected in the air samples and the predominant winds came across the sea from the north (Fig. 3). These results imply that sea ice is a major source for the High Arctic snowpack microflora and that sea ice microbiota are widely dispersed by the wind, not just by ocean currents.
One likely mechanism that could spread microbes from sea ice to the atmosphere is exposure of the ice community to the air by frost flowers. These surface structures are created under extreme cold and dry conditions by salt exclusion of the ice matrix during freezing (46). They have been found to contain 3- to 6-fold-higher concentrations of bacteria than are found in the underlying ice and are thought to be important for the long-range transport of ice-nucleating particles in the atmosphere (10). In August 2008, we observed frost flowers on Quttinirpaaq Lagoon, a brackish water feature north of Ward Hunt Island, and they are also likely to form on the sea ice along the coast.
The circumpolar flaw lead occurs intermittently along the northern coast of Ellesmere Island throughout the year, and the resultant open water could also provide a microbial inoculum. Microbes are concentrated in the sea surface microlayer compared to levels in the subsurface waters, and this microlayer has been implicated in the enrichment of microbes in marine aerosols produced via wave action and bubble bursting. Marine aerosols can be up to 22-fold enriched in microbes compared to bulk seawater (2).
Cosmopolitanism and long-range transport.
Most of the microbes detected in the snow and air had best matches to sequences from other cold environments, including Antarctica (some with 100% similarity, e.g., Octadecabacter; see Table S1 in the supplemental material), the Tibetan Plateau, and alpine regions of Japan, Europe, and North America, including the Arctic. No clones were >98% similar to sequences isolated from more typically warm regions, with the exception of WHIb50, which was 98.1% similar in its 16S rRNA gene sequence to Pseudomonas PsI, isolated from a woodland soil of the arid southwest United States. Rodrigues et al. (50) reported a global distribution of the psychrophilic species of Psychrobacter. Similarly, Jungblut et al. (29) reported cyanobacterial ecotypes throughout the cold biosphere and the absence of any gene sequences from warmer regions that matched those of High Arctic strains. Our detection of these taxa in the High Arctic snowpack is consistent with the global dispersal of a set of cold-adapted ecotypes throughout the cold biosphere. Other sequences from High Arctic snow had no close matches in GenBank and might represent novel species particular to the Arctic. For example, clone DFCb18 in Disraeli Fjord snow showed only 94.7% similarity with its closest match, Deinococcus radiomollis strain PO-04-20-144; the latter strain was isolated from a Mexican alpine soil at 5,065 m and is psychrophilic (13).
Viability in the snow.
The snow microbial communities likely contained a mixture of nonliving and living cells. The presence of marine bacteria and eukaryotes implied that part of the microflora is from marine microbial aerosols that may have acted as nucleation agents for the subsequent snowfall. Given the obligate requirement of many of these taxa for marine salts, it is unlikely that these cells would grow in the low-conductivity waters of snowmelt, although they could briefly survive in microenvironments of boundary layers of concentrated salts surrounding ice and soil grains. The eventual lysis of such cells by osmotic stress during snow melting would contribute nutrients and organic carbon substrates for the growth of the nonhalophilic taxa.
Felip et al. (18) studied the microbial communities in the ice and snow cover of three high-altitude lakes in the Tyrolean Alps and found that algal and protozoan biomass and microbial activity were greater in the snow cover than in the underlying lakes. Tyrolean Alps genera (Gymnodinium, Ochromonas, and Chlamydomonas) were also detected in our High Arctic snow, suggesting that these are likely true snow algae. We successfully cultivated cyanobacteria from snow samples, providing evidence that these organisms remain viable in snow. If phototrophs are able to grow in the snow, it is likely that some of the heterotrophic bacteria may also be viable and active. Several of the bacteria identified in snow samples reportedly have low nutrient requirements and may grow on diverse substrates (e.g., see references 26, 43, 48, and 68). Notably, 11 of the genera we recovered contain psychrophilic representatives, with potential to grow in situ. Several heterotrophic bacteria isolated from Ellesmere Island ice shelf mats grow under low-nutrient and cold-temperature conditions (8), and many of the same OTUs from these mats were found in our snow samples. Environmental surveys based on RNA would provide additional insights into which taxa are metabolically active in the Arctic snowpack. Contrary to our initial expectation, no spore-forming bacteria were detected in the High Arctic snow samples. This absence might be the result of methodological bias, for example, the difficulty of extracting DNA from bacterial spores, and this negative result is inconclusive.
In summary, our snow and air samples from northern Canada contained a distinct assemblage of heterotrophic and phototrophic microbes, and such communities are likely distributed throughout the High Arctic. A proportion of the microbial community was likely derived from two disparate sources near our sampling sites, specifically freshwater microbial mats and Arctic Ocean sea ice. The heterogeneous distribution of microbes in the snow suggested that microbes were redistributed into patches via wind action and localized melting. The high similarity between some of the Ellesmere Island SSU rRNA gene sequences and those found in polar and alpine environments elsewhere in the world suggests that High Arctic microbes are globally distributed in cold regions. These results show the value of the Ward Hunt Island region as a set of extreme northern sites for the study of snow and air microbiology. Further studies are needed to test other phylogenetic markers with finer resolution to unravel the relationships between High Arctic microbes and those from elsewhere. Deeper sampling of the rarer taxa could also be achieved with next-generation sequencing technologies, such as pyrosequencing, as in the work of Galand et al. (21). However, the tradeoff is a loss of fine taxonomic resolution, and a combination of approaches is required. Arctic microbial ecosystems are currently experiencing accelerated climate change (62), and the microbiology of High Arctic snow will require ongoing surveillance. Our detection of close relatives of well-known ice-nucleating bacteria (e.g., Pseudomonas syringae) in the air also highlights the need to study the role of microbes in atmospheric chemistry in the rapidly changing north polar environment.
Supplementary Material
ACKNOWLEDGMENTS
This research is part of the long-term program Northern Ellesmere Island in the Global Environment (NEIGE). It was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC), the Networks of Centres of Excellence program ArcticNet, the Canada Research Chair program, the Canadian International Polar Year program Microbiological and Ecological Responses to Global Environmental change in the Polar Regions (MERGE), and the Fonds Québecois de la Recherche sur la Nature et les Technologies.
We thank Julie Veillette, Dermot Antoniades, Sébastien Bourget, and Sophie Charvet for field assistance and Denis Sarrazin for technical support. We thank Caroline Duchaine for kindly providing the air sampler for this study. We thank Ladd Johnson and three anonymous reviewers for their insightful comments on the manuscript. We also thank Parks Canada, the Polar Continental Shelf Project (PCSP), and the Northern Scientific Training Program (NSTP) for logistical and infrastructure support.
This is PCSP/EPCP contribution no. 00511.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 April 2011.
REFERENCES
- 1. Aljanabi S. M., Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aller J. Y., Kuznetsova M. R., Jahns C. J., Kemp P. F. 2005. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J. Aerosol Sci. 36:801–812 [Google Scholar]
- 3. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Amato P., et al. 2007. Bacterial characterization of the snow cover at Spitzberg, Svalbard. FEMS Microbiol. Ecol. 59:255–264 [DOI] [PubMed] [Google Scholar]
- 5. Amato P., et al. 2007. An important oceanic source of micro-organisms for cloud water at the Puy-de-Dôme (France). Atmos. Environ. 41:8253–8263 [Google Scholar]
- 6. Baas Becking L. G. M. 1934. Geobiologie of Inleiding Tot de Milieukunde. WP Van Stockum & Zoon, The Hague, Netherlands [Google Scholar]
- 7. Bonilla S., Villeneuve V., Vincent W. F. 2005. Benthic and planktonic algal communities in a High Arctic lake: pigment structure and contrasting responses to nutrient enrichment. J. Phycol. 41:1120–1130 [Google Scholar]
- 8. Bottos E. M., Vincent W. F., Greer C. W., Whyte L. G. 2008. Prokaryotic diversity of arctic ice shelf microbial mats. Environ. Microbiol. 10:950–966 [DOI] [PubMed] [Google Scholar]
- 9. Bowers R. M., et al. 2009. Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl. Environ. Microbiol. 75:5121–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bowman J. S., Deming J. W. 2010. Elevated bacterial abundance and exopolymers in saline frost flowers and implications for atmospheric chemistry and microbial dispersal. Geophys. Res. Lett. 37:L13501 [Google Scholar]
- 11. Brinkmeyer R., Knittel K., Jürgens J., Weyland H., Amann R., Helmke E. 2003. Diversity and structure of bacterial communities in arctic versus antarctic pack ice. Appl. Environ. Microbiol. 69:6610–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown M. V., Bowman J. P. 2001. A molecular phylogenetic survey of sea-ice microbial communities (SIMCO). FEMS Microbiol. Ecol. 35:267–275 [DOI] [PubMed] [Google Scholar]
- 13. Callegan R. P., et al. 2008. Description of four novel psychrophilic, ionizing radiation-sensitive Deinococcus species from alpine environments. Int. J. Evol. Microbiol. 58:1252–1258 [DOI] [PubMed] [Google Scholar]
- 14. Carpenter E. J., Lin S., Capone D. G. 2000. Bacterial activity in South Pole snow. Appl. Environ. Microbiol. 66:4514–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christner B. C., et al. 2008. Geographic, seasonal, and precipitation chemistry influence on the abundance and activity of biological ice nucleators in rain and snow. Proc. Natl. Acad. Sci. U. S. A. 105:18854–18859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diez B., Pedrós-Alió C., Massana R. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esteban G. F., Finlay B. J. 1996. Morphology and ecology of the cosmopolitan ciliate Prorodon viridis. Arch. Protistenkunde 147:181–188 [Google Scholar]
- 18. Felip M., Sattler B., Psenner R., Catalan J. 1995. Highly active microbial communities in the ice and snow cover of high mountain lakes. Appl. Environ. Microbiol. 61:2394–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finlay B. J., Fenchel T. 2004. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist 155:237–244 [DOI] [PubMed] [Google Scholar]
- 20. Galand P. E., Lovejoy C., Pouliot J., Garneau M. -È, Vincent W. F. 2008. Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: a stamukhi lake and its source waters. Limnol. Oceanogr. 53:813–823 [Google Scholar]
- 21. Galand P. E., Potvin M., Casamayor E. O., Lovejoy C. 2010. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 4:564–576 [DOI] [PubMed] [Google Scholar]
- 22. Genitsaris S., Moustaka-Gouni M., Kormas K. A. 2011. Airborne microeukaryote colonists in experimental water containers: diversity, succession, life histories and established food webs. Aquat. Microb. Ecol. 62:139–152 [Google Scholar]
- 23. Gómez F., Moreira D., López-Garcia P. 2010. Neoceratium gen. nov., a new genus for all marine species currently assigned to Ceratium (Dinophyceae). Protist 161:35–54 [DOI] [PubMed] [Google Scholar]
- 24. Griffin D. W., Kellogg C. A., Garrison V. H., Shinn E. A. 2002. The global transport of dust: an intercontinental river of dust, microorganisms and toxic chemicals flows through the Earth's atmosphere. Am. Sci. 90:228–235 [Google Scholar]
- 25. Hammer O., Harper D. A. T., Ryan P. D. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4:1–9 [Google Scholar]
- 26. Hiraishi A., Imhoff J. F. 2005. Genus IX. Rhodoferax Hiraishi, Hoshino, and Satoh 1992a, 192VP, p. 727–732 In Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part C. Springer, New York, NY [Google Scholar]
- 27. Juhnke M. E., Mathre D. E., Sands D. C. 1987. Identification and characterization of rhizosphere-competent bacteria of wheat. Appl. Environ. Microbiol. 53:2793–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jungblut A.-D., et al. 2005. Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol. 7:519–529 [DOI] [PubMed] [Google Scholar]
- 29. Jungblut A. D., Lovejoy C., Vincent W. F. 2010. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 4:191–202 [DOI] [PubMed] [Google Scholar]
- 30. Lemmen D. S. 1990. Glaciomarine sedimentation in Disraeli Fjord, High Arctic Canada. Mar. Geol. 94:9–22 [Google Scholar]
- 31. Leslie A. 1879. The arctic voyages of Adolf Erik Nordenskiöld. MacMillan and Co., London, United Kingdom [Google Scholar]
- 32. Liu Y. Q., et al. 2009. Bacterial diversity in the snow over Tibetan Plateau Glaciers. Extremophiles 13:411–423 [DOI] [PubMed] [Google Scholar]
- 33. Logares R., et al. 2009. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 17:414–422 [DOI] [PubMed] [Google Scholar]
- 34. Logares R., Bråte J., Heinrich F., Shalchian-Tabrizi K., Bertilsson S. 2010. Infrequent transitions between saline and fresh waters in one of the most abundant microbial lineages (SAR11). Mol. Biol. Evol. 27:347–357 [DOI] [PubMed] [Google Scholar]
- 35. Lovejoy C., Potvin M. 2011. Microbial eukaryotic distribution in a dynamic Beaufort Sea and the Arctic Ocean. J. Plankton Res. 33:431–444 [Google Scholar]
- 36. Lovejoy C., et al. 2007. Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in arctic seas. J. Phycol. 43:78–89 [Google Scholar]
- 37. Medlin L., Elwood H. J., Stickel S., Sogin M. L. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499 [DOI] [PubMed] [Google Scholar]
- 38. Montresor M., Lovejoy C., Orsini L., Procaccini G., Roy S. 2003. Bipolar distribution of the cyst-forming dinoflagellate Polarella glacialis. Polar Biol. 26:186–194 [Google Scholar]
- 39. Mueller D., Vincent W. F., Jeffries M. O. 2006. Environmental gradients, fragmented habitats, and microbiota of a northern ice shelf cryoecosystem, Ellesmere Island, Canada. Arct. Antarct. Alp. Res. 38:593–607 [Google Scholar]
- 40. Mueller D. R., Van Hove P., Antoniades D., Jeffries M. O., Vincent W. F. 2009. High arctic lakes as sentinel ecosystems: cascading regime shifts in climate, ice cover, and mixing. Limnol. Oceanogr. 54:2371–2385 [Google Scholar]
- 41. Mueller D. R., Vincent W. F. 2006. Microbial habitat dynamics and ablation control on the Ward Hunt Ice Shelf. Hydrol. Proc. 20:857–876 [Google Scholar]
- 42. Mueller D. R., Vincent W. F., Bonilla S., Laurion I. 2005. Extremotrophs, extremophiles and broadband pigmentation strategies in a high arctic ice shelf ecosystem. FEMS Microbiol. Ecol. 53:73–87 [DOI] [PubMed] [Google Scholar]
- 43. Palleroni N. J. 2005. Genus I. Burkholderia Yabuuchi, Kosako, Oyaizu, Yano, Hotta, Hashimoto, Ezaki and Arakawa 1993, 398VP, p. 575–600 In Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (ed.), Bergey's manual of systematic bacteriolgy, 2nd edition, vol. 2, part C. Springer, New York, NY [Google Scholar]
- 44. Papke R. T., Ramsing N. B., Bateson M. M., Ward D. M. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650–659 [DOI] [PubMed] [Google Scholar]
- 45. Pearce D. A., Hughes K. A., Lachlan-Cope T., Harangozo S. A., Jones A. E. 2010. Biodiversity of air-borne microorganisms at Halley Station, Antarctica. Extremophiles 14:145–159 [DOI] [PubMed] [Google Scholar]
- 46. Piot M., von Glasow R. 2008. The potential importance of frost flowers, recycling on snow, and open leads for ozone depletion events. Atmos. Chem. Phys. 8:2437–2467 [Google Scholar]
- 47. Porter K. G., Feig Y. S. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943–948 [Google Scholar]
- 48. Pot B., Gillis M. 2005. Genus III. Aquaspirillum Hylemon, Wells, Krieg and Jannasch 1973b, 36IAL, p. 801–823 In Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part C. Springer, New York, NY [Google Scholar]
- 49. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61 [Google Scholar]
- 50. Rodrigues D. F., et al. 2009. Biogeography of two cold-adapted genera: Psychrobacter and Exiguobacterium. ISME J. 3:658–665 [DOI] [PubMed] [Google Scholar]
- 51. Rosselló-Mora R., Amann R. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39–67 [DOI] [PubMed] [Google Scholar]
- 52. Sattler B., Puxbaum H., Psenner R. 2001. Bacterial growth in supercooled cloud droplets. Geophys. Res. Lett. 28:239–242 [Google Scholar]
- 53. Schindler D. W., Welch H. E., Kalff J., Brunskill G. J., Kritsch N. 1974. Physical and chemical limnology of Char Lake, Cornwallis Island (75 °N lat.). J. Fish. Res. Board Can. 31:585–607 [Google Scholar]
- 54. Schloss P. D., et al. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Staley J. T., Gosink J. J. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189–215 [DOI] [PubMed] [Google Scholar]
- 56. Stanier R. Y., Palleroni N. J., Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159–271 [DOI] [PubMed] [Google Scholar]
- 57. Tillett D., Neilan B. A. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251–258 [Google Scholar]
- 58. Van Houdt R., et al. 2009. Evaluation of the airborne bacterial population in the periodically confined Antarctic base Concordia. Microb. Ecol. 57:640–648 [DOI] [PubMed] [Google Scholar]
- 59. Van Hove P., Belzile C., Gibson J. A. E., Vincent W. F. 2006. Coupled landscape-lake evolution in High Arctic Canada. Can. J. Earth Sci. 43:533–546 [Google Scholar]
- 60. Varin T., Lovejoy C., Jungblut A. D., Vincent W. F., Corbeil J. 2010. Metagenomic profiling of Arctic microbial mat communities as nutrient scavenging and recycling systems. Limnol. Oceanogr. 55:1901–1911 [Google Scholar]
- 61. Vézina S., Vincent W. F. 1997. Arctic cyanobacteria and limnological properties of their environment: Bylot Island, Northwest Territories, Canada (73°N, 80°W). Polar Biol. 17:523–534 [Google Scholar]
- 62. Vincent W. F. 2010. Microbial ecosystem responses to rapid climate change in the Arctic. ISME J. 4:1089–1091 [DOI] [PubMed] [Google Scholar]
- 63. Vincent W. F., Mueller D. R., Bonilla S. 2004. Ecosystems on ice: the microbial ecology of Markham Ice Shelf in the high Arctic. Cryobiology 48:103–112 [DOI] [PubMed] [Google Scholar]
- 64. Vincent W. F., et al. 2009. Arctic microbial ecosystems and impacts of extreme warming during the International Polar Year. Polar Sci. 3:171–180 [Google Scholar]
- 65. Wainwright M., Wickramasinghe N. C., Narlikar J. V., Rajaratnam P. 2003. Microorganisms cultured from stratospheric air samples obtained at 41 km. FEMS Microbiol. Lett. 218:161–165 [DOI] [PubMed] [Google Scholar]
- 66. Whitaker R. J., Grogan D. W., Taylor J. W. 2003. Geographic barriers isolate endemic populations of hyperthermophilic Archaea. Science 301:976–978 [DOI] [PubMed] [Google Scholar]
- 67. Xiang S. R., Shang T. C., Chen Y., Yao T. D. 2009. Deposition and postdeposition mechanisms as possible drivers of microbial population variability in glacier ice. FEMS Microbiol. Ecol. 70:165–176 [DOI] [PubMed] [Google Scholar]
- 68. Yagi J. M., Sims D., Brettin T., Bruce D., Madsen E. L. 2009. The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ. Microbiol. 11:2253–2270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.