Abstract
The effects of avilamycin, zinc bacitracin, and flavophospholipol on broiler gut microbial community colonization and bird performance in the first 17 days posthatch were investigated. Significant differences in gut microbiota associated with gut section, dietary treatment, and age were identified by terminal restriction fragment length polymorphism (T-RFLP), although no performance-related differences between dietary treatments were detected. Similar age-related shifts in the gut microbiota were identified regardless of diet but varied between the ilea and ceca. Interbird variabilities in ileal bacterial communities were reduced (3 to 7 days posthatch) in chicks fed with feed containing antimicrobial agents. Avilamycin and flavophospholipol had the most consistent effect on gut microbial communities. Operational taxonomic units (OTU) linked to changes in gut microbiota in birds on antimicrobial-supplemented diets were characterized and identified. Some OTUs could be identified to the species level; however, the majority could be only tentatively classified to the genus, family, order, or domain level. OTUs 140 to 146 (Lachnospiraceae), OTU 186/188 (Lactobacillus johnsonii), OTU 220 (Lachnospiraceae), OTUs 284 to 288 (unclassified bacterial spp. or Ruminococcaceae), OTU 296/298 (unclassified bacterium or Clostridiales), and OTU 480/482 (Oxalobacteraceae) were less prevalent in the guts of chicks fed antimicrobial-supplemented diets. OTU 178/180 (Lactobacillus crispatus), OTU 152 (Lactobacillus reuteri or unclassified Clostridiales), OTU 198/200 (Subdoligranulum spp.), and OTU 490/492 (unclassified bacterium or Enterobacteriaceae) were less prevalent in the gut of chicks raised on the antimicrobial-free diet. The identification of key bacterial species influenced by antimicrobial-supplemented feed immediately posthatch may assist in the formulation of diets that facilitate beneficial gut microbial colonization and, hence, the development of alternatives to current antimicrobial agents in feed for sustainable poultry production.
INTRODUCTION
The initial microbiota to which chicks are exposed as well as the nutrient composition of their diet affect their commensal gut microbiota, host gene expression, and immune system development (50, 62). During the first week posthatch the growth of the chicken gastrointestinal system far exceeds that of other organs in the body and is essential if the bird is to achieve its genetic potential (42). The primary function of the gastrointestinal tract is to absorb nutrients from the diet and excrete waste products; however, it also contains a unique microbial ecosystem that is affected by dietary nutrients, host secretions, and the systemic responses of the host (31, 42, 48).
The gastrointestinal microbiota has one of the highest cell densities of any ecosystem and in poultry ranges from 107 to 1011 bacteria per gram of gut content (3). The collective microbial genome (microbiome) has a coding capacity that vastly exceeds that of the host's genome and encodes biochemical pathways which the host has not evolved (13). Dominant bacteria identified within the ileum of broilers are lactobacilli and related genera, while those in the cecum are related predominantly to clostridia (4, 22, 35). The complexity and composition of the gut microbiota have been shown to increase as birds age (21, 28, 58). The most dramatic changes are evident within the first 2 weeks posthatch (23, 35, 57).
Antimicrobial agents in feed have been widely used since the 1950s to improve feed efficiency and animal growth through the modulation of the gut microbiota and the host's immune response (41) as well as to reduce morbidity and mortality due to clinical and/or subclinical disease (15). Results of previous studies investigating the effects of antimicrobials in feed on broiler performance and gut microbiota have varied. Broiler performance-related differences in response to antimicrobials given in feed have not always been associated with differences in the gut microbiota (38, 39, 43). Conversely, reports of changes in the gut microbiota composition as a result of antimicrobial agents in feed have not always been accompanied by broiler performance differences or performance data (4, 5, 14, 20, 21, 25, 32, 44, 63). Since the ban on antibiotics for growth promotion within the European Union was established in 2006, much interest has focused on the role of the gut microbiota in animal health, production, and product safety. However, alternatives to antimicrobials in feed have been evaluated in poultry, with various results (19, 38, 39). Knowledge of changes in broiler gut microbiota colonization and succession immediately posthatch, and the impact that dietary modification has on this, is essential if we are to reduce our reliance on antimicrobials in feed for poultry production. Such knowledge will assist in manipulating the gut microbiota in a beneficial way via diet or additives or isolating organisms that might be used as probiotics.
The variable results among different studies of antimicrobial agents in feed may be due in part to the methods used to investigate the gut bacterial communities and the differences in the sensitivities and specificities of these techniques. Evaluations of the effects of antimicrobials in feed on intestinal bacteria have been done either in vitro (47) or in vivo using both culture-dependent (14, 25, 39, 43) and culture-independent (20, 44, 61) methods. Culture-dependent techniques are limited by the culture conditions and media utilized, while culture-independent microbial profiling techniques, such as denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP), are able to detect most bacteria within the community structure. Few studies have investigated the influence that antimicrobials in feed have on the development and overall gut microbial communities in poultry (20, 44), and even fewer studies actually identified the potential bacterial species changing within the community in response to these compounds (21, 63). Hence, little research has been conducted to systematically evaluate the potential effects that antimicrobials in feed may have on the dynamics of the overall gut microbiota in broiler chickens.
The three antimicrobials (avilamycin, flavophospholipol, and zinc bacitracin) investigated in this study were chosen because of their relevance to the poultry industry internationally (49) and because they were reported previously to have various modes of action against bacteria in vitro (8). These three antimicrobials all belong to different antibiotic groups and classes with various mechanisms of action. Flavophospholipol (also known as bambermycin, moenomycin, and flavomycin) is a glycolipid antibiotic (phosphoglycolipid) produced by Streptomyces species, while bacitracin is a polypeptide antibiotic (cyclic peptide) produced by Bacillus licheniformis (8, 46). Both antibiotics inhibit bacterial cell wall synthesis through slightly different mechanisms. Avilamycin is an oligosaccharide antibiotic (orthosomycin) produced by Streptomyces viridochromogenes that inhibits protein synthesis (8). All three antibiotics have been reported to be active mainly against Gram-positive bacteria (8).
The aims of this study were to characterize the normal gut microbiota development in the first 17 days posthatch and determine if three antimicrobials given in feed (avilamycin, flavophospholipol, and zinc bacitracin) influence gut microbiota development and succession. Knowledge of the impacts of antimicrobial agents on the intestinal microbial ecology may give insight into the development of non-antibiotic-based methods to improve growth performance and health in broilers.
MATERIALS AND METHODS
Birds and housing.
All experimental work with animals was done at the Pig and Poultry Production Institute (PPPI), Roseworthy Campus, University of Adelaide, with animal ethics approval from both the Department of Primary Industries and Resources of South Australia (PIRSA) and the University of Adelaide. Newly hatched (n = 640) feather-sexed male broiler chicks (Cobb 500) were obtained from a local hatchery (Baiada Hatchery, Gawler, South Australia). Upon arrival chicks were weighed in groups of 40 and allocated to 1 of 16 raised-floor pens (0.9 m by 1.8 m) within a climate-controlled room. The floor of each pen was covered with brown paper and spread with fresh pine sawdust. Each pen also had its own feeder, drinker, and brooding lamp for warmth. The experiment had a 4-by-4 randomized-block design with four replicate pens receiving one of four diets from hatch (n = 160/treatment). The four experimental diets were based on a standard commercial starter diet (Steg 600 starter; Ridley Agriproducts, Australia) without any coccidiostats added and included the following components: the commercial starter crumbles without the addition of an antibiotic (control diet), control diet with the addition of zinc bacitracin (50 ppm), control diet with the addition of flavophospholipol (2 ppm), and control diet with the addition of avilamycin (15 ppm).
The pen bird weight was measured on the day of hatching (day 1), and pen bird weights and amounts of feed consumed were then recorded at days 3, 5, 7, 10, 12, 14, and 17 posthatch to allow the weight gain and feed conversion ratio (FCR) to be calculated as follows: FCR = pen weight gain (live + dead chicks)/feed consumed. Live weight was also recorded for individual chicks that were euthanized for microbial profiling.
Microbial profiling. (i) Sample collection and nucleic acid extraction.
At 3 and 5 days posthatch, six chicks per pen were taken (n = 24 birds/dietary treatment), while at 7, 10, 12, 14, and 17 days posthatch, three chicks per pen were taken (n = 12 birds/dietary treatment) for microbial profiling. Chicks were euthanized by cervical dislocation. An approximately 2-cm section of the ileum, midway between the Meckel's diverticulum and cecal junction, as well as both ceca were collected from each chick. Gut samples (tissue and associated digesta) obtained from chicks aged 3 or 5 days were pooled due to the limited amount of material available from these young birds. From the six chicks taken per pen at 3 or 5 days of age, gut samples were pooled (n = 2) to give three pooled samples per pen. Samples collected from birds 7 to 17 days posthatch were not pooled. Care was taken to not cross-contaminate samples by cleaning dissection instruments with 70% ethanol and changing gloves between dissections. Following collection, samples were kept on ice until frozen at −20°C and then freeze-dried. Total nucleic acid was extracted from the chicken gut samples by a modification (56) of a proprietary extraction method developed by the South Australian Research and Development Institute (52).
(ii) T-RFLP.
T-RFLP analysis was done on individual samples according to a technique described previously by Torok et al. (56). The bacterial rRNA gene was amplified with universal 16S bacterial primers 27F (33) and 907R (40). The forward primer (27F) was 5′ labeled with 6-carboxyfluorescein (FAM) to enable the subsequent detection of terminal restriction fragments (T-RFs). PCRs were done in duplicate in 50-μl volumes according to methods described previously by Torok et al. (56). Following PCR, all amplification products were quantified by fluorometry, and duplicate PCRs, which varied by less than 20% in fluorescein counts, were pooled (56). The specificities of the PCR products were analyzed by gel electrophoresis on a 1.5% agarose gel and visualized after staining with ethidium bromide. Approximately 200 ng of PCR product was digested with 2 U MspI (Genesearch, Arundel, Australia) in duplicate according to the manufacturer's instructions. The lengths of fluorescently labeled T-RFs were determined by comparison with an internal size standard (GeneScan 1200 LIZ; Applied Biosystems, Australia) following separation by capillary electrophoresis on an ABI 3730 automated DNA sequencer (Applied Biosystems, Australia). Data were analyzed by using GeneMapper v3.7 software (Applied Biosystems, Australia). Data points generated by the GeneMapper software were further analyzed by using a custom-built database containing queries to validate data points and generate outputs for statistical analysis (56). T-RFs were defined as peaks with a size of x ± 2 bp within pseudoreplicates of samples and rounded to the nearest even number between samples to produce operational taxonomic units (OTUs).
(iii) Lac-PCR DGGE.
Lactobacillus-specific PCR (Lac-PCR) DGGE analysis was used to investigate the diversity of Lactobacillus species and related genera within the ilea. Group-specific Lactobacillus primers Lac1 and Lac2-GC (59) were used to amplify the V3 region of the 16S rRNA gene from total DNA by using a Cool Gradient Palm Cycler 9600 instrument (Corbett Research, Sydney, Australia). Pooled DNA was used as a template. The pooled samples were prepared by combining the same amounts of DNA from the ilea of birds taken from the same pen (n = 3). The PCR products were subjected to DGGE (Lac-PCR DGGE) using the Bio-Rad (Hercules, CA) DCode universal mutation detection system as outlined previously (59). The identification ladders for DGGE were prepared by combining the Lac-PCR products from DNA extracted from the reference strains (Lactobacillus acidophilus ATCC 4356, Lactobacillus crispatus ATCC 33820, Lactobacillus gasseri ATCC 33323, Lactobacillus johnsonii ATCC 33200, Lactobacillus reuteri ATCC 23272, Lactobacillus salivarius subsp. salivarius ATCC 11741, Pediococcus acidilactici ATCC 8042, and Pediococcus pentosaceus ATCC 43200). L. crispatus, Lactobacillus gallinarum, and Lactobacillus amylovorus belong to the group A L. acidophilus taxonomic group, which cannot be distinguished by using Lac-PCR DGGE (23), and will be referred to here collectively as LCGA. Gels were stained with ethidium bromide and viewed by UV transillumination. The Lac-PCR DGGE band mobility and presence or absence were determined with the BioNumerics software package (Applied Maths, Sint-Martens-Latem, Belgium).
Statistical analysis.
Performance data were analyzed with the SAS for Windows, version 9.1, software package (Base SAS software; SAS Institute, Inc., Cary, NC). A univariate analysis of variance (ANOVA) was used to determine the effects of block and antimicrobial agents in feed (fixed factors) on bird performance, as measured by the live weight, feed consumed, and FCR, by using the general linear model (GLM), with differences between treatments being determined by Duncan's multiple-range test. ANOVA was also used to determine the effects of antimicrobial agents in feed on live weights of individual chicks taken for microbial profiling. Pearson's chi-square test (51) was used to determine whether incidences of mortality differed among antibiotic treatment groups and the control group over the experimental period.
OTUs obtained from the ileal and cecal contents of 240 individual broiler chicks aged 7 to 17 days and 96 pooled (n = 2) samples from birds aged 3 and 5 days were analyzed by using multivariate statistical techniques (PRIMER 6; PRIMER-E Ltd., Plymouth, United Kingdom). These analyses were used to examine similarities in chicken ileal and cecal bacterial communities associated with age and treatment with antimicrobials in feed. Bray-Curtis measures of similarity (7) were calculated to examine similarities between gut microbial communities of birds from the T-RFLP data (following standardization and fourth-root transformation) and the presence or absence Lac-PCR DGGE data (scored against the reference Lactobacillus strains). A two-way crossed analysis of similarity (ANOSIM) (9) was used to test whether gut microbial communities were significantly different between gut sections and diets for each age group investigated. A one-way ANOSIM (9) was used to test if ileal or cecal microbial communities were significantly different between age groups for each dietary treatment and among dietary treatments for each age group. The R statistic value describes the extent of similarity between each pair in the ANOSIM, with values close to unity indicating that the two groups are entirely separate and a zero value indicating that there is no difference between the groups. Analysis of similarity percentages (SIMPER) (9) was done to determine the overall average similarity in ileal or cecal microbial community compositions among birds fed the same diet for each age group. SIMPER was also done to determine OTUs driving significant differences in bacterial community compositions between dietary treatments (9, 56). Hierarchical cluster analysis (CLUSTER) (9) using the group average cluster mode on Bray-Curtis similarity data was done to show changes in gut microbial communities associated with age.
Cloning and sequencing of OTUs. (i) Isolation of OTUs of interest.
T-RFs were isolated from OTUs significantly associated with dietary treatments. Multiple samples containing OTUs of interest were targeted. Where possible, samples which lacked other OTUs within ±10 bp were chosen. A combination of adapter ligation, fragment size selection, and reamplification with adapter-specific PCR was used to isolate T-RFs of interest as described previously by Widmer et al. (60). T-RFs were characterized by a specific PCR primer sequence (27F) at the 5′ end and a specific restriction site (MspI) at the 3′ end. As this structure does not allow the direct reamplification and further characterization of a T-RF, a specific adapter matching the restriction site at the 3′ end of the T-RF and containing a known PCR priming site was ligated into restriction digests from samples containing OTUs of interest. The double-stranded MspI-adapter structure was prepared and ligated into restriction fragments as described previously by Widmer et al. (60). The size selection of T-RFs of interest was done by gel electrophoresis with an SEA 2000 electrophoresis apparatus (Elchrom Scientific, Inc., Switzerland) using precast Spreadex gels (EL 400, 600, 800, or 1200; Elchrom Scientific, Inc., Switzerland). Spreadex gel types and electrophoresis conditions were chosen based on the desired T-RF size range and were calculated by using the Gel Selection Guide and Virtual Electrophoresis software (Elchrom Scientific, Inc., Switzerland). Ten-microliter ligation products were electrophoresed along with 50-bp DNA ladder (New England BioLabs-Genesearch, Australia) and GeneRuler 100-bp DNA ladder (MBI Fermentas-Quantum Scientific, Australia) size standards to allow size estimations. A size range of approximately ±50 bp of the T-RF of interest was excised from the gel. The gel slice was cut into equal pieces, with each piece corresponding to a size range of approximately 12 bp. DNA was eluted from the gels as described previously by Widmer et al. (60).
(ii) PCR amplification and cloning of isolated OTUs.
Eluted DNA was used as a template for PCR amplification with primer 27F and the MspI-adapter-primer construct (60). Six microliters of DNA template was amplified in a reaction volume of 30 μl containing 1× PCR buffer (Applied Biosystems, Scoresby, Australia), 0.2 μM each primer (Sigma-Aldrich, Castle Hill, Australia), 2 mM MgCl2, 0.8 mM deoxynucleoside triphosphates (dNTPs) (Invitrogen, Mulgrave, Australia), and 1 U AmpliTaq (Applied Biosystems, Scoresby, Australia). PCR amplification was done with an MJ Research PTC-225 Peltier thermal cycler (GeneWorks, Adelaide, Australia), with an initial denaturation step for 5 min at 94°C, followed by 30 cycles with denaturation at 94°C for 45 s, annealing at 60°C for 60 s, and extension at 72°C for 90 s and a final extension step of 72°C for 5 min. PCR products were analyzed on a 2% agarose gel and visualized following staining with ethidium bromide. Single amplification products of the expected size range were excised and purified by using the NucleoSpin Extract II Macherey-Nagel kit (Scientifix, Clayton, Australia) according to the manufacturer's instructions. Purified products were ligated into the pGEM-T vector (Promega, Australia) and transformed into competent JM109 cells (Promega, Australia) according to the manufacturer's recommendations. Recombinant clones were identified by blue-white color selection (46a) and examined for the presence of recombinant plasmids by PCR using standard T7 and SP6 vector primers. Recombinant clones were grown overnight in Luria broth containing 150 μg/ml ampicillin (46a). Plasmids were purified by using the NucleoSpin Plasmid Macherey-Nagel kit (Scientifix, Clayton, Australia) according to the manufacturer's instructions.
(iii) 16S rRNA gene sequence analysis.
Plasmids were sequenced by Macrogen, Inc. (Seoul, South Korea). The vector sequence was removed by using Staden Package Pregap4, version 1.5 (6). Sizes of T-RFs were predicted in silico by using WatCut (Michael Palmer, University of Waterloo, Canada [http://watcut.uwaterloo.ca/watcut/watcut/template.php?act=restriction_new]). The 16S rRNA gene sequence data generated were assigned to a bacterial taxonomic hierarchy using the Ribosomal Database Project (RDP) release 10 classifier (10). The classifier estimates the classification reliability using bootstrapping. For sequences shorter than 250 bp a bootstrap cutoff threshold of 50% was used, while for longer sequences the default cutoff of 80% was used. BLASTn with nucleotide collection (nr/nt) databases and the Megablast algorithm (National Center for Biotechnology Information [NCBI]) were used to identify similarities of T-RFs to sequences available in public genome sequence databases (1). Unrooted neighbor-joining trees of 16S rRNA T-RF sequences from OTUs of interest and related sequences identified in the NCBI database (95 to 100% identity) were constructed. T-RFs were assigned to OTUs based on the closest nucleotide-length match or within ±6 bp. Sequences were aligned with ClustalW (55), and a bootstrapped (n = 500) consensus tree was created with MEGA 4 (53). The evolutionary distances were computed by using the maximum composite likelihood method (54).
Nucleotide sequence accession numbers.
Representative 16S rRNA gene sequences were deposited in the GenBank database under accession numbers HQ704902 to HQ705169.
RESULTS
Bacterial profiling of gut microbiota. (i) Role of gut section, diet, and age on composition of gut microbiota.
Multivariate statistical analysis showed that the compositions of the overall gut bacterial community were significantly different between gut sections as well as among dietary treatments for each of the age groups investigated (see Table S1 in the supplemental material). The ileal and cecal bacterial communities were different from each other regardless of diet; however, some overlap of common OTUs was observed for the gut sections (Fig. S1).
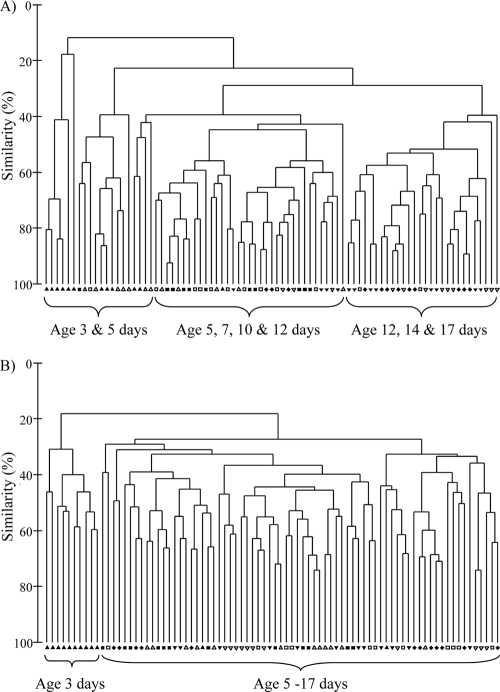
The effect of age on either the ileal or cecal bacterial community composition was also investigated for each of the dietary treatments. Within the ileum, significant age-related differences in gut microbiota compositions were detected for chicks on the control (global R = 0.274; P = 0.001), avilamycin-supplemented (global R = 0.329; P = 0.001), flavophospholipol-supplemented (global R = 0.415; P = 0.001), and zinc bacitracin-supplemented (global R = 0.360; P = 0.001) diets. Within the cecum, chick age also significantly influenced the gut bacterial community composition regardless of dietary treatment for chicks on control (global R = 0.362; P = 0.001), avilamycin-supplemented (global R = 0.500; P = 0.001), flavophospholipol-supplemented (global R = 0.572; P = 0.001), and zinc bacitracin-supplemented (global R = 0.542; P = 0.001) diets. Chicks showed a greater number of significant differences between age pairwise comparisons within the cecal bacterial communities than within the ileal bacterial communities. Overall, the temporal shifts in posthatch gut microbiota compositions were similar for all dietary treatments but varied between the ileum and cecum. Representative trends observed for chicks on the control diet are shown in Fig. 1. Three main clusters separated the ileal microbial communities for birds aged 3 to 5 days, 5 to 12 days, and 12 to 17 days (Fig. 1A). The cluster for birds aged 3 to 5 days was comprised of three subclusters, with levels of similarity in ileal microbial communities between these birds being generally lower than those for the older birds. Two main clusters separated the cecal microbial communities for birds aged 3 days and 5 to 17 days (Fig. 1B).
Fig. 1.
Dendrogram representing relationships between T-RFLP profiles of ileal (A) and cecal (B) bacterial communities from individual birds at 3, 5, 7, 10, 12, 14, and 17 days posthatch. All birds were raised on the control diet. ▴, 3 days posthatch; ▵, 5 days; ▪, 7 days; □, 10 days; ▾, 12 days; ▿, 14 days; ⧫, 17 days.
(ii) Identification of significant OTUs in the gut microbiota of chicks fed the control diet versus diets supplemented with antimicrobial agents.
Given the significance of the influence of age on the gut microbiota, the effects of antimicrobials in feed were further investigated for the ileum and ceca separately for each of the age groups. Significant (P < 0.05) diet-associated differences in bacterial community compositions were detected for both the ileum and ceca in the first 17 days posthatch, with the exception of the ceca at 14 days posthatch (Table 1). The influence of antimicrobials in feed was most evident for the ileal microbial communities. Ileal microbial communities of chicks fed diets containing flavophospholipol were significantly different from those of chicks fed the control diet for all seven age groups investigated. Zinc bacitracin had the least consistent effect on gut microbial communities compared with the control group, with differences detected only at 12 and 14 days posthatch within the ilea and 3 and 7 days posthatch within the ceca. Where significant differences were detected between the control diet and diets containing antimicrobials (Table 1), the OTUs contributing most to the dissimilarity between dietary treatments were identified for both the ileal and cecal microbial communities by SIMPER (Table 2). SIMPER identifies individual species (OTUs) contributing to the overall dissimilarity between treatments by calculating the average abundance of important OTUs for each dietary treatment. Diet-associated differences in gut microbiota were due to the presence or absence of specific OTUs as well as differences in the abundances of common OTUs.
Table 1.
One-way ANOSIM of the ileal or cecal microbial communities associated with dietary treatmentb
| Age (days) | Significance |
|
|---|---|---|
| Ileum | Cecum | |
| 3 | Global R = 0.182, P = 0.001 | Global R = 0.054, P = 0.039 |
| Control vs avilamycina | Control vs zinc bacitracin | |
| Control vs flavophospholipol | ||
| Avilamycin vs flavophospholipol | ||
| Flavophospholipol vs zinc bacitracin | ||
| 5 | Global R = 0.073, P = 0.036 | Global R = 0.272, P = 0.001 |
| Control vs flavophospholipol | Control vs avilamycin | |
| Avilamycin vs flavophospholipol | Control vs flavophospholipol | |
| Flavophospholipol vs zinc bacitracin | Avilamycin vs flavophospholipol | |
| Avilamycin vs zinc bacitracin | ||
| Flavophospholipol vs zinc bacitracin | ||
| 7 | Global R = 0.076, P = 0.008 | Global R = 0.117, P = 0.002 |
| Control vs flavophospholipol | Control vs flavophospholipol | |
| Flavophospholipol vs zinc bacitracin | Control vs zinc bacitracin | |
| Flavophospholipol vs zinc bacitracin | ||
| 10 | Global R = 0.067, P = 0.022 | Global R = 0.088, P = 0.014 |
| Control vs flavophospholipol | Avilamycin vs zinc bacitracin | |
| Avilamycin vs zinc bacitracin | Flavophospholipol vs zinc bacitracin | |
| Flavophospholipol vs zinc bacitracin | Avilamycin vs flavophospholipol | |
| 12 | Global R = 0.187, P = 0.001 | Global R = 0.087, P = 0.005 |
| Control vs avilamycin | Avilamycin vs flavophospholipol | |
| Control vs zinc bacitracin | Avilamycin vs zinc bacitracin | |
| Control vs flavophospholipol | Flavophospholipol vs zinc bacitracin | |
| Flavophospholipol vs zinc bacitracin | ||
| 14 | Global R = 0.153, P = 0.003 | Global R = 0.040, P = 0.102 |
| Control vs avilamycin | ||
| Control vs zinc bacitracin | ||
| Control vs flavophospholipol | ||
| Flavophospholipol vs zinc bacitracin | ||
| 17 | Global R = 0.222, P = 0.001 | Global R = 0.170, P = 0.001 |
| Control vs avilamycin | Control vs avilamycin | |
| Control vs flavophospholipol | Control vs flavophospholipol | |
| Avilamycin vs zinc bacitracin | Avilamycin vs flavophospholipol | |
| Flavophospholipol vs zinc bacitracin | Avilamycin vs zinc bacitracin | |
| Flavophospholipol vs zinc bacitracin | ||
Significant pairwise comparisons identified (P < 0.05).
For each age group the influence of dietary supplementation with antimicrobials on the gut microbiota was investigated. Where significant differences in the gut microbiota were identified, the significant pairwise differences between dietary treatments are listed.
Table 2.
OTUs contributing significantly to differences in microbial communities between chicks on control versus antimicrobial-supplemented diets
| Age of birda (days) | Treatment differing from control | Discriminating OTU(s) for groupb |
|
|---|---|---|---|
| Control | Antimicrobial treated | ||
| Ileum | |||
| 3 | Avilamycin | 220 | 60, 86, 492, 518, 536, 560/562 |
| Flavophospholipol | 220 | 86, 180, 186, 492, 518, 560, 566 | |
| 5 | Flavophospholipol | 186, 574 | 176, 180, 210/212, 566, 938 |
| 7 | Flavophospholipol | 186/188, 574 | 170, 178, 184, 210, 248 |
| 10 | Flavophospholipol | 186, 574 | 178/180, 212, 936 |
| 12 | Avilamycin | 186, 284/286, 936 | 152, 176, 212, 574 |
| Flavophospholipol | 186, 284/286, 894 | 152, 170, 176/178, 212, 936 | |
| Zinc bacitracin | 78, 86, 186, 284/286, 894 | 152, 176, 178/180, 212 | |
| 14 | Avilamycin | 86, 284/286, 574, 894 | 70, 180, 566 |
| Flavophospholipol | 86, 284/286, 894 | 70, 80, 180, 518, 566 | |
| Zinc bacitracin | 86, 284/286, 574, 894 | 70, 178/180, 566 | |
| 17 | Avilamycin | 86, 186/188, 284/286, 574, 894, 936 | 70, 178/180, 566 |
| Flavophospholipol | 86, 186/188, 284/286, 574, 894 | 70, 178/180, 566, 936 | |
| Ceca | |||
| 3 | Zinc bacitracin | 220 | 198, 910 |
| 5 | Avilamycin | 140/142, 180, 286/288, 296, 482 | |
| Flavophospholipol | 140/142, 286/288, 290, 296, 480/482, 564 | 178/180, 200, 216/218, 476 | |
| 7 | Flavophospholipol | 140/142 | 178/180, 198/200, 286, 476, 520, 564 |
| 17 | Avilamycin | 78, 144/146, 284/286, 296/298, 482, 536, 910 | 68, 140/142, 198, 216/218, 520 |
| Flavophospholipol | 78, 140/142, 144/146, 284/286, 536, 910 | 216, 294, 310, 490 | |
Age groups where significant differences between control and antimicrobial treatment groups were detected in gut microbial community compositions as determined by ANOSIM (Table 1).
OTUs identified by SIMPER as significantly discriminating between control and antimicrobial treatment groups. OTUs identified as being more abundant within a particular treatment group are indicated. OTUs in boldface type have been cloned and sequenced.
(iii) Interbird variation and representation of significant OTUs.
Similarities in gut bacterial communities among birds on the same diet were also calculated with SIMPER. The similarities in ileal microbial community compositions among chicks on the same dietary treatment ranged from 29 to 61% within the first 17 days posthatch, indicating high interchick variability in gut bacterial community compositions. The ileal bacterial community similarity was lower for the control group from 3 to 7 days posthatch (29 to 50%) than for the avilamycin-treated (43 to 55%), flavophospholipol-treated (45 to 67%), and zinc bacitracin-treated (38 to 58%) groups of the same age. Within the cecal microbial community, similarities among chicks on the same dietary treatment were lower than those for the ilea and ranged from 34 to 59% in the first 17 days posthatch. Cecal bacterial communities at 17 days posthatch were more complex (n = 94 OTUs) than the ileal bacterial communities (n = 58 OTUs). Chicks aged 3 to 5 days had lower cecal microbial community similarities within the avilamycin-treated (35 to 39%) and control (35 to 43%) groups than within the flavophospholipol-treated (50 to 52%) and zinc bacitracin-treated (50 to 59%) groups. However, by 17 days posthatch all treatments had comparable cecal microbial community similarities of 37 to 42%. Due to the interchick variability in both the ileal and cecal bacterial community compositions of birds on a particular diet, visual observations of treatment differences are highly subjective (see Fig. S2 and Fig. S3, respectively, in the supplemental material), hence supporting the need for a robust statistical analysis of such data.
(iv) Sequence identification of OTUs.
The targeted cloning and sequencing of OTUs generated 16S rRNA gene T-RF sequence information from 268 clones. Sixty percent of these sequences potentially represented treatment-specific OTUs, while the remainder represented nonsignificant OTUs contributing to the commensal gut microbiota. Targeted cloning and sequencing identified approximately one-third of all significant OTUs (Table 2). All 16S rRNA sequence information generated, representing both the significant diet-associated and other commensal bacteria, were classified according to the hierarchy of domain, phylum, class, order, family, and genus. Some of the cloned T-RF sequences could be classified only as unidentifiable bacteria. However, where sequences could be classified to the level of phyla, they belonged to the Firmicutes, Bacteroidetes, and Proteobacteria. Many sequences could be further classified to the level of class (Bacilli and Clostridia), order (Clostridiales and Lactobacillales), or even family (Lachnospiraceae, Lactobacillaceae, Enterobacteriaceae, Ruminococcaceae, and Oxalobacteraceae). In some cases the sequence could be classified to the level of genera and included Acinetobacter, Anaerotruncus, Clostridium, Dorea, Enterococcus, Erysipelotrichaceae incertae sedis, Escherichia, Faecalibacterium, Klebsiella, Lactobacillus, Lachnospiraceae incertae sedis, Oscillibacter, Pedobacter, Proteus, Ruminococcaceae incertae sedis, Shigella, Shuttleworthia, Subdoligranulum, and Syntrophococcus.
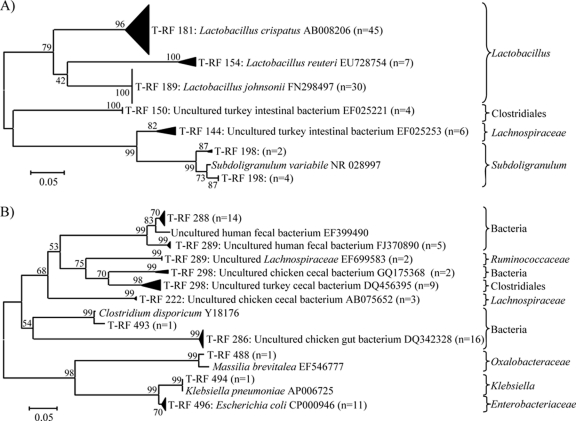
Only the 16S rRNA T-RF sequence data generated from significant OTUs associated with dietary treatment were used to construct phylogenetic trees (Fig. 2). Some T-RFs were shown to form several clades on the phylogenetic tree: T-RF 198 formed two clades related to Subdoligranulum spp. (Fig. 2A), while T-RF 289 and T-RF 298 each formed two clades related to an unclassified bacterium or the Ruminococcaceae and an unclassified bacterium or the Clostridiales, respectively (Fig. 2B). Significant OTUs that the T-RFs are likely to represent as well as the treatment with which they are predominantly associated are indicated in Table 3. Discrepancies between sequence-determined T-RFs and observed fragment sizes (T-RFLP-generated OTUs) were evident. Furthermore, some T-RFLP-generated OTUs represented several bacterial groups, such as OTUs 152, 198/200, 284 to 288, 296/298, and 490/492 (Table 3).
Fig. 2.
Unrooted neighbor-joining phylogenetic tree of 16S rRNA gene sequences (T-RFs) obtained by the targeted cloning and sequencing of OTUs identified as being significantly affected by dietary treatment. (A) Phylogenetic tree from OTUs of less than 200 bp. (B) Phylogenetic tree from OTUs of greater than 200 bp. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The line bars indicate evolutionary distances and are in units of the number of base substitutions per site. The values along the branches indicate percent confidence. The numbers in parentheses indicate numbers of sequences analyzed from multiple samples. The GenBank accession numbers with the closest identity to the T-RFs are indicated. The classifications of T-RF sequences according to the RDP classifier are indicated to the right of the tree.
Table 3.
Identities of 16S rRNA gene sequence-generated OTUs and the likely significant diet-associated T-RFLP-generated OTUs that they representb
| T-RF (bp) | Taxonomic assignment | Assigned OTU(s)a | Dietary treatment | Gut section | Age(s) of birds (days) |
|---|---|---|---|---|---|
| 144 | Lachnospiraceae | 140/142 and/or 144/146 | Control | Cecum | 5, 7, 17 |
| 150 | Clostridiales | 152 | Antimicrobial | Ileum | 12 |
| 154 | Lactobacillus reuteri | 152 | Antimicrobial | Ileum | 12 |
| 181 | Lactobacillus crispatus | 178/180 | Antimicrobial | Ileum/cecum | 3–17 |
| 189 | Lactobacillus johnsonii | 186/188 | Control | Ileum | 5–12, 17 |
| 198 | Subdoligranulum sp. | 198/200 | Antimicrobial | Cecum | 3–7 |
| 198 | Subdoligranulum sp. | 198/200 | Antimicrobial | Cecum | 3–7 |
| 222 | Lachnospiraceae | 220 | Control | Ileum/cecum | 3 |
| 286 | Unclassified bacteria | 284/286 and/or 286/288 | Control | Ileum/cecum | 5–17 |
| 288 | Unclassified bacteria | 284/286 and/or 286/288 | Control | Ileum/cecum | 5–17 |
| 289 | Unclassified bacteria | 284/286 and/or 286/288 | Control | Ileum/cecum | 5–17 |
| 289 | Ruminococcaceae | 284/286 and/or 286/288 | Control | Ileum/cecum | 5–17 |
| 298 | Unclassified bacteria | 296/298 | Control | Cecum | 5, 17 |
| 298 | Clostridiales | 296/298 | Control | Cecum | 5, 17 |
| 488 | Oxalobacteraceae | 480/482 | Control | Cecum | 5, 17 |
| 493 | Unclassified bacteria | 490/492 | Antimicrobial | Ileum | 3 |
| Cecum | 17 | ||||
| 494 | Klebsiella sp. | 490/492 | Antimicrobial | Ileum | 3 |
| Cecum | 17 | ||||
| 496 | Enterobacteriaceae | 490/492 | Antimicrobial | Ileum | 3 |
| Cecum | 17 |
T-RFs were assigned to OTUs based on the closest nucleotide-length match or within ±6 bp.
Associations of T-RFLP OTU with dietary treatment, gut section, and age are indicated.
Eighteen T-RFs were found to potentially represent 12 of the OTUs significantly associated with antimicrobial treatment (Table 3). A minority (28%) of these T-RFs represented unclassified bacteria, most of which (60%) represented OTUs 284/286 and/or 286/288. Bacterial groups identified as being more abundant in the gut of chicks fed the antimicrobial-supplemented diets were T-RFLP-generated OTU 152 (T-RF 154 [L. reuteri] or T-RF 150 [Clostridiales]), OTU 178/180 (T-RF 181 [L. crispatus]), OTU 198/200 (T-RF 198 [Subdoligranulum spp.]), and OTU 490/492 (T-RF 493 [unclassified bacterium], T-RF 494 [Klebsiella], and/or T-RF 496 [Enterobacteriaceae]) (Table 3), while bacteria more abundant in the gut of chicks on the antimicrobial-free control diet were T-RFLP-generated OTU 140/142 or 142/144 (T-RF 144 [Lachnospiraceae]), OTU 186/188 (T-RF 189 [L. johnsonii]), OTU 220 (T-RF 222 [Lachnospiraceae]), OTU 284/286 or 286/288 (T-RF 286, 288, or 289 [unclassified bacterial species] or T-RF 289 [Ruminococcaceae]), OTU 296/298 (T-RF 298 [unclassified bacterium] or T-RF 298 [Clostridiales]), and OTU 480/482 (T-RF 488 [Oxalobacteraceae]) (Table 3).
(v) DGGE analysis of the ileal Lactobacillus microbiota.
All eight lactic acid bacterial reference species investigated by Lac-PCR DGGE were detected in the ileum of chicks within this study. Analysis of ileal lactobacillus profiles by Lac-PCR DGGE showed that antimicrobials in feed did not significantly (global R = 0.039; P = 0.146) influence lactobacillus communities across age groups. However, the age of birds did significantly (global R = 0.200; P = 0.001) alter lactobacillus profiles across dietary treatments. Significant differences were detected between age groups for each of the dietary treatments, with the exception of the control dietary treatment groups (global R = 0.070 and P = 0.121 for the control group, global R = 0.286 and P = 0.001 for the avilamycin group, global R = 0.141 and P = 0.001 for the flavophospholipol group, and global R = 0.312 and P = 0.001 for the zinc bacitracin group). For the antimicrobial-supplemented groups, significant differences (P < 0.05) were detected between pairwise comparisons of age for 3 versus 10 and 3 versus 12 days posthatch regardless of diet. Fewer significant pairwise age differences were observed for the ileal lactic acid bacterial communities than for the overall ileal bacterial communities.
SIMPER of Lac-PCR DGGE results showed that as birds aged, the prevalence of L. johnsonii and L. reuteri increased. Pediococcus acidilactici was detectable in chicks at 3 days of age only. Birds of all age groups carried LCGA (group A L. acidophilus taxonomic group).
Chick performance.
Broiler chick performance, as measured by the FCR, live weight, and feed intake, was not significantly influenced (P > 0.05) by the addition of antimicrobials in feed in the first 17 days posthatch (see Table S2 in the supplemental material). Body weights of birds taken for microbial profiling also did not significantly differ (P > 0.05) among chicks on the various dietary treatments for each of the age groups investigated (data not shown). Overall chick mortality rates in the first 17 days posthatch were 4.2% (control diet), 1.9% (zinc bacitracin-supplemented diet), 5.6% (flavophospholipol-supplemented diet), and 3.1% (avilamycin-supplemented diet). No significant differences were observed for mortality, expressed as a percentage of newly hatched chicks, due to dietary treatment (χ2 = 2.07; P > 0.50). All mortalities observed occurred between 3 and 10 days posthatch.
DISCUSSION
In this study, T-RFLP was successfully used to compare gut microbiota developments in the first 17 days posthatch in chicks fed four different diets: a control diet (no antimicrobial agent) versus diets supplemented with three different antimicrobials in feed (avilamycin, flavophospholipol, and zinc bacitracin). T-RFLP analysis revealed that antimicrobial treatment, age, and gut section had significant impacts on the composition of the gut microbiota. A number of OTUs were identified as being significantly different between the gut microbiota of birds fed the control diet and those of birds fed antimicrobial-supplemented diets, and these OTUs were sequenced. A number of different taxa were represented, including Lactobacillus spp. These same Lactobacillus spp. were identified within the ileum of chicks by Lac-PCR DGGE but were not identified as being significantly different among dietary treatments.
The composition of the chick ileal and cecal microbiota was found to change with age during the period of 3 to 17 days posthatch. Increases in the complexities of gut bacterial communities (21, 28, 58) as well as changes in the gut bacterial community structure (23, 27, 61, 62, 64) as broilers age have both been reported. We found that the timing of age-related shifts in gut microbiota composition were similar regardless of the dietary treatment investigated but differed between gut sections. Within the ileum, three major shifts in the overall microbial community composition were observed and occurred at 3 to 5 days, 5 to 12 days, and 12 to 17 days posthatch. The similarities in ileal microbial communities between birds aged 3 to 5 days were generally lower than those for the older chicks, indicating that the initial gut microbiota colonizing chicks can be highly variable. This is in contrast to findings described previously by Pedroso et al. (45), who reported that day-old chicks carry common microorganisms regardless of origin.
We also found that ileal lactobacillus communities changed with age for chicks on each of the antimicrobial diets. Most of the lactic acid bacterial reference species investigated (L. acidophilus, LCGA, L. gasseri, L. johnsonii, L. reuteri, L. salivarius subsp. salivarius, P. acidilactici, and P. pentosaceus) were detectable in chicks 3 to 17 days of age by Lac-PCR DGGE. The LCGA group was dominant in all age groups, while P. acidilactici was detectable only in birds aged 3 days, and L. johnsonii and L. reuteri were more prevalent in the older chicks. OTUs 178/180 (T-RF 181 [L. crispatus]) and 186/188 (T-RF 189 [L. johnsonii]) were identified by T-RFLP and 16S rRNA gene sequence information as being present in high numbers within both the ilea and ceca for all chicks aged 3 to 17 days. L. reuteri (T-RF 154) was also confirmed by 16S rRNA gene sequencing as being present within these chicks but did not represent a dominant OTU as determined by T-RFLP (OTU 152 contributed less than 6% of the total in samples where it was detected). Dominant lactobacilli previously identified within the ileum belonged to the L. crispatus, L. salivarius, or L. johnsonii group (4). Lactobacilli have also been detected within the ceca but are less dominant (4, 22, 35). Our data support the autochthonous nature of these species in the chicken gastrointestinal tract, which were reported previously to be present in birds of various ages (21, 23, 27, 32, 35).
Within the ceca, two major shifts in the overall microbial community composition were observed at 3 days and 5 to 17 days posthatch. This may indicate that the cecal microbial communities are more stable than the ileal bacterial communities. A lack of successional changes within the cecal microbiota 5 to 20 days posthatch has also been reported previously by others (28, 62). Consistent with our current observations, Hume et al. (28) also previously showed shifts in the cecal microbiota at 2 days and 5 to 20 days of age. These observed age-related changes in the gut microbiota are likely to be partially related to the physiological changes occurring in the chick's gastrointestinal tract immediately posthatch. The size of the gastrointestinal tract increases more rapidly in relation to body weight than other organs and tissues of the chick, with a maximal relative size of digestive organs being reported at 3 to 8 days posthatch (42). However, the length and diameter of the intestine continue to further increase until 14 days posthatch (42). Early exposure to different environments, along with the host's physiology, was recently shown to have a direct impact on the development of the gut microbiota and the host's gene expression in the newly hatched chick (62).
Although some OTUs were common to both the ilea and ceca, the cecal microbial communities were generally different from those of the ilea from as early as 3 days posthatch. In contrast, Lu et al. (35) found previously that the ileal and cecal microbiota were not significantly different at 3 days posthatch and that the cecal microbiota was a subset of the ileal microbial community for the first 14 days posthatch. Bacterial communities along the gut of 4-day-old chicks were also reported previously to not differ (58). These contradictions to our findings could be due to the different techniques used to investigate the gut microbiota in those previous studies. In support of our findings Yin et al. (62) showed previously that the ileal and cecal gut microbiota already differed in day-old chicks. Those authors also showed that the cecal microbial inoculum produced from adult chickens colonized the cecum in preference to the ileum of posthatch chicks, suggesting that the gut physiology favors particular bacterial communities.
In addition to the influence that age and the gut environment had on posthatch ileal and cecal microbiota development, significant differences were detected in response to the various antimicrobials examined in this study. Antibiotic treatment has been shown to alter gut and fecal bacterial species composition rather than species richness (number of bacterial species) or evenness (relative distribution) (21, 37, 44). Two of the antimicrobials (avilamycin and flavophospholipol) had significant effects on both the ileal and cecal microbial communities although more consistently within the ileum. The proximal gut microbiota was reported previously to be more susceptible to antibiotics than the distal gut (12, 61). Furthermore, interchick variability in the ileal bacterial community composition was reduced for birds fed antimicrobials (38 to 67% similarity) than for the control group (29 to 50% similarity) at 3 to 7 days posthatch. In this study zinc bacitracin was not as consistent in its effects on either the ileal or cecal microbial communities. Why a greater effect was not detected with zinc bacitracin is not known; however, a possibility could be that the bacteria colonizing the young chick's gut may have been carrying resistance genes against this particular antibiotic and/or are inherently resistant. Bacterial resistance to antimicrobials was previously reported (17, 47). Resistant Enterococcus faecalis, Enterococcus faecium, staphylococci, and lactobacilli have been identified in broiler flocks never fed zinc bacitracin but originating from hens which had received zinc bacitracin as a growth promotant (17).
Lac-PCR DDGE did not detect differences in lactobacillus communities associated with the antimicrobial agents investigated in this study. However, T-RFLP not only identified differences in overall gut microbial communities associated with antimicrobial treatment but also identified species of the lactobacilli driving these differences: OTU 186/188 (T-RF 189 [L. johnsonii]), OTU 178/180 (T-RF 181 [L. crispatus]), and OTU 152 (T-RF 154 [L. reuteri]). Both techniques have the ability to detect these particular Lactobacillus species; however, the power of statistical analysis on the Lac-PCR DGGE data was likely reduced, as samples were pooled (n = 3), with only 4 replicates per treatment. Indeed, zinc bacitracin was previously shown to significantly alter lactobacillus communities in the ileum of birds aged 25 days, as determined by Lac-PCR DGGE of individual samples (n = 12/treatment) (20). We found that L. johnsonii (OTU 186/188) was less prevalent in the ileum of chicks fed antimicrobials, while L. reuteri (possibly OTU 152) and L. crispatus (OTU 178/180) were more abundant in the gut of groups fed antimicrobials. A differential response to antimicrobials in lactobacilli was also reported previously, with lactobacilli (14, 61) and L. salivarius (26, 63) being less abundant in the ileum of chickens fed diets containing antimicrobials, while others reported that levels of lactobacilli (including L. crispatus) increase in the gut in response to antimicrobials in the diet (12, 20).
Despite the high interchick variability (40 to 70%) observed for gut microbial communities of chicks reared on the same diet and under the same environmental conditions, T-RFLP in conjunction with multivariate statistical methods was able to identify significant differences related to age, gut environment, and diet. Such interbird variation previously led to the inability to successfully detect treatment differences (43). The variation in gut microbial profiles among birds raised and fed under the same conditions indicates that undefined host-related factors affect the establishment of the dominant bacterial community (58). For example, maternal flock age, egg size, and chick size all influence posthatch growth (42), and these factors may also have some bearing on the between-chick variability observed for the gut microbiota. One advantage of T-RFLP is that the interbird variability can be investigated.
Not only have we been able to characterize changes occurring in gut microbial communities using T-RFLP, but we have also been able to identify particular OTUs driving these differences via targeted cloning and sequencing. One limitation identified was the discrepancies between sequence-determined T-RFs and capillary electrophoresis-determined fragment sizes (T-RFLP-generated OTUs), which were previously reported to occur due to G+C nucleotide contents and secondary-structure melting temperatures (29, 30). Furthermore, in some cases numerous bacterial species were found to potentially represent a particular T-RFLP-generated OTU, e.g., OTU 152 (L. reuteri and/or Clostridiales), OTU 284/286 and/or OTU 286/288 (three unclassified bacterial species and/or Ruminococcaceae), OTU 296/298 (unclassified bacterium and/or Clostridiales), and OTU 490/492 (unclassified bacterium, Klebsiella sp., and/or Enterobacteriaceae). This makes it impossible to predict exactly which of these bacterial species may be responsible for the shifts in the bacterial community structure associated with the investigated antimicrobials. However, the 16S rRNA genome sequence information generated in this study could be invaluable for developing targeted diagnostic approaches for gaining a better understanding of the gut microbiota in poultry health and production. If some of these unknown but potentially culturable bacteria could be linked to immune responses, the competitive exclusion of pathogens, or production differences, they could be developed as poultry probiotics, which could be a viable alternative to antimicrobials in feed.
Many of the 16S rRNA gene sequences generated, including the treatment-specific OTUs, belong to unclassified bacteria; however, where phyla could be identified, the sequences belonged to the Firmicutes, Bacteroidetes, and Proteobacteria. Culture-independent techniques have estimated that only 50 to 80% of bacterial sequences identified belong to known bacterial species (43, 63, 64). Although many of the diet-associated bacteria detected in this study were unidentifiable, they did show sequence similarity to sequences of other bacteria inhabiting the gastrointestinal tract available in public genome sequence databases.
Other than the difference already described for three Lactobacillus spp. (L. crispatus, L. johnsonii, and possibly L. reuteri), some less-well-classified bacteria were also found to be influenced by antimicrobial treatment. The antimicrobials investigated potentially influenced members of the Clostridiales (unclassified Clostridiales, Lachnospiraceae, Ruminococcaceae, and Subdoligranulum spp.), members of the Enterobacteriaceae (unclassified Enterobacteriaceae and Klebsiella sp.), unclassified Oxalobacteraceae, as well as unclassified bacteria. Some bacterial groups were found to be more abundant in the gut of chicks fed antimicrobial-supplemented diets, while others were more abundant in the gut of chicks fed antimicrobial-free diets. Escherichia (family Enterobacteriaceae) was reported previously to be more prevalent in the ilea of 3-day-old chicks fed zinc bacitracin (21), which supports our observation that OTU 490/492 (Enterobacteriaceae) was more abundant in the ileum of chicks on antimicrobial-supplemented diets at 3 days posthatch. Faecalibacterium prausnitzii and Subdoligranulum variabile-like bacteria (Ruminococcaceae) were recently shown to contribute significantly to the cecal microbiota of chickens (4, 36). These bacteria produce short-chain fatty acids such as formic acid and butyric acid, which have important functions in growth performance (18) and protection against pathogens in poultry (16), respectively. Dietary supplementation with formic acid was shown previously to result in improvements in broiler growth performance traits similar to those for avilamycin supplementation (18). Our finding showing that dietary supplementation with avilamycin increased the abundance of OTU 198/200 (Subdoligranulum spp.) in the ceca of chicks could indirectly support the observations reported previously by Garcia et al. (18). The 16S rRNA gene sequences that we obtained from T-RFLP-generated OTU 198/200 were closely related to Subdoligranulum variabile (GenBank accession number NR_028997).
None of the antimicrobials evaluated in this study resulted in significant improvements in broiler chick performance, as measured by live body weight, feed consumption, or the FCR, or a significant reduction in overall chick mortality within the first 17 days posthatch. A growth promotion response in poultry due to dietary supplementation with antimicrobials in feed is not always evident (11, 12, 25, 44), particularly in a highly sanitized environment (12). A lack of significant performance differences in response to antimicrobials in feed was noted previously for birds raised in battery cages (44) or pens with wire floors (25). In such environments birds are not exposed to litter or coprophagy, which suggests that the environment is important for the performance-related response to antimicrobials. Had this study been conducted under commercial production conditions, performance differences in response to these antimicrobials in feed may have become evident.
Although this study did not detect changes in chick performance, it did identify and characterize changes in posthatch gut microbiota development in chicks in response to antimicrobial agents in feed. Some of these bacterial changes may have performance-related implications under certain rearing environmental conditions. Higher numbers of lactobacilli were previously implicated in broiler growth depression due to competition for nutrient uptake or impaired fat absorption (14). In particular, a decrease in numbers of L. salivarius bacteria has been linked with improved broiler performance (14, 24, 26). However, some probiotics containing either multiple bacterial species or a single bacterial species, including lactobacilli, have been shown to improve feed efficiency and weight gain in chickens (2, 38, 39), suggesting that perhaps strain-specific characteristics and/or host selective pressures determining the process of colonization (34, 62) could influence performance outcomes.
In conclusion, we have shown that both age and dietary supplementation with antimicrobials affect the overall gut bacterial and lactobacillus communities of broiler chicks. Furthermore, the influence of various antimicrobials was greatest on the ileal microbial communities. Although this and the influence of antimicrobials in feed on broiler gut microbiota were described previously for birds older than 2 weeks posthatch, this is the first detailed report of their influence on gut microbiota development immediately posthatch. We have also identified and characterized candidate bacterial species affected by antimicrobial treatment. This information could inform the design of new nutritional strategies to promote broiler health and facilitate the beneficial microbial colonization of the gastrointestinal tract.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Australian Poultry Cooperative Research Centre (project 06-25).
We thank Mark Geier, Martine Boulianne, Kylie Swanson, Evelyn Daniels, and Derek Schultz for help with animal handling and sample collection. We also thank Teresa Mammone, Rohit Philip, Chris Munday, and Fleur Roberts for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Angelakis E., Raoult D. 2010. The increase of Lactobacillus species in the gut flora of newborn broiler chicks and ducks is associated with weight gain. PLoS One 5:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Apajalahti J., Kettunen A., Graham H. 2004. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. Worlds Poult. Sci. J. 60:223–232 [Google Scholar]
- 4. Bjerrum L., et al. 2006. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and cellular-based techniques. Poult. Sci. 85:1151–1164 [DOI] [PubMed] [Google Scholar]
- 5. Bolder N., Wagenaar J., Putirulan F., Veldman K., Sommer M. 1999. The effect of flavophospholipol (Flavomycin) and salinomycin sodium (Sacox) on the excretion of Clostridium perfringens, Salmonella enteritidis and Campylobacter jejuni in broilers after experimental infection. Poult. Sci. 78:1681–1689 [DOI] [PubMed] [Google Scholar]
- 6. Bonfield J. K., Staden R. 1996. Experiment files and their application during large-scale sequencing projects. DNA Seq. 6:109–117 [DOI] [PubMed] [Google Scholar]
- 7. Bray J. R., Curtis K. R. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325–349 [Google Scholar]
- 8. Butaye P., Devriese L. A., Haesebrouck F. 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 16:175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clarke K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117–143 [Google Scholar]
- 10. Cole J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diarra M., et al. 2007. Impact of feed supplementation with antimicrobial agents on growth performance of broiler chickens, Clostridium perfringens and Enterococcus counts, and antibiotic resistance phenotypes and distribution of antimicrobial resistance determinants in Escherichia coli isolates. Appl. Environ. Microbiol. 73:6566–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dumonceaux T. J., Hill J. E., Hemmingsen S. M., Van Kessel A. G. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egert M., De Graaf A. A., Smidt H., De Vos W. M., Venema K. 2006. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 14:86–91 [DOI] [PubMed] [Google Scholar]
- 14. Engberg R. M., Hedemann M. S., Leser T. D., Jensen B. B. 2000. Effect of zinc bacitracin and salinomycin on intestinal microflora and performance of broilers. Poult. Sci. 79:1311–1319 [DOI] [PubMed] [Google Scholar]
- 15. Feighner S. D., Dashkevicz M. P. 1987. Subtherapeutic levels of antibiotics in poultry feed and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl. Environ. Microbiol. 53:331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernández-Rubio C., et al. 2009. Butyric acid-based feed additives help protect broiler chickens from Salmonella Enteritidis infection. Poult. Sci. 88:943–948 [DOI] [PubMed] [Google Scholar]
- 17. Frei A., Goldenberger D., Teuber M. 2001. Antimicrobial susceptibility of intestinal bacteria from Swiss poultry flocks before the ban of antimicrobial growth promoters. Syst. Appl. Microbiol. 24:116–121 [DOI] [PubMed] [Google Scholar]
- 18. Garcia V., Catala-Gregori P., Hernandez F., Megias M. D., Madrid J. 2007. Effect of formic acid and plant extracts on growth, nutrient digestibility, intestine mucosa morphology and meat yield of broilers. J. Appl. Poult. Res. 16:555–562 [Google Scholar]
- 19. Geier M. S., et al. 2010. Comparison of alternatives to in-feed antimicrobials for the prevention of clinical necrotic enteritis. J. Appl. Microbiol. 109:1329–1338 [DOI] [PubMed] [Google Scholar]
- 20. Geier M. S., Torok V. A., Allison G. E., Ophel-Keller K., Hughes R. J. 2009. Indigestible carbohydrates alter the intestinal microbiota but do not influence the performance of broiler chickens. J. Appl. Microbiol. 106:1540–1548 [DOI] [PubMed] [Google Scholar]
- 21. Gong J., et al. 2008. Effects of zinc bacitracin, bird age and access to range on bacterial microbiota in the ileum and caeca of broiler chickens. J. Appl. Microbiol. 104:1372–1382 [DOI] [PubMed] [Google Scholar]
- 22. Gong J., et al. 2007. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts; from crops to ceca. FEMS Microbiol. Ecol. 59:147–157 [DOI] [PubMed] [Google Scholar]
- 23. Guan L. L., et al. 2003. Detection and identification of Lactobacillus species in crops of broilers of different ages by using PCR-denaturing gradient gel electrophoresis and amplified ribosomal DNA restriction analysis. Appl. Environ. Microbiol. 69:6750–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guban J., Korver D., Allison G., Tannock G. 2006. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens. Poult. Sci. 85:2186–2194 [DOI] [PubMed] [Google Scholar]
- 25. Gunal M., Yayli G., Kaya O., Karahan N., Sulak O. 2006. The effects of antibiotic growth promoter, probiotic or organic acid supplementation on performance, intestinal microflora and tissue of broilers. Int. J. Poult. Sci. 5:149–155 [Google Scholar]
- 26. Harrow S., Ravindran V., Butler R., Marshall J., Tannock G. 2007. Real-time quantitative PCR measurement of ileal Lactobacillus salivarius populations from broiler chickens to determine the influence of farming practices. Appl. Environ. Microbiol. 73:7123–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hilmi H. T. A., Surakka A., Apajalahti J., Saris P. E. 2007. Identification of the most abundant Lactobacillus species in the crop of 1- and 5-week-old broiler chickens. Appl. Environ. Microbiol. 73:7867–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hume M. E., et al. 2003. Poultry digestive microflora biodiversity as indicated by denaturing gradient gel electrophoresis. Poult. Sci. 82:1100–1107 [DOI] [PubMed] [Google Scholar]
- 29. Kaplan C. W., Kitts C. L. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54:121–125 [DOI] [PubMed] [Google Scholar]
- 30. Kent A. D., Smith D. J., Benson B. J., Triplett E. W. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klasing K. C. 2007. Nutrition and the immune system. Br. Poult. Sci. 48:525–537 [DOI] [PubMed] [Google Scholar]
- 32. Knarreborg A., Simon M. A., Engberg R. M., Jensen B. B., Tannock G. W. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 34. Lu J., Domingo J. S., Shanks O. C. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41:3561–3574 [DOI] [PubMed] [Google Scholar]
- 35. Lu J., et al. 2003. Diversity and succession of the intestinal bacterial communities of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lund M., Bjerrum L., Pedersen K. 2010. Quantification of Faecalibacterium prausnitzii- and Subdoligranulum variabile-like bacteria in the cecum of chickens by real-time PCR. Poult. Sci. 89:1217–1224 [DOI] [PubMed] [Google Scholar]
- 37. McCracken V. J., Simpson J. M., Mackie R. I., Gaskins H. R. 2001. Molecular ecological analysis of dietary and antibiotic-induced alterations of the mouse intestinal microbiota. J. Nutr. 131:1862–1870 [DOI] [PubMed] [Google Scholar]
- 38. Mountzouris K. C., et al. 2010. Effects of probiotic inclusion levels in broiler nutrition on growth performance, nutrient digestibility, plasma immunoglobulins, and cecal microflora composition. Poult. Sci. 89:58–67 [DOI] [PubMed] [Google Scholar]
- 39. Mountzouris K. C., et al. 2007. Evaluation of the efficacy of a probiotic containing Lactobacillus, Bifidobacterium, Enterococcus, and Pediococcus strains in promoting broiler performance and modulating cecal microflora compositions and metabolic activities. Poult. Sci. 86:309–317 [DOI] [PubMed] [Google Scholar]
- 40. Muyzer G., Teske A., Wirsen C. O., Jannasch H. W. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165–172 [DOI] [PubMed] [Google Scholar]
- 41. Niewold T. 2007. The nonantibiotic anti-inflammatory effect of antimicrobial growth promoters, the real mode of action? A hypothesis. Poult. Sci. 86:605–609 [DOI] [PubMed] [Google Scholar]
- 42. Noy Y., Sklan D. 1997. Posthatch development in poultry. J. Appl. Poult. Res. 6:344–354 [Google Scholar]
- 43. Owens B., Tucker L. C. M. A., McCracken K. J. 2008. Effects of different feed additives alone or in combination on broiler performance, gut microflora and ileal histology. Br. Poult. Sci. 49:202–212 [DOI] [PubMed] [Google Scholar]
- 44. Pedroso A. A., et al. 2006. Intestinal bacterial community and growth performance of chickens fed diets containing antibiotics. Poult. Sci. 85:747–752 [DOI] [PubMed] [Google Scholar]
- 45. Pedroso A. A., Menten F. F. M., Lambais M. R. 2005. The structure of bacterial community in the intestine of newly hatched chicks. J. Appl. Poult. Res. 14:232–237 [Google Scholar]
- 46. Pfaller M. 2006. Flavophospholipol use in animals: positive implications for antimicrobial resistance based on its microbiologic properties. Diagn. Microbiol. Infect. Dis. 56:115–121 [DOI] [PubMed] [Google Scholar]
- 46a. Promega 2007. Technical manual TM042. Promega, Madison, WI [Google Scholar]
- 47. Rada V., Janesova J., Marounek M., Vorisek K. 1991. Susceptibility of chicken intestinal lactobacilli to antimicrobial compounds. Acta Vet. Brno 60:339–343 [Google Scholar]
- 48. Rehman H. U., Vanhjen W., Awad W. A., Zentek J. 2007. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch. Anim. Nutr. 61:319–335 [DOI] [PubMed] [Google Scholar]
- 49. Sarmah A. K., Meyer M. T., Boxall A. B. A. 2006. A global perspective on the use, sale, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725–759 [DOI] [PubMed] [Google Scholar]
- 50. Shira E. B., Sklan D., Friedman A. 2005. Impaired immune responses in broiler hatchling hindgut following delayed access to feed. Vet. Immunol. Immunopathol. 105:33–45 [DOI] [PubMed] [Google Scholar]
- 51. Snedecor G. W., Cochran W. G. 1980. Statistical methods, 7th ed. Iowa State University Press, Ames, IA [Google Scholar]
- 52. Stirling G. R., et al. 2004. Combining an initial risk assessment process with DNA assays to improve prediction of soilborne diseases caused by root-knot nematode (Meloidogyne spp.) and Fusarium oxysporum f. sp. lycopersici in the Queensland tomato industry. Aust. Plant Pathol. 33:285–293 [Google Scholar]
- 53. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 54. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Torok V. A., Ophel-Keller K., Loo M., Hughes R. J. 2008. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl. Environ. Microbiol. 74:783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Torok V. A., et al. 2007. Environment and age: impact on poultry gut microflora. Aust. Poult. Sci. Symp. 19:149–152 [Google Scholar]
- 58. van der Wielen P. W. J. J., Keuzenkamp D. A., Lipman L. J. A., van Knapen F., Biesterveld S. 2002. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44:286–293 [DOI] [PubMed] [Google Scholar]
- 59. Walter J., et al. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Widmer F., Hartmann M., Frey B., Kolliker R. 2006. A novel strategy to extract specific phylogenetic sequence information from community T-RFLP. J. Microbiol. Methods 66:512–520 [DOI] [PubMed] [Google Scholar]
- 61. Wise M., Siragusa G. 2007. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 102:1138–1149 [DOI] [PubMed] [Google Scholar]
- 62. Yin Y., et al. 2010. Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J. 4:367–376 [DOI] [PubMed] [Google Scholar]
- 63. Zhou H., et al. 2007. Appropriate chicken sample size for identifying the composition of broiler intestinal microbiota affected by dietary antibiotics, using the polymerase chain reaction-denaturing gradient gel electrophoresis technique. Poult. Sci. 86:2541–2549 [DOI] [PubMed] [Google Scholar]
- 64. Zhu X. Y., Joerger R. D. 2003. Composition of microbiota in content and mucus from ceca of broiler chickens as measured by fluorescent in situ hybridization with group-specific, 16S rRNA-targeted oligonucleotide probes. Poult. Sci. 82:1242–1249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.