Abstract
To longitudinally assess fecal shedding and animal-to-animal transmission of Clostridium difficile among finishing feedlot cattle as a risk for beef carcass contamination, we tested 186 ± 12 steers (mean ± standard deviation; 1,369 samples) in an experimental feedlot facility during the finishing period and at harvest. Clostridium difficile was isolated from 12.9% of steers on arrival (24/186; 0 to 33% among five suppliers). Shedding decreased to undetectable levels a week later (0%; P < 0.001), and remained low (<3.6%) until immediately prior to shipment for harvest (1.2%). Antimicrobial use did not increase fecal shedding, despite treatment of 53% of animals for signs of respiratory disease. Animals shedding C. difficile on arrival, however, had 4.6 times higher odds of receiving antimicrobials for respiratory signs than nonshedders (95% confidence interval for the odds ratio, 1.4 to 14.8; P = 0.01). Neither the toxin genes nor toxin A or B was detected in most (39/42) isolates based on two complementary multiplex PCRs and enzyme-linked immunosorbent assay testing, respectively. Two linezolid- and clindamycin-resistant PCR ribotype 078 (tcdA+/tcdB+/cdtB+/39-bp-type deletion in tcdC) isolates were identified from two steers (at arrival and week 20), but these ribotypes did not become endemic. The other toxigenic isolate (tcdA+/tcdB+/cdtB+/classic tcdC; PCR ribotype 078-like) was identified in the cecum of one steer at harvest. Spatio-temporal analysis indicated transient shedding with no evidence of animal-to-animal transmission. The association between C. difficile shedding upon arrival and the subsequent need for antimicrobials for respiratory disease might indicate common predisposing factors. The isolation of toxigenic C. difficile from bovine intestines at harvest highlights the potential for food contamination in meat processing plants.
INTRODUCTION
Clostridium difficile is the leading cause of infectious diarrhea in hospitals worldwide, with more than double the incidence observed today compared to the early 2000s (28). In the United States, C. difficile is estimated to be associated with over 300,000 clinical cases each year, resulting in a major financial burden to the health care system (14). Moreover, the incidence rates of severe infections and deaths in the community, particularly in children and pregnant women, are also increasing (8). This change in the epidemiology of C. difficile is coincidental with the emergence of hypervirulent antimicrobial-resistant strains, namely, epidemic PCR ribotypes 027 and 078 (22). The presence of these strains in livestock and up to 42% of retail meat products (41, 44) indicates that livestock might serve as a reservoir for contamination of the food supply, making foods a plausible source for community-acquired C. difficile infections. Resistance to fluoroquinolones and clindamycin, antibiotics used in human medicine, is also common among C. difficile isolates of animal and food origin (39).
Most pathogens that contaminate meat originate from the bacterial flora harbored by the live animal (19). As such, preharvest interventions to control food-borne pathogens on the farm are predicted to decrease the frequency of carcass contamination, therefore enhancing food safety (33). Clostridium difficile spores of epidemic genotypes have been isolated from retail meats (36, 38, 44) and vegetables (2, 4, 29) and from neonatal and young cattle and pigs (32, 39, 42). However, the patterns of fecal C. difficile shedding among cattle during the finishing period, including the prevalence at the time of harvest, and the frequency of bovine carcass contamination have not been reported.
A better understanding of the epidemiology of C. difficile in food animal populations will help to elucidate the sources of food contamination and determine if preharvest interventions for this pathogen may be beneficial. The goal of this study was to determine the prevalence of C. difficile in cattle upon entry to a feedlot, the incidence of new infections (assessed as new fecal shedders), and the spatio-temporal distribution of C. difficile subtypes in a group of beef cattle up until and at the time of harvest.
MATERIALS AND METHODS
Experimental unit and source of animals.
This study was conducted as part of a study evaluating body weight and dietary supplementation in a finishing feedlot facility at The Ohio State University, Wooster, OH. Purchased Angus-cross steers were maintained as a closed population; animals entered the feedlot in early fall (October 2007) and were harvested between 7 and 8.5 months later in the spring. Prior to animal arrival, disinfection of the facility included pressure washing of floors, feeders, and waterers and fresh painting of pen divisions and fences. The waterers were stainless steel, automatically refillable, and shared between two pens. The pens used had concrete slatted floors and an underground pit to accumulate fecal matter.
The steers originated from four university-owned operations and one commercial livestock auction. University-owned cattle originated from cow-calf farms that had the steers (∼7 to 8 months old) alongside their dams in permanent pasture with no transitional grain-based diets offered prior to shipment. Animals from the auction were of unknown origin and management history. Multiple trucks were used for transporting animals from the source to the feedlot facility. Farms A, B, C, and D and the auction market were located 151, 283, 70, 155, and 265 km away, respectively (transport time, 1 to 4 h).
Animal allocation, sampling, and antimicrobial therapy.
Upon arrival to the feedlot, fecal samples were collected from the rectum by using individual plastic sleeves (occasionally, some animals had no feces available for testing). Animals were also weighed, ear tagged, and systematically assigned to 24 pens based on order of arrival (8 to 10 animals/pen). Fecal samples were also collected from each animal on weeks 1, 4, 12, 20, and the day prior to shipment to the meat processing plant. During the first month, the steers remained in the originally assigned pens while they were fed a transitional diet. This diet consisted of a stepwise increase from corn silage, soy hulls, and high-moisture corn (45, 20, and 10% of the diet, respectively) to a finishing diet of mostly high-moisture corn (65%) and corn silage (10%). The remaining 25% of the diet was a pelleted supplement containing protein (soybean meal and urea), vitamins, and minerals to meet the requirements of growing cattle (31). No grass or hay was offered to the animals during the study.
Signs of bovine respiratory disease complex (ranging from fever to fatal pneumonia) are common in beef steers entering feedlots. A protocol based on antiinflamatory and antimicrobial drugs approved for use in cattle in the United States was used to treat affected animals. Flunixin meglumine (1.1 mg/kg of body weight; intravenous; single dose; Banamine; Schering-Plough) and tulathromycin (2.5 mg/kg; subcutaneous; one dose; Draxxin; Pfizer) were initially administered to animals with fever (>39.7°C), anorexia, and depression. If no clinical response was observed within 24 to 48 h, as indicated by persistently elevated rectal temperature, florfenicol was given (40 mg/kg; subcutaneous; one dose; Nuflor; Schering-Plough). Ceftiofur (1 mg/kg; subcutaneous; 1 dose/day for 3 days; Excenel; Pharmacia & Upjohn), and oxytetracycline (20 mg/kg; subcutaneous; one dose; Liquamycin LA-200; Pfizer) were the third and last antimicrobial options, respectively, used in nonresponsive cases. Due to removals associated with respiratory disease, sudden death, or injury, animals were systematically reallocated on weeks 4 and 6 to ensure a final animal density of 7 steers per pen (of comparable body weights), providing the space needed at the end of the finishing period.
Animals were harvested at two commercial meat processing plants in groups of 26 to 28 animals (4 pens per week) over a period of 6 weeks. Following evisceration, two additional samples were collected: cecal content (2 to 4 ml by transmural puncture of cecal apex with a 16-gauge needle) and a composite external carcass swab obtained using a single hydrated sponge (10 ml of buffered peptone water broth [HS10BPW]; 3M) per animal. Swabbing occurred after carcass trimming on both front quarters and the thoracic area (40 by 40 cm).
Laboratory analysis.
Eight samples per animal (six fecal samples at the feedlot and one cecal sample and one carcass swab at harvest) were cultured for C. difficile. Bacterial isolation was based on a described enrichment protocol (3, 38). Briefly, 1 gram of feces, cecal contents, and the carcass swabs were inoculated into 10, 10, and 35 ml of broth, respectively. This broth was prepared and supplemented with sodium taurocholate (0.1%; Sigma-Aldrich, St. Louis, MO) and antimicrobials d-cycloserine (250 mg/liter) and cefoxitin (8 mg/liter; SR0096; Oxoid) as previously described (3, 38). The broths were anaerobically incubated (CO2-H2-N2 at 10/10/80%) at 37°C for 10 days and then centrifuged (7,000 × g for 10 min). The sediments were treated with 96% ethanol (1:1 [vol/vol] for 30 min) to reduce contaminants, centrifuged again, and stored at −80°C without supernatant. Thawed sediments were inoculated onto prereduced cycloserine cefoxitin fructose C. difficile agar supplemented with 7% defibrinated horse blood. Agar plates were inspected after 5 days of anaerobic incubation at 37°C for suspect colonies (3). Up to five C. difficile-suspect (i.e., morphology and 365-nm fluorescence) colonies from each plate were subcultured onto tryptic soy agar (Acumedia, Lansing, MI) supplemented with 5% defibrinated sheep blood. Nonhemolytic colonies were biochemically confirmed as C. difficile via l-proline aminopeptidase activity (16). Five days later, spores were harvested and stored in selective broth with 30% glycerol at −80°C. All samples were recoded and blindly processed in composite batches. Positive controls included C. difficile strain ATCC 9569.
Molecular subtyping.
Thawed C. difficile isolates were revived on blood agar for crude DNA extraction. Single colonies resuspended in 200 μl of distilled water were boiled for 15 min and centrifuged (16,000 × g for 1 min) to harvest the supernatant, which was stored at −20°C. DNA concentrations ranged between 17 and 70 ng μl−1 (mean, 40.7; ND-1000; Nanodrop, Wilmington, DE). Recovered isolates were molecularly assessed using three multiplex PCRs (27, 34, 45). The gene combinations tested were tpi/tcdA/tcdB for multiplex 1, 16S rRNA gene/tcdA/tcdB/cdtA/cdtB for multiplex 2, and tcdE/tcdC for multiplex 3 (26, 34, 45). PCR ribotyping was also conducted to assess strain similarities (6). Cluster analysis of fingerprint patterns was conducted using the unweighted pair group method with median and Pearson coefficients (Bionumerics, version 5.1; Applied Maths Inc., Austin, TX). International PCR ribotype designations are provided when available; otherwise, the descriptive nomenclature for the first representative isolate was used for its respective cluster.
Verification of toxin production.
Two commercial enzyme-linked immunosorbent assays (ELISAs) were used to verify toxin production in the first two isolates recovered from each PCR ribotype cluster (18, 46), using 72-hour-old colonies grown on blood agar. One ELISA detected toxins A and B (Wampole-C. difficile TOX A/B II; TechLab, Blacksburg, VA), and the other detected toxin A only (Clearview C diff A; Inverness Medical Innovations, Princeton, NJ). ELISA testing was performed in duplicate. A results for the strains were defined as follows: (i) A− B− if no toxins were detected with the ELISA TOX A/B kit; (ii) A− B+ if the ELISA TOX A/B was positive but the ELISA C diff A was negative; (iii) A+ B+ if both ELISAs were positive. Clostridium difficile ATCC 9569 was used as a control. Binary toxin production was not evaluated.
Toxinotyping and antimicrobial susceptibility.
Restriction fragment length polymorphism of the pathogenicity locus (toxinotyping) was conducted on select isolates (one per major phylogenetic cluster) as described by Rupnik (toxin gene fragments A3 and B1) (http://www.mf.uni-mb.si/mikro/tox/). MICs against six antimicrobial classes relevant for treatment or induction of C. difficile infections in humans were also determined. Metronidazole, vancomycin, moxifloxacin, clindamycin, linezolid (first compound of oxazolidinones), and tigecycline (first compound of glycylcyclines) were assessed using the Etest method as described in a previous study (AB Biodisk; bioMérieux, France) using brucella agar (Acumedia, Lansing, MI) (36). Reference break points from the Clinical and Laboratory Standards Institute or reported ranges for vancomycin, linezolid, and tigecycline were used for interpretation (1, 20, 23, 30).
Statistical analyses.
Based on reported C. difficile prevalence rates in calves (39), a one-sample size estimation indicated that 171 animals would be sufficient to test a hypothetical shedding prevalence of 7% (alternate, 13%; power, 0.8; α = 0.05). As a closed population cohort study, variables were assessed using a risk-based cumulative incidence analysis (13). Binary and continuous data on arrival were analyzed using a chi-square test, Fisher's exact test, t test, or analysis of variance (ANOVA) statistics, or their nonparametric options, accordingly (35). Adjusting for events with a probability close to zero, 95% confidence intervals (CIs) were calculated using binomial exact statistics (7, 11). Analyses were conducted using the STATA software package (version 10.1; College Station, TX). To quantify the risk of animal-to-animal transmission for C. difficile, the effective reproduction number [Rt; the number of secondary shedders produced by each primary shedder during the first week after arrival) was estimated (9). No reproduction numbers were calculated after week 1 due to larger sampling intervals (3 to 8 weeks). We also compared the cumulative risk of becoming a shedder when exposed to shedders and nonshedders (13). For binary data at pen level, a join-count analysis using the Rook's case was computed as an index of spatial autocorrelation (5) to differentiate clustered from dispersed and random distributions of C. difficile-positive pens.
RESULTS
In total, 197 steers arrived at the feedlot, 116 directly from cow-calf farms and 81 from the livestock auction. Upon arrival, fecal samples were obtained from 186 animals (95.4% of 197; 105 from farms and 81 from auction; 11 steers had no feces in the rectum at arrival). A total of 1,369 samples were tested for C. difficile.
Shedding at arrival.
Clostridium difficile was isolated from 12.9% of steers on arrival (24/186; 95% CI, 8.4 to 18.5). At the source level, the prevalence of C. difficile ranged from 0 to 33% (Fig. 1 A), but there was no statistical difference between animals from farms and the livestock auction (chi-square, P > 0.5). Univariate and logistic regression analyses controlling for farm showed no association between body weight and C. difficile shedding at arrival (P > 0.2; odds ratio, 1); C. difficile was isolated from steers of all body weight ranges (Fig. 1B), indicating no effect of body weight (and possibly age) on shedding in young finishing steers.
Fig. 1.
Clostridium difficile in steers at arrival to the feedlot. (A) Prevalence of fecal shedding of C. difficile by source. The combined prevalence was 12.9%. (B) C. difficile shedding by body weight. Controlling for farm, there was no association between shedding status and body weight (F-test, P > 0.1). Lines with solid squares represent group means.
Shedding over time and at harvest.
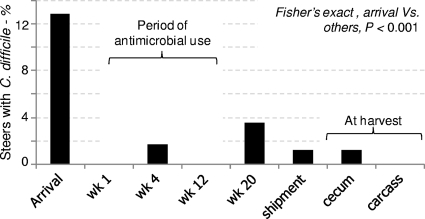
The proportion of animals shedding C. difficile at the feedlot facility decreased significantly from 12.9% (24/186) at arrival to 0% (0/176; 95% CI, 0 to 2.1) a week later and to 1.7% (3/176; 95% CI, 0.4 to 4.9) by week 4 (Fisher's exact test; P < 0.0001). Clostridium difficile was not detected at week 12 (0%; 0/168; 95% CI, 0 to 2.2), but the prevalence at week 20 was 3.6% (5/168; 95% CI, 1.3 to 7.6). Clostridium difficile was also isolated from the feces of two animals (1.2%; 2/167; 95% CI, 0.1 to 4.2) prior to shipment for harvest and from the intestinal content of two other animals at the time of harvest (cecum: 1.2%; 2/168; 95% CI, 0.1 to 4.2). One of these two steers that was C. difficile positive prior to shipment for harvest was also positive when it entered the feedlot. Likewise, one of the two steers that had C. difficile at harvest was positive at week 4. Neither of these two animals shed the same strain twice. All carcass swabs were negative (0/168; 95% CI, 0 to 2.2) (Fig. 2).
Fig. 2.
Longitudinal study of the prevalence of C. difficile shedding in cattle in the feedlot and at harvest. Cecum and carcass samples were collected at harvest, 24 h after the last on-farm sampling (shipment).
Antimicrobial use and C. difficile shedding.
Clinical signs of respiratory disease (i.e., fever and increased respiratory rate) prompted the use of antimicrobials in 53% (99/186) of the animals; most of them were treated between weeks 1 and 5. One, two, three, and four courses of the selected antibiotics were necessary in 63, 24, 8, and 7% of the 99 animals treated, respectively. Tulathromycin was the most commonly used antibiotic, followed by ceftiofur, florfenicol, and oxytetracycline (60, 22, 17, and 1% of 184 total single doses, respectively). The use of antimicrobials during the first month of the study (3 single doses on day 6, plus 156 doses between days 8 and 31) did not enhance C. difficile shedding (Fig. 2). By the end of the study, cumulative data analysis of the 12 steers that became new shedders at the feedlot indicated that antimicrobial use remained unassociated with C. difficile shedding (7/12 treated versus 5/12 nontreated; P > 0.1). Although no signs of enteric disease (i.e., watery diarrhea) were observed, logistic regression analysis controlling for farm effect indicated that animals shedding C. difficile on arrival had higher odds of subsequently receiving antimicrobials at the feedlot (odds ratio, 4.6; 95% CI, 1.4 to 14.8; P = 0.01) than nonshedders. Compared to the source of origin, this effect was driven by farm B (P = 0.003), whose steers also had the longest transport distance to the feedlot facility (283 km, ∼3.2 h, directly from cow-calf farm).
Reallocation and animal-to-animal transmission.
A total of 29 steers were removed (25 had moderate respiratory signs, 2 had leg injuries, and 2 died) early in the study, leaving 168 animals by week 6. The body weights of animals removed were not different from those of the remaining animals (t test, P = 0.132). Three of the 29 removed steers (10.3%) were positive for C. difficile on arrival. To adjust for withdrawals associated with disease or injuries and to balance body weights in each pen, animals were reallocated after having completed the dietary adjustment, on weeks 4 and 6. At the individual animal level, spatial statistics showed that the ordered allocation of incoming steers resulted in a random distribution of shedders within pens (join-count, P > 0.4) (Fig. 3). Regarding animal-to-animal transmission, the reproduction number derived from the data obtained within 1 week of arrival was zero (i.e., no new shedders arose from contact with shedding animals detected at arrival). No spatial statistics were done at individual sampling points after week 1, due to the limited number of shedders detected at each point (n, <5).
Fig. 3.
Spatio-temporal distribution of steers shedding C. difficile at the feedlot over time (n = 1,369 samples tested). Panels represent two separate rows of 12 contiguous pens (6 to 7 animals/pen). The spatial analysis indicated that pens holding incoming shedders followed a random distribution (join count, P > 0.4). Note the absence of new infections in association with PCR ribotype 078. Arrows and capital letters in the left panel indicate the order of arrival and farm of origin.
Molecular subtyping.
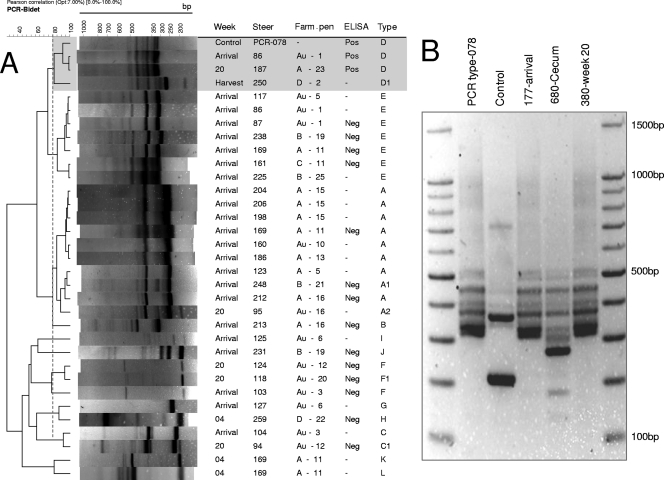
In total, 42 isolates recovered from 34 steers were examined. Molecular typing with one of the multiplex PCR methods (26) showed most strains were tcdA negative/tcdB positive, but the second multiplex method used (34) indicated they were tcdA negative/tcd negative (Table 1). A variant genotype (tcdA+/tcdB+/cdtB+/39-bp-type deletion in tcdC) identical to human and animal epidemic PCR ribotype 078 was also identified in two steers (n = 1 at arrival and n = 1 at week 20) by using the two multiplex methods. At harvest, one of the two cecal isolates identified was a toxigenic tcdA+/tcdB+/cdtB+/classic tcdC strain. PCR ribotyping clearly resulted in distinguishable ribotypes recovered from the same animal on different dates. In addition, some animals shed distinct ribotypes on the same date (Fig. 4). The two animals that shed PCR ribotype 078 in this feedlot never had direct nose-to-nose contact, never shared a common pen, and never had indirect contact via their penmates.
Table 1.
Toxin gene profiles of C. difficile PCR ribotypes from beef cattle in the feedlota
| PCR ribotype(s) | Toxin gene profile |
ELISA result | ||
|---|---|---|---|---|
| Multiplex 1 | Multiplex 2 | tcdE/tcdC | ||
| A, A1, and A2 | tpi+/tcdA negative/tcdB+ | 16S+/tcdA negative/tcdB negative/cdtA negative/cdtB negative | −/− | Negative |
| B | tpi+/tcdA negative/tcdB+ | 16S+/tcdA negative/tcdB negative/cdtA negative/cdtB negative | −/− | Negative |
| C, C1 | tpi+/tcdA negative/tcdB+ | 16S+/tcdA negative/tcdB negative/cdtA negative/cdtB negative | −/− | Negative |
| D (PCR 078) | tpi+/tcdA+/tcdB+ | 16S+/tcdA+/tcdB+/cdtA+/cdtB+ | +/39-bp deletion | A+ B+ |
| D1 | tpi+/tcdA+/tcdB+ | 16S+/tcdA+/tcdB+/cdtA+/cdtB+ | +/classic | ND |
| E | tpi+/tcdA negative/tcdB+ | 16S+/tcdA negative/tcdB negative/cdtA negative/cdtB negative | −/− | Negative |
| F to L | tpi+/tcdA negative/tcdB+ | 16S+/tcdA negative/tcdB negative/cdtA negative/cdtB negative | −/− | Negative |
| Others | tpi+/tcdA negative/tcdB negative | 16S+/tcdA negative/tcdB negative/cdtA negative/cdtB negative | ND | ND |
Subscripts in PCR ribotype letters indicate that extra bands were observed, compared to representative isolates in cluster (Fig. 4). Isolates were tested by using two complementary PCR multiplex methods for C. difficile (26, 34). The discrepancy between tcdB in both multiplexes for some PCR ribotypes (e.g., PCR ribotype A) indicates differential performance for the tcdB gene. Isolate type D1 was not available for further testing. ND, not determined.
Fig. 4.
PCR ribotypes of C. difficile from beef cattle in the feedlot. (A) Cluster analysis of representative isolates with at least one positive toxin gene. Note the similarities at arrival. The upper shaded rectangle highlights the similarity of three toxigenic isolates to PCR ribotype 078. ELISA tests for toxins A and B were conducted. −, not tested. Type, arbitrary letter designation; subscripts indicate that extra bands were observed compared to other isolates in the cluster. See Table 1 for toxin gene profiles of the PCR types (Pearson correlation similarity, >80%). Steers 86 and 169 carried two different strains. (B) PCR ribotypes highlighted in cluster type D, illustrating similarity to PCR ribotype 078.
Toxin production, toxinotyping, and antibiotic sensitivity.
ELISA testing of 20 isolates confirmed that tcdA-negative/tcdB-negative/cdtB-negative strains produced no toxins and that tcdA+/tcdB+/cdtB+ toxigenic strains produced both toxins. The multiplex-discrepant (tcdA-negative/tcdB-positive and tcdA-negative/tcdB-negative) strains had no detectable tcdE and tcdC genes, no toxin gene fragments A3 and B1, and no detectable toxins A and B. Cytotoxicity neutralization assays were not conducted. Toxinotyping and antimicrobial susceptibility testing showed that the two PCR ribotype 078 strains, identified at arrival and on week 20, were toxinotype V and were resistant to clindamycin and linezolid (MICs were 256 μg/ml and 8 to 12 μg/ml, respectively). All isolates were susceptible to metronidazole, vancomycin, tigecycline, and moxifloxacin.
DISCUSSION
This study provides new insights into the on-farm epidemiology of C. difficile in food animals, particularly during the final stages of beef production, as well as the associated potential for food contamination. Although young steers entering the feedlot harbored C. difficile, fecal carriage of this bacterium, irrespective of toxigenic genotype, was transient, and the transmission to other animals within the feedlot was neither temporally nor spatially clustered. The prevalence of C. difficile in purchased steers arriving from cow-calf farms was comparable to the reported prevalence for young calves (i.e., 10 to 13%) (39). However, most isolates were negative upon ELISA testing. The reasons for the discrepancy observed between the multiplex PCR performance regarding the detection of gene tcdB are unclear but may be in part due to differences in primer design and specificity.
Irrespective of toxin genotype, we did not observe molecular or temporal evidence to indicate that shedding persisted over time, either when animals were sampled 1 week apart after arrival to the feedlot or when testing occurred 24 h apart prior to harvest. These two short sampling intervals were considered critical to assess shedding persistence upon entry and prior to leaving the feedlot, but sparser sampling was chosen during the rest of the study (every 4 and 8 weeks) to reflect transition periods typical of feedlots (i.e., dietary adjustment, susceptibility to bovine respiratory disease, and reallocations). More frequent sampling could provide information about C. difficile shedding on a more precise time scale, but it may yield limited information with regard to food safety risks.
In the feedlot, the cause of the marked reduction of C. difficile during the first week was not determined but may be attributable to dietary changes (i.e., animals shifted from grass- and milk-based diets to grain-based diets in the feedlot). In humans, diet appears to modulate the growth of C. difficile in the intestinal tract (21), but similar effects are unknown for food animals. Alternatively, the move of cattle to the feedlot may have eliminated exposure to potential sources of C. difficile spores present on the farms of origin. At the feedlot, the slatted floors could have also reduced the number of spores to which the animals were exposed.
In humans and other animal species, antimicrobials enhance shedding, transmission of C. difficile, and the induction of C. difficile-associated disease (10, 28). For example, under experimental conditions, antimicrobials given to asymptomatic shedding mice resulted in “supershedder” states and increased transmission to previously C. difficile-negative immunocompetent cohoused cohorts (25). In contrast, in this study, we found that antimicrobial use was not the cause of the decrease in C. difficile shedding, because most animals were treated only after the significant shedding reduction to 0.6% was identified on week 1. Only 1.6% of all antimicrobial doses were given (i.e., two animals) prior to sampling the feedlot on week 1.
In feedlot steers, bovine respiratory disease complex often develops as an indicator of increased stress-related immune suppression following transportation and entry to feedlots (49). In our study, the association of C. difficile shedding at arrival with higher odds of receiving antimicrobials for respiratory signs in the feedlot indicates that C. difficile shedding and the bovine respiratory disease complex may share common predisposing factors at the farm of origin. However, the direct association between transportation time and C. difficile shedding was not measured in this study.
In neonatal calves, C. difficile shedding has been documented to last for at least 6 days (40). Fecal excretion of C. difficile was detected on only one occasion in most shedding steers, and those animals with two positive samples were shedding distinct C. difficile genotypes at nonconsecutive sampling points. The fecal shedding pattern was therefore considered transient. It is uncertain if detectable C. difficile shedding resulted from short-term successful bacterial colonization and proliferation or from intestinal passage of dormant spores ingested from the environment in food or water.
Despite the low incidence of new shedders in this study, we identified two animals with C. difficile immediately prior to shipment for slaughter and two different animals carrying C. difficile in the cecum at harvest. One of the cecal isolates was fully toxigenic (tcdA+/tcdB+/cdtB+/classic tcdC) and had 83% fingerprint pattern similarity to PCR ribotype 078, but it was clearly different, as it had additional bands of lower molecular weights (Fig. 4). Epidemic PCR ribotype 078 (tcdA+/tcdB+/cdtB+/39-bp deletion in putative toxin negative regulator tcdC/toxinotype V) was identified on two occasions in this cattle population (one animal at arrival and one in week 20) but it did not become endemic. PCR ribotype 078/toxinotype V is emerging as a cause of disease in hospitalized people (12) and the community (27), and it is a predominant type in swine operations (24), retail meats (44), and young dairy cattle (39). It was also recently isolated from clustered white-tailed deer farms in Ohio (17), in geographical proximity to where this study was conducted.
The presence of toxigenic C. difficile in feces and intestinal contents of cattle at the time of harvest indicates that contamination of transport vehicles, lariage areas, and meat processing plants is possible. For other food-borne pathogens, for instance Escherichia coli O157:H7, the prevalence of carcass contamination is strongly associated with the prevalence and load of fecal shedding by live animals at the time of harvest (15). The extent of such an association for C. difficile remains unknown. In this study, C difficile was not identified in sponges of carcass swabs. To date, there are no conclusive reports of food-borne C. difficile infections. However, the increasing isolation of human epidemic strains from retail meats and food animals, including finishing beef cattle in the present study, and the thermal resistance of C. difficile to minimum recommended cooking temperatures (37) underscore the potential for food-borne transmission.
In contrast to recent studies in which PCR ribotype 078 was clearly prevalent in young food animals (up to 94% in calves) and retail meats (73% of identified genotypes) (12, 24, 42, 44), this longitudinal study showed that such an epidemic genotype did not become endemic in our facility. The transient shedding observed in this feedlot indicates that cattle are unlikely persistent reservoirs of C. difficile during the finishing period. However, the isolation of one tcdA+/tcdB+/cdtB+ C. difficile strain from the cecum at harvest (although all carcass swabs were negative) demonstrated that meat contamination with clinically relevant isolates could occur at harvest. In addition, as with other clostridia (36, 43, 47, 48), spore intestinal translocation to deeper muscle tissues could also occur in vivo. Because bacterial proliferation during processing or in the final products is plausible, identifying and applying multiple barrier approaches, including those that emphasize control of cross-contamination, and decontamination during processing and food preparation could reduce the risk of food contamination with this emerging pathogen.
ACKNOWLEDGMENTS
This study was supported by an Ohio Agricultural Research and Development Center (OARDC) SEED grant, The Ohio State University, and state and federal funds allocated to the OARDC. A.R.P. is a research fellow of the Public Health Preparedness for Infectious Diseases program Targeted Investments in Excellence, The Ohio State University.
We are grateful to Mike Kauffman, Rose Schleppi, Jennifer Schrock, Laura Harpster, and Pamela Schlegel for technical assistance during sample collection to Juliette Hansen for veterinary assistance, and to Michele Williams for thorough comments during final preparation of the manuscript. We give special thanks to J. Scott Wesse (University of Guelph, Canada) for kindly donating reference strains and to the reviewers and editors for helpful suggestions.
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Ackermann G., Adler D., Rodloff A. C. 2003. In vitro activity of linezolid against Clostridium difficile. J. Antimicrob. Chemother. 51:743–745 [DOI] [PubMed] [Google Scholar]
- 2. al Saif N., Brazier J. S. 1996. The distribution of Clostridium difficile in the environment of South Wales. J. Med. Microbiol. 45:133–137 [DOI] [PubMed] [Google Scholar]
- 3. Arroyo L. G., et al. 2005. Use of a selective enrichment broth to recover Clostridium difficile from stool swabs stored under different conditions. J. Clin. Microbiol. 43:5341–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bakri M. M., Brown D. J., Butcher J. P., Sutherland A. D. 2009. Clostridium difficile in ready-to-eat salads, Scotland. Emerg. Infect. Dis. 15:817–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bell N., Schuurman N., Hameed S. M. 2008. Are injuries spatially related? Join-count spatial autocorrelation for small-area injury analysis. Injury Prev. 14:346–353 [DOI] [PubMed] [Google Scholar]
- 6. Bidet P., Barbut F., Lalande V., Burghoffer B., Petit J. C. 1999. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett. 175:261–266 [DOI] [PubMed] [Google Scholar]
- 7. Brown L. D., Cai T. T., DasGupta A. 2001. Interval estimation for a binomial proportion. Stat. Sci. 16:101–117 [Google Scholar]
- 8. Centers for Disease Control and Prevention 2008. Surveillance for community-associated Clostridium difficile—Connecticut, 2006. MMWR Morb. Mortal. Wkly. Rep. 57:340–343 [PubMed] [Google Scholar]
- 9. Chowell G., Hyman J. M., Bettencourt L. M. A., Castillo-Chavez C., Nishiura H. 2009. The effective reproduction number as a prelude to statistical estimation of time-dependent epidemic trends, p. 103–121 In Chowell G., Hayman J. M., Bettencourt L. M. A., Castillo-Chavez C. (ed.), Mathematical and statistical estimation approaches in epidemiology. Springer, Dordrecht, Netherlands [Google Scholar]
- 10. Clooten J., Kruth S., Arroyo L., Weese J. S. 2008. Prevalence and risk factors for Clostridium difficile colonization in dogs and cats hospitalized in an intensive care unit. Vet. Microbiol. 129:209–214 [DOI] [PubMed] [Google Scholar]
- 11. Clopper C. J., Pearson E. S. 1934. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413 [Google Scholar]
- 12. Debast S. B., et al. 2009. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ. Microbiol. 11:505–511 [DOI] [PubMed] [Google Scholar]
- 13. Dohoo I., Martin W., Stryhn H. 2003. Cohort studies, p. 69, 151–162 In McPike S. M. (ed.), Veterinary epidemiologic research. AVC, Inc., Charlottetown, Prince Edward Island, Canada [Google Scholar]
- 14. Dubberke E. R., Wertheimer A. I. 2009. Review of current literature on the economic burden of Clostridium difficile infection. Infect. Control Hosp. Epidemiol. 30:57–66 [DOI] [PubMed] [Google Scholar]
- 15. Elder R. O., et al. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. U. S. A. 97:2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fedorko D. P., Williams E. C. 1997. Use of cycloserine-cefoxitin-fructose agar and L-proline-aminopeptidase (PRO discs) in the rapid identification of Clostridium difficile. J. Clin. Microbiol. 35:1258–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. French E., Rodriguez-Palacios A., LeJeune J. T. 2010. Enteric bacterial pathogens with zoonotic potential isolated from farm-raised deer. Foodborne Pathog. Dis. 7:1031–1037 [DOI] [PubMed] [Google Scholar]
- 18. Garcia A., et al. 2000. Evaluation of four rapid methods for the investigation of the toxigenic capacity of Clostridium difficile strains isolated in a selective medium. Enferm. Infecc. Microbiol. Clin. 18:109–112 [PubMed] [Google Scholar]
- 19. Gill C. O. 2007. Microbiological conditions of meats from large game animals and birds. Meat Sci. 77:149–160 [DOI] [PubMed] [Google Scholar]
- 20. Hecht D. W., Citron D. 2007. Standard M11-A7. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 7th ed. Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 21. Iizuka M., et al. 2004. Elemental diet modulates the growth of Clostridium difficile in the gut flora. Aliment. Pharmacol. Ther. 20(Suppl. 1):151–157 [DOI] [PubMed] [Google Scholar]
- 22. Jhung M. A., et al. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg. Infect. Dis. 14:1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones R. N., Kohno S., Ono Y., Ross J. E., Yanagihara K. 2009. ZAAPS International Surveillance Program (2007) for linezolid resistance: results from 5591 Gram-positive clinical isolates in 23 countries. Diagn. Microbiol. Infect. Dis. 64:191–201 [DOI] [PubMed] [Google Scholar]
- 24. Keel K., Brazier J. S., Post K. W., Weese S., Songer J. G. 2007. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J. Clin. Microbiol. 45:1963–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawley T. D., et al. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemee L., et al. 2004. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 42:5710–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Limbago B. M., et al. 2009. Clostridium difficile strains from community-associated infections. J. Clin. Microbiol. 47:3004–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McFarland L. V. 2008. Update on the changing epidemiology of Clostridium difficile-associated disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 5:40–48 [DOI] [PubMed] [Google Scholar]
- 29. Metcalf D. S., Costa M. C., Dew W. M., Weese J. S. 2010. Clostridium difficile in vegetables, Canada. Lett. Appl. Microbiol. 51:600–602 [DOI] [PubMed] [Google Scholar]
- 30. Nagy E., Dowzicky M. J. 2010. In vitro activity of tigecycline and comparators against a European compilation of anaerobes collected as part of the Tigecycline Evaluation and Surveillance Trial (TEST). Scand. J. Infect. Dis. 42:33–38 [DOI] [PubMed] [Google Scholar]
- 31. National Research Council 2000. Nutrient requirements of beef cattle, 7th rev. ed. National Academy of Sciences, Washington, DC [Google Scholar]
- 32. Norman K. N., et al. 2009. Varied prevalence of Clostridium difficile in an integrated swine operation. Anaerobe 15:256–260 [DOI] [PubMed] [Google Scholar]
- 33. Oliver S. P., Patel D. A., Callaway T. R., Torrence M. E. 2009. ASAS centennial paper: developments and future outlook for preharvest food safety. J. Anim. Sci. 87:419–437 [DOI] [PubMed] [Google Scholar]
- 34. Persson S., Torpdahl M., Olsen K. E. 2008. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin. Microbiol. Infect. 14:1057–1064 [DOI] [PubMed] [Google Scholar]
- 35. Petrie A., Watson P. 1999. Statistics for veterinary and animal sciences, 1st ed. Blackwell Science, Oxford, United Kingdom [Google Scholar]
- 36. Rodriguez-Palacios A., et al. 2009. Possible seasonality of Clostridium difficile in retail meat, Canada. Emerg. Infect. Dis. 15:802–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez-Palacios A., Reid-Smith R. J., Staempfli H. R., Weese J. S. 2010. Clostridium difficile survives minimal temperature recommended for cooking ground meats. Anaerobe 16:540–542 [DOI] [PubMed] [Google Scholar]
- 38. Rodriguez-Palacios A., Staempfli H. R., Duffield T., Weese J. S. 2007. Clostridium difficile in retail ground meat, Canada. Emerg. Infect. Dis. 13:485–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodriguez-Palacios A., et al. 2006. Clostridium difficile PCR ribotypes in calves, Canada. Emerg. Infect. Dis. 12:1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodriguez-Palacios A., Stampfli H. R., Stalker M., Duffield T., Weese J. S. 2007. Natural and experimental infection of neonatal calves with Clostridium difficile. Vet. Microbiol. 124:166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rupnik M. 2007. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease?. Clin. Microbiol. Infect. 13:457–459 [DOI] [PubMed] [Google Scholar]
- 42. Rupnik M., Widmer A., Zimmermann O., Eckert C., Barbut F. 2008. Clostridium difficile toxinotype V, ribotype 078, in animals and humans. J. Clin. Microbiol. 46:2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sathish S., Swaminathan K. 2008. Molecular characterization of the diversity of Clostridium chauvoei isolates collected from two bovine slaughterhouses: analysis of cross-contamination. Anaerobe 14:190–199 [DOI] [PubMed] [Google Scholar]
- 44. Songer J. G., et al. 2009. Clostridium difficile in retail meat products, U.S.A., 2007. Emerg. Infect. Dis. 15:819–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spigaglia P., Mastrantonio P. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thonnard J., Carreer F., Avesani V., Delmee M. 1996. Toxin A detection on Clostridium difficile colonies from 24-h cultures. Clin. Microbiol. Infect. 2:50–54 [DOI] [PubMed] [Google Scholar]
- 47. Turcsan J., Varga L., Turcsan Z., Szigeti J., Farkas L. 2001. Occurrence of anaerobic bacterial, clostridial, and Clostridium perfringens spores in raw goose livers from a poultry processing plant in Hungary. J. Food Prot. 64:1252–1254 [DOI] [PubMed] [Google Scholar]
- 48. Vengust M., Arroyo L. G., Weese J. S., Baird H. R., Baird J. D. 2003. Preliminary evidence for dormant clostridial spores in equine skeletal muscle. Equine Vet. J. 35:514–516 [DOI] [PubMed] [Google Scholar]
- 49. Yates W. D. 1982. A review of infectious bovine rhinotracheitis, shipping fever pneumonia and viral-bacterial synergism in respiratory disease of cattle. Can. J. Comp. Med. 46:225–263 [PMC free article] [PubMed] [Google Scholar]