Abstract
The methods used to concentrate enteric viruses from water have remained largely unchanged for nearly 30 years, with the most common technique being the use of 1MDS Virozorb filters followed by organic flocculation for secondary concentration. Recently, a few studies have investigated alternatives; however, many of these methods are impractical for use in the field or share some of the limitations of this traditional method. In the present study, the NanoCeram virus sampler, an electropositive pleated microporous filter composed of microglass filaments coated with nanoalumina fibers, was evaluated. Test viruses were first concentrated by passage of 20 liters of seeded water through the filter (average filter retention efficiency was ≥99.8%), and then the viruses were recovered using various salt-based or proteinaceous eluting solutions. A 1.0% sodium polyphosphate solution with 0.05 M glycine was determined to be the most effective. The recovered viruses were then further concentrated using Centricon Plus-70 centrifugal ultrafilters to a final volume of 3.3 (±0.3 [standard deviation]) ml; this volume compares quite favorably to that of previously described methods, such as organic flocculation (∼15 to 40 ml). The overall virus recovery efficiencies were 66% for poliovirus 1, 83% for echovirus 1, 77% for coxsackievirus B5, 14% for adenovirus 2, and 56% for MS2 coliphage. In addition, this method appears to be compatible with both cell culture and PCR assays. This new approach for the recovery of viruses from water is therefore a viable alternative to currently used methods when small volumes of final concentrate are an advantage.
INTRODUCTION
Waterborne viruses are transmitted by the fecal-oral route upon ingestion of water contaminated with the excreta of humans and other animals. There are many human pathogenic viruses that are associated with waterborne illnesses, including the enteroviruses (polioviruses, echoviruses, and coxsackieviruses), adenoviruses, and caliciviruses (noroviruses) (29). Although enteric viruses are usually present in low densities in water, they may still pose a significant health risk, since the infectious dose for many viruses is quite low (∼10 to 100 virus particles) (24). Such low numbers are difficult to detect in water; therefore, it is necessary to concentrate viruses to higher densities from large volumes of water so that they may be successfully detected using cultural or molecular assays.
The VIRADEL (virus adsorption and elution) procedure has been the most commonly used method to recover and concentrate enteric viruses from water (3) for decades. It includes a primary step to concentrate viruses from large volumes of water using adsorptive media (filters), followed by virus elution from the filters. During the primary concentration step, water samples are passed through and the viruses are adsorbed to a microporous filter. The electropositive 1MDS filter (CUNO Inc., Meriden, CT) is the most commonly used filter for the concentration of viruses from water.
The positively charged NanoCeram virus sampler (Argonide Corporation, Sanford, FL) is a less expensive alternative. This is a nonwoven pleated cartridge depth filter that is composed of microglass filaments (0.6 μm in length) coated with nanoalumina (AlOOH) fibers (∼2 nm in diameter by ∼250 nm in length) derived from the mineral boehmite. NanoCeram filters have an extensive surface area (∼500 m2/g) due to the nanoalumina coating and an effective pore size of approximately 2.0 μm. The high surface area coupled with the relatively high isoelectric point (pI range = 8 to 9) confers a strong electropositivity to the filter surface. Karim et al. (21) found that the NanoCeram filter was able to concentrate poliovirus 1 from large volumes of water with an efficiency comparable to those of several currently available electropositive filters, including 1MDS (both cartridge and disc configurations) and Seitz filters.
The most commonly used eluting solution for the recovery of viruses from filters is beef extract (approximate pH of 9.0 to 9.5). It may be used at a range of concentrations (e.g., 1.5% to 3.0%) (4, 7, 8, 17, 21, 23, 25, 26, 27) and with glycine, an amphoteric amino acid (exhibiting both acidic and basic properties due to the presence of carboxylic acid and amine functional groups), to provide buffering (3, 14, 21, 23, 26, 27, 38). Beef extract solutions have been used to elute viruses from both electronegative (10, 32, 34) and electropositive (8, 21, 26, 37) cartridge filters, in addition to glass wool filters (15, 23, 28). Nonetheless, beef extract contains substances that can interfere with molecular assays, such as reverse transcriptase PCR (RT-PCR) (1). An eluent which is compatible with both cell culture and molecular assays is therefore desirable.
A number of inorganic chemicals have also been evaluated for their ability to elute viruses from filtration media. Tween 80 is a nonionic detergent that decreases the surface tension of water, allowing for the accommodation of nonpolar functional groups in the aqueous environment. It has been used as a component of eluting solutions to enhance virus desorption from microporous filters (12, 34, 39) and ultrafilters (18, 19, 31). Inorganic salt-based solutions can be used to disrupt the hydrophobic interactions between viruses and filter surfaces. Chaotropic salts are able to elute poliovirus 1 (12) and MS2 coliphage (9) from filters more efficiently than antichaotropic salts. The addition of a nonionic detergent, such as Tween 80, to chaotropic salt solutions further increases poliovirus 1 elution (36).
Sodium polyphosphates (NaPP) are inorganic polyanionic salts that are highly electronegative due in large part to the numerous phosphate groups comprising these molecules. This acts to reduce the zeta potential of surfaces exposed to the NaPP solution, resulting in an increase in the net negative charge of the surfaces. This in turn increases electrostatic repulsion, thus dispersing microorganisms and other particle types and facilitating virus desorption (35). NaPP are available in polymer chains ranging from two (pyrophosphates) to 17 or more units. Commercially available NaPP is heterogeneous and contains a mixture of chain lengths. Recently, NaPP salts coupled with Tween 80 have been used to elute viruses, bacteria, and protozoa from hollow-fiber ultrafilters (18, 19, 31).
Depending on the filter utilized, the volume of the eluate can range from ∼420 ml (e.g., with ultrafilter membranes, NanoCeram filters) to upward of 1.6 liters (e.g., with 1MDS filters) (8, 18, 21). Therefore, a secondary concentration step may be necessary to reduce the volume prior to assay. Organic flocculation is used for the secondary concentration step following elution with beef extract (22), typically yielding a virus concentrate with a volume of 15 to 40 ml. Alternatively, Centricon Plus-70 centrifugal filters (30-kDa cutoff; Millipore, Billerica, MA) have been used to concentrate ΦX174 and MS2 coliphages from 70 ml to approximately 2 ml (18).
In the current study, a positively charged filter was used in combination with a polyphosphate-based elution buffer followed by secondary concentration using an ultrafiltration method in order to minimize the final concentrate volume.
MATERIALS AND METHODS
MS2 coliphage propagation and assay.
MS2 bacteriophage (ATCC 15597-B1) and its host, Escherichia coli (ATCC 15597), were obtained from the American Type Culture Collection (ATCC; Manassas, VA). The MS2 coliphage was propagated monthly as described previously (40). This technique generally results in an MS2 stock culture of ≥1010 PFU/ml. No variations were observed in the MS2 cultures over the course of the study. The MS2 was assayed on tryptic soy agar (TSA; EMD Chemicals, Darmstadt, Germany) plates using the double overlay plaque-forming method (2).
Virus propagation and assay.
Adenovirus 2 (AV-2; ATCC VR-846), echovirus 1 (EV-1; ATCC VR-31), and coxsackievirus B5 (CV-B5; Faulkner, ATCC VR-185) were also obtained from the American Type Culture Collection (Manassas, VA). Poliovirus 1 (PV-1; strain LSc-2ab) was obtained from the Department of Virology and Epidemiology at the Baylor College of Medicine (Houston, TX). Adenovirus 2 was maintained on PLC/PRF/5 (primary liver carcinoma; ATCC CRL-8024) cell line monolayers with minimal essential medium (MEM; modified with Earle's salts; Irvine Scientific, Santa Ana, CA) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT), 100 mM sodium pyruvate (Mediatech Inc., Manassas, VA), and 1 mM glucose (Mediatech Inc., Manassas, VA) at an incubation temperature of 37°C with 5% CO2. Echovirus 1, coxsackievirus B5, and poliovirus 1 were maintained on BGM (Buffalo green monkey kidney; obtained from Dan Dahling at the United States Environmental Protection Agency, Cincinnati, OH) cell line monolayers with MEM containing 5% calf serum (CS; HyClone Laboratories, Logan, UT) at an incubation temperature of 37°C with 5% CO2.
Viruses were propagated by inoculating cell monolayers. Following the observation of ≥90% destruction of the monolayer, the cell culture flasks were frozen (at −20°C) and thawed (at 37°C) three successive times to release the viruses from the host cells. The suspension was then centrifuged (1,000 × g for 10 min) to remove cell debris, followed by precipitation with polyethylene glycol (PEG; 9% [wt/vol]) and sodium chloride (5.8% [wt/vol]) performed overnight at 4°C (6). The viruses were then centrifuged (15,300 × g for 30 min at 4°C). After resuspension of the virus pellet in phosphate-buffered saline (pH 7.4; Sigma-Aldrich, St. Louis, MO), a Vertrel XF extraction was performed at a 1:1 ratio to promote monodispersion of the virus and the removal of lipids (centrifugation at 1,900 × g for 15 min at 4°C) (6). The top aqueous layer containing the virus was carefully removed using a pipette and then filtered using a syringe filter (prewetted with 2 ml of 1.5% beef extract) with a pore size of 0.22 μm (Millex; Millipore, Bedford, MA). The viruses passed through the filter and were collected in sterile tubes and stored at −80°C until use.
Viral titrations for PV-1, EV-1, and CV-B5 were performed using 10-fold serial dilution plaque-forming assays described by Bidawid et al. (5). Host cell monolayers in 6-well tissue culture plates (Corning Inc., Corning, NY) were rinsed twice with 0.025 M Tris-buffered saline (0.32 liters TBS-1 [31.6 g/liter Trizma base, 81.8 g/liter NaCl, 3.73 g/liter KCl, 0.57 g/liter Na2HPO4-anhydrous] in 3.68 liters ultrapure H2O) and then inoculated with 0.1-ml volumes of 10-fold serial dilutions (in duplicate) of the virus stock and incubated at 37°C for 1 h to allow for virus adsorption to the cells. Following this incubation period, 3 ml of a molten solution of MEM containing 1.5% Bacto agar (Becton, Dickenson and Co., Sparks, MD), 2% FBS (HyClone Laboratories, Logan, UT), 1 M HEPES buffer (Mediatech Inc., Manassas, VA), 7.5% sodium bicarbonate (Fisher Scientific, Fair Lawn, NJ), 10 mg/ml kanamycin (HyClone Laboratories, Logan, UT), 100× antimycotic (HyClone Laboratories, Logan, UT), and 200 mM glutamine (Glutamax; HyClone Laboratories, Logan, UT) was added as an overlay to each well and allowed to solidify. The plates were then incubated at 37°C with 5% CO2 for 2 days for PV-1 and EV-1 and 5 days for CV-B5. Following this incubation, the agar overlays were removed and the cell monolayers were stained with 0.5% (wt/vol) crystal violet (Sigma-Aldrich, St. Louis, MO) dissolved in ultrapure water and mixed 1:1 with 95% ethanol. The plaques (clearings in the cell monolayer) were counted to enumerate infectious viruses.
AV-2 titrations were performed using the Reed-Muench method (30) to determine the tissue culture infectious dose that affected 50% of the wells (TCID50). Serial 10-fold dilutions of the virus sample were assayed in 96-well tissue culture plates (Nunclon, Roskilde, Denmark) containing monolayers of PLC cells and 100 μl of MEM containing 2% CS with incubation at 37°C with 5% CO2, as before. Ten wells were inoculated with 50 μl of each dilution. This number of wells was used to ensure adequate precision of the assay. Maintenance medium (25 μl of MEM containing 2% CS) was added to each well every third day for the duration of the assay to maintain the integrity of the monolayer. Each well was checked every day for 12 days for viral cytopathogenic effects (CPE). The greatest dilution in which more than 50% of the wells were positive was used to determine the virus TCID50/ml.
Primary virus concentration using NanoCeram cartridge filters.
Presterilized electropositive NanoCeram VS2.5-5 virus filters (Argonide Corporation, Sanford, FL) were used during the course of the method development. Tucson municipal tap water was dechlorinated via passage through an Amway activated carbon block filter (Amway, Ada, MI) prior to its use in this study and analyzed for pH and total dissolved solids (TDS). The average values were pH 7.65 (± 0.24) and 337 mg/liter (±25 mg/liter). Total organic carbon was less than 1 mg/liter. Initially, 19 liters of water was added to a stainless steel pressure vessel (Alloy Products, Waukesha, WI). Approximately 108 PFU (for PV-1, EV-1, and CV-B5) or 108 TCID50 (for AV-2) of each virus (in separate experiments) was added to a separate liter in a polypropylene beaker and mixed using a stir bar for 15 min. The 1-liter suspension was then added to the pressure vessel to bring the final test water volume to 20 liters, which was mixed thoroughly for an additional 15 min. To determine the influent titer, three 15-ml samples were collected from the 20-liter volume for assay.
Positive pressure was applied to the vessel using N2 gas at approximately 2.5 lb/in2 to achieve a flow rate of 2.5 liters/min for the seeded water through the NanoCeram filter. Effluent samples (1 liter) were collected after the passage of 5, 10, and 15 liters of the test water through the filter to determine the amount of virus retained by the filter.
Elution of viruses from NanoCeram filters.
Several test eluting solutions (see Table 2) were used in this study, including both salt-based (e.g., phosphate salts, chaotropic salts such as NaI) and proteinaceous (e.g., beef extract) solutions. Buffering agents (e.g., glycine) and dispersants (e.g., Tween 80, NaPP) were also components of some of the solutions tested. The pH of the eluting solutions was adjusted to either 7.5 or 9.3, and then the solutions were autoclaved prior to the experiments, with the exception of solutions containing 1.0% (wt/vol) Na-polyphosphate (Sigma-Aldrich, St. Louis, MO), which were autoclaved first (to prevent the formation of precipitates). MS2 coliphage was used as a surrogate for enteric viruses in the elution evaluation experiments. To assess the adequacy of MS2 as a surrogate, PV-1 was used for comparison in a smaller subset of elution experiments. Following the primary virus concentration on the filter, the test eluting solution was added to the housing unit until the filter was completely immersed (∼420 ml). The unit was then resealed and inverted 10 times, followed by a hold time of 15 min at room temperature. The unit was inverted 10 additional times, followed by another 15-min hold at room temperature. After the unit was inverted another 10 times, the eluting solution was passed through the filter under positive pressure (N2 gas) into a sterile polypropylene beaker. The eluate was then added again to the filter housing unit with a 1-min hold time (14) and then passed through the filter and collected in the same beaker. The pH of the final eluate was immediately adjusted to 7.0 to 7.2 using 1 M HCl (dropwise), and the volume was measured.
Table 2.
Recovery of MS2 coliphage from NanoCeram filters using various eluting solutions
| Eluting solutiona | pH | No. of trials | Influent titer (log10/20 liters ± SD) | Eluate titer (log10/420 ml ± SD) | Elution efficiency (% ± SD) |
|---|---|---|---|---|---|
| A (3% beef extract) | 9.3 | 2 | 7.99 ± 0.17 | 7.49 ± 0.07 | 34 ± 18 |
| B (Gly) | 9.3 | 3 | 8.12 ± 1.57 | 5.37 ± 2.35 | 0.4 ± 0.5 |
| C (3% beef extract + Gly) | 9.3 | 2 | 7.57 ± 0.33 | 6.63 ± 0.31 | 12 ± 1 |
| D (PB + Gly) | 9.3 | 2 | 7.67 ± 0.07 | 7.07 ± 0.01 | 26 ± 5 |
| D (PB + Gly) | 7.5 | 3 | 7.93 ± 1.23 | 7.31 ± 1.13 | 24 ± 7 |
| E (PB + Gly + 0.3% Tween 80) | 9.3 | 2 | 7.48 ± 1.55 | 7.06 ± 1.58 | 37 ± 2 |
| F (1.0% NaPP + PB + Gly) | 9.3 | 5 | 8.14b ± 0.46 | 7.90 ± 0.45 | 57 ± 3 |
| F (1.0% NaPP + PB + Gly) | 7.5 | 2 | 7.83b ± 0.02 | 7.23 ± 0.09 | 26 ± 4 |
| F (1.0% NaPP + PB + Gly) | 9.3 | 3 | 4.58c ± 0.02 | 4.51 ± 0.06 | 86 ± 9 |
| G (0.1% NaPP + PB + Gly) | 9.3 | 3 | 7.82 ± 0.16 | 7.41 ± 0.21 | 40 ± 7 |
| H (0.6 M NaI + PB + Gly) | 9.3 | 2 | 8.17 ± 0.38 | 6.59 ± 0.67 | 3 ± 2 |
Gly, 0.05 M glycine; PB, phosphate buffer (3.8 mM Na2HPO4, 6.5 mM KH2PO4); NaPP, sodium polyphosphate.
High (∼108 PFU) influent titer.
Low (∼104 PFU) influent titer.
Secondary virus concentration using Centricon Plus-70 centrifugal filters.
The Centricon Plus-70 ultrafilter (30-kDa cutoff; Millipore, Billerica, MA) was utilized to further concentrate the eluted viruses. The device was prewetted by adding 70 ml of Nanopure H2O, followed by centrifugation (1,900 × g for 8 min). The unit was inverted and centrifuged (800 × g for 2 min) to collect the remaining water, which was then discarded. A 70-ml volume of the NanoCeram filter eluate was added to the Centricon filter and concentrated via centrifugation (1,900 × g for 8 min). The viral concentrate was collected via inversion of the filter and further centrifugation for 2 min at 800 × g. An additional 70-ml volume of Nanopure H2O was processed through the same ultrafilter (1,900 × g for 8 min), and then the concentrated viruses were extracted by inversion of the filter and centrifugation at 800 × g for 2 min. The average volume of secondary concentrates measured 3.3 ± 0.3 ml. The 350-ml volume remaining from the primary concentration step was not concentrated further. An additional one or two 70-ml volumes can be processed using the same Centricon filter, yielding a comparable recovery efficiency (data not shown). However, since the approximate 3.3-ml volume is more than enough for subsequent assays, one concentration step was chosen to reduce the amount of time and labor required.
When 1.0% (wt/vol) NaPP was used as the eluting solution, the secondary concentrates of AV-2 required a desalting step (to remove ∼99% of salts) due to toxicity observed with the PLC cell line (this was not necessary for the BGM cell line monolayers). The original Centricon concentrate was collected from the filter and resuspended in 70 ml of Nanopure H2O and then processed through the same filter (1,900 × g for 8 min) and collected once more via centrifugation (800 × g for 2 min). Following this, the viruses on the filter were eluted further using an additional 70-ml volume of H2O, as before.
Sample preparation and assay.
All nonviral contaminants were removed using a preblocked (with 2 ml of 1.5% beef extract) Millex syringe filter with a pore size of 0.22 μm (Millipore, Bedford, MA). The samples were then aliquoted into 1.5-ml volumes in cryogenic vials and stored at either −80°C (human viruses) or 4°C (MS2 coliphage) until quantitative infectivity assays were performed.
MS2 coliphage assays were performed using 10-fold serial dilutions and the double agar overlay method (2). Human viruses were quantified using either the 10-fold serial dilution plaque-forming assay on BGM cells (for PV-1, EV-1, and CV-B5) or the Reed-Muench (TCID50) method on PLC cells (for AV-2) as described previously.
PCR detection of poliovirus.
One-step reverse transcriptase PCR (RT-PCR) followed by nested PCR as described by Rodriguez et al. (33) was used to determine if the primary concentrate and the secondary concentrate contained any PCR-inhibiting substances. The PCR products (bands of 195 and 105 bp) were visualized via gel electrophoresis (33). The 195-bp RT-PCR product is not always observed with PV-1-positive samples; therefore, the more sensitive nested PCR assay (105-bp product) was performed following the RT-PCR.
Data analysis and statistics.
The ability of the NanoCeram filter to effectively concentrate (the filter retention efficiency) (Table 1) each of the viruses studied was calculated by comparing the total number of viruses measured in the effluent samples by the total number present in the influent samples, i.e., 100 × [1 − (number of viruses in effluent samples/number of viruses in influent samples)]. The elution (virus recovery) efficiency from the filter for each eluting solution (Table 2) was determined by comparing the number of viruses recovered from the filter (the primary concentrate) to the number of viruses originally adsorbed (the influent titer minus the effluent titer) to the filter, i.e., 100 × (number of viruses in eluate/number of viruses adsorbed to the filter) (see also Table 3). The secondary concentration efficiency (Table 4) was determined by comparing the number of viruses in the Centricon concentrate to the number of viruses found in 70 ml of the primary concentrate (the 420-ml filter eluate), i.e., 100 × (number of viruses in secondary concentrate/number of viruses in 70 ml primary concentrate).
Table 1.
Virus retention efficiencies for the NanoCeram filters
| Virus | No. of trials | Influent titer (log10/20 liters ± SD) | Effluent titer (log10/20 liters ± SD) | Virus retention (% ± SD) |
|---|---|---|---|---|
| MS2 coliphage | 31 | 8.08 ± 0.36 | <4.38 ± 0.93 | >99.8 ± 0.59 |
| Poliovirus 1 (PV-1) | 4 | 7.32 ± 0.03 | <4.22a ± 0.00 | >99.92 ± 0.01 |
| Echovirus 1 (EV-1) | 5 | 7.87 ± 0.08 | <4.22a ± 0.00 | >99.98 ± 0.00 |
| Coxsackievirus B5 (CV-B5) | 4 | 8.29 ± 0.07 | <4.43 ± 0.41 | >99.991 ± 0.00 |
| Adenovirus 2 (AV-2) | 4 | 8.31 ± 0.12 | <3.82a ± 0.00 | >99.997 ± 0.00 |
Detection limit.
Table 3.
Virus elution efficiencies (primary concentration) from NanoCeram filters using the 1.0% NaPP elution buffera
| Virus | No. of trials | Influent titer (log10/20 liters ± SD) | Eluate titer (log10/420 ml ± SD) | Elution efficiency (% ± SD) |
|---|---|---|---|---|
| MS2 coliphage (∼108 PFU) | 5 | 8.14 ± 0.46 | 7.90 ± 0.45 | 57 ± 3 |
| MS2 coliphage (∼104 PFU) | 3 | 4.58 ± 0.02 | 4.51 ± 0.06 | 86 ± 9 |
| Poliovirus 1 (PV-1) | 4 | 7.32 ± 0.03 | 7.15 ± 0.05 | 69 ± 8 |
| Echovirus 1 (EV-1) | 5 | 7.87 ± 0.08 | 7.99 ± 0.14 | 134 ± 27 |
| Coxsackievirus B5 (CV-B5) | 4 | 8.29 ± 0.07 | 8.14 ± 0.04 | 72 ± 13 |
| Adenovirus 2 (AV-2) | 4 | 8.31 ± 0.12 | 7.88 ± 0.05 | 39 ± 13 |
Solution F from Table 2 at pH 9.3.
Table 4.
Virus secondary concentration efficiencies using Centricon ultrafilters
| Virus | No. of trials | Secondary concn efficiency (% ± SD) |
|---|---|---|
| MS2 coliphage (∼108 PFU) | 2 | 75 ± 21 |
| MS2 coliphage (∼104 PFU) | 3 | 65 ± 6 |
| Poliovirus 1 (PV-1) | 4 | 95 ± 5 |
| Echovirus 1 (EV-1) | 3 | 61 ± 18 |
| Coxsackievirus B5 (CV-B5) | 4 | 109 ± 11 |
| Adenovirus 2 (AV-2) | 4 | 33 ± 14 |
In order to determine the overall method efficiency for each of the viruses tested (Table 5), the number of viruses in the secondary concentrate was compared with an equivalent volume from the original 20-liter influent sample (i.e., 70 ml/420 ml total eluate concentrated using Centricon filter ≅ 3.3 liter/20 liter total influent). The number of viruses in the secondary concentrate was then divided by the number of viruses from the equivalent volume of the influent and then multiplied by 100. The standard deviations for all of the efficiencies were also calculated for each set of experiments.
Table 5.
Overall method efficiencies for virus concentration and recovery using NanoCeram filters along with Centricon ultrafilters
| Virus | No. of trials | Overall method efficiency (% ± SD) |
|---|---|---|
| MS2 coliphage (∼108 PFU) | 2 | 45 ± 15 |
| MS2 coliphage (∼104 PFU) | 3 | 56 ± 9 |
| Poliovirus 1 (PV-1) | 4 | 66 ± 6 |
| Echovirus 1 (EV-1) | 3 | 83 ± 14 |
| Coxsackievirus B5 (CV-B5) | 4 | 77 ± 11 |
| Adenovirus 2 (AV-2) | 4 | 14 ± 4 |
A two-tailed Student t test was used to compare the virus recovery efficiencies between experiments conducted with various elution buffers (Table 2). Differences were considered significant if the resultant P value was ≤0.05.
RESULTS
MS2 coliphage served as the enteric virus surrogate for the initial assessment of the NanoCeram filters. The mean virus retention efficiency for MS2 is shown in Table 1 (>99.8%). The NanoCeram filters were subsequently evaluated using PV-1, EV-1, CV-B5, and AV-2. The ability of the filters to concentrate these viruses was similarly high, with >99.9% retention (Table 1). The filter efficiencies are reported as >99.8% for all of the viruses tested, because either no virus was recovered from the effluent in any of the experiments (i.e., with PV-1, EV-1, and AV-2, for which the detection limit was used to calculate the filter efficiency) or the virus was not detected in the effluent in at least one of the experiments (i.e., with MS2 and CV-B5, for which the detection limit was used in the calculation for some of the filters).
Various eluting solutions (Table 2) were assessed for their ability to recover MS2 coliphage from the NanoCeram filters in preliminary tests. The average volume of the filter eluates following the pH adjustment to neutral was 420 ml. Of the eluting solutions tested, those containing the NaPP dispersant along with phosphate buffer (3.8 mM Na2HPO4, 6.5 mM KH2PO4; pH 7.5) and glycine (0.05 M) were the most effective, with the 1.0% NaPP (solution F in Table 2) solution yielding a significantly greater (P ≤ 0.05) MS2 recovery than the 0.1% NaPP solution (solution G). An alkaline 1.0% NaPP eluting solution (pH 9.3) was more effective than the same solution at pH 7.5 (57% versus 26%). A lower influent MS2 seed titer (∼104 versus ∼108 PFU) also resulted in a higher elution efficiency when using the 1.0% NaPP eluting solution (86% versus 57%). The comparison experiments performed with PV-1 yielded similar results (data not shown), indicating that MS2 was a reasonable surrogate for enteric viruses. The 1.0% NaPP (solution F) at pH 9.3 was therefore used in all subsequent experiments with PV-1, EV-1, CV-B5, and AV-2 (Table 3). The elution efficiencies observed using this elution buffer ranged from 39% for AV-2 to 134% for EV-1. Greater variation in the elution efficiency was observed between trials for EV-1 (standard deviation of 27%) than for the other human viruses tested (standard deviations ranging from 8 to 13%).
The viruses that were recovered from the NanoCeram filters using the 1.0% NaPP eluting buffer were concentrated further (to 3.3 ml ± 0.3 ml) using Centricon Plus-70 ultrafilters. MS2 coliphage, PV-1, EV-1, and CV-B5 were concentrated by the Centricon ultrafilters with virus recoveries of ≥61% (Table 4). As with the virus elution from the NanoCeram filters, the recovery of AV-2 was also lower for the secondary concentration step (33%).
The overall method efficiencies for the recovery of each virus were determined by comparing the virus titers measured in the secondary concentrates to the numbers of viruses originally used to seed the 20 liters of dechlorinated tap water. The method efficiency for each of the viruses tested is shown in Table 5. Concentration efficiencies of ≥66% were achieved for all of the human viruses tested with the exception of AV-2 (14%). Similar method efficiencies were found for both high-seed-titer (∼108 PFU) and low-seed-titer (∼104 PFU) trials with MS2 coliphage (45% and 56%, respectively).
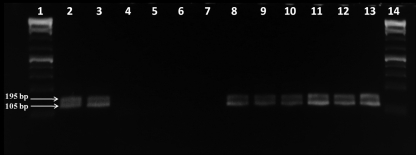
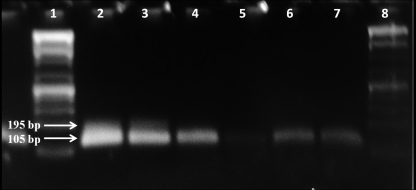
The amplification results from the RT-PCR and nested PCR assays are shown in Fig. 1. Although the primary PV-1 concentrate in the NaPP eluting solution did inhibit the PCR, this inhibition was eliminated during the secondary concentration step. To determine if the PCR was able to detect lower numbers of PV-1 in the presence of inhibition from NaPP, tests were performed in which lower initial PV-1 titers (10 PFU and 103 PFU/liter versus 106 PFU/liter) were added to the NaPP prior to the secondary concentration steps with the centrifugal filters (Fig. 2). The seminested PCR product (105 bp) was observed even with an initial virus titer of 10 PFU/liter of eluting solution.
Fig. 1.
Amplification using seminested reverse transcriptase PCR of poliovirus in primary and secondary concentrates. Lanes: 1 and 14, 1-kb ladder; 2 and 3, poliovirus-positive control in nuclease-free water (seeded with 108 PFU/ml); 4 and 5, negative control (no virus); 6 and 7, poliovirus in NaPP eluting buffer (seeded with 106 PFU/liter); 8 and 9, poliovirus recovered from the Centricon filter; 10 and 11, poliovirus recovered from the Centricon filter after the water rinse (end product); 12 and 13, poliovirus recovered from the Centricon filter after a desalting step (necessary for use with some cell culture lines).
Fig. 2.
Effect of virus concentration on amplification using seminested reverse transcriptase PCR. Lanes: 1 and 8, 1-kb ladder; 2 and 3, poliovirus-positive control in nuclease-free water (seeded with 108 PFU/ml); 4 and 5, poliovirus from secondary concentrate (originally seeded with 103 PFU/liter in NaPP eluting buffer); 6 and 7, poliovirus from secondary concentrate (originally seeded with 10 PFU/liter in NaPP eluting buffer).
DISCUSSION
The VIRADEL method has been refined and modified over several decades in an effort to enhance the concentration, recovery, and detection of enteric viruses in water. Nonetheless, this method remains far from ideal due to variable virus recovery efficiencies (20), fairly high final concentrate volumes (∼15 to 40 ml), and the presence of PCR-inhibiting substances in the recovered virus concentrates (1).
In the present study, the recently available and less expensive NanoCeram filter (Argonide, Sanford, FL) was evaluated for its ability to effectively concentrate viruses from water using a new VIRADEL approach. The NanoCeram filter retained ≥99.8% of all the viruses tested. Karim et al. (21) likewise studied the retention efficiency of the NanoCeram filter by passing 100 liters of dechlorinated tap water (flow rate of 5.6 ± 0.17 liter/min) seeded with PV-1 (105 PFU). The virus was retained by the filter with an efficiency of 84% (±9%). The reason for the difference between the results of these two studies is unclear. Higher flow rates often lead to a decrease in the virus adsorption potential of filters (41); nevertheless, in separate experiments with PV-1 using a higher flow rate (5.7 liters/min) and a higher volume of dechlorinated tap water (100 liters), retention efficiencies similar to those presented in Table 1 were observed in our laboratory (96% ± 2%; n = 4 trials). Gibbons et al. (16) also found relatively high adsorption efficiencies for adenovirus 41 (97%) and Qβ coliphage (>99%) by NanoCeram filters from 40 liters of dechlorinated finished waters processed at a high flow rate of 25 liters/min. In addition, both viruses were recovered from seeded seawaters and fresh source waters (40 liters at 25 liters/min) with an efficiency of >99% for seawater and approximately 80% for source waters. Nevertheless, the experiments described by Karim et al. (21) involved a lower initial poliovirus seed titer (∼105 PFU) than that of the present study (∼108 PFU), which might account for the difference in filter efficiencies.
The elution (recovery) of viruses from adsorptive media has presented several unique challenges for researchers. Adsorption kinetics are affected by numerous factors, including the virus surface properties (e.g., virus pI, the presence of a lipid envelope), the properties of the surrounding aqueous environment (e.g., ionic strength, pH), the presence of organic compounds, and the zeta potential of abiotic surfaces in a given system. Manipulation of one or more of these factors can lead to the successful desorption (elution) of viruses from the filters. Viruses that are more electronegative adsorb more strongly to positively charged filter surfaces. This may subsequently impact the efficiency of their elution. Solutions consisting of various amino acids (e.g., glycine) and complex proteinaceous solutions (e.g., beef extract) have been used to elute viruses from filters.
Beef extract has been used successfully for the elution of viruses from other filtration media; however, solutions with 3.0% beef extract (both nonbuffered and glycine buffered) evaluated in the current study yielded relatively low recoveries of MS2 coliphage from NanoCeram filters in comparison to the NaPP-based buffers (Table 2). The aforementioned published studies evaluating NanoCeram filters have both used beef extract solutions for the virus elution step. Karim et al. (21) reported elution efficiencies for PV-1 (54% ± 8%), CV-B5 (27% ± 17%), and echovirus 7 (32% ± 8%) from NanoCeram filters using 1.5% beef extract buffered with 0.05 M glycine (pH 9.0) with a primary concentrate volume of approximately 1 liter (2 elutions × 500 ml) and a secondary concentrate volume of 80 ml (2 elutions × 40 ml). Gibbons et al. (16) utilized 3.0% beef extract with 0.1 M glycine (pH 9.5) for the recovery of adenovirus 41 (1.4% ± 0.6%) and Qβ coliphage (91% ± 53%) from the NanoCeram filters with a primary concentrate volume of approximately 500 ml.
In the present study, the most effective recovery solutions were comprised of either 1.0% or 0.1% NaPP with phosphate buffer and glycine, recovering 57% and 40% of MS2 coliphage, respectively. Hill et al. (19) also reported greater recoveries (82% to 106%) using 0.01% or 0.1% NaPP to elute MS2 from ultrafiltration membranes. The 1.0% NaPP solution (pH 9.3) used in the present study was successful in eluting MS2 coliphage from the NanoCeram filters (57%). This recovery was significantly greater (P ≤ 0.05) than those achieved by any of the other eluting solutions tested, including the 1.0% NaPP solution at pH 7.5 (P = 8 × 10−5), the 0.1% NaPP solution (P = 0.002), and 3% beef extract (P = 0.02). When a lower concentration of MS2 (∼104 versus ∼108 PFU) was adsorbed to the filters, a significantly greater (P = 0.0004) elution efficiency was observed with the 1.0% NaPP solution (86% versus 57%). The number of viruses present in real water samples (from various sources) would likely be more reflective of this lower influent titer.
The recovery of human viruses from the NanoCeram filters was even more successful using the 1.0% NaPP solution, with ≥69% recovery for all viruses tested with the exception of AV-2 (39%). This included a greater recovery of 134% (±27%) for EV-1, likely due to the disruption of viral aggregates and a lack of precision of the assay. Sobsey et al. (38) found recovery efficiencies of EV-1 seeded in tap water and adsorbed to 1MDS filter media in small-volume (1.7-liter) and high-volume (1,000-liter) experiments of 53% and 9%, respectively, when eluting with 0.3% beef extract plus 0.05 M glycine at pH 9.5. Recovery efficiencies of echoviruses and coxsackieviruses have also been reported in adsorption-elution studies utilizing charge-modified filters. Cationic polymer-modified filters eluted with 3% beef extract (pH 9.5) yielded 99% of adsorbed EV-1 and 104% of CV-B5 (32). The same eluting solution used in combination with cellulose filters coated with ferric and aluminum hydroxide precipitates gave an average recovery of 34% of echovirus 5 and 37% of CV-B5 (11). Recovery efficiencies of CV-B5 from glass wool filters using 3% beef extract buffered with glycine (0.5 M) for elution ranged from 5% to 32% (23). The wide range of recovery values published for CV-B5 demonstrates that the compositions of both the adsorbent filter medium and the eluting solution impact recovery.
The recovery of AV-2 (39%) in this study, though lower than the recovery of the other viruses tested, was still much higher than that measured by Gibbons et al. (16) for adenovirus 41. Adenovirus 2 and adenovirus 41 exhibit different physicochemical properties (e.g., capsid protein isoelectric point values) (13) which may affect their concentration and elution from filters.
A secondary concentration step was used in this study to further concentrate viruses eluted from the NanoCeram filters. The process of organic flocculation used for concentrating beef extract (proteinaceous) eluates typically results in secondary concentrate volumes ranging from 15 to 40 ml. Centricon Plus-70 ultrafilters were used in the current study to reduce the eluate volume from 70 ml to <3.5 ml. In high-titer and low-titer MS2 coliphage experiments, 75% (±21%) and 65% (±6%) of viruses applied to the Centricon ultrafilters were recovered in the final concentrate volume, respectively (Table 4). Hill et al. (18) reported a secondary concentration efficiency of 82% (±26%) for MS2 using the same method. The secondary concentration of PV-1 and CV-B5 was highly effective (≥95%), as most of the viruses applied to the Centricon ultrafilter were recovered in the concentrates (Table 4). It is interesting to note that EV-1 had the highest primary elution efficiency yet was recovered less capably from the Centricon ultrafilters than the other enteroviruses tested. This may be attributable to intermolecular interactions between EV-1 and the material comprising the ultrafilter. AV-2, the largest of the test viruses, displayed the lowest secondary recovery efficiency. AV-2 has pentons (protein spikes) which extrude from the 12 vertices of the virus capsid (42). These may allow for further interactions with the filter material and inhibit the recovery of the virus.
The overall method efficiencies were ≥50% for PV-1 (66%), EV-1 (83%), CV-B5 (77%), and MS2 coliphage (low influent titer) (56%) but not for AV-2 (14%) (Table 5). The small secondary concentrate volume (<3.5 ml) achieved in the current study compares quite favorably against previously described methods in which similar efficiencies have been observed, but with much larger secondary concentrate volumes (from 5- to up to >12-fold higher). Also, the secondary concentration step appears to be effective at removing any PCR-inhibiting substances found in the NaPP eluting solution from the viral concentrates. This is true even with virus concentrations as low as 10 PFU/liter in the primary effluent (Fig. 2). The NanoCeram virus sampler, along with the elution and secondary concentration methods developed in the present study, is a new, less expensive concentration method for viruses from water. This method is able to recover viral pathogens from water with efficiencies at least equal to currently available methods used for pleated microporous filters, with much lower secondary concentrate volumes. This lower volume should help to increase the efficacy of current detection methods by allowing for the assay of very small volumes. In addition, this method appears to be compatible with both virus cell culture and PCR assays.
ACKNOWLEDGMENTS
This work was supported by a Science to Achieve Results (STAR) grant (R833009) from the United States Environmental Protection Agency.
Although the research described in this article has been funded wholly or in part by the United States Environmental Protection Agency through grant/cooperative agreement R833009 to Kelly R. Bright, it has not been subjected to the Agency's required peer and policy review and therefore does not necessarily reflect the views of the Agency, and no official endorsement should be inferred.
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Abbaszadegan M., Huber M. S., Gerba C. P., Pepper I. L. 1993. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl. Environ. Microbiol. 59:1318–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams M. H. 1959. Bacteriophages. Interscience Publishers, Inc., New York, NY [Google Scholar]
- 3. American Public Health Association (APHA) 2005. 9510 C. Virus concentration from large sample volumes by adsorption to and elution from microporous filters, p. 9-141–9-145 In Eaton A. D., Clesceri L. S., Rice E. W., Greenberg A. E. (ed.), Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, American Water Works Association, Water Environment Federation Publication, Washington, DC [Google Scholar]
- 4. Berg G., Dahling D., Berman D. 1971. Recovery of small quantities of viruses from clean waters on cellulose nitrate membrane filters. Appl. Microbiol. 22:608–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bidawid S., Malik N., Adegbunrin O., Sattar S. S., Farber J. M. 2003. A feline kidney cell line-based plaque assay for feline calicivirus, a surrogate for Norwalk virus. J. Virol. Methods 107:163–167 [DOI] [PubMed] [Google Scholar]
- 6. Black S., Thurston J. A., Gerba C. P. 2009. Determination of Ct values for chlorine resistant enteroviruses. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 44:336–339 [DOI] [PubMed] [Google Scholar]
- 7. Cashdollar J. L., Dahling D. R. 2006. Evaluation of a method to re-use electropositive cartridge filters for concentrating viruses from tap and river water. J. Virol. Methods 132:13–17 [DOI] [PubMed] [Google Scholar]
- 8. Dahling D. 2002. An improved filter elution and cell culture assay procedure for evaluating public groundwater systems for culturable enteroviruses. Water Environ. Res. 74:564–568 [DOI] [PubMed] [Google Scholar]
- 9. Farrah S. R. 1982. Chemical factors influencing adsorption of bacteriophage MS2 to membrane filter. Appl. Environ. Microbiol. 43:659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farrah S. R., Gerba C. P., Wallis C., Melnick J. L. 1976. Concentration of viruses from large volumes of tap water using pleated membrane filters. Appl. Environ. Microbiol. 31:221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farrah S., Preston D. 1985. Concentration of viruses from water by using cellulose filters modified by in situ precipitation of ferric and aluminum hydroxides. Appl. Environ. Microbiol. 50:1502–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farrah S. R., Shah D. O., Ingram L. O. 1981. Effects of chaotropic and antichaotropic agents on elution of poliovirus adsorbed to membrane filters. Proc. Natl. Acad. Sci. U. S. A. 78:1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Favier A. L., Burmeister W. P., Chroboczek J. 2004. Unique physicochemical properties of human enteric Ad41 responsible for its survival and replication in the gastrointestinal tract. Virology 322:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fout G. S., Schaefer F. W., III, Messer J. W., Dahling D. R., Stetler R. E. 1996. Information collection rule (ICR) microbial laboratory manual. EPA/600/R-95/178. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 15. Gassilloud B., Duval M., Schwartzbrod L., Gantzer C. 2003. Recovery of feline calicivirus infectious particles and genome from water: comparison of two concentration techniques. Water Sci. Technol. 47:97–101 [PubMed] [Google Scholar]
- 16. Gibbons C. D., Rodriguez R. A., Tallon L., Sobsey M. D. 2010. Evaluation of positively charged alumina nanofibre cartridge filters for the primary concentration of noroviruses, adenoviruses, and male-specific coliphages from seawater. J. Appl. Microbiol. 109:635–641 [DOI] [PubMed] [Google Scholar]
- 17. Goyal S. M., Zerda K. S., Gerba C. P. 1980. Concentration of coliphages from large volumes of water and wastewater. Appl. Environ. Microbiol. 39:85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hill V. R., et al. 2007. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl. Environ. Microbiol. 73:4218–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hill V. R., et al. 2005. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 71:6878–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hill V. R., et al. 2009. Comparison of hollow-fiber ultrfiltration to the USEPA VIRADEL technique and USEPA method 1623. J. Environ. Qual. 38:822–825 [DOI] [PubMed] [Google Scholar]
- 21. Karim M. R., Rhodes E. R., Brinkman N., Wymer L., Fout G. S. 2009. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl. Environ. Microbiol. 75:2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katzenelson E., Fattal B., Hostovesky T. 1976. Organic flocculation: an efficient second-step concentration method for the detection of viruses in water. Appl. Environ. Microbiol. 32:638–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lambertini E., et al. 2008. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 74:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LeBaron C. W., et al. 1990. Viral agents of gastroenteritis public health importance and outbreak management. Morb. Mortal. Wkly. Rep. 39:1–24 [PubMed] [Google Scholar]
- 25. Logan K. B., Rees G. E., Seeley N. D., Primrose S. B. 1980. Rapid concentration of bacteriophages from large volumes of freshwater: evaluation of positively charged, microporous filters. J. Virol. Methods 1:87–97 [DOI] [PubMed] [Google Scholar]
- 26. Ma J. F., Naranjo J., Gerba C. P. 1994. Evaluation of MK filters for recovery of enteroviruses from tap water. Appl. Environ. Microbiol. 60:1974–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melnick J. L., et al. 1984. Round robin investigation of methods for the recovery of poliovirus from drinking water. Appl. Environ. Microbiol. 47:144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Menut C., Beril C., Schwartzbrod L. 1993. Poliovirus recovery from tap water after concentration over glass powder and glass wool. Water Sci. Technol. 27:291–294 [Google Scholar]
- 29. Moe C. L. 2002. Waterborne transmission of infectious agents, p. 184–204 In Hurst C. J. (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 30. Payment P., Trudel M. 1993. Isolation and identification of viruses, p. 32–33 In Payment P., Trudel M. (ed.), Methods and techniques in virology. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 31. Polaczyk A. L., et al. 2008. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-liter tap water samples. J. Microbiol. Methods 73:92–99 [DOI] [PubMed] [Google Scholar]
- 32. Preston D. R., Vasudevan T. V., Bitton G., Farrah S. R., Morel J. L. 1988. Novel approach for modifying microporous filters for virus concentration from water. Appl. Environ. Microbiol. 54:1325–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodríguez R. A., Gundy P. M., Gerba C. P. 2008. Comparison of BGM and PLC/PRC/5 cell lines for total culturable viral assay of treated sewage. Appl. Environ. Microbiol. 74:2583–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scott T. M., Lukasik J., Farrah S. R. 2002. Improved method for recovery of bacteriophages from large volumes of water using negatively charged microporous filters. Can. J. Microbiol. 48:305–310 [DOI] [PubMed] [Google Scholar]
- 35. Sharma M. M., Chang Y. I., Yen T. F. 1985. Reversible and irreversible surface charge modification of bacteria for facilitating transport through porous media. Colloids Surf. 16:193–206 [Google Scholar]
- 36. Shields P. A., Farrah S. R. 1983. Influence of salts on electrostatic interactions between poliovirus and membrane filters. Appl. Environ. Microbiol. 45:526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sobsey M. D., Glass J. S. 1980. Poliovirus concentration from tap water with electropositive adsorbent filters. Appl. Environ. Microbiol. 40:201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sobsey M. D., Moore R. S., Glass J. S. 1981. Evaluating adsorbent filter performance for enteric virus concentrations in tap water. J. Am. Water Works Assoc. 73:542–548 [Google Scholar]
- 39. Sobsey M. D., et al. 2004. Development and evaluation of methods to detect coliphages in large volumes of water. Water Sci. Technol. 50:211–217 [PubMed] [Google Scholar]
- 40. Straub T. M., Pepper I. L., Gerba C. P. 1992. Persistence of viruses in desert soils amended with anaerobically digested sewage sludge. Appl. Environ. Microbiol. 58:626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wallis C., Melnick J. L., Gerba C. P. 1979. Concentration of viruses from water by membrane chromatography. Annu. Rev. Microbiol. 33:413–437 [DOI] [PubMed] [Google Scholar]
- 42. Zubieta C., Schoehn G., Chroboczek J., Cusack S. 2005. The structure of the human adenovirus 2 penton. Mol. Cell 17:121–135 [DOI] [PubMed] [Google Scholar]