Abstract
We report that the bacterial transposon Tn7 selects targets by recognizing features associated with DNA replication using the transposon-encoded DNA-binding protein TnsE. We show that Tn7 transposition directed by TnsE occurs in one orientation with respect to chromosomal DNA replication, indicating that a structure or complex involved in DNA replication is likely to be a critical determinant of TnsE insertion. We find that mutant TnsE proteins that allow higher levels of transposition also bind DNA better than the wild-type protein. The increased binding affinity displayed by the TnsE high-activity mutants indicates that DNA binding is relevant to transposition activity and suggests that TnsE interacts directly with target DNAs. In vitro, TnsE interacts preferentially with certain DNA structures, indicating a mechanism for the TnsE-mediated orientation and insertion preference. The pattern of TnsE-mediated insertion events around the Escherichia coli chromosome provides insight into how DNA replication forks proceed in vivo.
Keywords: Transposition, target selection, DNA replication
Mobile DNA elements called transposons are nearly ubiquitous in living organisms. Most often, transposons move with little apparent specificity for target sites. There are, however, examples of transposable elements that recognize various features in insertion sites (Craig 1997). The ability to recognize specific target sites could facilitate transposition and/or minimize the chance of inserting into essential host genes. Target features that could be recognized by a transposon include a specific DNA sequence, a DNA structure, or a protein–DNA complex. Tn7 is a bacterial transposon that has evolved an elaborate system of target site selection involving two different target selection pathways. We show here that one of Tn7's insertion pathways identifies target sites by recognizing a structure or complex associated with DNA replication using a transposon-encoded DNA-binding protein, TnsE.
Tn7 encodes five proteins to perform its two pathways of transposition: TnsA, TnsB, TnsC, TnsD, and TnsE (TnsABCDE) (Rogers et al. 1986; Waddell and Craig 1988; Kubo and Craig 1990). TnsABC make up the core transposition machinery that is incapable of transposition without one of two target site selecting proteins, TnsD or TnsE (Waddell and Craig 1988; Bainton et al. 1993). The TnsA and TnsB proteins collaborate to form the transposase responsible for the DNA breakage and joining activities that underlie transposition (May and Craig 1996; Sarnovsky et al. 1996; Biery et al. 2000; Lu and Craig 2000). TnsC is a regulator of transposase activity, communicating between the TnsAB transposase and the target selecting proteins, TnsD and TnsE (Stellwagen and Craig 1998). One transposition pathway, TnsABC+E, allows Tn7 to disseminate efficiently between host bacteria, whereas a second pathway, TnsABC+D, ensures that Tn7 can quickly find a safe insertion site called attTn7 that is found in many diverse hosts (Craig 1996). This paper focuses on the transposition pathway that uses the Tn7-encoded protein TnsE. In the TnsABC+E pathway, Tn7 preferentially inserts into plasmid DNAs that are transferred between cells in a process called conjugation (Wolkow et al. 1996). The TnsABC+E pathway has also been found to direct transposition into the Escherichia coli chromosome in regions where DNA replication terminates and proximal to DNA double-strand breaks (Peters and Craig 2000). The molecular basis for the target site selection displayed in TnsABC+E transposition and the role of TnsE in transposition has been unknown.
We show here that TnsE is a DNA-binding protein that recognizes a feature of DNA replication to direct Tn7 transposition. We describe the isolation and analysis of mutant TnsE proteins that promote increased frequencies of transposition. We also show that purified TnsE protein binds DNA. The mutant TnsE proteins that are more active in transposition in vivo bind DNA better than the wild-type protein, suggesting that TnsE's DNA-binding activity is involved in its role as a target site selector. Additionally, we show that TnsE selectively binds certain DNA structures that are likely to occur during DNA replication, indicating a mechanism for the TnsE-mediated insertion preference. The profile of TnsE-mediated Tn7 insertions in the chromosome provides insight into how DNA replication proceeds throughout the entire E. coli chromosome.
Results
Identification of the TnsE coding region
Determination of the N terminus of TnsE was necessary because it was uncertain if TnsE initiated with a valine (GTG) start codon or a methionine (ATG) codon located 37 codons downstream (Fig. 1). We examined TnsE-dependent transposition in the presence of two TnsE derivatives, one with and one without the N-terminal 36 amino acids, pJP118 and pJP117 (Fig. 1; Guzman et al. 1995). Transposition was tested using a “lambda hop” assay in which a defective lambda phage that cannot integrate or recombine into the host genome is used to deliver a kanamycin-resistant miniTn7 element (McKown et al. 1988). The Tn7 transposition proteins are provided in trans on compatible plasmids, and the frequency of transposition is scored as kanamycin-resistant colonies per infective lambda phage measured in plaque forming units (colonies/pfu). When the full-length TnsE construct was tested, we found that increasing levels of TnsE expression resulted in a greater frequency of transposition (Table 1). The N-truncated TnsE derivative did not allow the high level of transposition found with the full-length construct, nor did the vector-only control (Table 1).
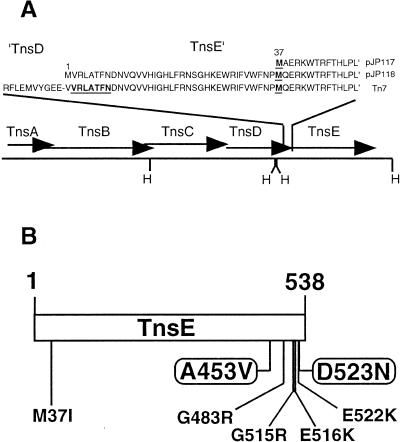
Figure 1.
The order of the Tns coding regions of Tn7 and the position of the mutations isolated in TnsE. (A) Schematic representation of the Tn7 Tns coding regions and the relative positions of HindIII sites (H). The amino acid sequences of the C terminus of TnsD and the N terminus of TnsE are shown in the context of native Tn7 (Flores et al. 1990). The N-terminal protein sequences of two TnsE expression constructs (pJP117 and pJP118) are also shown. The peptide sequence obtained by sequencing purified TnsE and amino acid M37 are shown in bold and underlined (see text for details). (B) Schematic representation of the 538-amino-acid TnsE protein showing the approximate position of the high-activity mutations. (Circled residues) Mutations isolated as single mutations (see text for details).
Table 1.
Transposition frequencies comparing two TnsE derivatives
| Inductiona
|
Transposition frequencyb
|
||
|---|---|---|---|
| Vector only
|
N-truncated TnsE
|
Full-length TnsE
|
|
| − | N.D. | <0.1 | 0.2 (+/−0.2) |
| + | N.D. | 0.4 (+/−0.7) | 4.6 (+/−2.4) |
| ++ | N.D. | <0.1 | 430 (+/−38) |
| +++ | <0.1 | 0.8 (+/−0.2) | 340 (+/−46) |
TnsE expression was modulated using the pBAD24 arabinose vector (Guzman et al. 1995). Expression was repressed with 0.2% glucose (−). TnsE expression was induced with increasing concentrations of arabinose: 0.00002% (+), 0.002% (++), 0.2% (+++) (see Materials and Methods).
The frequency of transposition was determined using the lambda hop assay and is expressed as Kanamycin-resistant colonies/plaque forming unit of phage (see Materials and Methods). Frequency represents the average from three experiments where the final value is multiplied by 106. TnsABC were expressed in all strains from pCW15, N-truncated TnsE was expressed from pJP117, and full-length TnsE was expressed from pJP118 (see Materials and Methods). The standard deviation from the mean is indicated in parentheses. (N.D.) not determined.
We also sequenced the TnsE protein isolated using a synthetic histidine affinity tag on the C terminus of the protein that possesses the native 5′ translation signals (see Materials and Methods). The purified protein was subjected to peptide sequencing and found to initiate with the amino acids V-R-L-A-T-F-N, a sequence found only at the N terminus of TnsE (Fig. 1). The histidine-tagged TnsE derivative is functional in vivo for transposition via multiple transposition assays (data not shown).
Isolation of high-activity TnsE proteins
We isolated TnsE mutants that allow high frequency transposition to gain a better understanding of how TnsE works. TnsE-expressing plasmids were subjected to in vitro hydroxylamine mutagenesis and transformed into a strain expressing TnsABC in which transposition was monitored using a visual screen based on promoter capture called the “papillation” assay (see Materials and Methods; Stellwagen and Craig 1997). Early mutagenesis experiments using a high-copy pUC18-based plasmid construct yielded 18 mutants in a screen of ∼23,000 colonies. DNA sequencing indicated that all 18 mutants corresponded to one of two coding changes in the 538amino-acid protein, TnsEA453V (10 isolates) and TnsED523N (8 isolates).
We were concerned that the TnsE expression levels in the high-copy pUC18-based construct could limit the spectrum of mutants obtained; therefore, we expressed TnsE in a very low-copy pSC101 replicon vector to facilitate the detection of TnsE mutants. We screened ∼1300 colonies for high-activity mutants in the low-copy plasmid and found one isolate, TnsEE522K/D523N. Given that we identified a double mutant allowing high levels of transposition, we actively sought double TnsE mutants that promote higher transposition levels by remutagenizing plasmids encoding the TnsEA453V and TnsED523N alleles. Two double mutants were obtained by screening ∼600 colonies with TnsEA453V: TnsEA453V/G483R and TnsEA453V/G515R. Five double mutants were obtained by screening ∼1200 colonies with the TnsE D523N-encoding plasmid: TnsE M37I/D523N (2 isolates), TnsE E516K/D523N, TnsEE522K/D523N, and TnsEA453V/D523N (Fig. 1). That two of the double mutants were isolated twice suggests that the mutagenesis procedure was approaching saturation. Almost all of the mutations occurred in the C-terminal 15% of the TnsE protein (Fig. 1).
In vivo analysis of the mutant TnsE proteins
TnsABC+E transposition levels promoted by the various TnsE mutants were quantitatively determined using the lambda hop transposition assay (Table 2). The single mutants TnsEA453V and TnsED523N promote transposition at about a 10-fold higher frequency then levels found with the wild-type protein. The TnsE double mutants displayed a wide range of transposition frequencies. The TnsEA453V/D523N mutant was the most active mutant, catalyzing a transposition frequency within twofold of the levels normally found only with the robust TnsABC+D transposition reaction (Kubo and Craig 1990).
Table 2.
TnsABC+E transposition frequency promoted by various TnsE constructs
| TnsE allele
|
Expressiona
|
Frequencyb
|
Frequencyc
|
|
|---|---|---|---|---|
| Chromosome
|
pOX38-Gen
|
|||
| None | <0.1 | |||
| Wild-type | Low | 0.18 (+/−0.08) | 0.02 | 0.16 |
| A453V | Low | 1.9 (+/−0.31) | 1.0 | 0.9 |
| D523N | Low | 1.9 (+/−0.7) | 0.83 | 1.1 |
| A453V/G483R | Low | 63.3 (+/−14.0) | — | — |
| A453V/G515R | Low | 43.3 (+/−10.8) | 40.8 | 2.5 |
| A453V/D523N | Low | 170.0 (+/−17.3) | 170.0 | <8.9 |
| E516K/D523N | Low | 45.7 (+/−4.5) | — | — |
| E522K/D523N | Low | 9.0 (+/−0.6) | — | — |
| M37I/D523N | Low | 40.3 (+/−6.4) | — | — |
| Wild-type | High | 16.3 (+/−2.3) | — | — |
TnsABC were expressed in all strains from pCW15 (see Materials and Methods). TnsE was expressed from pJP103 (low TnsE expression) or pJP104 (high TnsE expression).
The frequency of transposition was determined using the lambda hop assay and is expressed as Kanamycin-resistant colonies/plaque forming unit of phage (see Materials and Methods). Frequency represents the average from three experiments where the final value is multiplied by 106. The standard deviation from the mean is indicated in parentheses.
The frequency of transposition into the chromosome and pOX38-Gen was calculated from the data in Fig. 2, by multiplying the proportion of insertions into each replicon by the total transposition frequency.
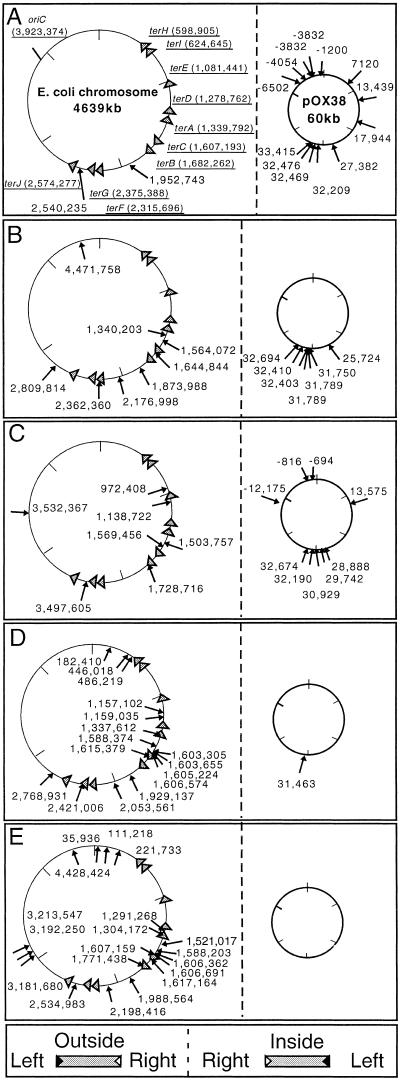
We also examined the insertion specificity of the various TnsE mutants. Previous work has shown that TnsE transposition events occur preferentially into conjugable plasmids rather than the chromosome (Wolkow et al. 1996). The position of TnsE-mediated transposition events was determined in an E. coli strain containing a conjugable F plasmid derivative, pOX38-Gen. When wild-type TnsE was tested, the results were consistent with previous work: The majority of the transposition events occurred into the conjugable plasmid (13/15) and in one orientation (Fig. 2A). With the two single mutants, we found that about half of the insertions occurred into the chromosome, 8/15 with TnsEA453V and 7/16 with TnsED523N (Fig. 2B,C). The profile of insertions was also examined with two of the TnsE double mutants. Nearly all of the insertions that were catalyzed by the double mutants were found in the chromosome, 16/17 with TnsEA453V/G515R and 19/19 with TnsEA453V/D523N (Fig. 2D,E). The high proportion of insertions isolated from the chromosome when examining the TnsE mutants could indicate that the mutants have lost the target specificity displayed by wild-type TnsE. However, based on other results presented in this paper, we argue that the mutants likely recognize the same target as the wild-type protein (see below).
Figure 2.
The location and orientation of TnsABC+E-mediated transposition events in the E. coli chromosome and the conjugable F plasmid derivative pOX38-Gen. All of the transposition events were collected in NLC51 pOX38-Gen pCW15 (TnsABC) pJP103 (TnsE). Wild-type or mutant TnsE were expressed in pJP103; TnsEWT (A), TnsEA453V (B), TnsED523N (C), TnsEA453V/ G515R (D), TnsEA453V/ D523N (E). Arrows indicate the position of insertion events; numbering is based on the central base pair of the 5-bp duplication found with Tn7 transposition. Numbering of the chromosome and the pOX38-Gen plasmid follow the previously established conventions (Frost et al. 1994; Blattner et al. 1997). The orientation of insertion events is indicated by placement of the arrow inside or outside the circle according to the key. (Triangles) The chromosomal ter sites (terA-terJ); replisomes approaching from the flat side of the triangle will be halted. The position of the terminators is indicated by the central base pair of the 23-bp consensus ter binding site (Coskun-Ari and Hill 1997). The location of the bidirectional origin of chromosomal DNA replication, oriC, is indicated.
Replication bias in TnsE-mediated transposition
Analysis of the insertion events promoted by the TnsE mutants reveals that TnsE-mediated transposition occurs with a distinct orientation bias with respect to DNA replication in the E. coli chromosome. Although Tn7 transposition events in the chromosome had been analyzed previously, the orientation bias with chromosomal DNA replication was not apparent (Peters and Craig 2000). Previous work with the TnsABC+E pathway used the wild-type TnsE protein, which preferentially directs transposition into the terminus region where DNA replication can occur from either direction (Peters and Craig 2000). Because transposition events found with the TnsE mutants occur more liberally throughout the chromosome, we can now realize a relationship between the direction of chromosomal DNA replication and the orientation of TnsABC+E transposition events.
Transposition events promoted by the TnsE mutants occurred in opposite orientations on either side of oriC, the bidirectional origin of chromosomal DNA replication (Fig. 3). The orientation bias that was found on either side of oriC continued around the circular E. coli chromosome until the terminus region. Interestingly, TnsABC+E transposition events were found in both orientations in the terminus region, where in theory DNA replication can occur via replisomes progressing from either orientation (Fig. 3). The results show clearly that although the mutant TnsE proteins seem to be less restricted in where insertions occur, many, if not all, of the insertion events orient in one direction with DNA replication. That transposition events mediated by the TnsE mutants still occur with an orientation bias indicates they continue to recognize the same structure as the wild-type protein.
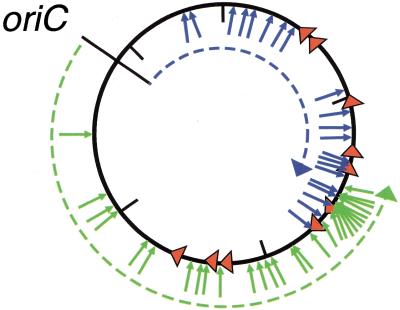
Figure 3.
The location and orientation of transposition events mediated by mutant TnsE and the direction of DNA replication in the E. coli chromosome. The 50 insertion events promoted by the TnsE mutants that mapped to the E. coli chromosome in Fig. 2B–E are compiled. The position and orientation of oriC, the ter sites, and the TnsABC+E-mediated transposition events are as described in Fig. 2. (Blue dashed line) Replication events initiated at oriC in a clockwise orientation that terminate at the first properly oriented ter site, terC; (green dashed line) replication events initiated at oriC in a counter-clockwise orientation that terminate at the first properly oriented ter site, terA.
It is important to note that the orientation of TnsABC+E transposition events in the chromosome is not determined by target site transcription or recognition of a DNA sequence that is distributed in a polar fashion, but instead stems from a more global polarity set by the direction of DNA replication. Of the 50 chromosomal insertion events shown in Figure 3, 46% (23/50) occurred in genes in which transcription occurred from right to left across the transposon, 38% (19/50) occurred in the opposite orientation, and 16% (8/50) fell between genes. Although there are DNA sequences that are distributed in a polar fashion in the chromosome, examination of a 200-bp window of DNA sequence surrounding the wild-type and mutant TnsE-derived insertion sites did not reveal a preferred target sequence (data not shown).
TnsE mutants: higher affinity for a limited target?
Recall that although wild-type TnsE directs transposition events preferentially into conjugable plasmids, transposition events promoted by the TnsE mutants were often found in the chromosome. However, we do not believe that the TnsE mutants have lost the ability to recognize the TnsEWT target found on conjugating DNA. Rather, our results indicate that the TnsE target found during conjugation is limited in the cell during the time window examined in our experiment and that the TnsE mutants are better able to recognize the same or similar target when it occurs in the chromosome.
Previous work has shown that the target structure recognized by TnsE must be limiting in the cell at low levels of TnsE, based on the observation that stimulating conjugation or DNA double-strand breaks stimulates TnsE-mediated transposition into the respective DNAs (Wolkow et al. 1996; Peters and Craig 2000). Consistent with this idea, we have also found that at high levels of wild-type TnsE, transposition is no longer stimulated by conjugation or by inducing DNA double-strand breaks (J.E. Peters and N.L. Craig, unpubl.). The results shown in Table 2 indicate there is a maximum observed frequency for transposition into the conjugable plasmid around 1 × 10−6 transposition events per Tn7 element in our experiments for wild-type or mutant TnsE (Table 2, last column). For example, with the single mutants in which the frequency of transposition is about 2 × 10−6, approximately half the insertions occur into the chromosome (Table 2; Fig. 2). When the transposition frequency is higher, as in the case of the mutant TnsEA453V/G515R (43 × 10−6), the frequency of transposition into the conjugable plasmid does not occur much more than 1 × 10−6 transposition events per Tn7 element. At the highest transposition frequency of 170 × 10−6, no insertions were found in the conjugable plasmid when 19 insertions were mapped (Table 2; Fig. 2). Thus, although the total frequency of transposition events increases with the mutants, the frequency of insertions into the conjugable plasmid remains relatively constant. We hypothesize that the TnsE mutants continue to recognize the preferred target found on conjugable plasmids, but that they can also recognize a related signal in the chromosome.
To further test the idea that a target structure recognized by wild-type and mutant TnsE and occurrs on conjugal plasmids can be limiting, we performed “mating-out” assays. The mating-out assay monitors the frequency of transposition only into a conjugable plasmid, not the chromosome. The frequency of transposition into a conjugable plasmid is determined by finding the number of transconjugants with Tn7; that is, recipient cells that have received the conjugable plasmid from a donor strain in which transposition can occur (see Materials and Methods). Consistent with there being a limited number of target structures occurring on conjugable plasmids, there appears to be a plateau in the frequency of transposition events into the conjugable plasmid (Table 3). Thus, whereas TnsABC+E transposition can be stimulated >100-fold by increasing the expression of TnsE or by using the TnsE mutants in the lambda hop assay (Table 2), the same TnsE constructs stimulate transposition only 8- to 10-fold in the mating-out assay (Table 3). These results indicate that the TnsE mutants continue to use a target found during conjugation like the wild-type protein, but must use a target found in the chromosome to promote very high transposition frequencies. The target found in the chromosome is likely to be very similar to the one recognized in conjugating DNA because wild-type and mutant TnsE both display the same orientation bias with DNA replication.
Table 3.
TnsABC+E transposition frequency quantified by the “mating-out” assay
| TnsEa
|
Expressionb
|
Frequencyc
|
|---|---|---|
| None | 0.16 | |
| Wild-type | Low | 25.7 (+/−5.1) |
| A453V | Low | 116.7 (+/−15.3) |
| A453V/G483R | Low | 256.7 (+/−15.3) |
| A453V/G515R | Low | 196.7 (+/−30.6) |
| Wild-type | High | 223.3 (+/−25.2) |
TnsABC were expressed in all strains from pCW15 (see Materials and Methods).
TnsE was expressed from pJP103 (low TnsE expression) or pJP104 (high TnsE expression).
Transposition frequency was determined using a “mating-out” assay (see Materials and Methods). Frequency represents the average from three experiments where the final value is multiplied by 106. The standard deviation from the mean is indicated in parentheses.
DNA binding of the TnsE protein in vitro
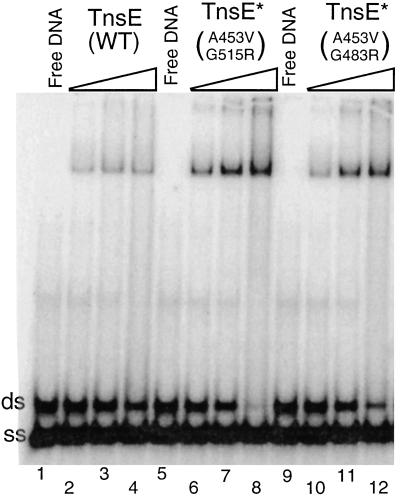
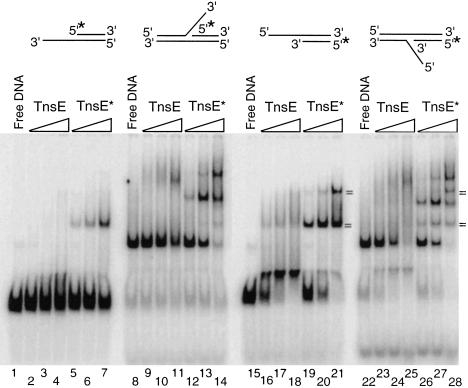
Given that the other Tn7 target-selecting protein, TnsD, is a DNA-binding protein, we explored the DNA-binding ability of TnsE. Wild-type and mutant TnsE derivatives with C-terminal histidine affinity tags were purified (see Materials and Methods). We analyzed the ability of TnsE to bind double- and single-stranded linear DNA by having both DNA species in the same binding reaction. Gel mobility-shift assays indicated that TnsE is able to bind double-stranded DNA (Fig. 4). The mutant TnsEA453V/G483R and TnsEA435V/G515R proteins that allow higher in vivo transposition frequencies also bind DNA better than the wild-type protein, indicating that the DNA-binding activity is likely relevant to transposition. TnsE appears to have a preference for double-stranded DNA over single-stranded DNA; this is most apparent with the mutant proteins (Fig. 4, lanes 8,12).
Figure 4.
Gel mobility-shift assays indicate TnsE does not interact preferentially with single-stranded DNA. Gel mobility-shift assay is shown comparing TnsEWT (lanes 2–4), TnsEA453V/G515R (lanes 6–8), and TnsEA453V/G483R (lanes 10–12). Increasing amounts of TnsE (0.06, 0.18, and 0.54 pmoles) were incubated with 0.05 pmoles of DNA substrate consisting of both single-stranded (ss) and double-stranded (ds) DNA for 30 min at 30°C and run on a 15% polyacrylamide gel (see Materials and Methods).
We also tested the ability of TnsE to bind various DNA structures produced from annealed oligonucleotides. We find that the TnsEWT protein interacts preferentially with structures containing 3′ recessed ends over those with 5′ recessed ends (Fig. 5, cf. lanes 2–4 and 16–18). Like the wild-type protein, the TnsEA453V/G515R mutant protein also interacts preferentially with the structure containing 3′ recessed ends (Fig. 5, cf. lanes 5–7 and 19–21). TnsEA453V/G515R forms predominantly slower migrating structures that are found only as a minority product with the wild-type protein (Fig. 5, cf. lanes 16–18 and 19–21). The binding preference of TnsE is especially intriguing given the known targeting preference of TnsE-mediated transposition for conjugating DNA in recipient cells where lagging-strand DNA synthesis is the exclusive mode of replication (Wolkow et al. 1996); during lagging-strand DNA synthesis, many 3′ recessed ends would be expected on Okazaki fragments.
Figure 5.
Certain DNA structures are preferred binding substrates for TnsE. Gel mobility-shift assay is shown comparing the ability of TnsEWT and TnsEA543V/G515R to bind various DNA structures. Each structure (0.08 pmoles) was incubated with no protein (Free DNA), or with 0.08, 0.24, or 0.72 pmoles of TnsEWT (TnsE) or TnsEA453V/G484R (TnsE*) for 30 min at 30°C and then run on a 6% polyacrylamide gel. The binding reactions contained a 50-fold excess of competitor DNA (see Materials and Methods). (=) Products found to comigrate when run in adjacent lanes; (*) the DNA strand that was 5′ end-labeled.
The mutant TnsEA453V/G515R protein interacts with replication fork-like structures and often gave multiple distinct altered migration products (Fig. 5 and data not shown). The wild-type TnsE protein also altered the mobility of DNA structures resembling a replication fork, but failed to give distinct bands, indicating the DNA–protein complexes are falling apart (Fig. 5, lanes 9–11,23–25). Two of the four altered mobility species found when TnsEA453V/G515R binds one of the fork-like structures may result from displacement of one DNA strand (Fig. 5, lane 28). When the fork structure used in Figure 5, lanes 22–28 was incubated with increasing concentrations of TnsEA453V/G515R, two of the altered mobility species comigrated with the species identified when the two-strand derivative was used in Figure 5, lanes 19–21.
Discussion
We report that the bacterial transposon Tn7 recognizes structures associated with DNA replication to identify target sites using the transposon-encoded DNA-binding protein TnsE. We find that TnsE-mediated transposition events in the chromosome occur with an orientation bias consistent with the direction of DNA replication. We isolated TnsE mutants that promote transposition levels that are much higher than levels found with the wild-type protein. In vivo, the TnsE mutants direct transposition events into the E. coli chromosome, without necessarily losing the ability to use conjugable plasmids as a preferred target. We have also found that TnsE has a DNA-binding activity that is likely involved in transposition, given that the high-activity TnsE mutants bind DNA better than the wild-type protein. TnsE's binding preference for certain oligonucleotide structures could help explain the target selection bias found with TnsE-mediated transposition events. These findings greatly extend our understanding of target site selection in the TnsE-mediated pathway of Tn7 transposition.
TnsE-mediated transposition and DNA replication
A striking observation found with TnsE-mediated insertion events is their orientation bias with DNA replication. The Tn7 orientation bias with chromosomal DNA replication was not apparent in previous work because TnsABC+E transposition events with wild-type TnsE occur in the terminus region, where replication can occur from either orientation (Peters and Craig 2000). DNA replication initiates bidirectionally at oriC in the E. coli chromosome and stops at one of a series of unidirectional terminators of DNA replication called ter sites (Hill 1996). Replisomes progressing in a clockwise direction will first encounter terC; counter-clockwise progressing replisomes will first encounter terA (Fig. 3). However, it is unclear how often individual ter sites are actually used in vivo. Because the ter sites stop replisomes approaching from only one direction, DNA replication between terA and terC could occur from either direction. Chromosomal transposition events mediated by wild-type TnsE occur preferentially in the region between terA and terC, where replication could occur from replisomes approaching from either direction (Peters and Craig 2000).
In the work presented here, we mapped a large number of chromosomal insertion events using TnsE mutants (Fig. 3). Because transposition events that occur with the TnsE mutants occur more liberally throughout the chromosome, we can now realize the relationship between the direction of DNA replication and the orientation of TnsABC+E transposition events. Of the 50 mutant TnsE-derived insertion events that were mapped to the chromosome, 37 occurred outside of the terA–terC terminators. Ninety-two percent (34/37) of the insertion events that occurred outside the terA–terC region were in a single orientation with respect to the direction of DNA replication initiated from oriC. It is possible that an even higher percentage of the transposition events orient with DNA replication; the ter sites are not totally effective at halting all replisomes, probably allowing some replication forks to progress outside the terA–terC region (Hill 1996). The region of the chromosome where transposition events are found in both orientations may experimentally define a region where replication can occur in either direction. Although the bidirectional initiation of DNA replication at oriC and the unidirectional termination of DNA replication at ter sites have been established previously (Kornberg and Baker 1992; Hill 1996), our studies address how replication proceeds throughout the bacterial chromosome. Differences in the rate of progression of replication forks in the two halves of the chromosome or differences in the activity of the ter sites at various positions in the chromosome could have resulted in any of a number of DNA replication profiles. The pattern of TnsE-mediated insertion events around the E. coli chromosome provides new insight into how DNA replication forks proceed in vivo.
Previous work has shown that in vivo the TnsABC+E proteins direct transposition into DNAs that are being actively transferred between cells in a process called conjugation (Wolkow et al. 1996). In previous work, it was unclear what aspect of conjugation specifically attracted Tn7 transposition events. Given that TnsE-mediated transposition events directed into conjugal plasmids occur almost exclusively in one orientation, an attractive explanation was that TnsE might recognize a feature of the unidirectional process of conjugation such as transfer or DNA replication (Wolkow et al. 1996). By analyzing the orientation of transposition events in the chromosome, we can now realize that DNA replication, rather than transfer or some other process, is the signal recognized in TnsE-mediated transposition. Interestingly, the orientation bias found in the chromosome matches the bias found with lagging-strand DNA synthesis identified in conjugating DNA (see below).
TnsE is a DNA-binding protein
We have also shown here that purified TnsE is a DNA-binding protein. Moreover, based on results from the mutant TnsE proteins, DNA binding is relevant to Tn7 transposition: Mutants that promote higher transposition frequencies bind DNA better. Our results show that TnsE interacts directly at target DNA molecules, like TnsD, the other Tn7 target site selecting protein (Bainton et al. 1993). Previously, it was unclear if TnsE acted as an allosteric effector of transposition by interacting only with the TnsABC core machinery to activate transposition or by interacting with DNA directly (Craig 1996). A role for a TnsE–DNA interaction does not rule out the possibility that host proteins are also involved in TnsE-mediated transposition; host proteins are known to be important in the TnsD-mediated transposition pathway (Sharpe and Craig 1998).
Because TnsE-mediated insertions do not occur randomly into all DNAs, TnsE must recognize a specific DNA structure or protein–DNA complex when choosing target sites. We examined the ability of TnsE to interact with various DNA structures (Figs. 4,5). TnsEWT has a very strong binding preference for structures with 3′ recessed ends, compared with structures containing 5′ recessed ends (Fig. 5). The mutant TnsEA453V/G515R protein also shows a marked preference for 3′ recessed ends, but forms predominantly supershifted species. Although supershifted species also occur with the TnsEWT protein, considerably more are found with the mutant TnsEA453V/G515R; forming the super-shifted complexes may be an important step in making an active target complex.
TnsE also binds DNA structures resembling replication forks in preference to linear duplex DNA (Fig. 5). The mutant TnsEA453V/G515R protein was able to interact with the replication fork-like derivatives and gave discrete altered mobility products (Fig. 5). Further research will be required to determine the stoichiometry and architecture of the slowed mobility products, although some of the products may stem from the displacement of DNA strands (Fig. 5). Wild-type TnsE also appears to interact preferentially with DNA three-way junction structures; however, the absence of a discrete slowed mobility product indicates this interaction is less stable than the interaction found with TnsEA453V/G515R (Fig. 5). Replication fork structures are likely always associated with multiple host proteins that could also be part of the signal recognized by TnsE in vivo.
Protein homology searches with the TnsE protein sequence are not informative in determining how TnsE operates. The only intriguing homolog of TnsE is found in a close Tn7 relative, Tn5468, identified in Thiobacillus ferrooxidans (Oppon et al. 1998). Like Tn7, Tn5468 resides in an attachment site downstream from the glmS gene in Thiobacillus ferrooxidans, and the tnsABCD genes occur in the same gene order as in Tn7 (Fig. 1). An open reading frame downstream from the Tn5468 tnsD gene, where a TnsE homolog would be expected, encodes a protein with limited homology with TnsE (Altschul et al. 1997).
The TnsE mutations TnsEA453V and TnsED523N are very important in allowing high-frequency TnsE-mediated transposition. The TnsEA453V and TnsED523N mutations were the only alleles isolated in our original screen. In addition, mutagenesis of the TnsEA453V and TnsED523N proteins allowed the isolation of higher-activity double mutants. The most active TnsE mutant was the result of the combination of the A453V and D523N alleles, where the increase was much greater than expected for an additive effect of these two alleles (Table 2). All of the secondary mutations in the C terminus of TnsE (G483R, G515R, E516K, and E522K) resulted from changes to basic amino acids that could favor an interaction with DNA in this region.
TnsE and lagging-strand DNA synthesis
It is tempting to speculate on how the ability of TnsE to interact preferentially with 3′ recessed ends could help explain TnsABC+E transposition (Fig. 5). TnsABC+E transposition must preferentially recognize something associated with only certain forms of DNA replication, given that the chromosome and both low- and high-copy nonconjugal plasmids are not preferred insertion targets (Wolkow et al. 1996). The preferred target for transposition mediated by wild-type TnsE is actively conjugating DNA in recipient cells in which lagging-strand DNA synthesis is the exclusive mode of DNA replication (Wolkow et al. 1996). The preference for conjugal replication processes in recipient cells could result from the abundance of 3′ recessed ends found during lagging-strand synthesis; multiple 3′ ends would reside in the plasmid DNA strand replicated by lagging-strand DNA synthesis because replication must be primed continually (Wilkins and Lanka 1993). Unlike conjugal DNA replication, leading-strand and lagging-strand DNA synthesis occur in concert during chromosomal DNA replication, which could eclipse structures on the lagging-strand from recognition by wild-type TnsE (Kornberg and Baker 1992). The TnsE mutants may promote higher transposition into the chromosome because the mutant proteins can recognize the 3′ recessed ends associated with lagging-strand DNA synthesis even in the context of a full replication fork. As expected from this hypothesis, mobility-shift assays indicate that the TnsE mutants have an increased ability to interact with DNA (Fig. 5). The same structure is likely recognized in both conjugal plasmids and the chromosome because the orientation is the same with respect to lagging-strand synthesis; the left and right ends of the transposon align in the same direction as the 5′ to 3′ orientation of Okazaki fragments.
Coordination of DNA metabolism in the bacterial chromosome
The link between Tn7 transposition and DNA replication joins a growing list of cell activities that are coordinated with DNA replication. The bacterial genome has evolved a polarity in its structure as a consequence of initiating bidirectional DNA replication from a single site on the circular chromosome. Features such as the orientation of operons and the orientation of the χ sites recognized by the recombination machinery are biased with the flow or replication from oriC to the terminus region (Blattner et al. 1997). Presumably, the orientation of operons with the direction of DNA replication allows the replisome to travel more efficiently. The orientation of χ sites in the chromosome likely directs replication toward the terminus following the use of DNA replication to repair lethal DNA double-strand breaks (for reviews, see Kuzminov 1995; Cox et al. 2000). The χ site is an 8-bp sequence recognized by the RecBCD recombination machinery when processing DNA double-strand breaks. The recombination machinery allows DNA replication to be templated off a sister chromosome to repair a DNA double-strand break and reactivate stalled replication forks. Given that the χ sequence is functional only in one orientation, the orientation bias found in the chromosome would direct DNA break-induced replication toward the terminus.
Why does TnsABC+E insertion also occur proximal to DNA double-strand breaks and in regions where DNA replication terminates? When DNA double-strand breaks are induced at different locations in the chromosome, TnsABC+E transposition is stimulated specifically and transposition events are found in the region around the break site (Peters and Craig 2000). An exciting possibility is that TnsE actually recognizes DNA replication structures involved in the repair of DNA double-strand breaks. Future dissection of the TnsABC+E transposition pathway of Tn7 will likely provide new information into how DNA replication is harnessed for the repair of lethal DNA double-strand breaks in bacteria. Multiple explanations could account for the propensity of TnsABC+E transposition events to occur in the terminus region. Given that double-strand breaks are associated with the termination of DNA replication, DNA breaks could explain why the terminus is a preferred region for TnsABC+E. transposition. Alternatively, a replication structure recognized by TnsE could become accessible when the progression of a replication fork is prohibited in the terminus region.
Why insert into replicating DNAs?
Other transposons also coordinate transposition with DNA replication of the host. However, in these cases transposons appear to move following replication of the transposon (Roberts et al. 1985; Chen et al. 1987, 1992). Tn10 transposition is stimulated following replication stemming from a methylation-sensitive transposase promoter and methylation-sensitive transposon ends (Roberts et al. 1985). The Ac element of maize transposes with replication but inserts into both replicated and unreplicated target sites (Chen et al. 1992). Tn7 is different than the previous examples in that transposition is not stimulated by replication of the transposon itself, but instead target DNAs become activated as transposition targets by DNA replication. Given that Tn7 preferentially recognizes DNA replication associated with plasmid transfer, the targeting preference could provide a mechanism for dissemination to new host organisms.
Materials and methods
Strains and plasmids
NLC28 is F− araD139Δ(argF−lac)U169 rspL150 relA1 flbB5301 deoC1 ptsF25 ValR, a valine-resistant derivative of MC4100 (McKown et al. 1987). CW51 is F− ara− lac−proXIII recA56 Nalidixic acid (NalR) Rifampicin-resistant (RifR) and was used as a recipient to select conjugable plasmids from NLC28 derivatives (Waddell and Craig 1988). NLC51 is a RecA− (recA56) derivative of NLC28 constructed as described previously (Peters and Craig 2000). A derivative of NLC28 unable to use arabinose was constructed by P1 transduction using a leu::Tn10 linked to the Δara714 allele, with strains kindly provided by Spencer Benson (University of Maryland, College Park; Miller 1992).
The plasmid pCW30 is a pUC18 derivative expressing TnsE, and pCW15 is a pACYC184 derivative expressing TnsABC (Waddell and Craig 1988). pTA106 is an ampicillin-resistant (AmpR) pSC101-based vector, kindly provided by Nancy Trun (Duquesne University, Pittsburgh, PA). pJP103 was constructed by cloning the Tn7 HindIII fragment encoding TnsE (Fig. 1A) into the HindIII site of pTA106 in an orientation in which it is not transcribed by the vector-encoded lac promoter. pJP104 is similar to pJP103, but the TnsE-encoding HindIII fragment is oriented such that it is transcribed by the vector-encoded lac promoter (Peters and Craig 2000). pCAW11 is a derivative of pET22b (Novagen), encoding TnsE with a C-terminal six histidine affinity tag that maintains the TnsE 5′ translation signals (Wolkow 1997). Mutant TnsE-encoding pCAW11 plasmids were constructed using PCR. A full-length 538-amino acid TnsE (pJP118), and an N-truncated TnsE derivative (pJP117) were constructed into the NcoI and XbaI sites of the arabinose-inducible expression vector pBAD24 using PCR (Fig. 1; Guzman et al. 1995). Western blots confirmed that the proteins were expressed in an arabinose induction system and that the derivatives had the expected difference in mobility (data not shown). In all cases we ensured that no Taq DNA polymerase mutations were introduced during PCR-based constructions by DNA sequencing. pOX38-Gen is an F plasmid derivative resistant to gentamycin (Johnson and Reznikoff 1984).
Mutant TnsE isolation
The TnsE-encoding plasmids pCW30 and pJP103 were mutagenized using 1 M hydroxylamine hydrochloride (in NaOH) for 24 h at 37°C (Rose et al. 1990). The hydroxylamine was removed by multiple ethanol precipitations, and the plasmid was transformed by electroporation into strain JP623 [NLC51 attTn7::miniTn7(Lac−) pCW15 (TnsABC)], in which transposition could be monitored visually using the papillation assay (DeBoy and Craig 1996). The papillation assay provides a visual screen for transposition levels in colonies of bacteria on indicator media (Huisman and Kleckner 1987; Stellwagen and Craig 1997). A miniTn7 element encoding the genes for lactose utilization, but which does not have the requisite lac promoter, miniTn7(Lac−), is located in a position and orientation in the chromosome where it is not transcribed, yielding a cell that is phenotypically Lac−. If the transposition proteins are provided in trans, the transposon can move, introducing the miniTn7(Lac−) element to positions behind actively transcribed promoters, allowing a cell that is now phenotypically Lac+. On the MacConkeys lactose indicators plates used in the experiment, red Lac+ papillae arise from the otherwise white colony of cells. The number of red Lac+ papillae on a colony provides a measure of transposition frequency. Following the preliminary screen, plasmids showing high papillation levels were retransformed and tested again in the papillation assay. Subcloning and DNA sequencing identified the actual coding change that conferred the mutant phenotype.
Western blots showed that similar levels of protein resulted when the TnsEWT, TnsEA453V, and TnsED523N alleles were expressed in the pUC18 backbone (pCW30) (data not shown). We could not visualize any of the TnsE proteins, wild-type or mutant, when expressed in the low-copy pJP103 vector using Western blots. We were unable to subclone the high-activity TnsE double mutants into high expression constructs that allow protein levels detectable by Western blot assays with our anti-TnsE antibody. Given that TnsE protein cannot be visualized in the low-expressing pJP103 constructs using Western blots, it remains formally possible that alterations in the stability of the TnsE double mutants may account for differences in the activity between the double mutants. However, it is clear that an increase in protein stability alone is not responsible for the increase in TnsE-mediated transposition based on comparisons with other higher TnsE expressing constructs; for example, pJP104 (data not shown).
Transposition assays
In vivo transposition levels were determined with the “lambda hop” or “mating-out” assay using miniTn7(KanR) elements comprised of the cis-acting Tn7 ends flanking a Kanamycin-resistance determinant. In the lambda hop assay, the miniTn7(KanR) resides on a defective lambda phage that cannot replicate or integrate in NLC51 (McKown et al. 1988). The phage is introduced into a strain where the Tns proteins are provided in trans. Transposition frequency was determined by scoring the number of kanamycin-resistant colonies per infectious phage particle as measured by plaque forming units.
The mating-out assay was performed as described previously (Waddell and Craig 1988). In the mating-out assay a miniTn7(KanR) resides on the chromosome of NLC51 pOX38-Gen with the Tn7 transposition proteins provided in trans. Transposition frequency is determined by finding the proportion of Gentamycin-resistant (GenR) pOX38-Gen plasmids that contain the miniTn7(KanR) element after selecting exconjugants in a NalR RifR recipient strain using the appropriate antibiotic selection (KanR NalR RifR/GenR NalR RifR).
Sequencing Tn7 insertion sites by PCR
The position of Tn7 insertions was determined by sequencing on an ABI automatic sequencer as described previously (Chun et al. 1997; Peters and Craig 2000). The DNA sequence flanking the transposon was determined using sequencing templates produced by PCR. The PCR templates were produced using primers specific to the transposon in conjunction with primers encoding random sequences to anneal to the region flanking the transposon. Insertions were localized using the E. coli MG1655 chromosome sequence, version M52 (http://www.genetics.wisc.edu/), or the pOX38-Gen plasmid using multiple sequence sources, accession numbers AF106329, ECPTRAX3, and U01159, assembled using regions of overlap. The entire pOX38-Gen plasmid sequence is not known; however, all insertions occurred in the available sequence.
TnsE protein isolation and gel mobility-shift assay
Wild-type and mutant TnsE protein were purified in parallel using a C-terminal six histidine affinity tag in the vector pET22b derivative pCAW11 according to the manufacturer's specifications. Protein concentrations were determined using BioRad assays. TnsE–DNA interactions were examined using mobility-shift assays. All gel mobility-shift assays were performed with 50-μL reaction volumes. DNA-binding substrates were constructed from oligonucleotide structures. One of the oligonucleotides was 5′ [γ-32P]ATP end-labeled before annealing. Mobility-shift gels were dried and subjected to phosphoimagery for analysis.
When analyzing the preference for single- verses double-stranded DNA, labeled oligonucleotide NLC95, 5′-ATA ATC CTT AAA AAC TCC ATT TCC ATT TCC ACC CCT-3′ was annealed with a limiting concentration (20%) of unlabeled NLC97, 5′-AGG GGT GGA AAT GGA GTT TTT AAG GAT TAT-3′ to give a mixture of DNA species. Target substrates (0.05 pmoles) were incubated in a 50-μL reaction of 25 mM HEPES (pH 7.6), 3.0 mM Tris-HCl (pH 7.6), 50 μg/mL BSA, 2 mM ATP, 4 mM DTT, 0.02 mM EDTA, 4 mM KCl, 0.2% Glycerol for 30 min at 30°C. Glycerol concentration was adjusted to 6% and loaded on a 15% polyacrylamide gel, 0.5× TBE at room temperature.
Larger oligonucleotide structures were constructed as described by Jones and Nakai (Jones and Nakai 1999) from HPLC-purified oligonucleotides where one was labeled, annealed to the appropriate oligos, and the structure was gel purified. Linear duplex structures were constructed from the 65-mer NLC707, 5′-CCA TTA GCA AGG CCG GAA ACG TCA CCA ATG CAA CGA TCA GCC AAC TAA ACT AGG ACA TCT TTG CC-3′ and the 65-mer NLC713, 5′-GGC AAA GAT GTC CTA GTT TAG TTG GCT GAT CGT TGC ATT GGT GAC GTT TCC GGC CTT GCT AAT GG-3′. The linear structures with 3′ and 5′ recessed ends were constructed by annealing the 65-mer NLC707 and the 30-mer NLC710, 5′-GGC AAA GAT GTC CTA GTT TAG TTG GCT GAT-3′ or the 60-mer NLC708, 5′-TAC TGA TTA CGG TGC TGC TAT CGA TGG TTT CAT TGG TGA CGT TTC CGG CCT TGC TAA TGG-3′ and 30-mer NLC709, 5′-AAA CCA TCG ATA GCA GCA CCG TAA TCA GTA. The replication fork-like structures were constructed by annealing NLC707, NLC708, NLC709, and NLC710. Greater than 90% of the structures remained intact after 60 min incubation in binding buffer (data not shown). In binding studies, 0.08 pmoles (4.4 ng in nucleotides) of structure and 220 ng of HincII-linearized pRM2 as a linear competitor (a 50-fold excess based on moles of base pairs) were incubated in a 50 μL reaction of 25 mM HEPES (pH 7.6), 3.0 mM Tris-HCl (pH 7.6), 50 μg/mL BSA, 2 mM ATP, 4 mM DTT, 0.02 mM EDTA, 4 mM KCl, 6.2% glycerol for 30 min at 30°C. Samples were run on a 6% polyacrylamide gel, 0.5× TBE at room temperature for analysis.
Acknowledgments
We are grateful to Spencer Benson, Nancy Trun, Jessica Jones, and Hiroshi Nakai for providing strains, plasmids, and advice. We thank Jeff Boeke and the members of the Craig laboratory for comments on the manuscript. This work was supported by NIH training grant 5T32CA09139 (J.E.P.), by the NSF under grant DBI-9750072 (J.E.P.), and by the NIH under grants GM53824 and PO1CA16519 to N.L.C. N.L.C. is an investigator in the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ncraig@jhmi.edu; FAX (410) 955-0831.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.870201.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton RJ, Kubo KM, Feng J-N, Craig NL. Tn7 transposition: Target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- Biery M, Lopata M, Craig NL. A minimal system for Tn7 transposition: The transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J Mol Biol. 2000;297:25–37. doi: 10.1006/jmbi.2000.3558. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Chen J, Greenblatt IM, Dellaporta SL. Transposition of Ac from the P locus of maize into unreplicated chromosomal. Genetics. 1987;117:109–116. doi: 10.1093/genetics/117.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Molecular analysis of Ac transposition and DNA replication. Genetics. 1992;130:665–676. doi: 10.1093/genetics/130.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun KT, Edenberg HJ, Kelley MR, Goebl MG. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast. 1997;13:233–400. doi: 10.1002/(SICI)1097-0061(19970315)13:3<233::AID-YEA88>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Coskun-Ari FF, Hill TM. Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J Biol Chem. 1997;272:26448–26456. doi: 10.1074/jbc.272.42.26448. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Craig NL. Transposon Tn7. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- ————— Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- DeBoy R, Craig NL. Tn7 transposition as a probe of cis interactions between widely separated (190 kilobases apart) DNA sites in the Escherichia coli chromosome. J Bacteriol. 1996;178:6184–6191. doi: 10.1128/jb.178.21.6184-6191.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C, Qadri MI, Lichtenstein C. DNA sequence analysis of five genes; tnsA, B, C, D and E, required for Tn7 transposition. Nucleic Acids Res. 1990;18:901–911. doi: 10.1093/nar/18.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Ippen-Ihler K, Skurray RA. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiolog Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and highlevel expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TM. Features of the chromosomal terminus region. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC.: ASM Press; 1996. pp. 1602–1614. [Google Scholar]
- Huisman O, Kleckner N. A new generalizable test for detection of mutations affecting Tn10 transposition. Genetics. 1987;116:185–189. doi: 10.1093/genetics/116.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Reznikoff WS. Copy number control of Tn5 transposition. Genetics. 1984;107:9–18. doi: 10.1093/genetics/107.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Nakai H. Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate. J Mol Biol. 1999;289:503–515. doi: 10.1006/jmbi.1999.2783. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA replication. New York: W.H. Freeman; 1992. [Google Scholar]
- Kubo KM, Craig NL. Bacterial transposon Tn7 utilizes two classes of target sites. J Bacteriol. 1990;172:2774–2778. doi: 10.1128/jb.172.5.2774-2778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- Lu F, Craig NL. Isolation and characterization of Tn7 transposase gain-of-function mutants: A model for transposase activation. EMBO J. 2000;19:3446–3457. doi: 10.1093/emboj/19.13.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May EW, Craig NL. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996;272:401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- McKown RL, Waddell CS, Arciszewska LA, Craig NL. Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. Proc Natl Acad Sci. 1987;84:7807–7811. doi: 10.1073/pnas.84.22.7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown RL, Orle KA, Chen T, Craig NL. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J Bacteriol. 1988;170:352–358. doi: 10.1128/jb.170.1.352-358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- Oppon JC, Sarnovsky RJ, Craig NL, Rawlings DE. A Tn7-like transposon is present in the glmUS region of the obligately chemoautolithotrophic bacterium Thiobacillus ferrooxidans. J Bacteriol. 1998;180:3007–3012. doi: 10.1128/jb.180.11.3007-3012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol Cell. 2000;6:573–582. doi: 10.1016/s1097-2765(00)00056-3. [DOI] [PubMed] [Google Scholar]
- Roberts D, Hoopes BC, McClure WR, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Rogers M, Ekaterinaki N, Nimmo E, Sherratt D. Analysis of Tn7 transposition. Mol Gen Genet. 1986;205:550–556. doi: 10.1007/BF00338097. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sarnovsky R, May EW, Craig NL. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- Sharpe P, Craig NL. Host proteins can stimulate Tn7 transposition: A novel role for the ribosomal protein L29 and the acyl carrier protein. EMBO J. 1998;17:5822–5831. doi: 10.1093/emboj/17.19.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A, Craig NL. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics. 1997;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Mobile DNA elements: Controlling transposition with ATP-dependent molecular switches. Trends Biochem Sci. 1998;23:486–490. doi: 10.1016/s0968-0004(98)01325-5. [DOI] [PubMed] [Google Scholar]
- Waddell CS, Craig NL. Tn7 transposition: Two transposition pathways directed by five Tn7-encoded genes. Genes & Dev. 1988;2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- Wilkins B, Lanka E. DNA processing and replication during plasmid transfer between gram-negative bacteria. In: Clewell DB, editor. Bacterial conjugation. New York: Plenum; 1993. pp. 105–136. [Google Scholar]
- Wolkow CA. Target site selection by the TnsABC+E pathway for Tn7 transposition in E. coli. Baltimore: Johns Hopkins University; 1997. [Google Scholar]
- Wolkow CA, DeBoy RT, Craig NL. Conjugating plasmids are preferred targets for Tn7. Genes & Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]