Abstract
Vesicular stomatitis virus (VSV) has been widely used to characterize cellular processes, viral resistance, and cytopathogenicity. Recently, VSV has also been used for oncolytic virotherapy due to its capacity to selectively lyse tumor cells. Mutants of the matrix (M) protein of VSV have generally been preferred to the wild-type virus for oncolysis because of their ability to induce type I interferon (IFN) despite causing weaker cytopathic effects. However, due to the large variability of tumor types, it is quite clear that various approaches and combinations of multiple oncolytic viruses will be needed to effectively treat most cancers. With this in mind, our work focused on characterizing the cytopathogenic profiles of four replicative envelope glycoprotein (G) VSV mutants. In contrast to the prototypic M mutant, VSV G mutants are as efficient as wild-type virus at inhibiting cellular transcription and host protein translation. Despite being highly cytopathic, the mutant G6R triggers type I interferon secretion as efficiently as the M mutant. Importantly, most VSV G mutants are more effective at killing B16 and MC57 tumor cells in vitro than the M mutant or wild-type virus through apoptosis induction. Taken together, our results demonstrate that VSV G mutants retain the high cytopathogenicity of wild-type VSV, with G6R inducing type I IFN secretion at levels similar to that of the M mutant. VSV G protein mutants could therefore prove to be highly valuable for the development of novel oncolytic virotherapy strategies that are both safe and efficient for the treatment of various types of cancer.
INTRODUCTION
Vesicular stomatitis virus (VSV) is an extensively studied virus for a large number of applications. A member of the Rhabdoviridae family, VSV is an enveloped virus containing an 11,161-kb single-strand RNA genome of negative polarity encoding five proteins: a nucleocapsid (N), a phosphoprotein (P), a matrix (M) protein, an envelope glycoprotein (G), and a polymerase (L) (30, 37, 47). VSV is not endemic to North America, and infection in humans is generally asymptomatic or may induce mild flu-like symptoms (29, 37).
VSV M protein is the smallest but the most abundant protein, with around 1,800 molecules per virion (32). Considered the key protein for assembly and budding, M is found mainly in the cytoplasm (80%) and sometimes is linked to the plasma membrane (10 to 20%) (51). The M protein can also be found in the nucleus (31), where it can inhibit transcription by interacting with the RNA polymerase II TFIID complex (1, 6, 13–14) and with NUP98, resulting in an inhibition of the nucleocytoplasmic transport of host mRNAs (20, 34). It also inhibits cellular translation (15) by modifying the initiation complex eIF4F through dephosphorylation of the initiation factors eIF4E and 4E-BP1 (10). Finally, it was also shown to participate in apoptosis induction (12, 23–25).
Trimeric VSV G is responsible for attachment to the cellular receptor and for fusion to the cell membrane. Binding to the still-controversial cellular receptor induces clathrin-mediated endocytosis (28, 44). As the pH drops in early endosomes, G changes conformation to allow fusion between the viral envelope and the endosomal membrane (36). The pH at which G is exposed during infection will determine three different structures: the prefusion state occurring at pH 7, the active hydrophobic state optimal at pH 6, which initiates fusion, and the postfusion state (35–36). Interactions between the G and M proteins have also been shown to increase budding efficiency (45). Apart from roles in fusion and particle assembly, G has recently been shown to also participate in cytotoxicity (22) and, under certain conditions, oncolysis (49).
VSV carrying mutant M proteins has been extensively studied (30). When mutated at methionine 51, the M protein fails to block host gene expression, thereby allowing the cell to secrete type I interferons (IFN) (16, 43). Consequently, VSV harboring an M protein containing the M51R mutation is not able to efficiently spread in normal tissue, whereas cancer cells, often deficient in their ability to mount an effective antiviral response due to deficiencies in the IFN pathway, are readily infected (4–5, 42). This confers on VSV its exquisite oncolytic properties (4–5, 11, 30, 41).
In the last decade, multiple viruses have been tested as oncolytic agents, with some of these already in clinical trials. One conclusion emerging from these studies is the need for developing a variety of different oncolytic agents if the successful treatment of a wide spectrum of cancer types is to be achieved. Moreover, the development of agents with diverse oncolytic characteristics could contribute to increasing our knowledge of the mechanisms involved in virus-mediated tumor regression, as well as providing new tools for the treatment of specific tumor types.
In light of this, we characterized the cytopathic profiles of four VSV G mutants. G5, G6, G5R, and G6R were previously shown to possess a wild-type M protein (11) while still being able to inhibit cellular protein translation (15) without persisting in infected cells (12). These properties render them attractive candidates for oncolytic virotherapy studies. We therefore investigated the cytopathic properties of these G mutants by analyzing their capacity to inhibit host cell transcription and protein synthesis. We also defined their ability to induce cell death in several tumor lines and their capacity to trigger type I IFN secretion in murine fibroblasts. Finally, we demonstrated that, as for other VSV variants, virus-induced cell death closely correlates with induction of apoptosis.
MATERIALS AND METHODS
Cells and viruses.
L929 mouse fibroblasts were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). B16 melanoma, 3LL carcinoma, and MC57 fibrosarcoma cells were obtained from A. Ochsenbein (Bern, Switzerland) and also were grown in DMEM supplemented with 10% FBS. The origin of VSV G mutants studied here was described previously (15). G5, G5R, G6, and G6R are referred to, respectively, as TP5, TP5R1, TP6, and TP6R1 in other studies (11, 15). G5 and G6 were isolated from nonmutagenized VSV HR strain stocks and thus share the same background. G5R and G6R are thermorevertants of G5 and G6, respectively (see Results for more detailed information). HR (here designated WT [wild type]) is a heat-resistant variant of the San Juan isolate of the Indiana serotype. The MM51R mutant, originally named T1026 (15), or AV1 in more recent publications (43), is derived from mutagenized HR stocks and harbors the M51R mutation in the M protein. All viruses were propagated and titrated on Vero cells.
Viral genome sequencing and analysis.
Vero cells were infected at a multiplicity of infection (MOI) of 10. Total RNA was isolated with the EZNA extraction kit (total RNA kit; Omega Bio-Tek, Norcross, GA) according to the manufacturer's instructions. A cDNA copy of the viral RNA was first obtained by reverse transcription using an oligonucleotide primer complementary to the very 3′ end of the genome. The cDNA copy was then subjected to PCR with 5 different pairs of primers, yielding 5 overlapping PCR products that covered the entire genome. Primers were derived from the VSV San Juan genome sequence (GI 9627229), and amplifications were done using Fidelitaq DNA polymerase (USB Corporation, Santa Clara, CA). After size verification and purification on agarose gels, amplicons were sequenced by Génome Québec (Montreal, Canada) with the Applied Biosystems 3730xl DNA analyzer using the Dye Terminator technique (Applied Biosystems, Streetsville, Ontario, Canada). Sequence alignments of G mutants with the HR parental strain, San Juan wild type, and MM51R mutant were performed using CLUSTALW2 (27). To identify mutations associated with the heat-resistant phenotype in our mutants, a modified in silico quantitative trait locus analysis was performed (21). A correlation between differences in phenotype (ΔP) and genotype (ΔG) was established. A matrix of phenotypic similarity was created for the mutants G5, G5R, G6, G6R, and MM51R, parental HR, and wild-type San Juan (values ranged from 0 to 1) and was defined as ΔP (phenotypic differences). For every amino acid position, a matrix of amino acid sequence differences between every mutant was created and defined as ΔG (genetic differences). A Pearson correlation coefficient was then calculated for every ΔG and ΔP value. This raw correlation coefficient (R), ranging from −1 to 1, indicates either a perfect or an inverse correlation between a mutation and the observed phenotype. To obtain a scaled value, the mean coefficient of correlation was subtracted from every coefficient of correlation, and this difference was divided by the standard deviation (SD) of all coefficients of correlation. The scaled R value obtained defines the number of SD above or below the mean R value.
Structural analysis.
Homology-based structural modeling of mutants was based on the crystal structure of VSV glycoprotein G (PDB identifier 2J6J) in its prefusion form (36), which shares between 97.5 and 98.5% sequence homology with mutants (7). Structures of generated models were regularized by steepest descent energy minimization using the GROMOS96 force field (39). Structural comparisons of proteins were performed using the SSM algorithm (26). Figures were prepared using the PyMOL molecular graphics system, version 1.3 (Schrödinger, LLC, Germany).
Viral replication.
L929 cells were infected at an MOI of 0.1 or 20 in DMEM containing 2% FBS. Forty-five minutes postinfection, medium was removed and cells were washed with phosphate-buffered saline (PBS), and fresh DMEM–2% FBS was added. At each time point, supernatants were harvested and titers were determined by plaque assay on Vero cells as described previously (11).
Analysis of viral mRNA synthesis.
L929 cells were infected at an MOI of 20 or mock infected. Forty-five minutes was allowed for adsorption of viral particles, and DMEM containing 2% FBS was added. Parallel samples were incubated in the presence of actinomycin D (5 μg/ml). At 2, 4, 6, and 8 h postinfection, cells were labeled with 10 μCi/ml of [3H]uridine ([5,6-3H]uridine; Perkin Elmer, Woodbridge, Ontario, Canada) for 30 min and harvested. Samples were then precipitated with 10% trichloroacetic acid on ice and washed twice with 5% trichloroacetic acid. Acid-precipitable radioactivity was measured by scintillation counting, subtracting counts per minute (cpm) of viral transcription (in the presence of actinomycin D) from cpm of total transcription. Results of host cell transcription are presented as percentages of mock-infected cells.
Analysis of viral protein production.
L929 cells were infected at an MOI of 20 or mock infected as described above. At 4, 8, and 12 h postinfection, cells were washed with PBS and pulsed for 20 min with 50 μCi/ml of 35S-labeled methionine-cysteine (Expres35S protein labeling mix; Perkin Elmer, Woodbridge, Canada) in Met-Cys-deficient medium (Invitrogen, Burlington, Ontario, Canada) supplemented with 2% FBS. Cells were then rinsed with PBS, harvested, and lysed in radioimmunoprecipitation assay (RIPA) buffer (0.15 M NaCl, 1% deoxycholic acid, 1% Triton, 10 mM Tris-HCl [pH 7.4], 0.1% SDS). Lysates were boiled, and 30 μg of labeled proteins were separated by migration on a 12% SDS-polyacrylamide gel. The gel was incubated for 15 min in 15% glycerol and 30 min in 5-times-gel-volume Enhancer solution (Perkin Elmer, Woodbridge, Canada), dried, transposed on Kodak film overnight at −80°C, and analyzed using the AlphaImager 3400 software program. For total protein synthesis determination, samples were precipitated with 10% trichloroacetic acid on ice and washed twice with 5% trichloroacetic acid. Acid-precipitable radioactivity was measured by scintillation counting.
Interferon induction.
Beta interferon (IFN-β) quantification was performed by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (Mouse IFN-Beta ELISA; PBL Biomedical Laboratories, New Brunswick, NJ) on supernatants from L929 cells infected with an MOI of 0.1 or 20 collected at 6, 12, 24, or 36 h postinfection as described above.
Oncolysis.
Mouse Lewis lung 3LL carcinoma, mouse B16 melanoma, mouse EL4 lymphoma, human Jurkat T cell lymphoma, human HepG2 hepatocarcinoma, mouse MC57 fibrosarcoma, and mouse L929 fibroblast cells were infected at an MOI of 20 or mock infected as described above. Cell viability was measured by mitochondrial activity determination at 6, 12, 24, 36, 48, and 60 h postinfection using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay (cell proliferation kit I MTT; Roche Diagnostics, Mississauga, Ontario, Canada). Briefly, in 96-well plates, 10 μl/well of MTT was added at each time point and cells were incubated for 1 h at 37°C in a 5%-CO2 atmosphere. Then, 100 μl/well of solubilization solution was added, and formazan production was detected after an overnight incubation at 37°C by spectrophotometric analysis at 570 nm.
Apoptosis induction.
L929 and B16 cells were infected at an MOI of 20 or mock infected as described above. At 6, 12, 18, and 24 h postinfection, CaspGLOW reagent (BioVision, Mountain View, CA) was added, and cells were then washed with PBS and harvested. After 5 min of centrifugation at 800 × g, cells were resuspended in 500 μl of the provided washing buffer. Cells were centrifuged again and fixed in washing buffer containing 2% paraformaldehyde. Finally, cells were kept at 4°C until they were analyzed by flow cytometry using a FACSCalibur instrument (BD Biosciences) and the FlowJo software program.
Statistical analysis.
An analysis of variance (ANOVA) with Tukey's test, using the SPSS software program, was used to determine the statistical significance of data wherever indicated, and P values of less than 0.05 were considered significant.
Nucleotide sequence accession numbers.
DNA sequences for the glycoproteins of mutants G5, G5R, G6, and G6R were deposited in GenBank under accession numbers HQ588113, HQ588114, HQ588115 and HQ588116, respectively. Those for the HR parental strain (GenBank accession no. HQ588112), San Juan wild type (GenBank accession no. M35219), and MM51R mutant (GenBank accession no. HQ593628) are also available.
RESULTS
Sequence analysis of VSV G mutants.
In order to characterize the potential of VSV G protein mutants as novel oncolytic agents, we took advantage of previously isolated mutants derived from the HR variant of the San Juan isolate of VSV (Indiana serotype) (15). Mutants G5, G5R, G6, and G6R were isolated, without prior mutagenesis by virtue of their small-plaque-size phenotype on interferon (IFN)-inducible cells but normal plaque size on Vero cells. Furthermore, G5 and G6 were shown to be thermosensitive, since their growth in a plaque assay on Vero cells was equal to that of HR at 34°C but slightly reduced at 37°C (15). The revertants G5R and G6R were isolated by plating G5 and G6 at 39 to 41°C on Vero cells (15). The MM51R (T1026) mutant was isolated in a different set of experiments from a mutagenized stock of HR (38). It is known to harbor an M51R amino acid replacement in the M protein (11), but the remainder of its genome had not been sequenced previously. We therefore set out to sequence the entire genome of all four G protein mutants and compared them to those of the parental HR virus and the MM51R mutant. Sequence analysis revealed that amino acid changes in these mutants were restricted to the G glycoprotein (Table 1 and data not shown). Even though the MM51R mutant shows the same D216G substitution in its G protein as mutants G5 and G6 (Table 1), previous studies using viral recombinants containing the mutant M protein on a wild-type background or M protein transfection showed that the M protein on its own confers the attenuated phenotype, suggesting that this mutation does not affect viral replication (2, 6, 13).
Table 1.
Amino acid sequences of the glycoproteins of G mutantsa
| VSV strain | Amino acid at position: |
|||||
|---|---|---|---|---|---|---|
| 100 | 216 | 226 | 238 | 415 | 471 | |
| WT | K | D | R | E | S | Y |
| G5 | R | G | Q | |||
| G5R | R | Q | ||||
| G6 | G | G | ||||
| G6R | G | |||||
| MM51R | G | H | A | H | ||
Amino acid differences between the glycoproteins of G mutants and that of the WT VSV. Each amino acid is numbered according to its position on the mature protein.
VSV G mutants replicate efficiently.
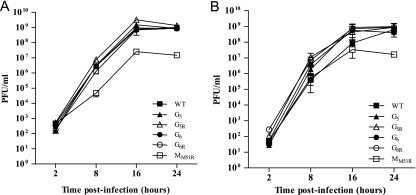
If VSV G mutants are to be considered for oncolytic virotherapy, they should first be able to replicate efficiently. VSV G mutants were therefore tested for their ability to replicate on L929 cells (Fig. 1). Despite mutations in the surface glycoprotein, all mutants replicated as efficiently as wild-type VSV regardless of the multiplicity of infection (MOI). We also tested replication of the M protein mutant MM51R and observed that it could also establish a productive infection, although this was reduced by 100-fold compared to that of G mutants.
Fig. 1.
Viral replication rates of VSV G mutants. L929 cells were infected at an MOI of 0.1 (A) or 20 (B). At the indicated time points, supernatants were harvested and viral titers were determined by plaque assay on Vero cells. Data are the means ± SEM for three independent experiments.
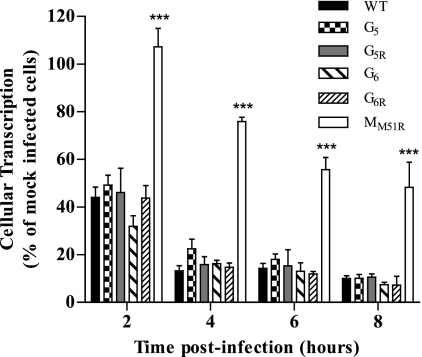
G mutants inhibit cellular transcription as efficiently as WT VSV.
Inhibition of cellular transcription by VSV is well documented. We next aimed at determining if mutations in the G protein influenced this capacity. Figure 2 shows that all G mutants efficiently inhibited cellular transcription as early as 2 h postinfection, with a reduction in RNA synthesis of more than 50% of that of mock-infected cells. By 8 h postinfection, cellular transcription was almost completely abolished. However, the matrix mutant MM51R failed to efficiently inhibit cellular transcription (P < 0.001 compared to results for all mutants), confirming results obtained previously (2).
Fig. 2.
Effect of VSV G mutants on cellular transcription rates. L929 cells were infected at an MOI of 20 or mock infected. At the indicated time points, cells were labeled with [3H]uridine (10 μCi/ml) and harvested. Acid-precipitable radioactivity was measured by scintillation counting. Data are the means ± SEM for three independent experiments in duplicate; ***, P < 0.001.
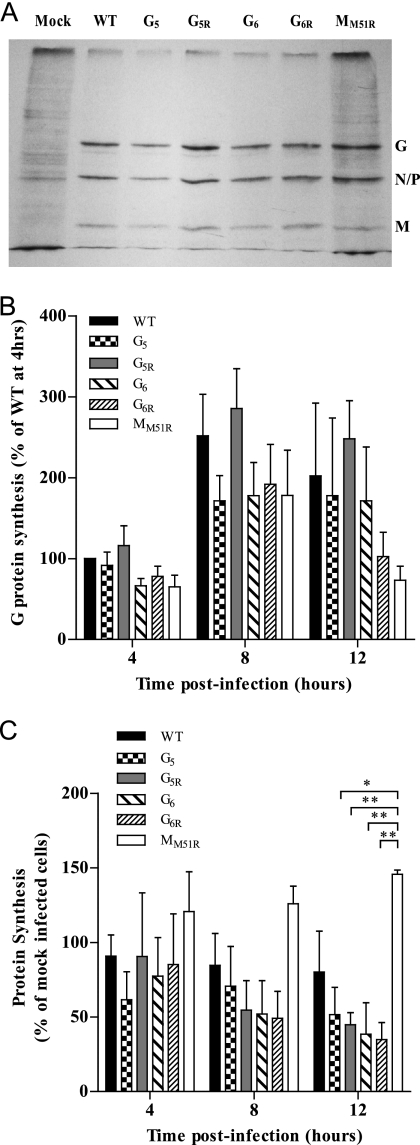
G mutants efficiently inhibit host protein synthesis.
Having showed that VSV glycoprotein mutants could rapidly inhibit cellular transcription upon infection, we next assessed if this would correlate with their ability to inhibit cellular translation. We first determined that all G mutants efficiently synthesized their viral proteins in a pattern similar to that of wild-type VSV (Fig. 3A and B). We then analyzed cellular protein synthesis during infection and observed an inhibition by all G mutants compared to results for wild-type VSV, although these differences did not reach statistical significance (Fig. 3C). In remarkable contrast, the MM51R mutant showed only transient G protein synthesis and an inability to shut off host protein synthesis compared to G mutants (P < 0.05 compared to results for G5; P < 0.01 compared to results for other G mutants).
Fig. 3.
Effect of VSV G mutants on cellular protein synthesis rates. L929 cells were infected at an MOI of 20 or mock infected. Cells were labeled with [35S]methionine/cysteine (50 μCi/ml) at 4, 8, and 12 h postinfection. Lysates were subjected to SDS-PAGE, and labeled proteins were quantified. (A) Representative image of an autoradiography film showing radiolabeled proteins at 8 h postinfection. Positions of viral proteins are indicated on the right. (B) The labeled G proteins in images similar to that of panel A were quantified using the AlphaImager 3400 software program. Results, corrected for the background level, are shown as a percentage of the WT-infected control at 4 h postinfection and are the means ± SEM for four independent experiments. (C) Acid-precipitable radioactivity was measured by scintillation counting for the determination of total protein synthesis. Results are shown as a percentage of those for the mock-infected control and are the means ± SEM for three independent experiments. *, P < 0.05; **, P < 0.01.
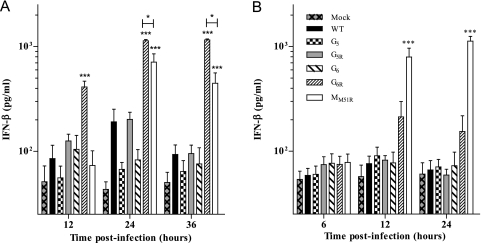
G6R induces type I interferon secretion in infected cells.
In order to develop oncolytic virotherapy as an effective and safe treatment for cancer, healthy cells should ideally be able to protect themselves against infection, while cancer cells should be sensitive to viral lysis. We therefore tested the capacity of G mutants to induce type I interferon secretion in L929 cells following infection at an MOI of 0.1 or 20 (Fig. 4). Mutants G5, G5R, and G6 did not induce significantly higher IFN secretion than wild-type VSV at either MOI. However, at an MOI of 0.1, mutant G6R showed rapid and strong IFN-β induction that was highly significant compared to that of WT or other G mutants (P < 0.001) and even superior to that of the MM51R mutant (P < 0.001 at 12 h; P < 0.05 at 24 h and 36 h). When an MOI of 20 was tested, G6R induced readily detectable levels of IFN-β; however, this did not reach statistical significance.
Fig. 4.
Interferon induction by VSV G mutants. L929 cells were infected at an MOI of 0.1 (A) or 20 (B). At the indicated time points, supernatants were collected and IFN-β quantification was performed by ELISA. Data are the means ± SEM for four independent experiments in duplicate. ***, P < 0.001.
G mutants induce death in several tumor cell lines.
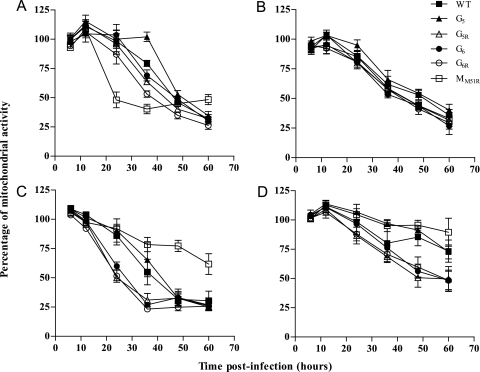
To investigate whether the strong inhibition of cellular transcription and translation observed for all G mutants would provide them with increased oncolytic properties, we assessed their abilities to kill various murine and human tumor cell lines in vitro (Fig. 5). Their capacity to induce death of murine 3LL pulmonary carcinoma (Fig. 5B), murine EL4 and human Jurkat lymphomas, or human HepG2 hepatocellular carcinoma (data not shown) was comparable to that of WT VSV or the M mutant. Importantly, however, all G mutants were more efficient than the M mutant at killing murine B16 melanoma cells (P < 0.001 at 48 h; P < 0.01 at 60 h), while G5R, G6, and G6R also showed a significantly faster induction of B16 cell death than wild-type VSV at 24 h (P < 0.01 for G5R and G6R; P < 0.05 for G6) (Fig. 5C). In addition, G5R and G6 showed increased killing of MC57 fibrosarcoma cells at 48 h postinfection compared to results for WT VSV (P = 0.01 for G5R; P < 0.05 for G6), while G5R, G6, and G6R also induced a significantly higher level of death than the M mutant (P < 0.01) (Fig. 5D). As a control, we analyzed cell death in L929 fibroblasts and showed that G mutants induced significantly slower death than the M mutant in these cells (P < 0.01 for G5, G5R, and G6; P < 0.05 for G6R at 24 h; P < 0.001 for G5, G5R, and G6 at 36 h) (Fig. 5A). Finally, G5R and G6R also induced increased death of L929 cells compared to WT results (P < 0.0001 for G6R; P < 0.05 for G5R). We confirmed these results using a trypan blue exclusion assay that showed the same trend for L929 and B16 cell lines (data not shown).
Fig. 5.
Cell viability modulation due to infection by VSV G mutants. L929 (A), 3LL (B), B16 (C), or MC57 (D) cells were infected at an MOI of 20. Cell viability was measured by mitochondrial activity using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. At the indicated time points, formazan production was detected by spectrophotometric analysis at 570 nm. Data are the means ± SEM for six replicates from three independent experiments.
Oncolysis correlates with apoptosis induction in cells infected with G mutants.
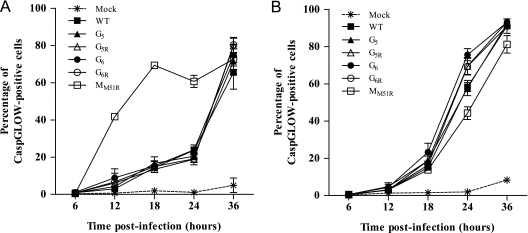
Finally, to determine if the observed cell death was due to increased apoptosis, caspase activation was analyzed by flow cytometry using a fluorescent irreversible inhibitor of caspase that binds to activated caspases (CaspGLOW assay). Infection of L929 cells with VSV induced similar increases in apoptosis over time for all G mutants and for WT VSV. In contrast, the MM51R mutant showed a dramatic increase of activated caspases as early as 12 h postinfection (P < 0.001) (Fig. 6A), correlating with the viability assay (Fig. 5). We also examined apoptosis induction in the B16 melanoma cell line (Fig. 6B) and demonstrated that contrary to what we observed with L929 cells, some G mutants induce significantly more programmed cell death than WT VSV or the MM51R mutant (for G6 compared to WT results, P < 0.05; for G5R, G6, and G6R compared to MM51R results, P < 0.01), also correlating with the viability assay (Fig. 5). We did not observe significant necrosis in any cell line tested or virus used (data not shown).
Fig. 6.
Caspase activation by VSV G mutants. L929 (A) or B16 (B) cells were infected at an MOI of 20 or mock infected. At the indicated time points, kinetics of caspase activity was determined by labeling the cells with the CaspGLOW reagent. Data are the means ± SEM for three independent experiments.
DISCUSSION
VSV is widely studied in the context of a growing interest for using viruses for vaccination (8–9, 18, 40) and the treatment of various cancers (4–5, 30). Glycoprotein G of VSV is responsible for viral fusion and entry into target cells. Both of these processes are critical for vaccination and oncolysis (48) and likely are involved in cytotoxicity and apoptosis induction (25). To develop such applications for an eventual use in humans, it is paramount to further study the multiple characteristics of VSV G and the various potential effects of G mutants on infected cells. Here we have presented sequence analyses of four new VSV G protein mutants and have characterized their cytopathic properties.
Mutations responsible for the phenotypes of mutants G5, G5R, G6, and G6R were found to map to the G glycoprotein of VSV, since comparison with the parental HR isolate revealed no amino acid change in any other viral genes (Table 1). To determine whether these mutations could be linked to known phenotypic differences between mutants, a modified quantitative trait locus analysis was conducted and revealed that mutation at position 216 best correlated with thermosensitivity (see Fig. S1A in the supplemental material). Comparison of structural models and predicted total energy between mutants and the HR parental virus revealed that the acquisition of the G216D mutation by G5R and G6R resulted in a lower total energy for G6R, a structure closer to that of the HR parental strain as well (see Fig. S1B). The presence of an aspartic acid at position 216 in each thermoresistant variant (HR, G5R, and G6R) is predicted to form an ionic bond (see Fig. S1C) that could have a stabilizing effect on the protein structure and could explain thermoresistance. It is tempting to speculate that such differences in protein stability and/or structure could also possibly explain other phenotypic differences between G mutants and their revertants, such as their capacity to induce interferon secretion; however, a detailed mutagenesis analysis will be required to directly assess this possibility. Interestingly, the E238Q or E238G mutation found in G mutants is proximal to the dominant neutralizing epitope of VSV, which could potentially modulate the kinetics of neutralizing antibody induction, leading to differences in the amount of virus reaching and infecting tumor cells (46). This aspect is beyond the scope of our in vitro studies but will eventually need to be tested in vivo. The similar replication rates observed between G mutants and wild-type VSV suggest that these mutations in G do not interfere with receptor binding or with other functions involved in viral replication (Fig. 1). Furthermore, cytopathic effects induced by G mutants in terms of transcription (Fig. 2) and translation (Fig. 3) inhibition were as strong as those induced by WT VSV. This is in agreement with previous results associating these properties with the M protein, unchanged in the G mutants studied here (1–2). Specific interactions between G and M proteins on the viral particle have been observed (17) and have been shown to be required for final assembly of the viral particle at budding sites (45) and for the release of nucleocapsids following endocytosis (33). Again, results showing normal replication rates of VSV G mutants argue against a modification of these interactions, although this was not tested directly.
Of central importance for the use of VSV for oncolytic virotherapy, and somewhat surprisingly in view of the wild-type M protein, the G6R mutant induced significant type I interferon secretion in L929 infected cells that was similar to or even higher than that with the MM51R mutant. This is particularly true at low MOI. This suggests that a different mechanism, not involving the M protein, is responsible for type I interferon secretion in infected cells between these two mutants. Such efficient IFN production will prime an antiviral state in tumor-surrounding healthy cells and should maintain the selectiveness of G6R for cancer cells deficient in the antiviral IFN pathway, presumably allowing the safe use of G6R in oncolytic virotherapy experiments. A further indirect indication for the safety of these G mutants was provided by our inability to detect any signs of toxicity following the inoculation of up to 1.5 × 109 PFU of each G mutant into C57BL/6 mice (data not shown). However, the complete innocuity of G mutants in comparison to WT VSV or the M mutant remains to be formally demonstrated in further in vivo studies. In addition, how G6R maintains the ability to inhibit cellular transcription and translation while allowing IFN production will also need to be determined. Previous studies have correlated IFN production and, more widely, primary immune responses following VSV infection with MyD88 stimulation (50) through either TLR4 (19) or TLR7 (3), depending on the cell type and viral strain considered. Mutations in G6R could therefore potentially modulate the triggering of such innate pattern recognition receptors, thus modifying the progression of the infection. This aspect is presently under study to explain differences observed between the cytopathogenicities of VSV G and M mutants.
Finally, for oncolytic virotherapy to be effective, cytotoxicity eventually needs to trigger cell death. We established that VSV G mutants induce cell death more efficiently in certain tumor cell lines than wild-type VSV or the M mutant (Fig. 6). We also determined that this killing efficiency differed dramatically between viral strains and tumor lines tested as previously shown in other models (24). We showed that these differences correlated with a potent induction of apoptosis by VSV mutants, as was previously observed with the TP6 (G6) mutant by Desforges et al. on neuroglioma cells (12). These results emphasize the need to develop a wide range of oncolytic viruses that could be customized for tumor types or origins in specific treatment protocols.
In conclusion, VSV G mutants represent promising oncolytic viruses due to their efficient replication and strong cytopathogenicity for various cancer cell lines. With the capacity to induce IFN in normal cells, conferring selectivity for cancer cells, and mutations proximal to the major VSV neutralizing epitopes, some of these mutants could be useful for further increasing our knowledge of the mechanisms involved in oncolysis. Further studies are under way to examine the in vivo oncolytic properties of these VSV G mutants using various murine tumor models. This will enable us to determine the full potential of VSV G mutants for oncolytic virotherapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Jeanne and J.-Louis Lévesque Chair in Immunovirology to A.L. from the J.-Louis Lévesque Foundation and by a grant from the Natural Sciences and Engineering Research Council of Canada to L.P. P.L. holds a Canadian Institutes of Health Research (CIHR)/Canadian Association for the Study of the Liver (CASL) hepatology fellowship.
We thank Omid Zahedi Niaki and Tanya Girard for their helpful comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Ahmed M., Lyles D. S. 1998. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J. Virol. 72:8413–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed M., et al. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 77:4646–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmed M., et al. 2009. Vesicular stomatitis virus M protein mutant stimulates maturation of Toll-like receptor 7 (TLR7)-positive dendritic cells through TLR-dependent and -independent mechanisms. J. Virol. 83:2962–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balachandran S., Barber G. N. 2004. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell 5:51–65 [DOI] [PubMed] [Google Scholar]
- 5. Balachandran S., Barber G. N. 2000. Vesicular stomatitis virus (VSV) therapy of tumors. IUBMB Life 50:135–138 [DOI] [PubMed] [Google Scholar]
- 6. Black B. L., Rhodes R. B., McKenzie M., Lyles D. S. 1993. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J. Virol. 67:4814–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bordoli L., et al. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4:1–13 [DOI] [PubMed] [Google Scholar]
- 8. Braxton C. L., Puckett S. H., Mizel S. B., Lyles D. S. 2010. Protection against lethal vaccinia virus challenge by using an attenuated matrix protein mutant vesicular stomatitis virus vaccine vector expressing poxvirus antigens. J. Virol. 84:3552–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cobleigh M. A., Buonocore L., Uprichard S. L., Rose J. K., Robek M. D. 2010. A vesicular stomatitis virus-based hepatitis B virus vaccine vector provides protection against challenge in a single dose. J. Virol. 84:7513–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Connor J. H., Lyles D. S. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 76:10177–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desforges M., et al. 2001. Different host-cell shutoff strategies related to the matrix protein lead to persistance of vesicular stomatitis virus mutants on fibroblast cells. Virus Res. 76:87–102 [DOI] [PubMed] [Google Scholar]
- 12. Desforges M., et al. 2002. Matrix protein mutations contribute to inefficient induction of apoptosis leading to persistent infection of human neural cells by vesicular stomatitis virus. Virology 295:63–73 [DOI] [PubMed] [Google Scholar]
- 13. Ferran M., Lucas-Lenard J. 1997. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J. Virol. 71:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flood E. A., McKenzie M. O., Lyles D. S. 2000. Role of M protein aggregation in defective assembly of temperature-sensitive M protein mutants of vesicular stomatitis virus. Virology 278:520–533 [DOI] [PubMed] [Google Scholar]
- 15. Francoeur M. A., Poliquin L., Stanners C. P. 1987. The isolation of interferon-inducing mutants of vesicular stomatitis virus with altered viral P function for the inhibition of total protein synthesis. Virology 160:236–245 [DOI] [PubMed] [Google Scholar]
- 16. Garcia-Sastre A., Biron C. A. 2006. Type 1 Interferons and the virus-host relationship: a lesson in detente. Science 312:879–882 [DOI] [PubMed] [Google Scholar]
- 17. Ge P., et al. 2010. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327:689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geisbert T. W., Bausch D. G., Feldmann H. 2010. Prospects for immunisation against Marburg and Ebola viruses. Rev. Med. Virol. 20:344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Georgel P., et al. 2007. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology 362:304–313 [DOI] [PubMed] [Google Scholar]
- 20. Glodowski D. R., Petersen J. M., Dahlberg J. E. 2002. Complex nuclear localization signals in the matrix protein of vesicular stomatitis virus. J. Biol. Chem. 277:46864–46870 [DOI] [PubMed] [Google Scholar]
- 21. Grupe A., et al. 2001. In silico mapping of complex disease-related traits in mice. Science 292:1915–1918 [DOI] [PubMed] [Google Scholar]
- 22. Hoffmann M., et al. 2010. Fusion-active glycoprotein G mediates the cytotoxicity of vesicular stomatitis virus M mutants lacking host shut-off activity. J. Gen. Virol. 91:2782–2793 [DOI] [PubMed] [Google Scholar]
- 23. Kopecky S. A., Lyles D. S. 2003. The cell-rounding activity of the vesicular stomatitis virus matrix protein is due to the induction of cell death. J. Virol. 77:5524–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kopecky S. A., Lyles D. S. 2003. Contrasting effects of matrix protein on apoptosis in HeLa and BHK cells infected with vesicular stomatitis virus are due to inhibition of host gene expression. J. Virol. 77:4658–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kopecky S. A., WIillingham M. C., Lyles D. S. 2001. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J. Virol. 75:12169–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krissinel E., Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60:2256–2268 [DOI] [PubMed] [Google Scholar]
- 27. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 28. Le Blanc I., et al. 2005. Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 7:653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Letchworth G. J., Rodriguez L. L., Del Cbarrera J. 1999. Vesicular stomatitis. Vet. J. 157:239–260 [DOI] [PubMed] [Google Scholar]
- 30. Lichty B. D., Power A. T., Stojdl D. F., Bell J. C. 2004. Vesicular stomatitis virus: re-inventing the bullet. Trends Mol. Med. 10:211–217 [DOI] [PubMed] [Google Scholar]
- 31. Lyles D. S., Puddington L., McCreedy B. J. J. 1988. Vesicular stomatitis virus M protein in the nuclei of infected cells. J. Virol. 62:4387–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCreedy B. J. J., Lyles D. S. 1989. Distribution of M protein and nucleocapsid protein of vesicular stomatitis virus in infected cell plasma membranes. Virus Res. 14:189–205 [DOI] [PubMed] [Google Scholar]
- 33. Mire C. E., Dube D., Delos S. E., White J. M., Whitt M. A. 2009. Glycoprotein-dependent acidification of vesicular stomatitis virus enhances release of matrix protein. J. Virol. 83:12139–12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petersen J. M., Her L. S., Varvel V., Lund E., Dahlberg J. E. 2000. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Cell. Biol. 20:8590–8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roche S., Bressanelli S., Rey F. A., Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187–191 [DOI] [PubMed] [Google Scholar]
- 36. Roche S., Rey F. A., Gaudin Y., Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848 [DOI] [PubMed] [Google Scholar]
- 37. Rose J. K., Whitt M. A. 2001. Rhabdoviridae: the viruses and their replication. In Knipe D. M., Howley P. M. (ed.), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 38. Schincariol A. L., Howatson A. F. 1970. Replication of vesicular stomatitis virus. I. Viral specific RNA and nucleoprotein in infected L cells. Virology 42:732–743 [DOI] [PubMed] [Google Scholar]
- 39. Schuler L., Daura X., van Gunsteren W. 2001. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J. Comput. Chem. 22:1205–1218 [Google Scholar]
- 40. Schwartz J. A., et al. 2010. Potent vesicular stomatitis virus-based avian influenza vaccines provide long-term sterilizing immunity against heterologous challenge. J. Virol. 84:4611–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stark G. R., Kerr I. M., Williams B. R. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264 [DOI] [PubMed] [Google Scholar]
- 42. Stojdl D. F., et al. 2000. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 6:821–825 [DOI] [PubMed] [Google Scholar]
- 43. Stojdl D. F., et al. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275 [DOI] [PubMed] [Google Scholar]
- 44. Sun X., Yau V. K., Briggs B. J., Whittaker G. R. 2005. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338:53–60 [DOI] [PubMed] [Google Scholar]
- 45. Swinteck B. D., Lyles D. S. 2008. Plasma membrane microdomains containing vesicular stomatitis virus M protein are separate from microdomains containing G protein and nucleocapsids. J. Virol. 82:5536–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vandepol S. B., Lefrançois L., Holland J. J. 1986. Sequences of the major antibody binding epitopes of the Indiana serotype of vesicular stomatitis virus. Virol. J. 148:312–325 [DOI] [PubMed] [Google Scholar]
- 47. Wagner E. K., Hewlett M. J. 2004. Basic virology, 2nd ed. Blackwell Science, Ames, IA [Google Scholar]
- 48. Whitt M. A. 2010. Generation of VSV pseudotypes using recombinant δG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol. Methods 169:365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wollmann G., Rogulin V., Simon I., Rose J. K., van den Pol A. N. 2010. Some attenuated variants of vesicular stomatitis virus show enhanced oncolytic activity against human glioblastoma cells relative to normal brain cells. J. Virol. 84:1563–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wongthida P., et al. 2011. VSV oncolytic virotherapy in the B16 model depends upon intact MyD88 signaling. Mol. Ther. 19:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ye Z., et al. 1994. Membrane-binding domains and cytopathogenesis of the matrix protein of vesicular stomatitis virus. J. Virol. 68:7386–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.