Abstract
Analysis of a large number of HIV-1 genomes at multiple time points after antiretroviral treatment (ART) interruption allows determination of the evolution of drug-resistant viruses and viral fitness in vivo in the absence of drug selection pressure. Using a parallel allele-specific sequencing (PASS) assay, potential primary drug-resistant mutations in five individual patients were studied by analyzing over 18,000 viral genomes. A three-phase evolution of drug-resistant viruses was observed after termination of ART. In the first phase, viruses carrying various combinations of multiple-drug-resistant (MDR) mutations predominated with each mutation persisting in relatively stable proportions while the overall number of resistant viruses gradually increased. In the second phase, viruses with linked MDR mutations rapidly became undetectable and single-drug-resistant (SDR) viruses emerged as minority populations while wild-type viruses quickly predominated. In the third phase, low-frequency SDR viruses remained detectable as long as 59 weeks after treatment interruption. Mathematical modeling showed that the loss in relative fitness increased with the number of mutations in each viral genome and that viruses with MDR mutations had lower fitness than viruses with SDR mutations. No single viral genome had seven or more drug resistance mutations, suggesting that such severely mutated viruses were too unfit to be detected or that the resistance gain offered by the seventh mutation did not outweigh its contribution to the overall fitness loss of the virus. These data provide a more comprehensive understanding of evolution and fitness of drug-resistant viruses in vivo and may lead to improved treatment strategies for ART-experienced patients.
INTRODUCTION
Despite significant advances in antiretroviral therapy (ART), some HIV-infected patients still experience treatment failure due to drug resistance, poor adherence, drug toxicity, and suboptimal drug metabolism (4, 35). Among these causes, the emergence of drug resistance mutations plays a central role in ART failure (14, 35). In addition, the presence of preexisting drug-resistant viruses correlates with poor responses to ART (18, 28, 36–39). Multiple-drug-resistant (MDR) mutations have been shown to exist in various complex linkage patterns in viral genomes (3, 26, 34), and these MDR viruses can play an important role in ART failure. After discontinuation of ART in patients who have developed drug resistance, drug resistance mutations disappear from the viral population within weeks and are replaced with wild-type (WT) viruses (7–9, 20, 40). However, a sensitive allele-specific PCR (ASPCR) method detects minority drug-resistant viruses (0.01% to 20%) in most patients months or even years following ART discontinuation (12, 33). The evolution of both majority and minority linked MDR virus populations after treatment interruption has been characterized in only a limited number of viral genomes in HIV-1-infected patients.
The fitness of drug-resistant viruses has been extensively studied in vivo and in vitro (6, 10, 11, 13, 15, 23–25, 27). A more comprehensive understanding of the viral fitness of various drug-resistant viruses could lead to improved treatment strategies and novel pathways in antiretroviral drug discovery. Previous studies have examined a limited set of mutations and a small number of viral genomes in HIV-1-infected individuals. A more thorough evaluation of drug resistance mutations in the absence of drug selection pressure can be accomplished by analyzing large viral populations in patients who discontinued failing ART. This can provide a unique opportunity to investigate the evolution of viral fitness and reservoirs of drug-resistant viruses, especially those with linked MDR mutations.
We recently developed a highly sensitive parallel allele-specific sequencing (PASS) assay that can detect minority drug resistance populations at frequencies as low as 0.01% to 0.1% of the viral population by simultaneously analyzing thousands of viral genomes in a single assay (3). In addition, this new technology allows identification of linkages among MDR mutations on individual viral genomes. In this study, we characterized the evolution of MDR mutations and their linkage relationships among a large viral population in longitudinal samples collected from individual patients following treatment interruption. These data were used to model the evolution and fitness of various drug-resistant viruses and to estimate the time to emergence of WT viruses following treatment interruption.
MATERIALS AND METHODS
Patient plasma samples.
The study population consisted of HIV-1-infected patients followed at the Duke University Medical Center HIV/AIDS Clinic between 2001 and 2003. Residual plasma samples remaining after clinical viral load testing were stored at −80°C. Patients selected for this study were identified from the HIV Patients Sample Repository database after reviewing characteristics of their treatment history. For inclusion in the study, patients had to have been followed in the Duke HIV/AIDS Clinic for at least 11 weeks, during which time they underwent ART interruption and thus had both on-treatment and off-treatment samples available at different time points to be studied. Based on patient responses to previous ART regimens and/or results from population-based genotypic drug resistance testing, samples were selected from patients experiencing treatment failure with viral load rebound. Patients were excluded if fewer than two blood samples after treatment interruption were available for analysis. Written informed consent was obtained from all individuals whose blood samples were collected. The study was approved by the Duke University Institutional Review Board.
vRNA extraction and cDNA synthesis.
One milliliter of each sample was concentrated by ultracentrifugation at 32,000 rpm for 3 h at 4°C. The virus pellet was resuspended with 400 μl of phosphate-buffered saline (PBS), and the viral RNA (vRNA) was extracted using the PureLink viral RNA/DNA minikit (Invitrogen, Carlsbad, CA). The vRNA was eluted into 17 μl of the elution buffer and used for cDNA synthesis using Superscript III (Invitrogen, Carlsbad, CA) and primer RTuni1 (5′-CCAATCCCCCCTTTTCTTTTAAAATTGTG-3′) in a 40-μl reaction mixture. An appropriate amount of cDNA was used for the PASS assay to obtain an optimal number of viral genomes in each assay.
Detection of drug resistance mutations by PASS.
The PASS assay was performed as previously described (3). Briefly, 20 μl of 6% acrylamide gel mix, containing 1 μM acrydite-modified reverse primer, 5′Acr-AATCCCTGCATAAATCTGACTTGCCCAAT-3′, cDNA template (5 μl to 18.5 μl), 0.3% diallyltartramide, 5% Rhinohide, 0.1% ammonium persulfate (APS), 0.1% TEMED (N,N,N′,N′-tetramethylethylenediamine), and 0.2% bovine serum albumin (BSA), was used to cast a gel on a bind-saline (Amersham Biosciences, Piscataway, NJ)-treated glass slide. The in-gel PCR amplification was then performed in a PTC-200 Thermal Cycler with a mix of 1 μM forward primer, 5′-TTAGCTTCCCTCAGATCACTCTTTGGCA-3′; 0.1% Tween 20; 0.2% BSA; 1× PCR buffer; 250 μM deoxynucleoside triphosphate (dNTP) mix, 3.3 units of Jumpstart Taq DNA polymerase (Sigma, St. Louis, MO), and H2O (up to 200 μl) under a sealed SecurSeal chamber (Grace Bio-Labs, Inc., Bend, OR). The PCR conditions were 94°C for 3 min; 65 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 3 min; and 72°C for 6 min.
After PCR amplification, single-base extension (SBE) was performed with mutant and wild-type (WT) bases distinctively labeled with Cy3 and Cy5, respectively, using the primers that annealed just upstream of the mutation sites of interest. To detect multiple drug resistance mutations on the same viral genome for linkage analysis, the amplified viral genomes (polonies) in each gel were then sequentially interrogated by 6 to 12 SBE reactions for targeted drug resistance mutations. Mutation sites were selected for analysis based on the treatment history of the patients and genotypic test results when they were available. Between 6 and 12 drug resistance mutation sites were assayed for the amplified viral genomes in each sample by repeating SBEs. After each SBE, the gel was scanned with a GenePix 4000B Microarray Scanner (Molecular Devices, Sunnyvale, CA) to acquire images.
PASS data analysis.
The two channel images (Cy5 for WT and Cy3 for mutant) acquired from each PASS assay were first cropped with Picture Window Pro3.5 to remove the edge area containing no signal. The cropped images were analyzed with the Progenesis PG200 (Nonlinear Dynamics, Durham, NC) software. After background subtraction, normalization, and spot filter setting, only the unambiguous spots at either channel were included for further analysis. The normalized pixel count data at multiple mutation sites for each spot were exported into an Excel file with a unique number. By comparing each spot's normalized values at the two channels, the position was classified as WT or mutant. Finally, the linkage pattern of all mutations on each viral genome was determined by compiling mutation information at all analyzed sites with the Linksys program (22).
Frequency of mutants in the population.
At each sampling time, a number of sequences, N, are analyzed. If, as in Fig. 1, we find that 9 out of 9 sequences all have the M184V mutation, the frequency of M184V mutants in the sample is 100%, but other sequences not containing M184V may be present in the population and just not sampled. For example, if the frequency of non-M184V sequences is 1/(N + 1), i.e., 10% in this case, then the probability of observing 9 out of 9 sequences with M184V, computed from the binomial distribution, Binom(9, 9, P = 0.9), is ∼0.39. One can also use the binomial distribution to determine with what frequency non-M184V sequences would need to be present in order to obtain with 95% probability no non-M184V mutants in a sample of size N. As discussed in more detail in reference 21, this is the solution of Binom(0, N, P) = 0.95. For N = 9, if non-M184V sequences were present at P = 0.5% or less, one would expect 9 out of 9 sequences to be M184V with >95% probability.
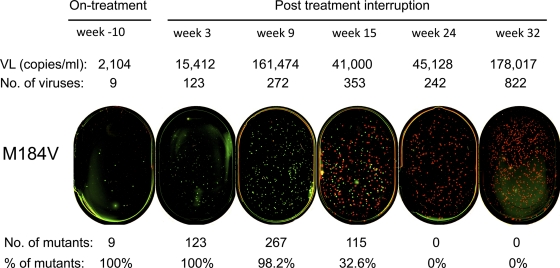
Fig. 1.
Detection of the M184V mutation after treatment interruption. The M184V mutation was determined by PASS in plasma samples collected on therapy (week −10) and off therapy (week 3 to week 32) in patient PID811. Green and red dots indicate mutant and WT bases detected in amplified individual viral genomes, respectively. Viral loads (VL), numbers of viral genomes detected, numbers of mutants in the viral population, and percentages of mutant viruses in the sequenced genomes are shown above and underneath the PASS images. Due to limited sampling of the viral population, the percentages of mutants observed may differ from the percentage of mutants in the population (see Materials and Methods).
Viral fitness comparison.
A conventional method (11, 15, 41) of analyzing the difference in growth rates (or fitness) between mutant and WT was used to evaluate the relative fitness between strains. In this method, one assumes the wild-type (W) and mutant (M) virus grow according to
Here the number of cells infected by WT and mutant virus was assumed to be proportional to the amount of wild-type and mutant virus, respectively (i.e., the virus and infected cells were assumed to be in quasi-steady state [31]). Cells infected with WT virus were assumed to grow at rate r and to be cleared at rate δ. Cells infected with mutant virus were assumed to grow at rate r′ and to be lost at rate δ′. The solution for these equations, which gives the evolution of the viral levels for each strain, between times t1 and t2 is
The ratio of mutant to WT virus at the two times was used to calculate d, the difference in the net growth rates, where d = (r′ − δ′) − (r − δ). This difference, d, has been called the log relative fitness (41) and is computed here by the formula
| (1) |
where f(t) = M(t)/W(t) and ln denotes the natural logarithm. For simplicity and to be consistent with much of the virology literature, d will simply be called the relative fitness rather than the more precise log relative fitness. The relative fitness can also be related to the selection coefficient, s, by the formula d = ln(1 + s) (41). Note that when s ≪ 1, d is approximately equal to s, and this approximation has been used to calculate s (11). Equation 1 is written such that we are assessing the fitness of the mutant relative to the WT. Since our measurements were made after interruption of therapy, we expect the drug resistance mutants to be less fit than WT and d will be negative, and its value can be viewed as the fitness loss of the mutant relative to the WT.
For each patient, the relative fitness was calculated over the time frame where both WT and mutant virus were observed. For this time frame, we write equation 1 as
| (2) |
which is the equation of a straight line with Δt = t2 − t1 as the independent variable. Using linear regression, one can then estimate d (41). Note that if the fitness of the mutant or WT changes over time, equation 2 can be interpreted as the mean relative fitness of the mutant averaged over the time interval, i.e.,
We analyzed the data by first grouping all sequences that had the same number of mutations to calculate the relative fitness of a “strain” with i mutations, i = 1, 2, 3… versus the WT. Since we had the most data for strains with one or two mutations in patient PID811, we also computed the relative fitness of these strains individually for this patient.
We calculated confidence intervals for the relative fitness estimates using a bootstrap method. In each case, we generated 1,000 new data sets, with the same number of total genomes as the data and with a distribution of WT and mutants as given by the data. That is, we used a multinomial distribution with the probabilities given by the data to calculate for each bootstrap replicate the new number of WT, 1-point, 2-point, etc., mutant genomes. In those few cases where the number of genomes for a given strain in the bootstrap data set was chosen as zero, we imputed 1 genome (note that equation 2 cannot be applied when one of the frequencies is zero). Implicitly, this is the same as assuming that the lowest frequency of detection is 1/N (where N is the total number of genomes analyzed at a given time point) and that the next genome sampled would be of that strain (1). We then used equation 2, as before, to calculate the relative fitness in these bootstrap data sets. We picked the 2.5 percentile and the 97.5 percentile of these estimates as the limits of the 95% confidence interval.
Estimation of the time for WT virus appearance after treatment interruption.
The time at which the wild-type strain emerged was estimated by a method introduced by Asquith and McLean (1) in the context of determining the time that cytotoxic T-lymphocyte (CTL) escape mutants arise. The fraction of the wild-type strain, p(t), is calculated as
| (3) |
where g = M(0)/W(0), and d is the difference of viral growth rates, as before. Equation 3 can be transformed to a linear equation as
| (4) |
where G = ln(g). To estimate the parameters d and G, we used the data after treatment interruption in Table S1 in the supplemental material. From these data we computed the fraction of WT virus, p(t), at various times and then used linear regression (equation 4) to estimate d and G. We then define the time when the WT strain emerges as the time that its frequency reaches 1% of the population. We can determine this time by choosing p(t) = 0.01 and calculating t in equation 4, using the best-fit parameters d and G from the linear regression described above.
RESULTS
Clinical characteristics of the study subjects.
Five patients who interrupted antiretroviral therapy, either per physician recommendation or by self-choice, were selected for analysis (Table 1). All were followed regularly in the HIV/AIDS clinic and had periodic viral load testing during the duration of their treatment interruption. In all cases, plasma viral loads quickly increased following treatment interruption. Multiple off-therapy samples and one on-therapy sample (0 to 61 weeks before treatment interruption) were available for assessment in each individual. All patients had received therapy with three or more antiretroviral agents over a period of 37 to >260 weeks prior to treatment interruption. Drug resistance mutations were identified in a clinical population-based HIV genotyping resistance test in two patients (PID811 and PID908) either while still on a failing ART regimen or just after treatment interruption commenced, confirming the presence of drug resistance mutations in both individuals. Clinical data, including prior ART regimens, duration of ART, dates of sample collection, viral load test results, and the results from clinical HIV resistance testing, are summarized in Table 1.
Table 1.
Demographic information for treatment interruption subjectsa
| Patient ID | ART before treatment interruption |
Sample collection date (mo/day/yr) | Viral load (copies/ml) | Wk from treatment interruption | Mutation analyzed by PASS |
Mutations detected by genotyping (date [mo-day-yr]) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Drugs | Duration (wk) | No. of viral genomes | Detected drug resistance mutationsc | Undetected drug resistance mutations | No. of mutations analyzed | |||||
| PID811 | AZT, 3TC, ABC | 48 | 8/1/2002 | 2,104 | −10 | 9 | M41L, K65R, D67N, K70R, | T215YF, L74V, Y115F, I84Vb | 12 | M41L, D67N, K70R, M184V, |
| 10/10/2002 | NA | 0 | NA | M184V, L210W, | L210W, T215Y, K219E; | |||||
| 11/1/2002 | 15,412 | 3 | 123 | K219QE, K103Nb | K103N, V118I (9-16-2002) | |||||
| 12/12/2002 | 161,474 | 9 | 272 | |||||||
| 1/23/2003 | 41,000 | 15 | 353 | |||||||
| 3/27/2003 | 45,128 | 24 | 242 | |||||||
| 5/22/2003 | 178,017 | 32 | 822 | |||||||
| PID908 | d4T, 3TC, NFV, EFV | 183 | 12/4/2002 | 55,541 | −2 | 124 | D67N, K70R, M184V; | None | 7 | M184V; L10I, L33F, G48V, |
| 12/20/2002 | NA | 0 | NA | M46I, G48V, V82A, | I54V, V82A, I84V | |||||
| 1/29/2003 | 353,899 | 6 | 1,458 | I84V | (1-29-2003) | |||||
| 3/5/2003 | 570,166 | 11 | 1,219 | |||||||
| PID268 | AZT, 3TC, LPVr | 37 | 6/28/2001 | 4,945 | 0 | 15 | D67N, K70R, M184V, | M41L, L210W, T215YF, | 11 | NA |
| 2/21/2002 | 89,440 | 34 | 553 | K219QE; V82A | K65R, V32I, I47V | |||||
| 4/11/2002 | 246,457 | 41 | 988 | |||||||
| 6/27/2002 | 181,134 | 52 | 1,167 | |||||||
| PID895 | AZT, 3TC, NFV | >260 | 6/13/2001 | 1,111 | −61 | 1 | M41L, D67N, K70R, | L210W, T215YF, K219QE, | 10 | NA |
| 8/17/2002 | NA | 0 | NA | M184V | K65R, D30N, L90M | |||||
| 10/16/2002 | 225,470 | 9 | 2,953 | |||||||
| 2/26/2003 | 129,546 | 28 | 1,772 | |||||||
| 8/6/2003 | 750,000 | 51 | 4,350 | |||||||
| 10/1/2003 | 96,115 | 59 | 1,159 | |||||||
| PID295 | 3TC, d4T, ddI, LPVr | 79 | 4/19/2001 | 213 | −39 | 1 | None | D67N, K70R, T215YF, | 6 | NA |
| 1/27/2002 | NA | 0 | NA | M184V, V32I, V82A | ||||||
| 4/18/2002 | 13,849 | 13 | 215 | |||||||
| 7/25/2002 | 108,052 | 27 | 175 | |||||||
| 10/31/2002 | 83,097 | 41 | 180 | |||||||
| 2/27/2003 | 179,242 | 58 | 300 | |||||||
Boldface indicates mutations that were not detected by a genotypic assay, underlining indicates mutations that were not tested by PASS, and italic indicates secondary mutations. NA, no data available. d4T, stavudine; NFV, nelfinavir; EFV, efavirenz; LPVr, lopinavir/ritonavir; ddI, didanosine.
Mutations that are not related to the drugs in the regimen when the treatment was terminated.
RTI mutations are listed first, and PI mutations are listed second (separated by a semicolon).
Dynamic population changes of individual drug resistance mutations.
Viral RNA was extracted from longitudinal samples in each patient, and drug resistance mutations were assayed by PASS. A total of 18,451 viral genomes (an average of 802 per time point) were analyzed. In general, samples collected during nonsuppressive ART that only partially suppressed viral replication had a low viral load, and only few or no viral genomes were detected in samples from four patients (PID811, PID908, PID268, and PID295). In each of two cases (PID895 and PID295) with low viral loads (1,111 copies/ml and 213 copies/ml, respectively), only one viral genome was detected. These samples were not included for analysis.
Six to 12 mutation sites in each patient were selected for analysis based on potential resistance patterns that were anticipated to have been selected by the particular agents being used during the time of treatment failure. When available, clinical genotypic resistance test results were also used to identify mutations of interest. Four to eight drug resistance mutations were detected in four of the five patients; no drug resistance mutations were detected in the fifth patient, PID295 (Table 1).
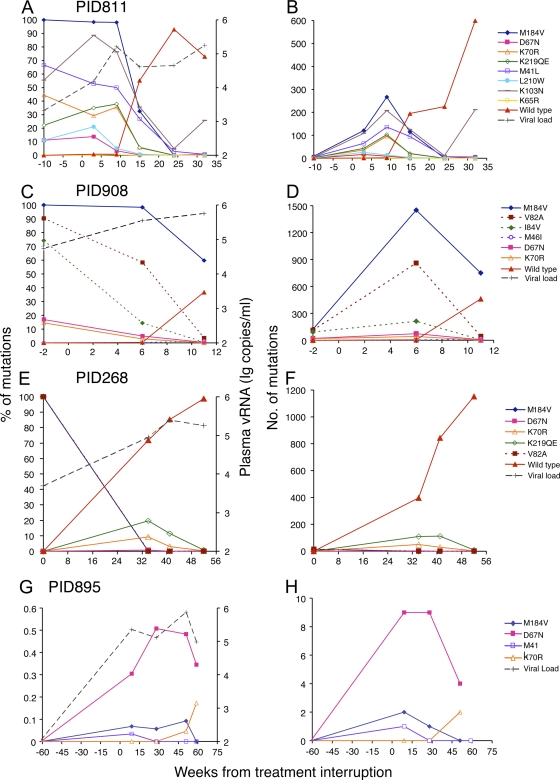
Drug resistance mutations were first analyzed individually for each patient. Patient PID811 was treated with zidovudine (AZT), lamivudine (3TC), and abacavir (ABC). The patient had also received nevirapine (NVP) in an earlier regimen. The viral load was 2,104 copies/ml in an on-therapy sample collected 10 weeks before treatment interruption, and it continuously increased to 178,017 copies/ml at week 32 after ART interruption. Eight primary drug resistance mutations and one secondary drug resistance mutation were detected by a clinical genotype resistance test in a sample obtained 3 weeks prior to treatment interruption (Table 1). The PASS assay was completed on one on-therapy sample and five off-therapy samples collected within 32 weeks after treatment interruption. On average, 304 viral genomes (range, 9 to 822) were analyzed at each time point. Based on the treatment history and available genotypic resistance tests, 11 primary drug resistance mutations to reverse transcriptase inhibitors (RTIs) and one to protease inhibitors (PIs) were assessed. Of these, eight (M41L, D67N, K65R, K70R, K103N, M184V, L210W, and K219Q/E) were detected and four (L74V, Y115F, T215Y/F, and I84V) were not (Table 1). K103N (RTI mutation) and I84V (PI mutation) were included for analysis based on the genotypic test result and the previous treatment history. While the patient was still on therapy, the viral load was low and only nine viral genomes were analyzed. At this time, all viruses had the M184V mutation (Fig. 1). The proportion of observed viruses with the M184V mutation was relatively stable (89% to 100%) for the first 9 weeks following treatment interruption and then dramatically declined over the next 23 weeks, during which time WT viruses predominated (Fig. 1 and Fig. 2A). After week 24, the M184V mutation became undetectable. Similar results were observed for the other seven identified mutations, although they were present in various proportions in the viral population. The exception to this pattern was the K103N mutation, which initially decreased to ∼5% at week 24 but then became a higher proportion (26%) at week 32 (Fig. 2A). The K013N substitution can be caused by either the AAC or AAT allele. Since the AAC allele is about 100-fold more common than the AAT allele in patients who have developed the K103N mutation (32), we analyzed only the AAC allele. Thus, the total percentage of the K103N mutation might be higher in this patient. The K103N mutation has been detected in patients months after treatment termination, and the proportion of this mutation in the viral population generally decreased over time (17, 33). The cause of a one-time increase in the proportion of K103N virus detected at one time point in patient PID811 is unclear.
Fig. 2.
Evolution of drug resistance mutations following treatment interruption. Detected individual drug resistance mutations were plotted as percentages (A, C, E, and G) and absolute numbers (B, D, F, and H) in the viral population at each time point in each individual. Drug resistance mutations are indicated by unique symbols as shown at the far right for each patient. The RTI mutations are shown as solid lines, and the PI mutations are shown as dotted lines. The WT virus and viral load data are indicated as red triangles and black crosses, respectively. WT viruses were not included in patient PID895 in order to show the differences among minority drug-resistant virus populations (G and H). The G48V mutation was not analyzed at week 11 and is not shown in the plots (C and D).
Fewer time point samples from three other patients were available for analysis. However, the evolution of drug resistance mutations in these patients was similar to what was observed in patient PID811. Two samples in the early weeks after treatment interruption were analyzed in patient PID908 (Table 1 and Fig. 2C). Three mutations in the reverse transcriptase gene (M184V, D67N, and K70R) and four in the protease gene (M46I, G48V, V82A, and I84V) were detected. M184V was present in all detected viral genomes, while other mutations accounted for various fractions of the viral population when the patient was on therapy. Similar to the pattern seen in patient PID811, the proportions of the various mutations either persisted at the same level (M184V) or modestly decreased (others) at week 6 but then all declined substantially by week 11 when WT viruses began to emerge (Fig. 2C).
In patient PID268, the first sample was collected on the day the patient discontinued ART. Fifteen viral genomes were analyzed at this time point, and all had the RTI resistance mutation M184V and the PI resistance mutation V82A (Fig. 2E). Interestingly, neither the M184V mutation nor the V82A mutation was detected in the other three samples obtained later during the course of treatment interruption (34 to 52 weeks). Instead, three RTI resistance mutations (D67N, K70R, and K219QE) were detected at week 32 and decreased to very low levels by week 52.
Samples were available only 61 or 39 weeks before treatment interruption in patients PID895 and PID295, respectively (Table 1). Both samples had low viral loads (1,111 copies/ml in PID895 and 213 copies/ml in PID295), suggesting that viral replication was relatively well controlled when ART was stopped. Four drug resistance mutations were detected in patient PID895, and all were present at very low frequencies (<0.6%) even 9 weeks after treatment interruption. The sum of all the detected resistant viruses accounted for less than 1% of the total viral population at any time point during treatment interruption (Fig. 2G). No mutations were detected at any time points between weeks 13 and 58 following treatment interruption in patient PID295 (Table 1). Of note, three RTI resistance mutations (D67N, K70R, and M184V) and one PI resistance mutation (V82A) were consistently detected in other patients (Table 1 and Fig. 2) but not in this patient. The infrequent detection of minority resistant mutations in these two patients suggests that selection of drug resistance mutations was limited at the time treatment was discontinued. It is also possible that the actual treatment termination dates might be earlier than the reported dates in both patients and the initial evolution phase of the drug resistance mutations was missed.
Linkage analysis of MDR mutations.
The ability of the PASS assay to detect different mutations present in each viral genome allows determination of the linkage patterns of MDR mutations among a large population of individual viral genomes. Viral genomes with linked MDR mutations were identified in three individuals (PID811, PID908, and PID268), all of whom had higher levels of viral replication at or shortly before treatment interruption. In all three patients, nearly all detected on-therapy viruses carried two or more drug resistance mutations (Tables 2 to 4). Linkage analysis of eight drug resistance mutations in patient PID811 revealed 35 different linkage patterns (Table 2). During the first 15 weeks following treatment interruption, viruses with as many as 6 linked MDR mutations were identified but at very low frequencies, while the majority of detected viruses had 2, 3, or 4 linked MDR mutations. Nearly all these MDR viruses became undetectable by weeks 24 and 32. In contrast, single-drug-resistant (SDR) viruses were rare in the first 9 weeks after treatment interruption (only M184V mutants were consistently detected). After week 15, nearly all drug-resistant viruses had only a single mutation, almost always the K103N or M41L mutation. Similar results were observed in patient PID908. A total of 28 linkage patterns were identified, and the MDR viruses predominated in the viral population from the on-therapy sample to the sample from week 6 after interruption (Table 3). These viruses then rapidly decreased in frequency and were replaced by WT viruses and minority SDR viral populations (<2%) by week 11. Interestingly, viruses with only the M184V mutation accounted for 39% of the viral population at week 6 and increased further to 57% of the viral population by week 11.
Table 2.
Detection of drug-resistant and WT viruses in treatment interruption patient PID811
| No. of mutations in linked MDR viruses | Linkage pattern | No. of viral genomes found at wk from treatment interruption: |
|||||
|---|---|---|---|---|---|---|---|
| −10 | 3 | 9 | 15 | 24 | 32 | ||
| ≥7 | |||||||
| 6 | M184V+K103N+D67N+K70R+M41L+L210W | 1 | |||||
| M184V+K103N+D67N+K70R+K219QE+M41L | 1 | ||||||
| 5 | M184V+K103N+D67N+K70R+K219QE | 3 | 3 | ||||
| M184V+K103N+D67N+K70R+M41L | 4 | ||||||
| M184V+K103N+D67N+K219QE+M41L | 1 | ||||||
| M184V+K103N+K70R+K219QE+M41L | 1 | 3 | |||||
| M184V+K103N+K70R+K219QE+L210W | 1 | ||||||
| M184V+K103N+D67N+M41L+K65R | 1 | ||||||
| 4 | M184V+K103N+D67N+K219QE | 5 | 1 | ||||
| M184V+K103N+D67N+M41L | 1 | ||||||
| M184V+K103N+K70R+M41L | 1 | 1 | |||||
| M184V+K103N+K70R+K219QE | 1 | 24 | 69 | 13 | |||
| M184V+K103N+K219QE+M41L | 1 | 5 | 2 | ||||
| M184V+K103N+M41L+L210W | 1 | 22 | 11 | 2 | |||
| M184V+D67N+K70R+K219QE | 2 | ||||||
| M184V+D67N+K70R+M41L | 1 | ||||||
| 3 | M184V+K103N+D67N | 1 | |||||
| M184V+K103N+K70R | 3 | 2 | 2 | ||||
| M184V+K103N+K219QE | 7 | 5 | 2 | ||||
| M184V+K103N+M41L | 2 | 32 | 73 | 66 | |||
| M184V+K103N+L210W | 2 | ||||||
| M184V+D67N+M41L | 1 | ||||||
| M184V+K70R+K219QE | 1 | 11 | |||||
| M184V+K70R+M41L | 1 | ||||||
| M184V+K219QE+M41L | 2 | ||||||
| M184V+M41L+L210W | 1 | ||||||
| K103N+K70R+K219QE | 1 | 1 | |||||
| 2 | M184V+K103N | 4 | 27 | 9 | |||
| M184V+D67N | 2 | ||||||
| M184V+K70R | 2 | 1 | |||||
| M184V+K219QE | 1 | 1 | |||||
| M184V+M41L | 1 | 36 | 6 | ||||
| K103N+D67N | 1 | ||||||
| K103N+K70R | 3 | ||||||
| K103N+M41L | 2 | 7 | 2 | ||||
| 1 | M41L | 1 | 12 | 5 | 6 | ||
| K65R | |||||||
| D67N | 5 | ||||||
| K70R | 1 | ||||||
| K103N | 18 | 10 | 211 | ||||
| M184V | 1 | 8 | 8 | 10 | |||
| L210W | 1 | ||||||
| K219QE | |||||||
| No. of wild-type genomes | 0 | 0 | 2 | 195 | 225 | 599 | |
| Total no. of viral genomes | 9 | 123 | 272 | 353 | 242 | 822 | |
Table 4.
Detection of drug-resistant and WT viruses in treatment interruption patient PID268
| No. of mutations in linked MDR viruses | Linkage pattern | No. of viral genomes found at wk from treatment interruption: |
|||
|---|---|---|---|---|---|
| 0 | 34 | 41 | 52 | ||
| ≥3 | |||||
| 2 | M184V+V82A | 15 | |||
| D67N+K219QE | 1 | ||||
| K70R+K219QE | 6 | ||||
| 1 | D67N | 3 | 1 | 2 | |
| K70R | 45 | 30 | 3 | ||
| K219QE | 101 | 113 | 10 | ||
| M184V | |||||
| V82A | |||||
| No. of wild-type genomes | 0 | 397 | 844 | 1,152 | |
| Total no. of viral genomes | 15 | 553 | 988 | 1,167 | |
Table 3.
Detection of drug-resistant and WT viruses in treatment interruption patient PID908
| No. of mutations in linked MDR viruses | Linkage pattern | No. of viral genomes found at wk from treatment interruption: |
||
|---|---|---|---|---|
| −2 | 6 | 11a | ||
| ≥7 | ||||
| 6 | M184V+V82A+G48V+I84V+D67N+K70R | 11 | 13 | |
| 5 | M184V+V82A+G48V+D67N+K70R | 5 | 5 | |
| M184V+V82A+G48V+I84V+D67N | 3 | 7 | ||
| M184V+G48V+I84V+D67N+K70R | 1 | |||
| M184V+V82A+I84V+D67N+K70R | 1 | |||
| M184V+V82A+G48V+I84V+K70R | 1 | |||
| M184V+V82A+G48V+I84V+M46I | 1 | |||
| 4 | M184V+V82A+G48V+I84V | 74 | 163 | |
| M184V+V82A+G48V+D67N | 6 | |||
| M184V+V82A+D67N+K70R | 15 | |||
| M184V+I84V+D67N+K70R | 1 | |||
| M184V+V82A+I84V+D67N | 2 | |||
| 3 | M184V+V82A+M46I | 10 | ||
| M184V+V82A+G48V | 18 | 263 | ||
| M184V+V82A+I84V | 1 | 21 | 2 | |
| M184V+G48V+D67N | 2 | |||
| M184V+I84V+D67N | 1 | |||
| M184V+D67N+K70R | 5 | |||
| V82A+G48V+I84V | 1 | |||
| M184V+V82A+K70R | 2 | |||
| M184V+V82A+D67N | 8 | |||
| 2 | M184V+M46I | 3 | 4 | |
| M184V+D67N | 9 | 2 | ||
| M184V+I84V | 1 | 1 | 4 | |
| M184V+G48V | 8 | |||
| M184V+V82A | 349 | 28 | ||
| V82A+G48V | 1 | |||
| V82A+M46I | 1 | |||
| 1 | D67N | 3 | ||
| K70R | ||||
| M184V | 9 | 564 | 700 | |
| N46I | ||||
| G48V | ND | |||
| V82A | 1 | 4 | ||
| I84V | ||||
| No. of wild-type genomes | 0 | 5 | 461 | |
| Total no. of viral genomes | 124 | 1,458 | 1,219 | |
G48V mutation was not determined at week 11.
In patient PID268, 15 viral genomes were analyzed at the time of treatment discontinuation, and all had both M184V and V82A mutations (Table 4). However, neither M184V nor V82A was detected in subsequent samples (weeks 34, 41, and 52), perhaps because no samples were available during the early time period following ART interruption. Instead, low frequencies of linked MDR mutations (D67N/K219QE and K70R/K219QE) were detected at week 34, but all viruses from subsequent time points had only SDR mutations, which continuously decreased over time. No viruses in patient PID895 were found to carry two or more mutations at any time points (Table 5).
Table 5.
Detection of drug-resistant and WT viruses in treatment interruption patient PID895
| No. of mutations in linked MDR viruses | Linkage pattern | No. of viral genomes found at wk from treatment interruption: |
||||
|---|---|---|---|---|---|---|
| −61 | 9 | 28 | 51 | 59 | ||
| ≥2 | ||||||
| 1 | M41L | 1 | ||||
| D67N | 9 | 9 | 21 | 4 | ||
| K70R | 2 | 2 | ||||
| M184V | 2 | 1 | 4 | |||
| No. of wild-type genomes | 1 | 2,941 | 1,762 | 4,323 | 1,153 | |
| Total no. of viral genomes | 1 | 2,953 | 1,772 | 4,350 | 1,159 | |
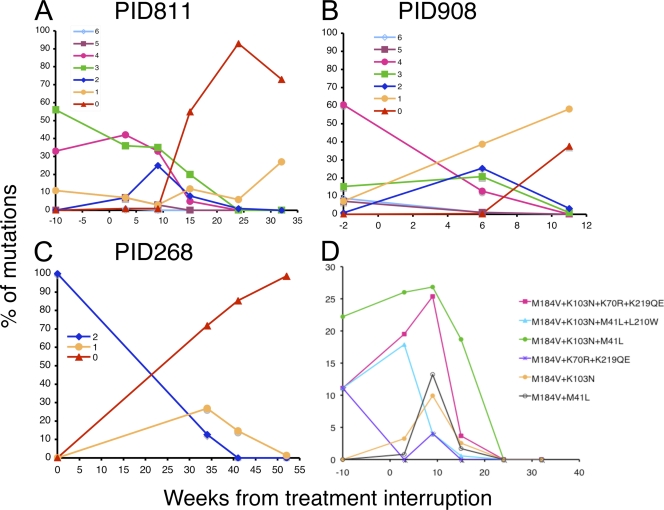
Most viruses with linked MDR mutation patterns were present at low frequencies (<2%), but a few linkage patterns were more common. For example, four linkage patterns (M184V/K103N/K70R/K219QE, M184V/K103N/M41L, M184V/K103N, and M184V/K219QE) in patient PID811 accounted for more than 25% of the population at weeks 3 and 9 (Table 2). Similarly, three linkage patterns (M184V/V82A/G48V/I84V, M184V/V82A/G48V, and M184V/V82A) accounted for much higher proportions of viruses than others in patient PID908 (Table 3). These results indicate that some drug-resistant viruses with particular linked MDR mutation patterns might have a significant fitness advantage. In all three patients, viruses with 7 or more mutations were not found at any time point, suggesting that such viruses are too unfit to survive, i.e., that the potential resistance advantage provided by the seventh mutation does not outweigh its contribution to the overall fitness loss of the virus or that such viruses do not exist in sufficient quantity for detection.
In the three patients who carried linked MDR viruses, those with combinations of four or five linked mutations were noted to gradually decrease over time after therapy interruption, but viruses with one or two mutations transiently increased during the first 6 to 10 weeks and then decreased thereafter (Fig. 3). Viruses with five or six mutations occurred too infrequently for conclusions to be drawn. However, some linkage patterns, for example, M184V/K103N/K70R/K219QE and M184V/K103N/M41L in patient PID811, continued to increase in the viral population during the first 9 weeks after ART interruption and persisted at relatively high frequency (19% and 4%, respectively) at week 15 (Fig. 3D and Table 2). Interestingly, predominant viruses with linked MDR mutations shared the same mutations (M184V, K103N, and M41L in patient PID811; M184V and V82A in both patients PID908 and PID268, who were treated with RTIs and PIs). The mutations present in the dominant viruses with linked MDR mutations were generally those present at higher frequencies as SDR mutations (Tables 2 and 3). These results suggest that some drug-resistant viruses with different combinations of MDR mutations exist in higher frequencies than others, probably because they are selected more easily as a result of these particular ART regimens, and that the fitness cost of these mutation patterns was less substantial.
Fig. 3.
Dynamic changes of linked MDR viruses following treatment interruption. The viral population changes over time were compared in patients PID811 (A), PID908 (B), and PID268 (C). The numbers of linked drug resistance mutations in viruses are indicated by different symbols and colors as indicated in each graph. The population changes of select predominant viruses with linked MDR mutations in patient PID811 were compared (D).
Three-phase evolution of drug resistance mutations during treatment interruption.
Three patients with higher viral loads (PID811, PID908, and PID268) had samples available at the time of or shortly before treatment interruption. When the combined data from these three patients were analyzed, a three-phase evolution of drug resistance mutations was observed following treatment interruption. In the first phase (the initial 9 weeks after treatment interruption), the proportions of drug-resistant viruses present were stable or slightly decreased (Fig. 2A and C; Tables 2 and 3) while the absolute number of viruses with resistance mutations increased (Fig. 2B and D). In this phase, the predominant viruses were those with MDR mutations, while viruses with only SDR mutations were uncommon and WT viruses were very rare or undetectable. In the second phase (9 to 32 weeks after treatment interruption), the proportion and number of drug-resistant viruses rapidly decreased and they were quickly replaced by WT viruses (Fig. 2A to D; Tables 2 and 3). In this phase, viruses with MDR mutations decreased markedly while viruses with minority SDR mutations became the major drug-resistant population. In the third phase (32 to 59 weeks after treatment interruption), all drug-resistant viruses became very rare (<1% of the viral populations) and only viruses with SDR mutations were detectable (Fig. 2E and G).
Fitness of drug-resistant viruses.
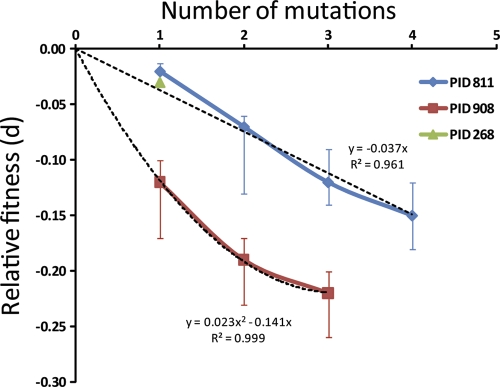
To determine the fitness of various groups of drug-resistant viruses, we first classified them into seven groups based on the number (0 through 6) of mutations that each virus carried (see Table S1 in the supplemental material). Each mutation was counted as one, so viruses in each group might contain different combinations of mutations. For each patient, t1 was chosen as the first time for which data were available for both WT and mutant strains and t2 was chosen as a subsequent time. Then, we estimated the relative fitness (d) by fitting equation 2 to the data. Viral genomes with 5 to 6 mutations were not included for analysis since there were too few for comparison. The estimated relative fitness (d) of viral strains with different numbers of mutations was computed, together with the respective 95% confidence intervals (Fig. 4). In patients PID811 and PID908, using this procedure, we compared the cost of increased number of mutations in relation to the WT virus (Fig. 4). The loss in relative fitness increased linearly in patient PID811 (P = 0.00016), whereas in PID908 the three-point mutant did not lose as much fitness as predicted by a linear loss (i.e., a positive quadratic term for fitness loss was significant, P = 0.02). Because the specific mutations that form the one-, two-, and three-mutation groups are different and appear at different times in these patients, it is difficult to compare the relative fitness of the mutation groups between the two patients.
Fig. 4.
Relationship between relative fitness loss of drug resistance mutants compared to the WT virus and the numbers of drug resistance mutations. The vertical bars indicate the 95% confidence interval range. Note that for patient PID268, the error bars are so small that they cannot be easily visualized. The dashed lines represent the best fit for the trend of fitness loss with higher numbers of mutations.
Various combinations of drug resistance mutations at different time points were observed in patient PID811 (Table 6). This allowed for calculation of the relative fitness by taking into account the different combinations of mutations. Equation 1 was applied to determine the relative fitness based on which drug resistance mutations were present in the viral genome. Among 41 strains of drug-resistant viruses detected in patient PID811, the majority of them carried MDR mutations (2 to 6) while only six had SDR mutations (Table 2). Over time, the frequency of the fittest strains increased (Table 6). For instance, from days 63 to 105, the M184V virus was present with low fitness (d = −0.104) relative to the WT; however, SDR mutants with M41L or K103N emerged between days 105 and 168, and they were more fit (d = −0.016 or −0.012) than the M184V mutant, which was lost in this time window. This also was seen for the double mutant population, where from days 105 to 168, all double mutants except K103N/M41L were lost, and the K103N/M41L double mutant was at least six times more fit than any other double mutants in the previous time period.
Table 6.
Relative fitness (d) of mutant strains compared to WT in patient PID811
| No. of mutations | Linkage pattern |
d at days from treatment interruption (95% confidence interval) |
||
|---|---|---|---|---|
| 63–105 | 105–168 | 168–224 | ||
| 4 | M184V+K103N+D67N+K219QE | |||
| M184V+K103N+D67N+M41L | ||||
| M184V+K103N+K70R+M41L | ||||
| M184V+K103N+K70R+K219QE | −0.149 (−0.18, −0.12) | |||
| M184V+K103N+K219QE+M41L | −0.131 (−0.17, −0.08) | |||
| M184V+K103N+M41L+L210W | −0.15 (−0.19, −0.11) | |||
| M184V+D67N+K70R+K219QE | ||||
| M184V+D67N+K70R+M41L | ||||
| 3 | M184V+K103N+D67N | |||
| M184V+K103N+K70R | −0.109 (−0.15, −0.06) | |||
| M184V+K103N+K219QE | −0.131 (−0.17, −0.08) | |||
| M184V+K103N+M41L | −0.111 (−0.13, −0.09) | |||
| M184V+K103N+L210W | ||||
| M184V+D67N+M41L | ||||
| M184V+K70R+K219QE | ||||
| M184V+K70R+M41L | ||||
| M184V+K219QE+M41L | ||||
| M184V+M41L+L210W | ||||
| K103N+K70R+K219QE | −0.109 (−0.14, −0.07) | |||
| 2 | M184V+K103N | −0.135 (−0.17, −0.11) | ||
| M184V+D67N | ||||
| M184V+K70R | −0.126 (−0.16, −0.07) | |||
| M184V+K219QE | ||||
| M184V+M41L | −0.152 (−0.19, −0.12) | |||
| K103N+D67N | ||||
| K103N+K70R | ||||
| K103N+M41L | −0.079 (−0.12, −0.04) | −0.022 (−0.04, 0.001) | ||
| 1 | M41L | −0.016 (−0.04, 0.0) | −0.014 (−0.04, 0.015) | |
| K65R | ||||
| D67N | ||||
| K70R | ||||
| K103N | −0.012 (−0.03, 0.0) | 0.037 (0.027, 0.053) | ||
| M184V | −0.104 (−0.14, −0.07) | |||
| L210W | ||||
| K219QE | ||||
Estimation of timing for WT virus emergence during treatment interruption.
WT viruses gradually increased and became dominant after treatment interruption in patients PID811, PID908, and PID268 (Tables 2 to 4). It would be informative to estimate the time at which the WT viruses emerged in each patient. Using the values of d and G in equation 4, the time for WT viruses to emerge after treatment was estimated. Here we define the time of emergence as the time that the WT virus reaches 1% of the population, when it was undetectable before. The time for WT virus to reach 1% of the population was ∼56 days in patient PID811 (based on data collected until week 24), ∼50 days in patient PID908, and ∼44 days in patient PID268. On average, WT viruses emerged 50 days after treatment interruption. However, these times depend on our definition of “emergence.” If instead of a frequency of 1%, we define it as the time that it takes the WT to reach 0.1%, then the estimated times are 23 and 35 for patients PID811 and PID908, respectively. For patient PID268, we cannot calculate the time that it takes WT to reach 0.1%, because the estimated proportion of WT at time zero is already 0.3%. Note that for patient PID268, we have no data between weeks 0 and 34 (i.e., day 238), which leads to high uncertainty in frequency estimates early after therapy interruption.
DISCUSSION
Interrupting antiretroviral therapy in HIV-infected persons whose treatment regimens are not achieving viral suppression provides an opportunity to study the evolution of drug resistance mutations and viral fitness in vivo during the period after drug selection pressure is removed. In this study, WT, SDR, and linked MDR mutations were analyzed from over 18,000 viral genomes in five treatment-interrupted patients at multiple time points using the PASS assay. These data demonstrated a three-phase evolution of drug resistance mutations and complicated patterns of linked MDR mutations. Mathematical modeling showed that the relative loss of viral fitness increased with the number of mutations in the viral genome and that WT viruses reached an estimated frequency of 1% after treatment was interrupted for 1 to 2 months.
Understanding of the evolution of viral populations following treatment interruption has been hindered by the inability to characterize a large number of viruses and the limited availability of samples from appropriate time points. Analysis of thousands of viral genomes by PASS from three patients at multiple time points (up to 59 weeks) following treatment discontinuation demonstrated a three-phase evolution pattern of drug resistance mutations. In the first phase, HIV viral load rebounded rapidly after ART interruption. These early rebound viruses were almost exclusively those with linked MDR mutations, although the proportion of drug-resistant viruses within the population remained stable or decreased slightly. WT viruses were generally undetectable during this time period. However, our model predicted that they had begun to accumulate. In the second phase, the proportions and the numbers of viruses with MDR mutations rapidly decreased while viruses with SDR mutations gradually increased. In this phase, WT viruses increased rapidly and became predominant. In the third phase, the vast majority of viruses were wild type and the only detected drug-resistant viruses were those with SDR mutations present at very low frequencies (<1%). This phase was observed to last more than a year.
Our data suggest that the relatively stable proportion of drug resistance mutations observed in the first phase is maintained by a continuous increase of drug-resistant viruses in blood following ART interruption, while the rapid disappearance of these drug-resistant viruses is a consequence of increases in more fit WT viruses and a possibly decreased release of drug-resistant viruses into the blood during the second phase. Similar evolution patterns of drug-resistant viruses have been reported previously (7, 8). In those studies, the drug susceptibility of the viral populations was determined by a phenotypic resistance assay after ART discontinuation. Viruses in those patients were fully resistant to ART over the first 15 weeks following ART discontinuation, but they were then quickly replaced by drug-susceptible viruses over a very short period, and viral populations remained fully drug susceptible thereafter. However, specific drug resistance mutations were not described in those studies.
Although viruses with linked MDR mutations are less fit than the WT virus, they continuously replicate early on when drug selection pressure has been removed, since initially they are the only detectable viruses. This results in the observed early increase in the number of drug-resistant viruses and the persistence of the proportion of drug-resistant viruses in the first phase. However, due to the impaired fitness of these viruses in the absence of continued drug selection pressure, they are subsequently outcompeted by WT viruses, which become the dominant population beginning about 10 weeks after treatment interruption. The drug selection pressure declines relatively quickly after ART interruption since the half-lives of antiretroviral drugs are generally only a few days even for long-lasting nonnucleoside RTIs (NNRTIs) (2, 5, 19, 29). Therefore, the presence of mostly drug-resistant viruses during the first 9 weeks following treatment interruption cannot be due to the continued suppression of WT viruses by residual antiretroviral effect in these patients but rather may reflect the fitness costs of resistance, as calculated here, and the low frequency of the WT virus when treatment is interrupted.
An area of considerable potential importance is the effect of linked MDR mutations present in a single viral genome as opposed to various drug resistance mutations being present as single mutations in different virions within a patient. This phenomenon has not been well studied due to the difficulty of assessing a large number of viral genomes within any one HIV-1-infected individual (26, 34). Using the recently developed PASS assay (3), we were able to perform linkage analysis of drug resistance mutations from thousands of viral genomes in each patient. Many different genetic variants with various combinations of linked MDR mutations were identified in patients in whom MDR mutations were detected. Viruses with 5 to 6 mutations were detected but were present at much lower frequencies, suggesting that they had impaired fitness. In contrast, viruses with 2 to 4 mutations were the majority of viruses at the time of ART failure and during the early stage following treatment interruption. Population genetic analysis also indicated that a few virus populations with specific combinations of some drug resistance mutations were more prevalent than others. The precise reason why some mutations predominate within certain combinations is not clear but is likely the result of differences in viral fitness, reflecting the interplay between mutations driven by particular drugs and the fitness consequences of the various combinations of possible mutations. Although hundreds of viral genomes at each time point were analyzed by PASS during treatment failure and treatment interruption, no viral genomes were found to have 7 or more drug resistance mutations. There may be two reasons why this was so. First, the resistance gain offered by the 7 or more mutations did not outweigh the seventh mutation's contribution to the overall fitness loss of the virus. The fitness cost for such viruses was too high for them to outcompete viruses with fewer mutations. Second, they might not develop because viruses did not need to obtain as many mutations in their genomes to be fully resistant to the combinations of drugs.
Analysis of a large number of virus genomes after removal of antiretroviral drug selection pressure allowed evaluation of viral fitness in vivo. Our mathematical model showed a positive correlation between the number of mutations in the viral genome and the loss of viral fitness. This is in agreement with our observation that viruses with 5 to 6 drug resistance mutations were rare and viruses with more than 6 mutations were not identified in any patients. While viruses with greater numbers of drug resistance mutations can theoretically be more advantageous in the presence of antiretroviral drug combinations, the severe fitness cost of multiple mutations renders such viruses incapable of replication or so impaired that they do not compete successfully against more fit viruses with fewer mutations. Our model also showed that the time to emergence of WT viruses varies from patient to patient, but the average time to reach 1% WT virus in the three most closely studied patients (PID811, PID908, and PID268) was 50 days.
Our study has limitations, primarily those of any cohort study. Treatment interruption is no longer considered an appropriate clinical treatment strategy. As a result, the total number of these patients who were available to be studied is limited. Patients were treated with a variety of antiretroviral drug combinations and had differing prior treatment histories, leading to differing resistance patterns at the time of treatment interruption. The samples were collected at different time points for each subject. The analysis was performed for select drug resistance mutations, and the effect of potential compensatory mutations (16, 30, 39) was not assessed. A well-controlled cohort in which a larger number of patients are treated with the same drugs and samples are collected at similar schedules would be ideal, but such a population is no longer feasible. However, the results from this comprehensive study of the dynamics of drug-resistant virus populations following treatment interruption provide new insights on the understanding of ART resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brooke Walker for editorial assistance.
This work was supported by grants from the National Institutes of Health (GM065057, AI64518, AI067854, AI28433, RR06555, P20-RR18754, and K24-AI01608) and the National Science Foundation (grant NSF PHY05-51164), and portions of the work were done under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Asquith B., McLean A. R. 2007. In vivo CD8+ T cell control of immunodeficiency virus infection in humans and macaques. Proc. Natl. Acad. Sci. U. S. A. 104:6365–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boffito M., et al. 2008. Pharmacokinetics of atazanavir/ritonavir once daily and lopinavir/ritonavir twice and once daily over 72 h following drug cessation. Antivir. Ther. 13:901–907 [PubMed] [Google Scholar]
- 3. Cai F., et al. 2007. Detection of minor drug-resistant populations by parallel allele-specific sequencing. Nat. Methods 4:123–125 [DOI] [PubMed] [Google Scholar]
- 4. Clavel F., Hance A. J. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023–1035 [DOI] [PubMed] [Google Scholar]
- 5. Cressey T. R., et al. 2005. Persistence of nevirapine exposure during the postpartum period after intrapartum single-dose nevirapine in addition to zidovudine prophylaxis for the prevention of mother-to-child transmission of HIV-1. J. Acquir. Immune Defic. Syndr. 38:283–288 [PubMed] [Google Scholar]
- 6. Croteau G., et al. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deeks S. G., et al. 2005. Interruption of treatment with individual therapeutic drug classes in adults with multidrug-resistant HIV-1 infection. J. Infect. Dis. 192:1537–1544 [DOI] [PubMed] [Google Scholar]
- 8. Deeks S. G., et al. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472–480 [DOI] [PubMed] [Google Scholar]
- 9. Devereux H. L., Youle M., Johnson M. A., Loveday C. 1999. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 13:F123–F127 [PubMed] [Google Scholar]
- 10. Goudsmit J., de Ronde A., de Rooij E., de Boer R. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goudsmit J., De Ronde A., Ho D. D., Perelson A. S. 1996. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J. Virol. 70:5662–5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hance A. J., et al. 2001. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J. Virol. 75:6410–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrigan P. R., Bloor S., Larder B. A. 1998. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J. Virol. 72:3773–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirsch M. S., et al. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 37:113–128 [DOI] [PubMed] [Google Scholar]
- 15. Holland J. J., de la Torre J. C., Clarke D. K., Duarte E. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huigen M. C., et al. 2008. Identification of a novel resistance (E40F) and compensatory (K43E) substitution in HIV-1 reverse transcriptase. Retrovirology 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iarikov D. E., et al. 2010. Use of HIV resistance testing after prolonged treatment interruption. J. Acquir. Immune Defic. Syndr 53:333–337 [DOI] [PubMed] [Google Scholar]
- 18. Johnson J. A., et al. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kikaire B., et al. 2007. Nevirapine clearance from plasma in African adults stopping therapy: a pharmacokinetic substudy. AIDS 21:733–737 [DOI] [PubMed] [Google Scholar]
- 20. Lawrence J., et al. 2003. Structured treatment interruption in patients with multidrug-resistant human immunodeficiency virus. N. Engl. J. Med. 349:837–846 [DOI] [PubMed] [Google Scholar]
- 21. Lee H. Y., et al. 2009. Modeling sequence evolution in acute HIV-1 infection. J. Theor. Biol. 261:341–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J., et al. 2011. Analysis of low-frequency mutations associated with drug resistance to raltegravir before antiretroviral treatment. Antimicrob. Agents Chemother. 55:1114–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mammano F., Petit C., Clavel F. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mammano F., Trouplin V., Zennou V., Clavel F. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Markowitz M. 2000. Resistance, fitness, adherence, and potency: mapping the paths to virologic failure. JAMA 283:250–251 [DOI] [PubMed] [Google Scholar]
- 26. Martinez-Picado J., et al. 2000. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 97:10948–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martinez-Picado J., Savara A. V., Sutton L., D'Aquila R. T. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Metzner K. J., et al. 2009. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin. Infect. Dis. 48:239–247 [DOI] [PubMed] [Google Scholar]
- 29. Moore K. H., et al. 1999. The pharmacokinetics of lamivudine phosphorylation in peripheral blood mononuclear cells from patients infected with HIV-1. AIDS 13:2239–2250 [DOI] [PubMed] [Google Scholar]
- 30. Nijhuis M., et al. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349–2359 [DOI] [PubMed] [Google Scholar]
- 31. Nowak M. A., May R. M. 2000. Virus dynamics: mathematical principles of immunology and virology. Oxford University Press, Oxford, England [Google Scholar]
- 32. Palmer S., et al. 2006. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS 20:701–710 [DOI] [PubMed] [Google Scholar]
- 33. Palmer S., et al. 2006. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc. Natl. Acad. Sci. U. S. A. 103:7094–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Palmer S., et al. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richman D. D. 2006. Antiviral drug resistance. Antiviral Res. 71:117–121 [DOI] [PubMed] [Google Scholar]
- 36. Richman D. D., Grimes J. M., Lagakos S. W. 1990. Effect of stage of disease and drug dose on zidovudine susceptibilities of isolates of human immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 3:743–746 [PubMed] [Google Scholar]
- 37. Shafer R. W. 2009. Low-abundance drug-resistant HIV-1 variants: finding significance in an era of abundant diagnostic and therapeutic options. J. Infect. Dis. 199:610–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simen B. B., et al. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693–701 [DOI] [PubMed] [Google Scholar]
- 39. Svarovskaia E. S., et al. 2008. The A62V and S68G mutations in HIV-1 reverse transcriptase partially restore the replication defect associated with the K65R mutation. J. Acquir. Immune Defic. Syndr. 48:428–436 [DOI] [PubMed] [Google Scholar]
- 40. Verhofstede C., Wanzeele F. V., Van Der Gucht B., De Cabooter N., Plum J. 1999. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS 13:2541–2546 [DOI] [PubMed] [Google Scholar]
- 41. Wu H., et al. 2006. Modeling and estimation of replication fitness of human immunodeficiency virus type 1 in vitro experiments by using a growth competition assay. J. Virol. 80:2380–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.