Abstract
Productive replication of Epstein-Barr virus occurs in discrete sites in nuclei, called replication compartments, where viral DNA replication proteins and host homologous recombinational repair (HRR) and mismatch repair (MMR) factors are recruited. Three-dimensional (3D) surface reconstruction imaging clarified the spatial arrangements of these factors within the replication compartments. Subnuclear domains, designated BMRF1 cores, which were highly enriched in viral polymerase processivity factor BMRF1 could be identified inside the replication compartments. Pulse-chase experiments revealed that newly synthesized viral genomes organized around the BMRF1 cores were transferred inward. HRR factors could be demonstrated mainly outside BMRF1 cores, where de novo synthesis of viral DNA was ongoing, whereas MMR factors were found predominantly inside. These results imply that de novo synthesis of viral DNA is coupled with HRR outside the cores, followed by MMR inside cores for quality control of replicated viral genomes. Thus, our approach unveiled a viral genome manufacturing plant.
INTRODUCTION
Epstein-Barr virus (EBV), a human lymphotropic herpesvirus with a linear double-stranded DNA 172 kb in length (2), infects resting B lymphocytes, inducing their continuous proliferation without production of virus particles, this being termed latent infection. Productive infection, which occurs spontaneously or can be induced artificially, is characterized by expression of lytic genes, leading to virus production. The EBV genome is amplified 100- to 1,000-fold by viral replication machinery composed of BALF5 DNA polymerase, BMRF1 polymerase processivity factor, BALF2 single-stranded-DNA (ssDNA) binding protein, and BBLF4-BSLF1-BBLF2/3 helicase-primase complex in discrete sites in nuclei, which are called replication compartments (9, 13). With progression of productive replication, the replication compartments become enlarged and fuse to form large globular structures that eventually fill the nucleus in late stages (9).
We have previously reported the architecture of the EBV replication compartments (9). The BZLF1 oriLyt binding proteins show a fine, diffuse pattern of distribution throughout the nuclei at immediate-early stages of induction and then become associated with the replicating EBV genome in the replication compartments during lytic infection. The BMRF1 proteins show a homogenous, not dot-like, distribution in the replication compartments, coinciding with the synthesized viral DNA. In contrast, the BALF5 Pol catalytic protein, the BALF2 single-stranded-DNA binding protein, and the BBLF2/3 protein, a component of the helicase-primase complex, were colocalized as distinct dots distributed within replication compartments, representing viral replication factories.
The BMRF1 protein is a major phosphoprotein abundantly expressed during EBV productive infection (7, 26), associating with the BALF5 polymerase catalytic subunit with one-to-one stoichiometry to enhance its polymerase processivity (41). Judging from immunostaining data, together with the finding that almost all expressed BMRF1 proteins bind to viral genome DNA, the factor has been assumed not only to act as a polymerase processivity factor but also to protect the viral genome after synthesis. In addition, it can transcriptionally activate the BHLF1 promoter (48) and enhance BZLF1-mediated transcription of the BALF2 promoter (33).
It has been suggested that DNA replication is coupled with DNA recombination to generate large branched head-to-tail concatemers of replication intermediates during herpesvirus genome replication (4, 44, 49). We previously showed that homologous recombinational repair (HRR) factors such as replication protein A (RPA), Rad51, Rad52, and the Mre11/Rad50/Nbs1 (MRN) complex are recruited and loaded onto the newly synthesized viral genome in replication compartments (23). HRR is an accurate repair process known to be mediated by the MRN complex, RPA, members of the RAD52 epistasis group of gene products such as Rad51, Rad52, and Rad54, and phosphorylated BRCA1 and BRCA2 (5, 24). Knockdown of RPA32 and Rad51 by RNA interference significantly prevents viral DNA synthesis (23), indicating an HRR involvement in viral DNA synthesis.
We have also previously demonstrated that proliferating cell nuclear antigen (PCNA), the PCNA loader complex (RF-C), and a series of mismatch repair (MMR) proteins such as MSH2, MSH6, MLH1, and PMS2 can be assembled to Epstein-Barr virus replication compartments (10). MMR works primarily to correct mutations by removing base-base and small insertion-deletion mismatches that arise during DNA replication, and it is mediated by MSH heterodimers (MSH2-MSH3 and MSH2-MSH6) and MLH heterodimers (MLH1-PMS2 and MLH1-MLH3) (18, 20). PCNA, which was originally characterized as a DNA sliding clamp for replicative DNA polymerases, interacts with MSH2-MSH6 or MSH2-MSH3 complexes, searching for mispairs on newly replicated DNA (8, 14, 19). RF-C recruits PCNA (the clamp) and loads it onto DNA in the presence of ATP (clamp loading), with this being required for MMR (47).
In other herpesviruses such as herpes simplex virus type 1 (HSV-1) and human cytomegalovirus (HCMV), viral replication compartments are also formed in the infected nuclei during the productive replication, and HRR factors, including the MRN complex and Rad51, are reported to be recruited to the replication compartments (27, 29, 36, 37, 46). Furthermore, Taylor and Knipe reported that the HSV-1-encoded single-stranded-DNA binding protein ICP8 interacts either directly or indirectly with HRR and MMR factors (37). Also, HSV-1 alkaline exonuclease UL12 has recently been shown to interact specifically with the MRN complex (3).
Here we examined the spatial arrangements of viral DNA replication factors and cellular HRR and MMR factors in the replication compartments by means of confocal laser scanning microscopy and three-dimensional (3D) surface reconstruction imaging. BMRF1-rich subnuclear domains, designated BMRF1 cores, could be identified inside the replication compartments. As a result, each replication compartment was partitioned into two subdomains, outside and inside the BMRF1 core. We here present data demonstrating that viral DNA replication and viral genome maturation are assigned to outside and inside subdomains, respectively.
MATERIALS AND METHODS
Cell culture.
Tet-BZLF1/B95-8 cells were maintained in RPMI 1640 medium supplemented with 1 μg/ml puromycin, 250 μg/ml hygromycin B, and 10% tetracycline-free fetal calf serum (Clontech) at 37°C in a humidified 5% CO2 atmosphere. To induce lytic EBV replication, the tetracycline derivative doxycycline was added to the culture medium at a final concentration of 4 μg/ml. When blocking lytic replication, phosphonoacetic acid (PAA), a herpesvirus DNA polymerase-specific inhibitor, was added to the culture medium at a final concentration of 400 μg/ml.
Antibodies.
Anti-BALF2 and anti-BMRF1 rabbit polyclonal antibodies were as previously prepared (10, 40, 42). Anti-BALF5 protein-specific rabbit antibodies (43) were affinity purified with BALF5 protein coupled-Sepharose 4B as described previously (15). An anti-EBV EA-D-p52/50 (BMRF1 gene product) protein-specific mouse monoclonal antibody, clone name R3, was purchased from Chemicon Inc. 5-Chloro-2′-deoxyuridine (CldU)-labeled DNAs were detected with anti-5-bromo-2′-deoxyuridine (anti-BrdU) rat monoclonal antibody clone BU1/75 (ICR1), purchased from Abcam. The anti-BrdU antibody clone BU1/75 (ICR1) does not cross-react with 5-iodo-2′-deoxyuridine (IdU). Anti-pRPA32 S4/S8, -BRCA1 S1524, and -Mre11 rabbit polyclonal antibodies were purchased from Abcam and anti-Rad52 antibodies from Cell Signaling. Anti-PCNA mouse monoclonal and rabbit polyclonal antibodies were purchased from Transduction Laboratories and Abcam, respectively, and anti-MSH2, -MSH3, and -MSH6 monoclonal antibodies were obtained from Transduction Laboratories and BD Biosciences. The secondary goat anti-rabbit, anti-rat, and anti-mouse IgG antibodies conjugated with Alexa 488 or 594, a Zenon mouse IgG labeling kit (Alexa 594), and a Zenon rabbit IgG labeling kit (Alexa 594) were obtained from Molecular Probes.
Immunofluorescence analysis.
All staining procedures except for extraction and incubation with primary antibodies were carried out at room temperature. For immunofluorescence experiments, cells were washed with ice-cold phosphate-buffered saline (PBS) and extracted with 0.5% Triton X-100–mCSK buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.8), 300 mM sucrose, 1 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.5% Triton X-100) on ice for 10 min. Multiple protease inhibitors (Sigma; 25 μl/ml), 200 μM Na3VO4, and 20 mM NaF were also added to the buffer. Cells were fixed with 70% ethanol for 24 h at −20°C, washed with PBS containing 0.1% normal goat serum and 0.01% Tween 20, permeabilized with 0.5% Triton X-100 in PBS for 15 min, blocked for 1 h in 10% normal goat serum in PBS, and then incubated overnight with the primary antibodies diluted in PBS containing 0.1% normal goat serum and 0.01% Tween 20. The samples were then incubated for 1 h with secondary goat anti-rabbit, anti-rat, and anti-mouse IgG antibodies conjugated with Alexa Fluor 488 or 594. For MSH2, MSH3, and MSH6 staining, anti-MSH2, -MSH3, and -MSH6 antibodies were directly labeled with a Zenon tricolor mouse IgG1 labeling kit purchased from Molecular Probes. Also, for BALF2, BRCA1 S1524, and Rad52 staining, anti-BALF2, -BRCA1 S1524, and -Rad52 antibodies were directly labeled with a Zenon tricolor rabbit IgG1 labeling kit purchased from Molecular Probes. Cells were incubated with Alexa Flour 594-labeled anti-MSH2, -MSH3, -MSH6, -BALF2, -BRCA1 S1524, and -Rad52 antibodies for 45 min at room temperature and washed three times with PBS, followed by a second fixation with 4% paraformaldehyde solution in phosphate buffer for 15 min at room temperature. All the primary antibodies were employed at a 1:100 dilution, and the secondary antibodies were employed at a 1:500 dilution. All washes after antibody incubation were performed with PBS containing 0.1% normal goat serum and 0.01% Tween 20. The slides were mounted in ProLong Gold antifade reagent with 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes) and analyzed by fluorescence confocal microscopy. Laser scanning confocal fluorescence microscopic images were captured and processed using an LSM510 Meta microscope (Carl Zeiss MicroImaging, Inc.) with a plan-Apochromat 100×/1.4-numerical-aperture oil immersion objective.
Pulse-chase experiments.
For CldU pulse-labeling, newly synthesized DNA was labeled by incubating lytic replication-induced Tet-BZLF1/B95-8 cells with 10 μM CldU added directly to the incubation medium for 10 min at 24 h postinduction. For the pulse-chase experiments, the cells were pulse-labeled with 10 μM CldU for 10 min at 24 h postinduction, and the CldU-containing medium was removed and replaced with new medium containing IdU to inhibit CldU incorporation to newly synthesized DNA. Cells were chased for 1 h prior to harvesting. Cells were washed with ice-cold PBS and extracted with 0.5% Triton X-100–mCSK buffer on ice for 10 min. Multiple protease inhibitors (Sigma; 25 μl/ml), 200 μM Na3VO4, and 20 mM NaF were also added to the buffer. Cells were then fixed with 70% ethanol for 24 h at −20°C and treated for 60 min with 2 N HCl containing 0.5% Triton X-100 to expose the incorporated CldU residues before blocking. The cells were washed twice with PBS and neutralized with 0.1 M sodium tetraborate, pH 9.0, for 5 min prior to immunofluorescence.
Fluorescence in situ hybridization.
EBV bacterial artificial chromosome (BAC) DNA was labeled with digoxigenin (DIG) nick translation mix (Sigma) and used as a probe. First, cells were fixed in 4% paraformaldehyde and 70% ethanol. After digestion with RNase, cells were treated with 50% formamide in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), air dried, and immediately covered with a probe mixture containing 60% formamide in 2× SSC containing probe DNA (4 ng/μl), 12% dextran sulfate, and sheared salmon DNA (0.1 μg/μl). Probes and cells were simultaneously heated at 85°C for 5 min and incubated overnight at 37°C. After hybridization, specimens were washed at 45°C with 50% formamide in 2× SSC (three times for 3 min each), at 45°C with 2× SSC (three times for 3 min each), and at 65°C with 0.1× SSC for 10 min. After washing, specimens were blocked for 1 h in 5% milk in 4× SCC containing 1% bovine serum albumin (BSA) and then stained with anti-DIG sheep monoclonal antibodies in 4× SSC containing 1% BSA at 37°C for 30 min. After staining, specimens were washed at room temperature with 4× SSC, 0.1% Triton X-100 in 4× SSC, 4× SSC, and PN buffer (0.5 M Na2HPO4, 0.5 M NaH2PO4, 0.5% NP-40) (three times for 3 min each). Finally, cells were stained with anti-BMRF1 mouse monoclonal antibodies, mounted in ProLong Gold antifade reagent with DAPI (Molecular Probes), and analyzed by fluorescence confocal microscopy. Images were captured and processed using an LSM510 Meta microscope (Carl Zeiss MicroImaging, Inc.) with a plan-Apochromat 100×/1.4-numerical-aperture oil immersion objective.
3D reconstruction with confocal laser scanning microscopy.
Images observed with a confocal laser scanning microscope (Carl Zeiss MicroImaging, Inc.) were computerized to automatically make 50 to 100 serial optical sections at intervals of around 0.26 μm. The 3D reconstruction was performed with Imaris software (Carl Zeiss MicroImaging, Inc.). Image files created by the LSM510 Meta microscope were opened with Imaris, and a 3D surface model was created based on the appropriate intensity threshold.
RESULTS
BMRF1-rich structures (cores) are observed in viral replication compartments.
We previously described a B95-8 derivative cell line in which we can trigger onset of viral lytic replication via tetracycline-inducible expression of viral immediate-early protein BZLF1 (Tet-BZLF1/B95-8 cells) (22). In this study, we utilized Tet-BZLF1/B95-8 cells to examine more details of replication compartments during viral productive replication. The lytic replication-induced Tet-BZLF1/B95-8 cells were harvested and extracted with 0.5% Triton X-100–mCSK buffer. It should be noted that the treatment extracts soluble viral or cellular proteins, permitting investigation of DNA-bound fractions of viral and cellular proteins. We tested for specificity of the secondary antibodies and for reliability of discrimination with the fluorescence microscopy filters. When cells were stained singly for either antigen with inappropriate combinations of first and second antibodies, no fluorescence was observed. Also, no immunofluorescence was observed with an alternate filter. We have previously defined EBV replication compartments as BMRF1- or BALF2-staining sites where viral DNA genomes are colocalized as judged by immunofluorescence and fluorescence in situ hybridization (FISH) analysis (9). A representative image of a viral replication compartment is illustrated in Fig. 1A, visualized by means of indirect immunofluorescence analyses. As shown in a merged image, BALF2 protein dots were distributed not only inside but also outside BMRF1-stained subnuclear domains. A correlative immunofluorescence microscopy and electron microscopic imaging (FM-EM) study found that BMRF1 proteins were localized within an electron-lucent region in the nuclear interior, in contrast with electron-dense chromatin regions (data not shown). At higher magnification, the area corresponding to the replication compartment contained diffuse noncondensed fibers and granules but lacked any specific structures.
Fig. 1.
The majority of viral genome DNA is localized inside BMRF1-rich structures. (A) Lytic replication-induced Tet-BZLF1/B95-8 cells were fixed, stained with anti-BMRF1 (green) and anti-BALF2 (red) antibodies, and observed by laser scanning confocal microscopy to locate viral replication compartments. The right panel is a merged image. (B) Lytic replication-induced Tet-BZLF1/B95-8 cells were fixed in 4% paraformaldehyde and 70% ethanol. After digestion with RNase, they were treated with 50% formamide, air dried, and immediately hybridized with a mixture containing the EBV BAC DNA probe labeled with DIG nick translation mix. Specimens were stained with fluorescein isothiocyanate (FITC)-conjugated anti-DIG sheep (green) and anti-BMRF1 mouse (red) monoclonal antibodies. The right panel is a merged image. (C) Newly synthesized DNAs were labeled by incubation with 10 μM CldU added directly to the culture medium of lytic replication-induced Tet-BZLF1/B95-8 cells for 10 min at 24 h postinduction in the presence (b) or absence (a) of PAA (400 μg/ml). Specimens were stained with anti-BALF2 (green), anti-CldU (yellow), and anti-BMRF1 (red) antibodies.
In an independent experiment, the localizations of viral genome DNA and BMRF1 protein were simultaneously examined by means of combinational FISH and immunofluorescence. As shown in a representative image (Fig. 1B), the majority of viral genome DNA was localized inside BMRF1-localized regions. To support FISH analysis, we performed pulse-labeling analysis combined with immunofluorescence staining. The labeling reagent CldU, a nucleotide analog like BrdU, was added to the culture medium at 24 h postinduction and left for 10 min so that newly synthesized viral DNAs were labeled with incorporated CldU. As shown in Fig. 1C, CldU staining coincided with BMRF1- and BALF2-localized regions. In the presence of PAA, a herpesvirus DNA polymerase-specific inhibitor, CldU and BMRF1 signals were not observed (Fig. 1C). In the presence of the inhibitor, viral replication compartments are not formed and the expressed BMRF1 proteins are solubilized by the detergent treatment, while BALF2 proteins are distributed throughout nuclei as distinct spots (9). Overall, these results indicate that viral genome DNA is synthesized in the replication compartments, confirming the previous observations (9).
Spatial localization of viral replication proteins within replication compartments.
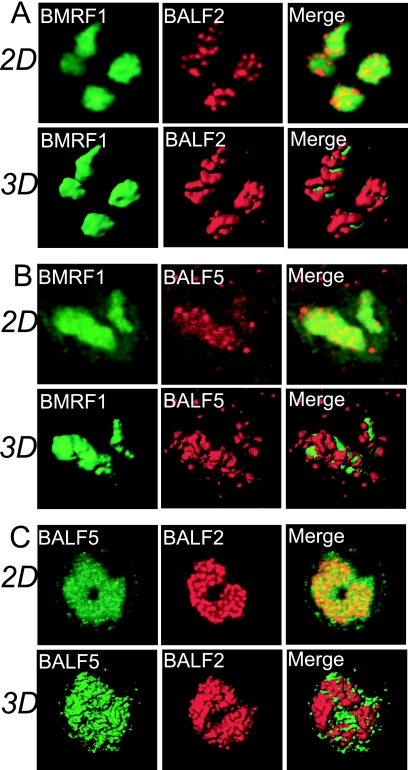
Confocal images are frequently presented as brightest point projections, but this format is not appropriate for demonstrating the spatial distribution of objects. To establish spatial relationships among viral replication proteins within replication compartments, 3D surface reconstruction imaging was employed. We used a 3D visualization and volume modeling software program, Imaris, to create images. We first examined the spatial distribution of BMRF1 relative to viral ssDNA binding protein BALF2 and viral DNA polymerase BALF5. Our previous work demonstrated colocalization of BALF5 with BALF2 as distinct spots within replication compartments, with the two proteins most likely cooperatively working at viral replication forks (9). As illustrated in a representative 3D image (Fig. 2A), each mass of BMRF1 protein appeared to be surrounded by BALF2 protein. We named the BMRF1-rich structures “BMRF1 cores.” It should be noted that BALF2 protein also localized inside the BMRF1 core, as observed in 2D images (Fig. 2A). Similarly, as shown in another representative 3D surface reconstruction image (Fig. 2B), each BMRF1 core was surrounded by BALF5 DNA polymerase, although BALF5 was also localized inside the BMRF1 core as shown in the 2D image (Fig. 2B). On the other hands, the BALF5 and BALF2 proteins colocalized well both outside and inside the BMRF1 core (Fig. 2). From the 3D surface reconstruction image it appeared that both proteins are mingled on their surfaces (Fig. 2C).
Fig. 2.
3D surface reconstruction imaging of viral replication compartments. (A) Laser scanning confocal images of BMRF1 and BALF2 proteins. Lytic replication-induced Tet-BZLF1/B95-8 cells were treated with 0.5% mCSK buffer, fixed with 70% ethanol, and stained with anti-BMRF1 (green) and anti-BALF2 (red) antibodies. The 2D images show brightest-point projections of 60 images collected at 0.26-μm steps in the z axis. The same data are displayed as 3D topographical reconstructions of BMRF1 and BALF2 proteins (left and middle panels, respectively). The right panel shows a 3D surface reconstruction image of both proteins showing the BMRF1 core covered by BALF2 proteins. (B) Laser scanning confocal images of BMRF1 (green) and BALF5 (red) proteins. The 2D images show brightest-point projections of 60 images collected at 0.26-μm steps in the z axis. The same data are displayed as 3D topographical reconstructions of BMRF1 and BALF5 proteins (left and middle panels, respectively). The right panel shows a 3D surface reconstruction image of both proteins showing the BMRF1 core covered by BALF5 Pol proteins. (C) Laser scanning confocal images of BALF5 (green) and BALF2 (red) proteins. The 2D images show brightest-point projections of 60 images collected at 0.26-μm steps in the z axis. The same data are displayed as 3D topographical reconstructions of BALF5 and BALF2 proteins (left and middle panels, respectively). The right panel shows a 3D surface reconstruction image of both proteins, showing that the BALF5 Pol proteins and BALF2 proteins are mingled.
Viral DNA genomes newly synthesized outside the BMRF1 core move to the inside.
We have previously demonstrated that the sites stained with anti-BMRF1 protein-specific antibodies coincided with the foci of 1-h-pulse-labeled viral DNA as judged by 5-bromodeoxyuridine (BrdU) incorporation and FISH analyses on confocal immunofluorescence analyses and that the BrdU-pulse-labeled DNA moved out of nucleus with time, clarifying that BrdU-labeled DNAs at 24 h postinduction are mostly viral and not cellular DNAs (10).
Our observation that the BMRF1 core was surrounded by a BALF2 ssDNA binding protein and the BALF5 DNA polymerase protein, whereas the majority of viral genomes were inside the core, let us hypothesize that viral DNAs are synthesized outside cores and then transported inside. This hypothesis fits well with the idea that BMRF1, a viral polymerase processivity factor, also protects newly synthesized viral genomes during lytic replication. To test the hypothesis, we performed pulse-chase labeling experiments to monitor the location of newly synthesized viral DNAs. The labeling reagent CldU, a nucleotide analog like BrdU, was added to cells at 24 h postinduction and left for 10 min so that newly synthesized viral DNAs were labeled with incorporated CldU. As shown in a representative image (Fig. 3A), pulse-labeled newly synthesized viral DNAs were localized mainly outside the BMRF1 core, although some existed inside. In contrast, when 1 h of chasing was included after CldU pulse-labeling, all of the labeled viral DNAs were localized inside the BMRF1 core (Fig. 3B). From the 3D surface reconstruction image it appeared that the BALF2 protein and pulse-labeled DNA were mingled (Fig. 3A, bottom panels). These results correspond well with the idea that BMRF1 protein assembles on newly synthesized DNAs to form the cores, and the cores progressively enlarge during the course of productive infection.
Fig. 3.
Newly synthesized viral DNA genomes localized outside the BMRF1 core move to the inside. Pulse-chase labeling experiments were performed with Tet-BZLF1/B95-8 cells at 24 h after induction. Outlines of the experimental protocol are given at the tops of panels A and B. (A) Tet-BZLF1/B95-8 cells were pulse-labeled with CldU for 10 min at 24 h postinduction. (B) The pulse-labeled cells were washed, and then chased for 1 h (B). For the 2D images, cells were treated with 0.5% mCSK buffer and stained with anti-BMRF1 or -BALF2 (green) and anti-CldU (red) antibodies. These images show brightest-point projections of 60 images collected at 0.26-μm steps in the z axis. The same data are displayed as 3D topographical reconstructions of the BMRF1 or BALF2 protein and CldU-labeled viral DNA (left and middle panels, respectively). The right panels show 3D surface reconstruction images.
HRR proteins are recruited to outside and inside BMRF1 cores.
We have previously reported that homologous recombinational repair (HRR) factors such as replication protein A (RPA), Rad51, Rad52, and the Mre11/Nbs1/Rad50 (MRN) complex are recruited and loaded onto the newly synthesized viral genome in replication compartments (23). Therefore, we determined the spatial localization of the HRR factors within these compartments. As shown in a representative image (Fig. 4A), Mre11, a component of the MRN complex, covered and also existed inside the BMRF1 cores. It is noteworthy that the spatial localization of Mre11 (Fig. 4A) resembles that of newly synthesized viral DNAs (Fig. 3A, pulse-labeling without chasing). Similarly, BRCA1, Rad52, and phosphorylated RPA32 at Ser-4/-8 covered the cores (Fig. 4B, C, and D), again resembling the localization of newly synthesized viral DNAs (Fig. 3A). Figure 4D shows relative localizations of BMRF1, BALF2, and phosphorylated RPA (pRPA) within the replication compartment of the same cell. BALF2 and pRPA were colocalized and surrounded the BMRF1 core. This observation corresponds well with our previous demonstration that knockdown of RPA32 and Rad51 by RNA interference significantly prevented viral DNA synthesis (23) and supports the idea that HRR somehow contributes to coordinated viral DNA replication.
Fig. 4.
Homologous recombinational repair proteins are located both outside and inside BMRF1 cores. (A to C) Lytic replication-induced Tet-BZLF1/B95-8 cells were treated with mCSK buffer, fixed with 70% ethanol, and stained with combinations of the indicated antibodies as follows: (A) anti-BMRF1 (green) and anti-Mre11 (red) antibodies; (B) anti-BMRF1 (green) and anti-BRCA1 ser-1524 (red) antibodies; (C) anti-BMRF1 (green) and anti-Rad52 (red) antibodies. The 2D images show brightest-point projections of 60 images collected at 0.26-μm steps in the z axis. The same data are displayed as 3D topographical reconstructions of each protein (left and middle panels, respectively). The right panels shows 3D surface reconstruction images of both proteins indicated, showing the BMRF1 core covered by HRR proteins. (D) The cells were stained with anti-BMRF1 (green), anti-BALF2 (blue), and anti-phosphorylated RPA32 Ser-4/Ser-8 (red) antibodies. Top panels, 2D images showing a brightest-point projection of 60 images collected at 0.26-μm steps in the z axis. Middle panels, each combination of the merged image of the top panels. Bottom panels, each combination of merged 3D surface reconstruction images.
MMR proteins are recruited and loaded inside BMRF1 cores.
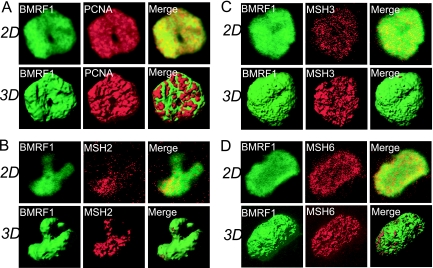
Next, the spatial localization of mismatch repair (MMR) proteins was determined. As shown in a representative image, the majority of PCNA was localized inside the BMRF1 cores, while some appeared on their surfaces (Fig. 5A). This is in contrast to the localization of HRR factors and MRN complexes (Fig. 4). Regarding the localization of MSH2, MSH3, and MSH6, unlike PCNA, they were almost completely localized inside the BMRF1 cores (Fig. 5B, C, and D). Such a difference in localization implies that MMR factors are last to be loaded onto viral DNAs, whereas the BMRF1 protein and PCNA are first loaded on newly synthesized viral DNA, resulting in enlargement of the BMRF1 core. Because MSH3 and MSH6 interact with PCNA, such loading might trigger transfer of a series of host MMR proteins to sites of viral DNA maturation. Taken together, the results indicate that MMR of the viral DNA genome might occur after BMRF1 proteins bind to a viral DNA and contribute to a mature intact viral genome.
Fig. 5.
Mismatch repair proteins such as PCNA, MSH2, MSH3, and MSH6 localize inside BMRF1 cores. Lytic replication-induced Tet-BZLF1/B95-8 cells were treated with mCSK buffer, fixed with 70% ethanol, and stained with combinations of antibodies as follows: (A) anti-BMRF1 (green) and anti-PCNA (red) antibodies; (B) anti-BMRF1 (green) and anti-MSH2 (red) antibodies; (C) anti-BMRF1 (green) and anti-MSH3 (red) antibodies; (D) anti-BMRF1 (green) and anti-MSH6 (red) antibodies. The 2D images show brightest-point projections of 60 images collected at 0.26-μm steps in the z axis. The same data are displayed as a 3D topographical reconstructions of each protein (left and middle panels, respectively). The right panels show 3D surface reconstructions image of both proteins indicated, showing MMR proteins located inside the BMRF1 core.
DISCUSSION
Previous cross-sectional studies of EBV replication compartments by confocal laser microscopy demonstrated EBV replication proteins to be localized at viral replication compartments, also revealing variation in staining pattern between individual proteins (9). Here, the BMRF1 protein, which is multifunctional, showed a homogenous, not dot-like, distribution. This is evidence of BMRF1-rich structures (BMRF1 cores) existing within viral replication compartments. Three-dimensional surface reconstruction imaging revealed that replication compartments are partitioned into two subdomains, inside and outside the BMRF1 core. This approach further revealed factories for viral genome synthesis and maturation mediated by HRR and MMR host factors. The BALF5 DNA polymerase and BALF2 ssDNA binding protein were almost colocalized outside the cores. Since both proteins are thought to form an item of the viral elongation machinery acting at replication forks on the replicating EBV genome, they would position at sites of viral genome synthesis. Pulse-chase experiments indicated that viral DNA genomes were synthesized mainly outside BMRF1 cores, where HRR factors were localized, with subsequent movement into the cores. On the other hand, MMR proteins were recruited to viral DNAs exclusively inside, indicating that loading of MMR proteins onto viral DNA is likely to occur at the step of viral genome maturation. These observations let us propose a new model of viral DNA synthesis and maturation as follows. First, viral DNA genomes are synthesized by viral replication machinery, which is coupled with homologous recombination with the help of host HRR factors. Next, BMRF1 proteins bound to the newly synthesized viral DNA are assembled to form cores. During the progression of viral replication, the size of the core increases. Inside, MMR factors are loaded through PCNA to repair and mature newly synthesized viral DNA. The observed sequential loading of HRR factors and MMR proteins on newly synthesized viral DNA implies that spatially and temporarily different repair mechanisms are working during EBV genome maturation.
From our recent resolution of the crystal structure of C-terminally truncated BMRF1 protein (32), the molecular structure shares structural similarity with other processivity factors, such as HSV-1 UL42, HCMV UL44, and human proliferating cell nuclear antigen (PCNA). Although the crystal structure of BMRF1 indicates an interesting tetramer ring formation, electrophoresis and sedimentation assay suggested that the main component of EBV BMRF1 in solution is a head-to-head dimer. Tail-to-tail association of the dimers forms the ring structure. Replication of a recombinant virus with a point mutation at C206E of BMRF1 (which is expected to impair tail-to-tail contact) is severely restricted (34), although the mutant protein possesses the same in vitro biochemical activities as the wild type (32), indicating that its tetrameric ring formation might be essential for EBV replication. PCNA proteins adopt a ring-shaped trimer conformation with the head-to-tail contacts, forming a central channel to accommodate the template DNA (6). The BMRF1 ring-shaped structure (30, 32) is almost twice as large as the previously reported PCNA ring structure. In contrast, HSV-1 UL42 stably exists as a monomer (35), whereas HCMV UL44 forms a head-to-head-contacting C-shaped dimer in the crystal structure (1). The BMRF1 dimer formation overlaps the UL44 dimer. Substitutions of positively charged residues on the concave surface of the BMRF1 C-shaped dimer reduce DNA binding affinity. Furthermore, an amino acid mutation disrupting the dimer formation, C95E, results in no DNA binding (32). Another study of dimerized processivity factor HCMV UL44 also indicates that DNA binding affinity is related to dimer formation (1). Thus, the basic concave surface is very important for DNA binding by the dimerized proteins BMRF1 and UL44. UL44 is consequently thought to bind to DNA like PCNA, which surrounds DNA by ring formation (21). The BMRF1 protein is abundantly expressed in the lytic infected cells and is distributed homogenously, not in a dot-like pattern, within the replication compartments. We speculate that the tetrameric ring form of BMRF1 might be involved in formation of the BMRF1 cores within the replication compartments to protect the synthesized viral DNA by occupying the surfaces of the DNA molecules.
In general, processivity factors are associated with their cognate DNA polymerases on the template during DNA replication. These proteins are also known as “sliding clamps,” including PCNA, which interacts with DNA polymerase δ or ε. However, the herpesvirus polymerase processivity factors display different molecular assemblies to cognate viral DNA polymerase. The HSV-1 UL42 form a heterodimer with the UL30 DNA polymerase (35). Mutational analyses of BMRF1 revealed that the monomer form of EBV BMRF1 can function as polymerase processivity factor in vitro (32), suggesting that BMRF1 interacts with BALF5 DNA polymerase to form a heterodimer like UL42. Thus, the BMRF1 protein adopts different subunit architecture during the replication process.
HRR is a repair system for double-strand breaks (DSBs), essential for cellular survival in eukaryotes, which relies on several proteins, including Rad-51, -52, and -54, the Mre11/Rad50/Nbs1 (MRN) complex, RPA, and BRCA1/2 (5, 28). First, the MRN complex searches DSBs and generates 3′ ssDNA by removing the 5′ ends of the DSB regions, followed by phosphorylated RPA binding to 3′ ssDNA strands. Rad52, BRCA1, and BRCA2 replace RPA with Rad51. Both Rad51 and Rad52 bind specifically to the terminal region of tailed duplex DNA, the substrate thought to initiate recombination by promoting homologous pairing and strand transfer reactions in vivo (12, 16, 38). HRR is also required to repair replication-associated DNA lesions and replication fork stalling or collapse (31, 39). In eukaryotic cells, HRR also occurs coupled with replication at replication forks to ensure proper replication and prevent genomic instability (11). Considering these results and our present study, the host HRR system might be used, being necessary for efficient viral replication coupled with viral genome replication.
It has recently been reported that the HSV-1 alkaline exonuclease UL12 and the single-stranded-DNA binding protein ICP8 interact with each other, are recruited to replication compartments, and together mediate strand exchange in vitro, suggesting a role as a two-component recombinase reminiscent of the lambda Red α/β recombination system (3). Further, RPA, Mre11, Rad50, Nbs1, and Rad51 are recruited to HSV-1 replication compartments as in the case of EBV (46). These viral and cellular proteins might together be involved in homologous recombinational repair of herpesvirus genome DNA maturation. However, unlike in the case of EBV (23), there is no induction of hyperphosphorylation of RPA upon productive HSV-1 infection. Instead, endogenous hyperphosphorylated RPA is sequestered away from replication compartments into discrete nuclear foci (VICE domains) that are enriched for cellular components involved in protein folding and degradation (45). Since those authors never observed either induction of RPA hyperphosphorylation or recruitment of phosphorylated RPA to HSV-1 replication compartments, they concluded that these signaling molecules are excluded from sites that contain viral DNA. Although the discrepancy remains unclear, hyperphosphorylated RPA32, Rad51, Rad52, and MRN complex were recruited to the EBV replication compartments and interacted with EBV genomic DNA (23). HRR represents an error-free subpathway of damage tolerance, allowing replicational bypass of lesions through a template switch. The EBV genome is amplified to several hundred copies during lytic infection, and further, the small interfering RNA (siRNA) depletion of Rad51 or RPA32 reduces new viral DNA synthesis (23). We propose that HRR occurs during EBV lytic genome replication with the aid of host cellular HRR factors and that the HRR factors might help not only to increase the fidelity of viral replication but also to facilitate viral genome replication.
On the other hand, MMR contributes to recognize and repair DNA mismatches that are generated during chromatin DNA replication in eukaryotic cells (20), correcting 99% of such lesions. Because MMR reduces the number of replication-associated errors, defects in MMR increase the spontaneous mutation rate. The MMR machinery works as a postreplication repair near a replication fork. The first step of MMR is that MSH2-MSH6 or MSH2-MSH3 searches for and recognizes a mismatch region through interaction with the β-clamp accessory protein PCNA (14, 18). A MLH1-PMS2 complex is then recruited, and the mismatch region is removed by endonuclease activity of MLH1-PMS2 and exonuclease (Exo1) (17, 25). Thus, this MMR machinery works as a postreplication repair near a replication fork. Our present data show that some MMR proteins contribute to EBV genome maturation after replication. High-copy replication with high fidelity might require the host MMR machinery to repair produced viral genomes.
ACKNOWLEDGMENTS
We thank Ayumi Kudoh (Salk Institute) and Tohru Daikoku (Toyama University) for discussions and Keizo Tomonaga, Yusuke Matsumoto, and Hiroko Omori (Osaka University) and Y. Tanaka (Nagoya University) for technical suggestions.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science, Sports, Culture and Technology of Japan (no. 20390137 and 21022055 to T.T.) and partly by the Uehara Memorial Research Fund (to T.T.).
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Appleton B. A., Loregian A., Filman D. J., Coen D. M., Hogle J. M. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233–244 [DOI] [PubMed] [Google Scholar]
- 2. Baer R., et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 [DOI] [PubMed] [Google Scholar]
- 3. Balasubramanian N., Bai P., Buchek G., Korza G., Weller S. K. 2010. Physical interaction between the herpes simplex virus type 1 exonuclease, UL12, and the DNA double-strand break-sensing MRN complex. J. Virol. 84:12504–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bataille D., Epstein A. 1994. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology 203:384–388 [DOI] [PubMed] [Google Scholar]
- 5. Benson F. E., Baumann P., West S. C. 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391:401–404 [DOI] [PubMed] [Google Scholar]
- 6. Bruck I., O'Donnell M. 2001. The ring-type polymerase sliding clamp family. Genome Biol. 2:REVIEWS3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho M. S., Milman G., Hayward S. D. 1985. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J. Virol. 56:860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark A. B., Valle F., Drotschmann K., Gary R. K., Kunkel T. A. 2000. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J. Biol. Chem. 275:36498–36501 [DOI] [PubMed] [Google Scholar]
- 9. Daikoku T., et al. 2005. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. J. Virol. 79:3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daikoku T., et al. 2006. Postreplicative mismatch repair factors are recruited to Epstein-Barr virus replication compartments. J. Biol. Chem. 281:11422–11430 [DOI] [PubMed] [Google Scholar]
- 11. Delacote F., Lopez B. S. 2008. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle 7:33–38 [DOI] [PubMed] [Google Scholar]
- 12. Errico A., Costanzo V. 2010. Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns. EMBO Rep. 11:270–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fixman E. D., Hayward G. S., Hayward S. D. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flores-Rozas H., Clark D., Kolodner R. D. 2000. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 26:375–378 [DOI] [PubMed] [Google Scholar]
- 15. Fujita N., et al. 1999. Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta 208:283–293 [DOI] [PubMed] [Google Scholar]
- 16. Gottipati P., Helleday T. 2009. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis 24:203–210 [DOI] [PubMed] [Google Scholar]
- 17. Hoeijmakers J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366–374 [DOI] [PubMed] [Google Scholar]
- 18. Jiricny J. 2000. Mediating mismatch repair. Nat. Genet. 24:6–8 [DOI] [PubMed] [Google Scholar]
- 19. Kleczkowska H. E., Marra G., Lettieri T., Jiricny J. 2001. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 15:724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolodner R. D., Marsischky G. T. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89–96 [DOI] [PubMed] [Google Scholar]
- 21. Komazin-Meredith G., et al. 2008. The human cytomegalovirus UL44 C clamp wraps around DNA. Structure 16:1214–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kudoh A., et al. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 77:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kudoh A., et al. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuzminov A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 98:8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li G. M. 2008. Mechanisms and functions of DNA mismatch repair. Cell Res. 18:85–98 [DOI] [PubMed] [Google Scholar]
- 26. Li J. S., et al. 1987. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J. Virol. 61:2947–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lilley C. E., Carson C. T., Muotri A. R., Gage F. H., Weitzman M. D. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lundin C., et al. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 22:5869–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo M. H., Rosenke K., Czornak K., Fortunato E. A. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Makhov A. M., Subramanian D., Holley-Guthrie E., Kenney S. C., Griffith J. D. 2004. The Epstein-Barr virus polymerase accessory factor BMRF1 adopts a ring-shaped structure as visualized by electron microscopy. J. Biol. Chem. 279:40358–40361 [DOI] [PubMed] [Google Scholar]
- 31. Michel B., et al. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 98:8181–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murayama K., et al. 2009. Crystal structure of Epstein-Barr virus DNA polymerase processivity factor BMRF1. J. Biol. Chem. 284:35896–35905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakayama S., et al. 2009. Epstein-Barr virus polymerase processivity factor enhances BALF2 promoter transcription as a coactivator for the BZLF1 immediate-early protein. J. Biol. Chem. 284:21557–21568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakayama S., et al. 2010. Tetrameric ring formation of Epstein-Barr virus polymerase processivity factor is crucial for viral replication. J. Virol. 84:12589–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Randell J. C., Coen D. M. 2004. The herpes simplex virus processivity factor, UL42, binds DNA as a monomer. J. Mol. Biol. 335:409–413 [DOI] [PubMed] [Google Scholar]
- 36. Shirata N., et al. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336–30341 [DOI] [PubMed] [Google Scholar]
- 37. Taylor T. J., Knipe D. M. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thompson L. H., Hinz J. M. 2009. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat. Res. 668:54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toueille M., Hubscher U. 2004. Regulation of the DNA replication fork: a way to fight genomic instability. Chromosoma 113:113–125 [DOI] [PubMed] [Google Scholar]
- 40. Tsurumi T. 1993. Purification and characterization of the DNA-binding activity of the Epstein-Barr virus DNA polymerase accessory protein BMRF1 gene products, as expressed in insect cells by using the baculovirus system. J. Virol. 67:1681–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsurumi T., Daikoku T., Kurachi R., Nishiyama Y. 1993. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J. Virol. 67:7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tsurumi T., et al. 1998. Overexpression, purification and helix-destabilizing properties of Epstein-Barr virus ssDNA-binding protein. J. Gen. Virol. 79:1257–1264 [DOI] [PubMed] [Google Scholar]
- 43. Tsurumi T., et al. 1993. Functional expression and characterization of the Epstein-Barr virus DNA polymerase catalytic subunit. J. Virol. 67:4651–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weber P. C., Challberg M. D., Nelson N. J., Levine M., Glorioso J. C. 1988. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell 54:369–381 [DOI] [PubMed] [Google Scholar]
- 45. Wilkinson D. E., Weller S. K. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilkinson D. E., Weller S. K. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie Y., Counter C., Alani E. 1999. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics 151:499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Q., et al. 1996. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J. Virol. 70:5131–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang X., Efstathiou S., Simmons A. 1994. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology 202:530–539 [DOI] [PubMed] [Google Scholar]