Abstract
Interferon is a principal component of the host antiviral defense system. In this study, abortive focus formation by Japanese encephalitis virus (JEV) in primate cells was accompanied by early interferon induction, while productive focus formation in porcine cells was associated with a late interferon response. Neutralization antibodies against interferon relieved the restricted infection in primate cells, and increasingly larger foci were generated as treatment with exogenous interferon was delayed, thereby establishing a solid correlation between interferon response and viral dissemination. However, delayed interferon induction in JEV-infected porcine cells occurred in the absence of active inhibition by the virus. We further demonstrated that JEV mediates interferon activation through double-stranded RNA and cytosolic pattern recognition receptors. Immunofluorescence and subcellular fractionation studies revealed that double-stranded RNA is concealed in intracellular membranes at an early phase of infection but eventually appears in the cytosol at later periods, which could then allow detection by cytosolic pattern recognition receptors. Interestingly, cytosolic exposure of double-stranded RNA was delayed in porcine cells compared to primate cells, independent of total double-stranded RNA levels and in correlation with the timing of the interferon response. Furthermore, when double-stranded RNA was artificially introduced into the cytosol of porcine cells, more rapid and robust interferon activation was triggered than in viral infection. Thus, cytosolic exposure of JEV double-stranded RNA is imperative for interferon induction, but in cell lines (e.g., porcine cells) with delayed emergence of cytosolic double-stranded RNA, the interferon response is late and viral dissemination is consequently enhanced.
INTRODUCTION
Japanese encephalitis virus (JEV) is a member of the genus Flavivirus of the family Flaviviridae. JEV is an enveloped virus that harbors a single-stranded positive-sense RNA but produces three types of viral RNA when replicating inside the host cell: the genomic RNA, the semi-double-stranded replicative intermediate (RI), and the double-stranded replicative form (RF) (23). The 11-kb viral genome encodes three structural and seven nonstructural proteins.
JEV is transmitted to humans and animals by mosquitoes, primarily the Culex tritaeniorhynchus group (26, 44). However, humans are dead-end hosts because of low-level and transient viremia. JEV infection in humans is mainly asymptomatic, although severe cases of the disease are the predominant cause of encephalitis incidence in eastern and southern Asia. In view of its low symptomatic-to-asymptomatic ratio, which ranges from 1:25 to 1:1,000, it is believed that adaptive immunity controls JEV replication before the virus invades the blood-brain barrier and establishes an infection in the central nervous system (26). Consistent with this, low-level IgM and IgG titers are associated with enhanced viremia and mortality (18). Hence, vaccination is considered the most reliable method for preventing Japanese encephalitis (9, 44). However, no antiviral therapeutic agent has been developed for the disease (9).
JEV infection among swine is characterized differently. Pigs are very susceptible to JEV, with a rate of natural infection that reaches 98 to 100% and high-titer viremia that lasts for 2 to 4 days (44). As such, pigs are considered to be the main amplification host of JEV. Considering that synchronous infection of pigs has led to significant transmission of JEV to humans (33), targeting the amplifying host is a rational strategy for control and prevention of human cases. The live-inactivated vaccine is not recommended for piglets less than 6 months of age due to neutralization by maternal antibodies, but most pigs live only up to 6 to 8 months before they are slaughtered (13). Hence, controlling JEV infection of swine requires a different approach.
The distinction between humans and pigs in terms of JEV susceptibility implicates various factors that are critical for viral propagation. Elucidating these factors could facilitate the development of alternative strategies for the prevention and treatment of Japanese encephalitis. Among these factors are virus-host interactions that modulate the innate immune system in order to direct the outcome of an infection. The interferon (IFN) system is the first and one of the most important antiviral defense mechanisms of the host. It is divided into two major pathways: (i) IFN activation and (ii) IFN signaling. The IFN activation pathway involves a group of viral RNA-sensing molecules called pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), RIG-I, and MDA5 (1, 32). When a PRR recognizes a specific type of viral RNA (e.g., single stranded versus double stranded, short versus long, or cytosolic versus endosomal), a cascade of signaling occurs to mediate the transcription of beta interferon (IFN-β) (1). IFN-β constitutes the primary wave of the IFN system and is critical for initiation of immune responses. During IFN signaling, IFN-β binds to type I IFN receptors and stimulates the JAK/STAT signal transduction pathway, leading to the expression of other antiviral genes (interferon-stimulated genes [ISGs]) and, eventually, IFN-α (32). This second wave amplifies the IFN response and is responsible for the establishment of an antiviral state. While the general pathway of the IFN system has already been completely laid out, virus-specific mechanisms are still being elucidated (14, 25). Dissecting these virus-specific pathways is crucial for investigating virus-host interactions.
The rapid response and efficiency of the host to establish and maintain an antiviral state puts viruses at a disadvantage. Nevertheless, viruses have developed various defense mechanisms to frustrate the host immune system. Subversion of IFN signaling by flaviviruses is a well-researched phenomenon (20, 21, 22, 29, 40), but modulation of IFN activation deserves equally profound investigation. In this study, we observed that JEV developed plaque-forming foci in porcine cells, but focus formation was abortive in primate cells. We therefore investigated IFN responses in these cells and their influence on the intercellular spread of JEV. We report for the first time a delayed activation of IFN in response to JEV infection of porcine cells, in contrast to the immediate response in primate cells. We further show that focus formation is regulated by IFN, and the timing of the IFN response is critical for this purpose. However, JEV did not actively suppress IFN induction in porcine cells. Examination of the IFN pathway confirmed cytosolic PRRs as the primary immune sensors for JEV and double-stranded RNA (dsRNA) as the major IFN-activating factor of JEV. Even though the majority of the dsRNA was concealed among intracellular membranes, its eventual exposure to the cytosol was crucial for initiating the antiviral response. JEV dsRNA emerged later in the cytosol of porcine cells than in that of primate cells, although total levels were similar in the two cell types. Hence, delayed cytosolic exposure of dsRNA is most likely responsible for the impaired IFN activation in porcine cells. This implies a role for cell/species-specific IFN response in differential host susceptibility, such as in pigs and humans, a hypothesis that should be evaluated in future studies.
MATERIALS AND METHODS
Cells and viruses.
Rhesus monkey kidney LLC-MK2 and Vero; human epithelial HeLa; porcine kidney PS, PK, and ESK; baby hamster kidney BHK-21; and mosquito C6/36 cells were maintained in minimum essential medium (MEM) supplemented with 10% fetal bovine serum and 0.2 mM nonessential amino acids. Meanwhile, porcine kidney LLC-PK1 cells were maintained in 199 medium (MP Biomedicals) with 5% fetal bovine serum. The cells were allowed to grow at 37°C with 5% CO2 (28°C with 5% CO2 for C6/36 cells). The Japanese encephalitis virus strain JaOArS982 was isolated from a Culex mosquito pool in Osaka, Japan, in 1982 (39). Japanese encephalitis virus and vesicular stomatitis virus (VSV) were propagated in C6/36 and BHK21 cells, respectively, to generate working stocks. Virus stocks were stored at −80°C.
Viral infection.
To allow equal infection among different cell types, the titer of viral stocks was determined in each cell line and used to calculate the multiplicity of infection (MOI). Monolayers of the indicated cells were adsorbed with JEV at specified MOIs for 90 min at 37°C. The unbound virus was removed from cells by gentle washing with medium, after which the cells were incubated in culture medium at 37°C with 5% CO2. The supernatants were harvested at the indicated times and stored at −80°C for future use (growth curve analysis and focus size reduction assay).
Focus formation and virus titration.
Focus formation was performed on various cells, while JEV focus titration and VSV plaque titration were done in C6/36 and BHK21 cells, respectively. A 10-fold dilution series of viral stocks was used for infection, and then an overlay of MEM with 2% fetal calf serum (FCS) and 1.25% methylcellulose was added after viral adsorption. The cells were incubated at 37°C with 5% CO2 (LLC-MK2, Vero, HeLa, PS, PK, LLC-PK1, and BHK-21 cells) or 28°C with 5% CO2 (C6/36 cells) until focus staining, which was performed as described previously (17). For plaque titration, cells were stained with crystal violet after fixation.
The antiviral activities of supernatants and recombinant human IFN-β 1a (PBL Interferon Source) was evaluated using the focus size reduction assay. Supernatants from virus-infected or uninfected cells were exposed for 5 min to 120 mJ/s UV light (UV) using GS Gene Linker (Bio-Rad) prior to the assay. After viral adsorption by the cellular monolayer in 24-well plates, the medium was replaced with the UV-inactivated supernatant or recombinant IFN-β and overlaid with MEM containing 2% FCS and 1.25% methylcellulose until focus staining was performed. For neutralization experiments, the UV-inactivated supernatant was incubated with human anti-IFN-β antibody (Abcam) overnight at 4°C prior to application. In other studies, cells seeded in 96-well plates were pretreated with an anti-IFN antibody cocktail containing 2,000 neutralization U/ml anti-human IFN-β (PBL Interferon Source) and 20 μg/ml anti-human IFN-α/βR2 (CD118) (PBL Interferon Source) for 1 h prior to viral challenge, maintained with the same concentrations throughout the infection, and then focus stained at the indicated number of days postinfection.
Real-time quantitative reverse transcription (RT)-PCR.
Total RNA was harvested from cells using the RNeasy Mini Kit (Qiagen). One microgram RNA in a total volume of 20 μl was reverse transcribed with Superscript III (Invitrogen) using the Oligo(dT)12-18 primer (Invitrogen) or random primers (Invitrogen) for cellular mRNA and JEV RNA, respectively. Three microliters of the product was used for SYBR green real-time PCR (Applied Biosystems), which was performed in triplicate. Primer sequences for the target genes are listed in Table 1. Cellular mRNA expression was measured via relative quantification using the Pfaffl method (35). The number of copies of JEV RNA was calculated using absolute quantification based on in vitro-transcribed JEV RNA standards. All values were normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin for primate and porcine cells, respectively.
Table 1.
Sequences of real-time RT-PCR primers
| Gene | Primer orientation | Primer sequence (5′ to 3′) |
|---|---|---|
| JEV NS5 | Forward | CAACATGATGGGAAAAAGAGAAAAGAAGC |
| Reverse | TGCTCCAAGCCACATGAACCAAAT | |
| Primate IFN-β | Forward | TGCCTCAAGGACAGGATGAAC |
| Reverse | GCGTCCTCCTTCTGGAACTG | |
| Primate GAPDH | Forward | AAATCAAGTGGGGCGATGCTG |
| Reverse | CAAATGAGCCCCAGCCTTCTC | |
| Porcine IFN-βa | Forward | AGTTGCCTGGGACTCCTCAA |
| Reverse | CCTCAGGGACCTCGAAGTTCAT | |
| Porcine β-actina | Forward | CATCACCATCGGCAACGA |
| Reverse | GCGTAGAGGTCCTTCCTGATGT |
Source, Moue et al. (28).
Transfection and quantitation of total RNA.
Total cellular RNA was obtained using the hot-phenol RNA extraction method in order to preserve dsRNA structures. Cells were lysed in TE buffer (10 mM Tris, pH 7.5, 10 mM EDTA) with 0.5% SDS, mixed with acid phenol-chloroform (5:1; Sigma-Aldrich), incubated at 65°C for 20 min, and then cooled at −80°C for 30 min. After centrifugation at 20,000 × g, RNA isolation was continued using phenol-chloroform extraction, and the RNA was subjected to DNase treatment (Qiagen). Finally, the RNA was purified by performing another round of phenol-chloroform extraction. Total RNA was subjected to viral RNA quantitation by real-time RT-PCR or transfected into cells using Lipofectamine LTX (Invitrogen). In other experiments, total RNA was incubated with RNase III or RNase R (Epicentre Biotech) at 37°C for 6 h prior to use.
Quantitation of double-stranded RNA.
Double-stranded RNA was quantified using a solid-phase immunosorbence-based assay, as described previously (16). Briefly, MaxiSorp Immuno 96-microwell solid plates (Nunc) were coated with K1 anti-dsRNA antibody (English and Scientific Consulting Bt.) diluted at 1.25 μg/ml in coating buffer (15 mM sodium carbonate, 35 mM sodium bicarbonate, and 3 mM sodium azide, pH 9.6) and incubated at 4°C overnight. After three washes with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST), RNA samples diluted in PBST with 1 U/μl RNasin (Promega) were loaded and incubated for 2 h at 37°C. A portion of each diluted RNA sample was reserved as the input RNA and left untreated. The plates were washed three times with PBST, and RNA was harvested by treatment with 50 μg/ml proteinase K in 0.5% sodium dodecyl sulfate for 1 h at 37°C. Finally, RNA was reextracted using the RNeasy Mini Kit and subjected to real-time quantitative RT-PCR using JEV-specific primers.
Subcellular fractionation.
Cytosolic fractions were isolated as described by Hardy et al. (10), with slight modification. Briefly, uninfected or JEV-infected cells were harvested, counted, and subjected to low-speed centrifugation (900 × g). The cell pellets were resuspended in 200 μl hypotonic buffer (10 mM Tris-HCl, pH 7.8, and 10 mM NaCl), allowed to swell on ice for 15 min, and then disrupted by passing them 20 times each through 21- and 26-gauge needles. The resulting homogenate was centrifuged at 900 × g and 4°C for 5 min to pellet the nuclei. The remaining supernatant was centrifuged again at 16,000 × g and 4°C for 10 min to pellet the intracellular membranes. RNA from the cytosolic supernatant (10 μl) was extracted using the RNeasy Mini Kit and used for quantitative real-time RT-PCR of JEV RNA. The number of viral RNA copies per 1 × 106 cells was calculated based on the original number of cells harvested. To perform the solid-phase immunosorbent assay for dsRNA quantitation, phenol-chloroform-extracted RNA from the cytosolic extract was used, and the number of viral RNA copies per 100 ng total RNA was calculated. In other studies, RNA was isolated from the intracellular membrane pellet of 4 × 106 cells using the hot-phenol method of RNA extraction, and its dsRNA content was evaluated by the solid-phase immunosorbent assay for dsRNA.
Immunoblotting.
Cells were resuspended in lysis buffer (0.5% Nonidet-P40, 1 mM CaCl2, and 2 mM MgCl2 in PBS) with HALT Protease Inhibitor Cocktail (Thermo Scientific) and incubated on ice for 30 min. After centrifugation at 20,000 × g for 30 min, the supernatant was mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiled at 95°C for 3 min. Equal volumes of samples were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with appropriate antibodies. The primary antibodies used were anti-RIG-I, anti-MDA5, anti-MAVS, and anti-β-actin antibodies (Santa Cruz Biotechnology). Protein bands were developed using DNA Thunder Chemiluminescent Reagent (Perkin Elmer LAS) and detected using a LAS-4000 mini-luminescent-image analyzer (Fujifilm).
Indirect immunofluorescence assay.
JEV infection was performed on a monolayer of cells on a 16-well chamber slide. At the indicated times, the cells were subjected to immunostaining. Briefly, the cells were fixed with 2% paraformaldehyde, permeabilized with 1% Nonidet-P40, blocked, and stained. Streptolysin O permeabilization was conducted as described previously (30). The primary antibodies used were as follows: K1 mouse anti-dsRNA antibody (English and Scientific Consulting Bt.), 12D11/7E8 mouse anti-flavivirus E monoclonal antibody (MAb), and goat anti-calregulin antibody (Santa Cruz Biotechnology). Alexa 488-conjugated anti-rabbit, Alexa 488-conjugated anti-goat, or Alexa 549-conjugated anti-mouse antibody (Invitrogen) was used for secondary labeling. For double staining using mouse antibodies, MAb 12D11/7E8 was converted to a rabbit antibody by coating it with a Fab fragment rabbit anti-mouse antibody (Jackson Immunoresearch Laboratories) prior to staining it with a secondary antibody according to the manufacturer's protocol. Finally, cells were incubated with DAPI (4′,6-diamidino-2-phenylindole) for nuclear counterstaining. Images were captured using the Leica TCS SP2 confocal microscope (Leica Microsystems). The percentage of cells positively staining for double-stranded RNA was calculated in five different fields.
RESULTS
Focus formation by JEV is associated with the timing of IFN expression.
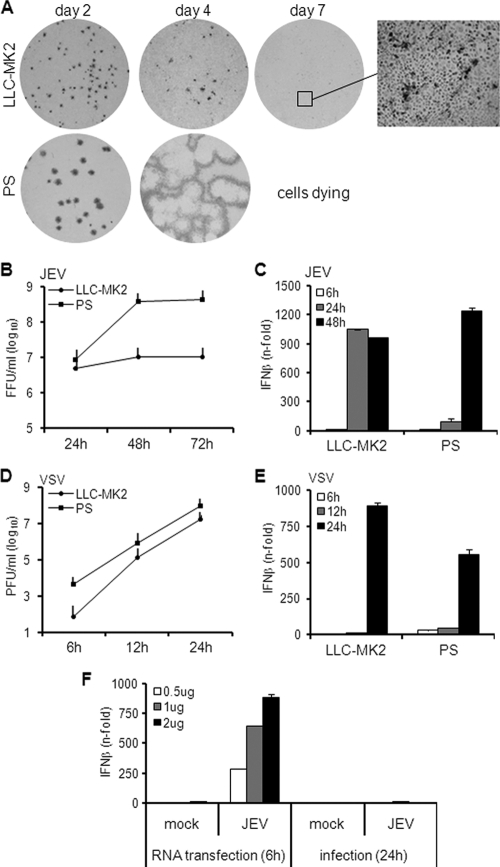
JEV typically develops large foci in several cell lines, but we noticed an abortive type of focus formation in the rhesus monkey LLC-MK2 cell line. Figure 1A demonstrates a minute focus size of JEV in LLC-MK2 cells, which was reduced to a single-cell infection level by day 7 postinfection (p.i.). On the other hand, JEV foci not only dramatically increased by day 4 p.i. in the porcine PS cell line (Fig. 1A), but also demonstrated plaque formation. Most of the cells were already dying by day 7 due to viral cytopathic effect (data not shown).
Fig. 1.
Focus formation, growth kinetics, and IFN activation by JEV in rhesus and porcine cells. (A) LLC-MK2 and PS cells were infected with JEV, and foci were detected by immunostaining them on the indicated days postinfection. The inset was obtained using ×100 magnification. (B) Viral titer of supernatants from JEV-infected cells (MOI, 1) at the indicated hours postinfection. The error bars indicate standard deviations of the means. (C) IFN-β mRNA expression in cells infected with JEV at an MOI of 1 was quantified via real-time quantitative RT-PCR. The results are expressed as fold increase over mock-treated cells and normalized to GAPDH and β-actin for LLC-MK2 and PS cells, respectively. The error bars indicate standard deviations of the means. (D) Viral titer of supernatants from VSV-infected cells (MOI, 0.1) at the indicated hours postinfection. The error bars indicate standard deviations of the means. (E) IFN-β mRNA expression in cells infected with VSV at an MOI of 0.1 was quantified via real-time quantitative RT-PCR. The results for IFN-β mRNA are expressed as the fold increase over mock-treated cells and normalized to GAPDH and β-actin for LLC-MK2 and PS, respectively. (F) Total RNA from mock-treated or JEV-infected LC-MK2 cells was transfected for 6 h in PS cells at 0.5, 1, or 2 μg. Another set of cells was mock treated or infected with JEV at an MOI of 1 for 24 h. IFN activation was measured by real-time quantitative RT-PCR. The results for IFN-β mRNA are expressed as the fold increase over mock-treated cells and normalized to β-actin. The error bars indicate standard deviations of the means.
Abortive focus formation by JEV in LLC-MK2 cells may be due to its inability to replicate in that cell line. Therefore, a viral growth curve was constructed to explore this possibility. In Fig. 1B, high yet steady levels of infectious particles were produced in LLC-MK2 cells. In contrast, JEV established a higher peak titer in PS cells by 48 h p.i. (Fig. 1B). Thus, JEV replication occurs in LLC-MK2 cells, although the viral titers are controlled. In PS cells, replication is less restricted, leading to higher titers. These results are consistent with the previous data (Fig. 1A), in which focus formation by JEV in LLC-MK2 cells is limited but is unhampered in PS cells. Furthermore, production of infectious particles in LLC-MK2 cells implies that other factors are responsible for the impaired development of foci.
This led us to the hypothesis that focus formation by JEV in LLC-MK2 cells is restrained due to the antiviral IFN response of the host cells. To test this hypothesis, the IFN response was monitored via real-time quantitative RT-PCR of IFN mRNA during JEV infection. As shown in Fig. 1C, JEV triggered a significant increase in IFN transcription in LLC-MK2 cells by 24 h p.i. When the IFN response in the PS cell line was evaluated, elevated levels of IFN mRNA were also observed, but the response was delayed until 48 h p.i. (Fig. 1C). Thus, JEV triggers IFN activation in LLC-MK2 and PS cells, but the timing of the host response is different for the two cell lines. The controlled replication status of JEV in LLC-MK2 cells may be associated with the early IFN response, while productive growth in PS cells is probably due to the late IFN response, as demonstrated in later experiments.
The IFN response of the cells to another virus, VSV, was checked, and VSV induced high levels of IFN expression in both LLC-MK2 and PS cells by 24 h p.i. (Fig. 1E), with fast growth kinetics (Fig. 1D). This suggests that impaired IFN activation in PS occurs specifically for JEV. Therefore, we tested the ability of PS cells to recognize JEV RNA. As shown in Fig. 1F, transfection of RNA from mock-infected cells did not induce IFN activation, but RNA from JEV-infected cells induced IFN expression to a significant degree within 6 h of treatment of PS cells. This is in stark contrast to a 24-hour infection by JEV, which induced only a minimal IFN response (Fig. 1F). Thus, PS cells are perfectly capable of a rapid and robust IFN induction but exhibited a late response exclusively during infection by JEV.
JEV stimulates early production of IFN in LLC-MK2 cells but late production in PS cells.
In earlier experiments, mRNA expression was quantified to monitor the IFN response. However, this does not reflect the actual amount of active IFNs released by infected cells. Hence, a focus size reduction assay was performed to confirm the IFN contents of supernatants from JEV-infected cells. IFN activity was assessed by the ability of supernatants to reduce or inhibit focus formation. As shown in Fig. 2A, UV-inactivated supernatants from JEV-infected LLC-MK2 cells harvested at all time points inhibited focus formation, unlike the supernatant from mock-infected cells. Supernatants from JEV-infected PS cells harvested at 48 and 72 h p.i. also exhibited antiviral activity, but the effect of the 24-h supernatant was not different from that of the mock supernatant (Fig. 2A). These data prove that LLC-MK2 cells respond to JEV infection by producing significant amounts of antiviral factors even during early infection. PS cells also produce sufficient levels of antiviral factors to inhibit cell-to-cell spread of JEV, albeit at a later time. This is consistent with our data from real-time quantitative RT-PCR of IFN mRNA, confirming that the antiviral response to JEV is early in LLC-MK2 cells but deferred in PS cells.
Fig. 2.
Antiviral activity of JEV supernatants and roles of IFN and viral proteins. Supernatants (supe) from mock-treated or JEV-infected cells were UV inactivated and tested for the ability to block focus formation. JEV-infected cells were treated with the supernatants immediately after the viral adsorption period, and focus staining was performed 2 days postinfection. (A) LLC-MK2 supernatants harvested at the indicated times were applied on infected Vero cells, while PS supernatants were applied on infected PS cells. (B) Supernatant from JEV-infected LLC-MK2 cells was preincubated with 0, 0.05, and 0.5 μg/ml anti-human IFN-β prior to application on infected Vero cells. Mock supernatant was used as a control. (C) LLC-MK2 supernatants were applied on infected LLC-MK2 or PS cells.
To verify that the antiviral activity of the supernatants is due to IFN, JEV-infected LLC-MK2 supernatants were incubated with an anti-IFN-β antibody prior to treatment. Supernatants in the absence of antibody treatment inhibited focus formation (Fig. 2B). However, pretreatment with 0.5 μg/ml of anti-IFN-β antibody partially restored focus formation (Fig. 2B), implying a role for IFN. Partial activity can be explained by the incomplete effect of the human antibody against a rhesus antigen. However, in addition to IFN, viral proteins may also mediate an antiviral effect. UV treatment of supernatants does not destroy viral proteins, and their presence in the supernatant may influence the IFN response. To clarify the role of viral proteins in this phenomenon, the antiviral activity of supernatants from virus-infected rhesus monkey cells was tested on porcine cells. Since rhesus monkey IFN does not cross-react with porcine cells, any antiviral effect exhibited by the supernatant should be attributed to factors other than IFN. Figure 2C shows that JEV-infected LLC-MK2 supernatants inhibited focus formation in the LLC-MK2 cell line, but not in the PS cell line. This confirms the incompatibility of rhesus monkey IFN with porcine cells. More importantly, it demonstrates that viral proteins included in the supernatant do not stimulate IFN responses. Therefore, the antiviral activity stimulated by JEV supernatant is mediated solely by IFN.
Other cell lines reinforce the relationship between the timing of IFN activation and focus formation.
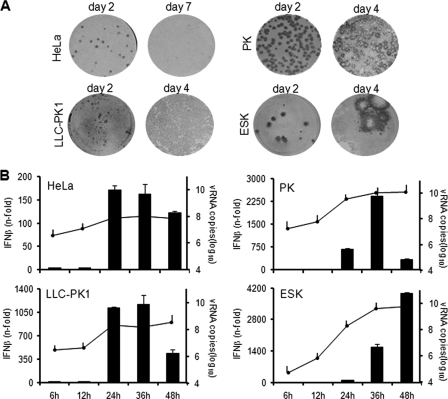
JEV focus formation and IFN response were tested in other primate and porcine cells. Figure 3A shows abortive focus formation by JEV in human HeLa and porcine LLC-PK1 cells, but plaque-forming foci were developed in porcine PK and ESK cells. Accordingly, IFN activation in HeLa and LLC-PK1 cells occurred early, at 24 h p.i., while in PK and ESK cells, the onset was delayed (Fig. 3B). All infections reached high viral RNA titers by 24 h p.i. (Fig. 3B), suggesting that the total level of viral RNA is not the sole determinant for the timing of IFN induction. These results establish a relationship between the timing of IFN activation and the propensity of the virus for cell-to-cell-spread in various cell lines.
Fig. 3.
Focus formation and IFN activation in other cell lines. (A) HeLa, LLC-PK1, PK, and ESK cells were infected with JEV, and foci were detected by immunostaining at the indicated days postinfection. (B) IFN activation (bar graph) and viral RNA titers (line graph) were measured in JEV-infected cells (MOI, 1) at the indicated times postinfection by real-time quantitative RT-PCR. The results for IFN-β mRNA are expressed as the fold increase over mock-treated cells. The results for JEV RNA are expressed as log10 copies of RNA per 1 μg total RNA. The values were normalized to GAPDH and β-actin for primate and porcine cells, respectively. The error bars indicate standard deviations of the means.
IFN restricts viral dissemination in primate cells, but its protective function diminishes upon delayed treatment.
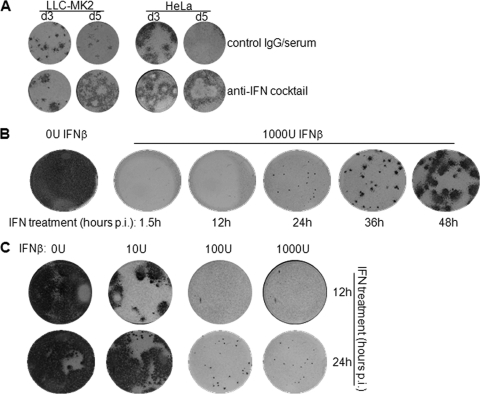
To prove the correlation between IFN response and viral dissemination, we first had to establish that IFN is the restrictive agent against JEV focus formation in primate cells. Thus, focus formation in LLC-MK2 and HeLa cells was assessed in the presence of neutralizing antibodies against IFN-β and IFN-α/βR2 (anti-IFN cocktail). Focus formation in both cell lines was abortive in the presence of a control IgG/serum cocktail (Fig. 4A). However, JEV produced larger foci in antibody-treated cells and even managed to form plaques (Fig. 4A). Thus, intercellular spread of the virus in primate cells is delimited by IFN.
Fig. 4.
Regulation of focus formation by IFN and timing of the IFN response. (A) JEV focus formation was assessed at 3 and 5 days (d) p.i. in LLC-MK2 and HeLa cells treated with an anti-IFN antibody cocktail or a control IgG/serum cocktail. (B and C) Vero cells were infected with JEV and then treated with recombinant human IFN-β at the indicated times postinfection. Focus staining was performed 3 days postinfection. (B) Cells were treated with 0 U IFN-β at 1.5 h postinfection or 1,000 U IFN-β at 1.5, 12, 24, 36, and 48 h postinfection. (C) Cells were treated with 0, 10, 100, and 1,000 U IFN-β at 12 and 24 h postinfection.
We also predicted that intercellular spread would be enhanced if the IFN response were delayed. To test this, Vero cells were treated with exogenous IFN-β at various times postinfection, after which the development of JEV foci was evaluated. IFN-deficient Vero cells were used for this purpose to eliminate the effect of endogenously produced IFNs. IFN treatment at all time points inhibited focus formation compared to the control well without IFN (Fig. 4B). However, JEV developed larger foci with late IFN treatment from 36 h p.i. onward, as opposed to the impaired focus development with early treatment at 1.5 h to 24 h p.i. (Fig. 4B), demonstrating enhanced cell-to-cell spread of JEV when IFN is applied at later periods of infection. When the concentration of IFN was varied, 10 U IFN afforded some protection when applied at 12 h but not at 24 h p.i. (Fig. 4C). On the other hand, higher doses of IFN (100 and 1,000 U) were effective when applied at both time points (Fig. 4C). This further implies that delaying the antiviral response demands higher concentrations of IFN in order to combat viral dissemination. Taken together, these results suggest an IFN-dependent restriction of JEV dissemination in primate cells, which is relieved in cells that develop a delayed IFN response.
The IFN activation pathway of PS cells remains intact and functional during JEV infection.
We were interested in elucidating the mechanism of IFN induction by JEV. To verify the role of cytosolic PRRs in JEV-mediated IFN response, we performed small interfering RNA (siRNA)-mediated knockdown of RIG-I and MDA5 in HeLa cells, for which reliable siRNAs are available. Gene-specific knockdown was demonstrated by evaluation of mRNA levels through RT-PCR (see Fig. S1A and B, bottom, in the supplemental material). Treatment with RIG-I siRNA radically prevented IFN activation by JEV infection, whereas MDA5 siRNA had an intermediate effect (see Fig. S1A in the supplemental material). Nucleic acid delivery via transfection occurs initially through the endosome (15), which houses TLRs that act as principal detectors of RNA viruses (1). To check if the IFN pathway utilized during viral RNA transfection is still mediated by cytosolic PRRs instead of endosomal TLRs, we repeated the experiment using transfected JEV RNA. Consistent with the previous result, RIG-I siRNA significantly reduced IFN activation by transfection of RNA from JEV-infected cells, while MDA5 siRNA had a median effect (see Fig. S1B in the supplemental material). Thus, in agreement with previous studies (2, 14), the IFN response to JEV is dependent on cytosolic PRRs, regardless of whether the viral RNA is introduced via infection or by transfection.
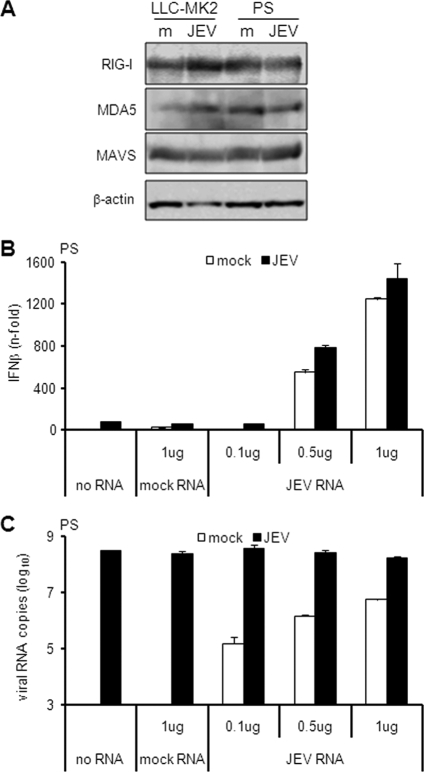
Next, we determined whether JEV has an immunosuppressive activity on the IFN pathway in PS cells. Some viruses inhibit the IFN system by degrading RIG-I or cleaving its downstream effector molecule, MAVS (3, 19, 31). To check if JEV employs a similar mechanism, protein levels of RIG-I, MDA5, and MAVS were determined by immunoblotting. Compared to uninfected controls, there was no significant reduction in the expression levels of all proteins tested during JEV infection (Fig. 5A), but even though these proteins are intact, they may be nonfunctional, possibly through immune suppression by JEV-encoded proteins. To test this hypothesis, we evaluated the response of JEV-infected cells to an exogenous activator of IFN. PS cells were infected for 18 h, after which IFN was induced for another 6 h using total RNA from JEV-infected cells. This RNA was chosen because it allows activation of the same IFN signal transduction pathway used by a live JEV infection (see Fig. S1A and B in the supplemental material). JEV infection alone triggered minimal IFN activation (Fig. 5B). Transfection of total RNA from mock-treated cells neither stimulated IFN activation nor altered the IFN response of infected cells (Fig. 5B). However, transfection of 0.5 and 1 μg JEV RNA stimulated significant levels of IFN expression in the absence of JEV infection and slightly enhanced levels during JEV infection (Fig. 5B). Viral RNA was detected among cells treated with JEV RNA (Fig. 5C), indicating that the transfection was successful. Furthermore, viral RNA levels did not vary among treatments for the naturally infected cells (Fig. 5C), indicating that the transfection had no effect on JEV replication. These results demonstrate that the IFN activation pathway is still inducible in JEV-infected PS cells. Hence, the delayed activation of IFN in PS cells is not the result of an antagonistic mechanism by JEV on the IFN activation pathway. Furthermore, although cells infected with JEV alone had at least 10-fold-higher intracellular viral RNA levels than cells transfected with JEV RNA alone (Fig. 5C), IFN activation was hardly stimulated in the former (Fig. 5B), suggesting that high intracellular titers of JEV RNA do not necessarily translate into a high-level IFN response.
Fig. 5.
Effect of JEV on the IFN activation pathway. (A) LLC-MK2 or PS cells were mock treated (m) or infected with JEV at an MOI of 1 for 24 h. Cellular extracts were subjected to immunoblotting for the proteins indicated on the left, with β-actin as the internal control. (B and C) PS cells were mock treated or infected with JEV at an MOI of 1 for 18 h, and then the indicated amounts of total RNA from mock-treated or JEV-infected LLC-MK2 cells were transfected for another 6 h. IFN activation (B) and viral RNA titers (C) were measured by real-time quantitative RT-PCR. The results for IFN-β mRNA are expressed as the fold increase over mock-treated cells. The results for JEV RNA are expressed as the log10 number of RNA copies per 1 μg total RNA. The values were normalized to β-actin, and the error bars indicate standard deviations of the means.
The total viral RNA and double-stranded RNA levels of JEV do not account for the delayed IFN response in PS cells.
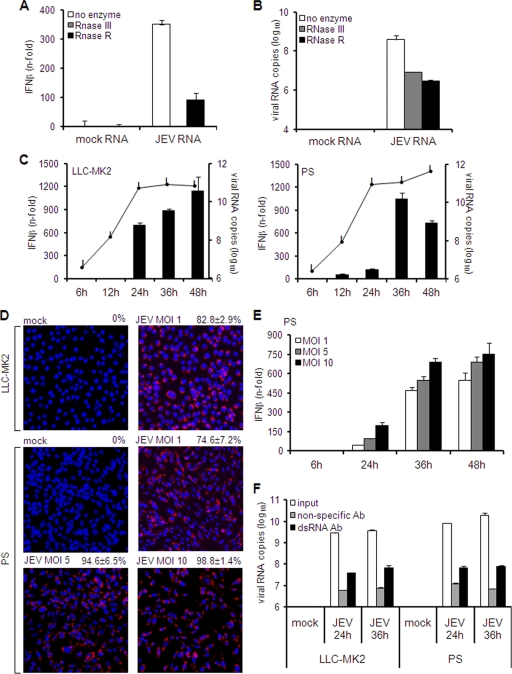
The lack of an inhibitory mechanism by JEV against IFN induction suggests that the virus is most likely not recognized by PS cells. In this case, limiting the expression of or concealing viral RNA would be an appropriate approach, since viral RNA is the instigator of IFN activation. JEV generates both single-stranded RNA (ssRNA) and dsRNA intracellularly, and hence, we initially determined which form is important for IFN response. Total RNA from JEV-infected cells was treated with RNase to remove ssRNA or dsRNA and then transfected into HeLa cells to measure IFN activation. RNA digestion with dsRNA-specific RNase III completely abolished the stimulatory activity, while ssRNA-specific RNase R left some residual stimulation (Fig. 6A). Quantification of viral RNA confirmed its degradation by RNase, resulting in similar titers for both RNase treatments (Fig. 6B). This implies that RNA structure accounts for the distinct IFN inductions in the two RNase treatments. Thus, dsRNA is mainly responsible for JEV-mediated IFN activation.
Fig. 6.
Role of double-stranded RNA in IFN activation and quantitation of intracellular viral RNA and double-stranded RNA. (A and B) Five hundred nanograms of total RNA from mock-treated or JEV-infected cells was incubated with 1 U RNase III, 1 U RNase R, or glycerol (no enzyme). (A) Pretreated RNA was transfected in HeLa cells for 6 h, and IFN activation was measured by real-time quantitative RT-PCR. The results are expressed as the fold increase over cells transfected with mock RNA and normalized to GAPDH. The error bars indicate standard deviations of the means. (B) A similar set of pretreated RNAs was subjected to JEV RNA quantitation by real-time RT-PCR. The results are expressed as the log10 number of RNA copies per 1 μg total RNA and normalized to GAPDH. The error bars indicate standard deviations of the means. (C and E) LLC-MK2 or PS cells were infected with JEV at an MOI of 1 (C) or PS cells were infected at MOIs of 1, 5, and 10 (E). IFN activation (bar graphs) and viral RNA titers (line graphs) were measured at the indicated times postinfection by real-time quantitative RT-PCR. The results for IFN-β mRNA are expressed as the fold increase over mock-treated cells. The results for JEV RNA are expressed as the log10 copies of RNA per 1 μg total RNA. The values were normalized to GAPDH and β-actin for LLC-MK2 and PS cells, respectively. The error bars indicate standard deviations of the means. (D) Immunodetection of dsRNA in LLC-MK2 or PS cells mock treated or infected with JEV at the indicated MOIs 24 h postinfection. The rate of dsRNA expression was obtained by calculating the mean percentage of positively stained cells in five fields. The values at the top right indicate the rates of dsRNA expression ± standard deviations. (F) Quantitation of double-stranded RNA via a solid-phase immunosorbent method. Hot-phenol-extracted total RNA (1 μg) from uninfected or JEV-infected cells was incubated with anti-dsRNA antibody (Ab) or nonspecific antibody on immunosorbent plates. Antibody-bound RNA was harvested via proteinase K/SDS treatment, reextracted, and quantified by real-time RT-PCR using primers for JEV. Untreated RNA was also quantified as the input viral RNA. The results represent the log10 copies of viral RNA. The error bars indicate standard deviations of the means.
We next examined if the deferred IFN response in PS cells is a consequence of low-level expression of viral RNA at early periods of infection. Therefore, we evaluated the kinetics of intracellular viral RNA expression (including single-stranded and double-stranded RNA) versus transcription levels of IFN. The levels of intracellular viral RNA in LLC-MK2 and PS cells were similar at 6 and 12 h p.i., with increasingly higher titers in PS cells starting at 24 h p.i. (Fig. 6C). Although there was a notable increase in IFN expression in LLC-MK2 cells by 24 h p.i., expression levels in PS cells were minimal for the same period (Fig. 6C). These data further validate the previous observations that total viral RNA titers are not entirely predictive of the level of IFN response (Fig. 3B and 5B and C). Considering its vital role in IFN activation, dsRNA expression in both cell lines was next evaluated through indirect immunofluorescence. For both cell lines, a significant number of cells infected at an MOI of 1 contained dsRNA by 24 h p.i. (83% in LLC-MK2 cells and 75% in PS cells) (Fig. 6D).
IFN activation was further examined when PS cells were infected at different MOIs. As illustrated in Fig. 6E, IFN transcription increased in a dose-dependent manner as the MOI was elevated from 1 to 5 and 10. Nonetheless, IFN expression was low at 24 h p.i. for all MOIs (Fig. 6E). Indirect immunofluorescence of dsRNA confirmed the enhanced rate of infection from 75% at an MOI of 1 to 95% and 99% at MOIs of 5 and 10, respectively (Fig. 6D).
Finally, we determined the intracellular dsRNA content using a solid-phase immunosorbence-based assay. Hot-phenol-extracted RNA was used for this purpose in order to preserve the dsRNA structure. Total RNA was incubated with anti-dsRNA antibodies on immunosorbent plates. Antibody-bound dsRNA was then harvested by proteinase K/SDS treatment, reextracted, and quantified by real-time RT-PCR. Consistent with previous results (Fig. 6C), total RNA from JEV-infected cells, i.e., input RNA, contained high levels of viral RNA, with slightly higher titers in PS cells (Fig. 6F). Nonspecific antibody was able to bind viral RNA, but the levels were significantly increased when dsRNA-specific antibody was applied (Fig. 6F). Lastly, the titers of anti-dsRNA antibody-bound viral RNA from JEV-infected cells were similar in all samples (Fig. 6F). To eliminate the possibility of dsRNA saturation in this immunosorbent assay, smaller amounts (10 and 100 ng) of total RNA were tested and gave similar results (see Fig. S2 in the supplemental material). Hence, equal levels of intracellular dsRNA are maintained during JEV infection of both LLC-MK2 and PS cells. Altogether, these results prove that the late IFN response in PS cells is not due to a deficiency of viral RNA or dsRNA expression, and improving the rate of infection does not accelerate the IFN response.
IFN activation is associated with the extent to which dsRNA is exposed in the cytosol.
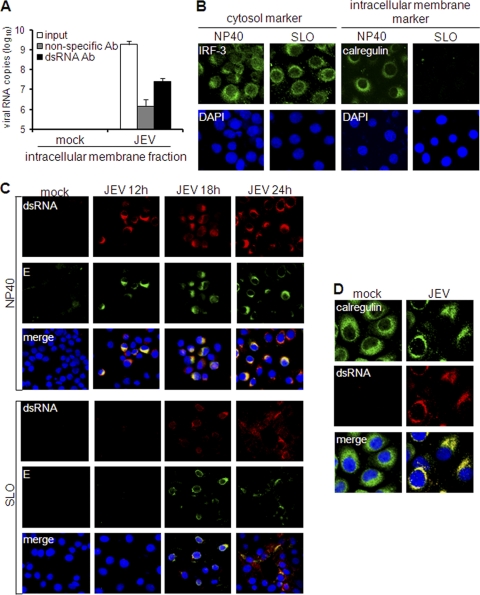
Since JEV triggers IFN through PRRs localized in the cytosol, we investigated the role of dsRNA localization in IFN activation. Several studies have proposed that flavivirus dsRNA, including that of JEV, is recruited inside intracellular membrane vesicles (8, 30, 43, 46, 47). Hence, we initially ascertained the intracellular membrane association of dsRNA through subcellular fractionation. Intracellular membrane fractions of LLC-MK2 cells were subjected to the hot-phenol method of RNA extraction in order to release membrane-associated RNA and preserve the dsRNA structure. Then, the dsRNA content of the extracted RNA was analyzed by solid-phase immunosorbent assay. As opposed to the nonspecific antibody, the anti-dsRNA antibody bound a more significant amount of viral RNA from the membrane fraction (Fig. 7A). Similar results were obtained using intracellular membranes from JEV-infected PS cells (see Fig. S3 in the supplemental material). Hence, JEV dsRNA is closely associated with intracellular membranes. This was further validated by immunostaining of cells that have been subjected to membrane-specific permeabilization. Nonidet P-40 (NP-40) permeabilizes all types of membranes, permitting immunostaining of all cellular regions. In contrast, streptolysin O (SLO) creates pores on the plasma membrane while keeping the intracellular membranes intact, allowing antibodies to pervade only the cytosolic region of the cell. This protocol was initially tested by immunostaining of differentially localized proteins: the cytosolic protein IRF-3 and the endoplasmic reticulum (ER) protein calregulin. Indeed, cytosolic staining is possible using SLO permeabilization, because IRF-3 was detected equally in both NP-40 and SLO treatments (Fig. 7B). In contrast, calregulin was detected by NP-40, but not by SLO permeabilization (Fig. 7B), proving that the internal contents of intracellular membranes are inaccessible during SLO permeabilization. When the method was applied to dsRNA immunostaining, JEV dsRNA was readily detected in LLC-MK2 cells by 12 h p.i. using NP-40 permeabilization, but staining was reduced upon SLO treatment (Fig. 7C). This confirms that dsRNA is unavailable in the cytoplasm and is concealed among intracellular membranes. Colocalization studies showed that JEV dsRNA completely colocalized with the ER marker calregulin (Fig. 7D), suggesting that it is encased particularly within the ER organelle.
Fig. 7.
Subcellular localization of double-stranded RNA. (A) Hot-phenol-extracted RNA from intracellular membrane fractions of mock-treated or JEV-infected LLC-MK2 cells was incubated with nonspecific or dsRNA-specific antibody on immunosorbent plates or left untreated (input). Antibody-bound RNA was harvested by proteinase K/SDS treatment, reextracted, and quantified by real-time RT-PCR using JEV-specific primers. The results are expressed as log10 copies of viral RNA. The error bars indicate standard deviations of the means. (B and C) Membrane-specific permeabilization of LLC-MK2 cells using NP-40 to permeabilize all membranes or SLO to permeabilize only the plasma membrane. Nuclear staining was achieved using DAPI. (B) Immunodetection of IRF-3 or calregulin in NP-40- or SLO-permeabilized LLC-MK2 cells. (C) Immunodetection of dsRNA and E protein in NP-40- or SLO-permeabilized LLC-MK2 cells at various times postinfection by JEV (MOI, 1). (D) Colocalization of dsRNA and calregulin in LLC-MK2 cells mock treated or infected with JEV (MOI, 1) 24 h postinfection. Nuclear staining was achieved using DAPI.
However, cytosolic PRR-mediated activation of IFN by JEV requires the cytosolic presence of dsRNA. To resolve this, we checked for cytosolic dsRNA at later periods of infection. Interestingly, some LLC-MK2 cells exhibited staining of cytosolic dsRNA by 18 h p.i. (Fig. 7C). To further study this cytosolic appearance of dsRNA, we monitored its localization, along with the viral protein E. When JEV E protein is synthesized, it remains in the ER and Golgi apparatus for processing and viral assembly before its final release from the host cell. Indeed, SLO treatment also reduced staining for E (Fig. 7C), validating its localization inside intracellular membranes. Interestingly, some E proteins were detected in the cytosol at 18 h p.i., and the presence of cytosolic E coincided with cytosolic dsRNA (Fig. 7C). Thus, the emergence of dsRNA in the cytosol appears to occur along with that of the viral protein E. Despite the localization of dsRNA inside membranes, its eventual exposure to the cytosol can explain how the host recognizes the virus to initiate the IFN response.
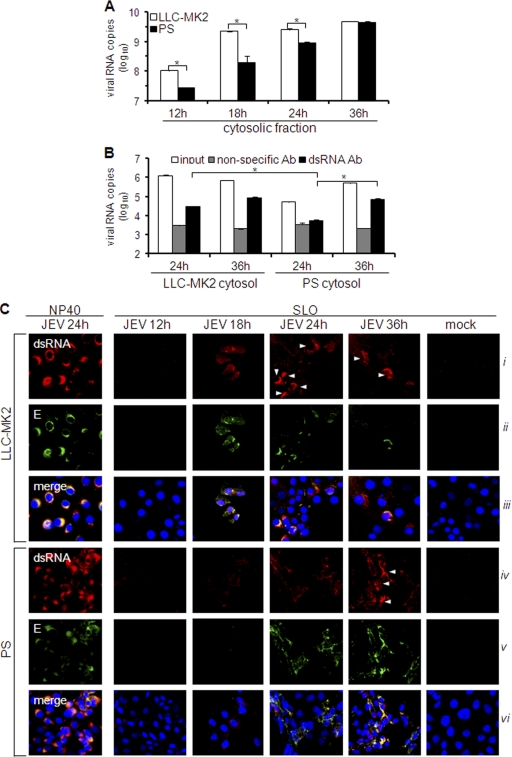
We next quantified and compared the cytosolic viral RNA content of LLC-MK2 and PS cells through subcellular fractionation. As illustrated in Fig. 8A, the titer of viral RNA obtained from the cytosolic fraction of LLC-MK2 cells had already peaked by 18 h p.i. A similar pattern was observed in LLC-PK1 cells (see Fig. S4A in the supplemental material), which display the same type of IFN response and virus permissivity as LLC-MK2 cells (Fig. 3A and B). On the other hand, the cytosolic viral RNA titer in PS cells was significantly lower than that in LLC-MK2 cells, particularly at 12 h to 24 h p.i., but slowly increased to high-level titer by 36 h p.i. (Fig. 8A). We proceeded to evaluate cytosolic dsRNA levels by performing the solid-phase immunosorbent assay for dsRNA quantitation. Crude cytosolic extracts could not be directly used for this purpose due to high nonspecific activity (data not shown). Instead, phenol-extracted RNA was utilized. The amount of cytosolic dsRNA in PS cells was significantly smaller than in LLC-MK2 cells at 24 h p.i. and than PS cells at 36 h p.i. (Fig. 8B). However, like LLC-MK2 cells, cytosolic dsRNA levels in LLC-PK1 cells were relatively high at early times of infection (see Fig. S4B in the supplemental material). Cytosolic exposure was further verified by assessing the dsRNA staining of NP-40- and SLO-treated cells. Total dsRNA staining did not vary among the infected cells, as demonstrated by the NP-40-permeabilized cells (Fig. 8C, i and iv). For cytosolic dsRNA of LLC-MK2 cells, positive staining occurred at 18 h p.i., followed by intense staining in some cells from 24 h to 36 h p.i. (Fig. 8C, i), but in PS cells, slight staining of cytosolic dsRNA appeared at 24 h p.i., and the staining was amplified by 36 h p.i. (Fig. 8C, iv). The appearance of cytosolic dsRNA is also concomitant with that of cytosolic E (Fig. 8C, ii and v). Thus, JEV maintains low levels of cytosolic dsRNA during early infection of PS cells, a phenomenon that is probably responsible for the late IFN response.
Fig. 8.
Quantitation of cytosolic viral RNA and double-stranded RNA. (A and B) LLC-MK2 or PS cells were infected with JEV (MOI, 1), and cytosolic fractions were obtained at various times postinfection. (A) RNA was isolated from cytosolic extracts using the spin column method and subjected to viral RNA quantification by real-time RT-PCR. The results are expressed as log10 copies of viral RNA per 1 × 106 cells. The error bars indicate standard deviations of the means. The asterisks indicate the statistical significance (P < 0.001) between the comparison groups. (B) Phenol-extracted RNA from the cytosolic extract was incubated with nonspecific or dsRNA-specific antibody on immunosorbent plates or left untreated (input). Antibody-bound RNA was harvested by proteinase K/SDS treatment, reextracted, and quantified by real-time RT-PCR using JEV-specific primers. The results are expressed as log10 copies of viral RNA per 100 ng total RNA. The error bars indicate standard deviations of the means. The asterisks indicate the statistical significance (P < 0.001) between the comparison groups. (C) Immunodetection of dsRNA and E protein in NP-40- or SLO-permeabilized cells at various times postinfection by JEV (MOI, 1). NP-40 was used to permeabilize all membranes or SLO to permeabilize only the plasma membrane. Nuclear staining was achieved using DAPI. The white arrowheads indicate cells with intense dsRNA staining.
DISCUSSION
This study demonstrates different IFN responses to JEV in primate and porcine cells, which were correlated with the ability of the virus for cellular dissemination. JEV did not possess an immunosuppressive activity against the IFN system. Instead, we demonstrated the role of dsRNA localization in regulating the host antiviral response.
IFN is an immediate and powerful defense mechanism of the host against viruses. Indeed, IFN has been noted to be a potent antiviral agent against JEV in vitro and in vivo (7, 11, 42, 45). However, in the IFN-producing porcine PS, PK, and ESK cell lines, JEV manages to propagate well, as indicated by its plaque-foming foci and high-titer replication (Fig. 1A and B and 3A and B). When IFN responses were monitored, we discovered a delayed induction of IFN in these porcine cells (Fig. 1C, 2A, and 3B). The delayed IFN response is specific to JEV, since PS cells responded to VSV infection with early and robust IFN induction (Fig. 1E). Interestingly, deferred activation of IFN has been demonstrated for the closely related West Nile virus (WNV) and tick-borne encephalitis virus (TBEV) (5, 6, 30). In connection with this, WNV developed large plaque size and produced infectious particles in a sustained manner when the IFN response was eliminated through IRF-3 knockout of cells (5). Recently, highly virulent avian influenza virus strains that replicated more efficiently were also reported to exhibit late production of IFN (49). Thus, there seems to be a positive correlation between delayed IFN activation and enhanced viral replication and/or dissemination. In agreement with this, delayed IFN production in mice due to MAVS/Cardif gene knockout resulted in elevated levels of dengue virus (DENV) RNA in serum and lymphoid tissues (34). In contrast, the IFN response to JEV in the rhesus monkey LLC-MK2, human HeLa, and porcine LLC-PK1 cell lines was early (Fig. 1C and 3B), and it was accompanied by abortive focus formation (Fig. 1A and 3A) in spite of productive infection at a high MOI (Fig. 1B). Restoration of plaque-forming foci through IFN-neutralizing antibodies in primate cells (Fig. 4A) highlights the restrictive function of IFN against viral intercellular movement, but we further propose that the timing of the IFN response is critical, as well. An early antiviral reaction adequately prevents intercellular spread of JEV, but delaying the IFN response creates a window of opportunity for the virus to disseminate ahead of the antiviral effect. Indeed, intercellular dissemination of JEV was inhibited to a lesser degree when IFN treatment was delivered late (Fig. 4B), and higher concentrations of the antiviral agent were necessary for protection as treatment was deferred (Fig. 4C). Accordingly, late IFN treatment implies more difficulty in inhibiting viral replication. This principle is reiterated in the in vitro and in vivo studies of Harinasuta et al. (11) and Taylor et al. (42), respectively. However, when Diamond and his group (4) performed a similar experiment on DENV, IFN treatment postinfection did not inhibit viral replication and dissemination, although IFN treatment prior to infection did. This discrepancy can be explained by the low concentration of IFN applied in the DENV study, emphasizing the importance of the IFN dosage in antiviral treatment. Thus, both the timing and the intensity of the host IFN response play crucial roles in influencing the outcome of an infection. This has strong implications for antiviral therapy. In a clinical study, IFN-α2a treatment of children with Japanese encephalitis yielded negative results (38). The authors rationalized that the doses of IFN used were not sufficient to improve the outcome of the patients. To complement those conclusions, our results can attribute the failure of the IFN dosage to the timing of the treatment, since the patients were already in the acute phase of infection. Hence, future work should consider both the aspects of timing and dosage for a successful antiviral therapy.
During IFN activation, PRRs function as the frontline sensors of viral infection (1). We confirmed previous reports (2, 14) that JEV-mediated IFN induction is dependent on the cytosolic PRRs RIG-I and MDA5 (see Fig. S1 in the supplemental material). Although it has been popularly assumed, we also provided for the first time direct evidence that JEV dsRNA is the major activating factor for IFN (Fig. 6A). Hence, dsRNA interaction with RIG-I and MDA5 (1) may serve as the IFN-initiating complexes in response to JEV infection, a function that is dictated by the cytosolic availability of dsRNA. However, according to Overby et al. (30), TBEV encloses its dsRNA inside intracellular membrane vesicles to hide it from the host immune system. Studies on Kunjin virus, WNV, DENV, and JEV have also shown that dsRNA is confined inside ER-derived membrane vesicles, along with the replication complex (8, 43, 46, 47). Alternatively, dsRNA may localize on the ER surface, but it is hidden among ER-bound proteins, as has been proposed for hepatitis C virus (41). In agreement with these reports, JEV dsRNA was closely associated with intracellular membranes (Fig. 7A; see Fig. S3 in the supplemental material), most probably derived from the ER (Fig. 7D). Membrane-specific permeabilization confirmed that the majority of this RNA is hidden among intracellular membranes at an early phase of infection (Fig. 7C). It is reasonable for the virus to recruit dsRNA away from cytosolic PRRs, and disruption of membrane structures should eliminate this protection by exposing the dsRNA, as has been demonstrated for WNV (12).
The ability of cells to activate the IFN system during JEV infection implies that dsRNA eventually becomes available in the cytosol. Indeed, cytosolic dsRNA was detected by 18 h p.i. in LLC-MK2 cells, and high levels were reached by 24 h p.i. (Fig. 7C and 8B and C). Early cytosolic exposure of dsRNA was also observed in LLC-PK1cells (see Fig. S4 in the supplemental material), in correlation with the timing of the IFN response (Fig. 3B). For PS cells, cytosolic dsRNA appeared only at 24 h p.i., and high levels were reached later at 36 h p.i. (Fig. 8B and C). Since most of the dsRNA is inaccessible during JEV infection, a threshold level for cytosolic dsRNA has to be reached before robust IFN induction sets in. Hence, early exposure of cytosolic dsRNA in LLC-MK2 and LLC-PK1 cells primes them in advance for IFN activation, whereas delayed cytosolic exposure of dsRNA in PS cells impedes the IFN response. Increasing the infection rate in PS cells to almost 100% did not accelerate the IFN response (Fig. 6D and E), most likely because a modest amount of dsRNA is maintained in the cytosol early in infection. In support of this, transfection of viral RNA (containing dsRNA) in PS cells, which directly introduces it into the cytosol (15), induced a much stronger IFN response than a live JEV infection (Fig. 1F and 5B).
It is interesting that while cytosolic levels of viral RNA in PS cells were lower than in LLC-MK2 cells during early infection (Fig. 8A), their total viral RNA levels were comparable (Fig. 6C). A similar pattern was also observed for dsRNA (Fig. 6F and 8B; see Fig. S2 in the supplemental material). Thus, the low cytosolic level of viral RNA in PS cells is most likely a reflection of the small quantity of cytosolic dsRNA. Moreover, this suggests that minimal exposure of dsRNA in the cytosol of porcine cells occurs independently of total viral RNA and dsRNA titers. Accordingly, the total RNA of JEV was not absolutely predictive of the intensity of the IFN response (Fig. 3B, 5B and C, and 6C). This is different from the case of WNV and TBEV, where IFN activation was proportional to the amount of total viral RNA or the replication level (5, 30). Thus, although the deferred IFN activation by WNV was attributed to insufficient levels of viral RNA (or other viral factors) at early times (5), these results emphasize that the quantity of dsRNA exposed in the cytosol is the determinant of the IFN response to JEV infection.
A few other studies have already demonstrated the cytosolic presence of flavivirus dsRNA. In DENV-infected cells, dsRNA has been observed inside membrane vesicles, as well as on their cytosolic surfaces, indicating its temporary confinement inside the membranes (46). Overby et al. (30) used SLO permeabilization to demonstrate the membrane localization of TBEV dsRNA, but cytosolic dsRNA was not detected, probably since it was monitored at only one time point. However, they were able to show, using indirect methods, that dsRNA is exposed in the cytosol at later periods of infection. The mechanism for cytosolic emergence of flaviviral dsRNA is currently unknown, but it appears to occur simultaneously with the liberation of viral E protein (Fig. 7C and 8C). This may be simply a physiological reaction to infection (e.g., by the rupture of ER membranes), or it may be a stage in the process of viral replication and assembly (e.g., active movement of viral products). Certainly, the level of cytosolic dsRNA during early infection also varies with the cell line, with primate cells containing high levels while porcine cells possess low levels. These differences can be explained by a cellular factor that influences the cytosolic appearance of dsRNA. Future investigations can focus on how flaviviral dsRNA becomes available in the cytosol and which viral or cellular factors are involved. Such factors are potential targets for antiviral therapy.
Our study indicates that JEV does not retain any immunosuppressive activity. First, IFN signaling is still active, since IFN treatment successfully abrogated JEV focus formation in PS cells (Fig. 2A). Moreover, unlike other viruses that degrade PRRs or cleave MAVs to inhibit the IFN response (3, 19, 31), the molecular facility of the IFN activation pathway is still intact in JEV-infected cells (Fig. 5A). JEV also failed to block IFN activation (Fig. 5B), indicating that the IFN activation pathway can still operate in the presence of the virus. This further implies that JEV-encoded proteins are not the major components that inhibit IFN induction, contrary to what has been reported for other flaviviral proteins (24, 27, 36, 48). In similar experiments, DENV was able to inhibit IFN activation using various inducers (36), but WNV and TBEV failed to inhibit IFN activation by poly(I · C) (6, 30), suggesting that not all flaviviruses possess an active mechanism to antagonize the host IFN pathway. The intact and functional IFN system of PS cells during JEV infection reinforces the idea that the virus (i.e., dsRNA) is simply not detected at early stages of infection. The delayed exposure of dsRNA in the cytosol explains this phenomenon. Hence, a regulated cytosolic exposure of dsRNA at an early phase of infection limits the IFN response and contributes to viral propagation in culture cells. To our knowledge, this is the first investigation of the tripartite relationship between cytosolic dsRNA, the IFN system, and viral dissemination among flaviviruses.
JEV establishes high-level viremia in pigs, but viral titers in humans are usually undetectable, and the virus can hardly be isolated (26, 44). A cell/species-specific IFN response could contribute to the differential susceptibility of humans and pigs, as demonstrated by the behavior of JEV in this study. The early IFN response to JEV in primate cells has been observed in other human cell lines (2) and could be a restraining factor for the virus. A prompt IFN response may control JEV replication and/or dissemination and contribute further by allowing the early development of adaptive immune responses, which play key roles in restricting JEV infection among humans (26). In support of this, Rokutanda (37) showed that a low-viremia strain of JEV triggered high-level IFN production in mice by 24 h postinfection, while a high-viremia variant did not elicit an IFN response. Likewise, an impaired IFN response could be responsible for high susceptibility of pigs to JEV. Three out of four porcine cell lines tested in this study exhibited late IFN activation. While not all porcine cells behaved similarly, if a majority of them could display late IFN response in vivo, this could certainly promote JEV propagation. An investigation of the JEV-mediated IFN response in other types of porcine cells (in vitro) or in pig models (in vivo) is required to validate this hypothesis. Nevertheless, this study suggests the importance of IFN in mediating protection against early infection with JEV, and an examination of the IFN response is encouraged to substantiate its role in host susceptibility.
Supplementary Material
ACKNOWLEDGMENTS
L.A.E.-M. is a recipient of the Monbukagakusho Scholarship from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). This study was supported by the 21st Century Centers of Excellence (COE) Program on Global Strategies for Control of Tropical and Emerging Infectious Diseases at Nagasaki University from MEXT; a U.S.-Japan Cooperative Medical Science Program and Research Grant (H21 Shinkou-ippan-005) from the Ministry of Health, Labor and Welfare of Japan; and the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, JST, Japan.
We thank Daisuke Hayasaka and Joanna L. Shisler for critical and helpful comments on the study and Fuxun Yu for technical advice.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Baum A., García-Sastre A. 2010. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids 38:1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang T. H., Liao C. L., Lin Y. L. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I dependent IRF-3 and PI3K-dependent NF-κB activation. Microb. Infect. 8:157–171 [DOI] [PubMed] [Google Scholar]
- 3. Chen Z., et al. 2007. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J. Virol. 81:964–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diamond M. S., et al. 2000. Modulation of dengue virus infection in human cells by alpha, beta and gamma interferons. J. Virol. 74:4957–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fredericksen B. L., Smith M., Katze M. G., Shi P. Y., Gale M., Jr 2004. The host response to West Nile Virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737–7747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fredericksen B. L., Gale M., Jr 2006. West Nile Virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 80:2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh S. N., et al. 1990. Protective effect of 6-MFA, a fungal interferon inducer against Japanese encephalitis virus in bonnet macaques. Indian J. Med. Res. 91:408–413 [PubMed] [Google Scholar]
- 8. Gillespie L. K., Hoenen A., Morgan G., Mackenzie J. M. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84:10438–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gould E. A., Solomon T., Mackenzie J. S. 2008. Does antiviral therapy have a role in the control of Japanese encephalitis? Antiviral Res. 78:140–149 [DOI] [PubMed] [Google Scholar]
- 10. Hardy R. W., Marcotrigiano J., Blight K. J., Majors J. E., Rice C. M. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 77:2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harinasuta C., Wasi C., Vithanomsat S. 1984. The effect of interferon on Japanese encephalitis virus in vitro. Southeast Asian J. Trop. Med. Public Health 15:564–568 [PubMed] [Google Scholar]
- 12. Hoenen A., Liu W., Kochs G., Khromykh A. A., Mackenzie J. M. 2007. West Nile virus-induced cytoplasmic membrane structures provide partial protection against the interferon-induced antiviral MxA protein. J. Gen. Virol. 88:3013–3017 [DOI] [PubMed] [Google Scholar]
- 13. Igarashi A. 2002. Control of Japanese encephalitis in Japan: immunization of humans and animals, and vector control. Curr. Top. Microbiol. Immunol. 267:139–152 [DOI] [PubMed] [Google Scholar]
- 14. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 15. Khalil I. A., Kogure K., Akita H., Harashima H. 2006. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 58:32–35 [DOI] [PubMed] [Google Scholar]
- 16. Kinney J. S., Viscidi R. P., Vonderfecht S. L., Eiden J. J., Yolken R. H. 1989. Monoclonal antibody assay for detection of double-stranded RNA and application for detection of group A and non-group A rotaviruses. J. Clin. Microbiol. 27:6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kinoshita H., et al. 2009. Isolation and characterization of two phenotypically distinct dengue type-2 virus isolates from the same dengue hemorrhagic fever patient. Jpn. J. Infect. Dis. 62:343–350 [PubMed] [Google Scholar]
- 18. Leake C. J., Burke D. S., Nisalak A., Hoke C. H. 1986. Isolation of Japanese encephalitis virus from clinical specimens using a continuous mosquito cell line. Am. J. Trop. Med. Hyg. 35:1045–1050 [DOI] [PubMed] [Google Scholar]
- 19. Li X. D., Sun L., Seth R. B., Pineda G., Chen Z. J. 2005. Hepatitis C virus protease NS34A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 102:17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin C. W., et al. 2008. Interferon antagonist function of Japanese encephalitis virus NS4A and its interaction with DEAD-box RNA helicase DDX42. Virus Res. 137:49–55 [DOI] [PubMed] [Google Scholar]
- 21. Lin R. J., Liao C. L., Lin E., Lin Y. L. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285–9294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin R. J., Chang B. L., Yu H. P., Liao C. L., Lin Y. L. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindenbach B. D., Rice C. M. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23–61 [DOI] [PubMed] [Google Scholar]
- 24. Liu W. J., Chen H. B., Wang X. J., Huang H., Khromykh A. A. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 78:12225–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loo Y. M., et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Misra U. K., Kalita J. 2010. Overview: Japanese encephalitis. Prog. Neurobiol. 91:108–120 [DOI] [PubMed] [Google Scholar]
- 27. Moriyama M., et al. 2007. Interferon-beta is activated by hepatitis C virus NS5B and inhibited by NS4A, NS4B, and NS5A. Hepatol. Int. 1:302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moue M., et al. 2008. Toll-like receptor 4 and cytokine expression involved in functional immune response in an originally established porcine intestinal epitheliocyte cell line. Biochim. Biophys. Acta 1780:134–144 [DOI] [PubMed] [Google Scholar]
- 29. Muñoz-Jordán J. L. 2010. Subversion of interferon by dengue virus. Curr. Top. Microbiol. Immunol. 338:35–44 [DOI] [PubMed] [Google Scholar]
- 30. Overby A. K., Popov V. L., Niedrig M., Weber F. 2010. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 84:8470–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Papon L., et al. 2009. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393:311–318 [DOI] [PubMed] [Google Scholar]
- 32. Paun A., Pitha P. M. 2007. The innate antiviral response: new insights into a continuing story. Adv. Virus Res. 69:1–66 [DOI] [PubMed] [Google Scholar]
- 33. Peiris J. S., et al. 1993. Japanese encephalitis in Sri Lanka: comparison of vector and virus ecology in different agro-climatic areas. Trans. R. Soc. Trop. Med. Hyg. 87:541–548 [DOI] [PubMed] [Google Scholar]
- 34. Perry S. T., Prestwood T. R., Lada S. M., Benedict C. A., Shresta S. 2009. Cardif-mediated signaling controls the initial innate response to dengue virus in vivo. J. Virol. 83:8276–8281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodriguez-Madoz J. R., et al. 2010. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J. Virol. 84:9760–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rokutanda H. K. 1969. Relationship between viremia and interferon production of Japanese encephalitis virus. J. Immunol. 102:662–670 [PubMed] [Google Scholar]
- 38. Solomon T., et al. 2003. Interferon alfa-2a in Japanese encephalitis: a randomized double-blind placebo-controlled trial. Lancet 361:821–826 [DOI] [PubMed] [Google Scholar]
- 39. Sumiyoshi H., et al. 1987. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology 161:497–510 [DOI] [PubMed] [Google Scholar]
- 40. Suthar M. S., Gale M., Jr., Owen D. M. 2009. Evasion and disruption of innate immune signalling by hepatitis C and West Nile viruses. Cell. Microbiol. 11:880–888 [DOI] [PubMed] [Google Scholar]
- 41. Targett-Adams P., Boulant S., McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taylor J. L., Schoenherr C., Grossberg S. E. 1980. Protection against Japanese encephalitis virus in mice and hamsters by treatment with carboxymethylacridanone, a potent interferon inducer. J. Infect. Dis. 142:394–399 [DOI] [PubMed] [Google Scholar]
- 43. Uchil P. D., Satchidanandam V. 2003. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 278:24388–24398 [DOI] [PubMed] [Google Scholar]
- 44. van den Hurk A. F., Ritchie S. A., Mackenzie J. S. 2009. Ecology and geographical expansion of Japanese encephalitis virus. Annu. Rev. Entomol. 54:17–35 [DOI] [PubMed] [Google Scholar]
- 45. Vithanomsat S., Wasi C., Harinasuta C., Thongcharoen P. 1984. The effect of interferon on flaviviruses in vitro: a preliminary study. Southeast Asian J. Trop. Med. Public Health 15:27–31 [PubMed] [Google Scholar]
- 46. Welsch S., et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Westaway E. G., Mackenzie J. M., Kenney M. T., Jones M. K., Khromykh A. A. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structure. J. Virol. 71:6650–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson J. R., de Sessions P. F., Leon M. A., Scholle F. 2008. West Nile Virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 82:8262–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeng H., et al. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J. Virol. 81:12439–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.