Abstract
Although hepatitis C virus (HCV) assembly remains incompletely understood, recent studies with the genotype 2a JFH-1 strain suggest that it is dependent upon the phosphorylation of Ser residues near the C terminus of NS5A, a multifunctional nonstructural protein. Since genotype 1 viruses account for most HCV disease yet differ substantially in sequence from that of JFH-1, we studied the role of NS5A in the production of the H77S virus. While less efficient than JFH-1, genotype 1a H77S RNA produces infectious virus when transfected into permissive Huh-7 cells. The exchange of complete NS5A sequences between these viruses was highly detrimental to replication, while exchanges of the C-terminal domain III sequence (46% amino acid sequence identity) were well tolerated, with little effect on RNA synthesis. Surprisingly, the placement of the H77S domain III sequence into JFH-1 resulted in increased virus yields; conversely, H77S yields were reduced by the introduction of domain III from JFH-1. These changes in infectious virus yield correlated well with changes in the abundance of NS5A in RNA-transfected cells but not with RNA replication or core protein expression levels. Alanine replacement mutagenesis of selected Ser and Thr residues in the C-terminal domain III sequence revealed no single residue to be essential for infectious H77S virus production. However, virus production was eliminated by Ala substitutions at multiple residues and could be restored by phosphomimetic Asp substitutions at these sites. Thus, despite low overall sequence homology, the production of infectious virus is regulated similarly in JFH-1 and H77S viruses by a conserved function associated with a C-terminal Ser/Thr cluster in domain III of NS5A.
INTRODUCTION
Infection with hepatitis C virus (HCV) is associated with chronic hepatitis, progressive hepatic fibrosis leading to cirrhosis, and hepatocellular carcinoma (for a review, see reference 21). The virus establishes lifelong persistent infection in most infected persons. It is currently thought to infect 130 to 170 million people worldwide, placing them at risk for potentially life-threatening liver disease. Available interferon-based treatment is limited in its efficacy, and immunization currently is not possible. To overcome these shortcomings in therapeutic and preventive measures, there is a need to increase our current understanding of HCV pathogenesis, including the molecular mechanisms involved at various steps in the virus life cycle and the role of virus-host interactions in virus persistence and disease progression.
HCV is an enveloped, positive-strand RNA virus classified within the genus Hepacivirus of the family Flaviviridae. Its 9.7-kb genome encodes a single polyprotein, which is co- and posttranslationally processed by both viral and cellular proteases (21). The N-terminal segment of the polyprotein comprises the structural proteins (core, E1, and E2), while the downstream proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) are nonstructural. Only the segment extending from NS3 to NS5B is required for genome replication (24), with NS5B, an RNA-dependent RNA polymerase, serving as the catalytic core of a membrane-bound, macromolecular viral replicase complex. p7 and NS2 are not required for RNA replication, and accumulating evidence supports important functions for both of these proteins in virus assembly and the release of virus from infected cells (18, 19, 26, 37, 45, 48). Emerging data also support essential roles for other nonstructural proteins, in particular NS5A (2, 16, 28, 32, 39) and NS3 (27, 32, 47), in the assembly and release of infectious particles.
The HCV NS5A protein is unique, with no known human or viral homologs other than NS5A in the closely related GB virus B (GBV-B). It appears to have multiple functions in the virus life cycle and to interact with numerous viral and host proteins (43), but little is known about NS5A at a mechanistic level. It is required for viral RNA replication and, as mentioned above, plays an essential role in virion production. It is a phosphoprotein (15, 20, 38), and its phosphorylation status was suggested to regulate genome replication and virus assembly (11, 12). It is also a frequent site of cell culture-adaptive mutations that promote the amplification of HCV replicon RNAs in Huh-7 hepatoma cells (3). At a structural level, there appear to be three distinct domains that are separated by segments of the protein that are relatively disordered. The N-terminal domain (domain I) contains a Zn-binding motif and is essential for viral RNA replication (41). Two high-resolution structural models have been developed for domain I of NS5A based on X-ray crystallography (25, 42). While they differ significantly, both models suggest a dimeric structure that associates with lipid bilayers in membrane-bound replicase complexes. Importantly, NS5A mutants that are deficient in the ability to support viral RNA replication can be trans-complemented in a cell culture system (1).
The middle domain II, which is involved in antagonizing innate immune responses, and C-terminal domain III of NS5A appear to play lesser roles in viral RNA replication (30, 40). Nonetheless, mutations within domain III may reduce the efficiency of genome amplification. The amino acid sequence of domain III is poorly conserved between different viral genotypes and has been considered to be relatively unstructured (14). Nonetheless, recent data suggest that it plays an important role in the assembly of infectious virus particles. This aspect of NS5A function has emerged from studies of the genotype 2a JFH-1 virus, a unique strain of HCV that replicates efficiently in cell culture. The transfection of synthetic JFH-1 genomic RNA produces relatively high titers of infectious virus in permissive Huh-7 hepatoma cells (22, 44, 52). During an early step in the process of particle assembly, the JFH-1 NS5A protein is recruited to the surface of cytoplasmic lipid droplets that are decorated with the viral core protein (6, 29). Genetic evidence also suggests that NS5A, in association with NS2, acts at a later step in virus production, following intracellular particle assembly but prior to the release of infectious virus (48). Several recent studies indicated that domain III of the JFH-1 NS5A protein is particularly important for the production of infectious virions and that the phosphorylation of Ser residues within this domain, possibly by cellular casein kinase II (CK II), is required for this process (2, 16, 28, 39).
We have found the construction and analysis of intergenotypic chimeric HCV genomes to be a productive approach to the characterization of functional interactions among HCV proteins that are important in the virus life cycle (27, 47). We adopted a similar approach to investigate the role of NS5A in the production of infectious virus, generating intergenotypic NS5A chimeras within the background of JFH-1 and a genotype 1a molecular clone, H77S, that also produces infectious particles when transfected into cells as RNA (49). H77S RNA produces substantially fewer infectious particles in transfected cells than does JFH-1 RNA, but like JFH-1 virus produced in cell culture (23), recent studies have shown that virus produced from H77S RNA (with an additional adaptive mutation in E2) is infectious for chimpanzees as well (data not shown).
We exchanged the entire NS5A sequences between these two genotypes, as well as only the domain III sequences, and observed the effects of these exchanges on intracellular genome amplification and infectious virus production. We also carried out a mutational analysis of potential phosphoacceptor sites within the C-terminal domain III sequence of the NS5A protein in the H77S virus and compared these results with results of previously reported studies of the JFH-1 virus. Despite limited sequence identity, our data indicate that domain III of the genotype 1a H77S NS5A protein shares conserved structural and functional elements with the JFH-1 protein that are essential for the production of infectious virions.
MATERIALS AND METHODS
Plasmids.
The cell culture-adapted pH77S infectious molecular clone of genotype 1a HCV was described previously (49). pH77S.3 is a modified version of this plasmid that has an enhanced capacity for the production of infectious virus in cell culture and that contains an additional N476D mutation in E2 and lacks the Q1067R adaptive mutation in NS3 (36). “No see'm” cloning (50) was used to generate NS5A chimeras in the pH77S (49), pJFH-1 (44), and chimeric intergenotypic pHJ3-5 (47) plasmid DNAs without altering the original genome sequences outside NS5A or NS5A domain III. The SapI restriction enzyme was employed for the deletion and insertion of heterologous sequences since its recognition site is nonpalindromic. For Ser/Thr-to-Ala or -Asp substitution mutations, the QuikChange mutagenesis kit (Stratagene) was used. DNA sequencing verified the integrity of the manipulated sequences and the presence of the intended substitution mutations.
Cells.
Huh-7.5 cells (4) were kindly provided by Charles Rice and Apath, LLC. The cells were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and 1× penicillin-streptomycin at 37°C in a 5% CO2 environment.
RNA transcription and transfection.
Plasmid DNAs were linearized by XbaI restriction digestion. Synthetic RNA was transcribed from linearized DNAs using the MEGAscript kit (Ambion). The concentration and integrity of the transcribed RNAs were confirmed by spectrophotometry and denaturing agarose gel electrophoresis. In vitro-transcribed RNAs (1.25 μg) were transfected into Huh-7.5 cells in six-well culture dishes (6 × 105 cells/well) using the TransIT-mRNA transfection reagent (Mirus Bio) as recommended by the manufacturer. Six hours after transfection, the culture medium was replaced with fresh medium. The cells were split at a 1:2 ratio before further incubation at 37°C in a 5% CO2 incubator.
DNA transfection.
For NS5A protein expression studies, 0.9 μg of plasmid DNA was mixed with 0.1 μg of the pCMV-GLuc plasmid (New England BioLabs) and transfected into Huh-7.5 cells in 12-well culture dishes (2 × 105 cells/well) using TransIT-2020 reagent (Mirus Bio) as recommended by the manufacturer. NS5A species were quantified in immunoblots using different antibody probes (see below), and the results were normalized to the luciferase activity present in supernatant fluids of transfected cultures.
Virus titration.
Culture supernatants were collected 3 days after transfection with the synthetic RNAs, and the titer of infectious virus was determined by the inoculation of serial 10-fold dilutions onto naïve Huh-7.5 cells seeded 1 day previously in 8-well chamber slides (Lab-Tek) (1 × 105 cells/well). Three days after inoculation, infected cells were fixed with methanol-acetone and labeled for the detection of the HCV core protein by immunofluorescence microscopy as described previously (27). The titer of infectious virus was determined from the number of foci of infected cells observed at each dilution and is expressed as focus-forming units (FFU) per ml. To quantify intracellular infectious virus, cell lysates were prepared by multiple freeze-thaw cycles as described previously (13) and similarly tested.
Immunoblots.
A standard immunoblotting procedure was employed (35). Protein samples transferred onto polyvinylidene difluoride (PVDF) membranes were probed with the following primary antibodies: anti-core (1:2,000) (MA1-080; Affinity BioReagents), 9E10 (kindly provided by Charles Rice and Tim Tellinghuisen), rabbit polyclonal anti-NS5A (kindly provided by Craig Cameron), anti-NS3 (217-A; Virogen), anti-actin (A5441; Sigma), anti-Myc (catalog number 2278; Cell Signaling), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (AM4300; Ambion). Proteins were visualized with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (catalog numbers 103005 and 405005; Southern Biotech) and enhanced chemiluminescence detection (Amersham ECL-GE Healthcare). For the quantitative assessment of immobilized proteins, proteins were visualized with IRDye 800CW goat anti-mouse IgG or IRDye 680 goat anti-rabbit IgG, and images were collected with an Odyssey infrared imaging system (Li-Cor Biosciences).
HCV core ELISA.
The amount of soluble HCV core protein released into the cell culture medium by transfected cells was quantified by using the Ortho Diagnostics Trak-C enzyme-linked immunosorbent assay (ELISA) kit (Ortho-Clinical Diagnostics) according to the manufacturer's instructions.
RT-PCR.
For the quantitative measurement of the HCV RNA replication capacity, Huh-7.5 cells were transfected by electroporation (46), and total cellular RNA was isolated at the time of harvest by using the RNeasy minikit (Qiagen). Real-time quantitative reverse transcription (RT)-PCRs (qRT-PCRs) were carried out with the iQ5 real-time PCR detection system (Bio-Rad) as described previously (27).
CK II inhibitor treatment.
Cell cultures were split as described above 6 h after HCV RNA transfection and refed with medium containing 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT), a specific CK II inhibitor (Calbiochem). The cells were incubated for 48 h, followed by the refeeding of the cells with fresh medium (no DMAT). Culture supernatant fluids were collected 1 day later and assayed for infectious virus as described above. The cytotoxic effects of DMAT were assessed by using the WST-1 cellular proliferation assay (Roche Applied Sciences) as recommended by the manufacturer.
RESULTS
Intergenotypic NS5A domain exchanges and genome replication.
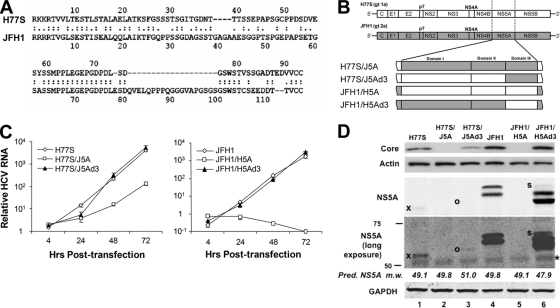
Genome-length HCV RNAs transcribed from genotype 2a plasmid pJFH-1 and genotype 1a plasmid pH77S produce infectious virus particles when transfected into Huh-7 hepatoma cells (44, 49, 52). As described above, several previous studies suggested that C-terminal domain III of the NS5A protein is essential for the assembly of infectious JFH-1 virus particles and that the phosphorylation of this domain may regulate assembly (2, 16, 28, 39). While it is likely that this is also true for the genotype 1a H77S virus, the genotype 1a NS5A protein has not been similarly investigated. Since genotype 1 infections account for the large majority of infections in most geographic regions (31), it is important to study these events in the context of genotype 1 viruses. This is especially important since the H77S NS5A protein shares only 58% amino acid identity with the JFH-1 protein overall and only 46% identity within domain III (Fig. 1A). To determine whether these NS5A proteins can function interchangeably, despite the differences in amino acid sequences, we exchanged sequences encoding the entire NS5A protein, or the NS5A domain III only, within the background of these full-length constructs (Fig. 1B). To avoid any changes in the HCV sequence outside the exchanged domains, we employed “no see'm” cloning (50) to generate the mutants. The chimeric sequences were verified by sequencing of the plasmid DNAs.
Fig. 1.
Construction and evaluation of replication properties of NS5A chimeras. (A) Amino acid sequences of NS5A domain III in H77S and JFH-1 viruses. The amino acid sequence identity between these two domains is 46.2%. (B) Schematic diagram of the NS5A chimeras constructed for these studies. The entire NS5A sequence or NS5A domain III was exchanged between H77S and JFH-1 to assess the impact on infectious virus production and viral RNA replication. (C) Replication properties of transfected H77S (left) and JFH-1 (right) chimeric RNAs. The abundance of in vitro-transcribed genomic RNA relative to that of a replication-defective mutant with a GND mutation in NS5B was determined at various times following transfection by using a real-time RT-PCR assay. Shown are mean values ± standard errors (SE) from triplicate RT-PCR assays. (D) Immunoblots of lysates of HCV RNA-transfected cells showing the abundance of core and NS5A in comparison to actin and GAPDH loading controls, respectively. Faint bands are indicated to their left as follows: “x,” H77S NS5A; “o,” H77S/J5Ad3 NS5A; “s,” additional hyperphosphorylated NS5A species (see the text); “*,” nonspecific band. The predicted molecular masses of the H77S, JFH-1, and chimeric NS5A proteins, prior to any posttranslational modification, are shown between the NS5A and GAPDH blots. Pred. NS5A m.w., predicted NS5A molecular weight (in thousands).
The impact of these NS5A domain swaps on RNA replication was determined following their transfection into Huh-7.5 cells by using a real-time, quantitative RT-PCR assay to compare the RNA abundance relative to the abundance of a replication-defective control RNA containing a GND substitution in the GDD motif of NS5B (Fig. 1C). These results demonstrated similar increases in the relative intracellular abundances of the parental H77S and JFH-1 RNAs over the 72 h following transfection (Fig. 1C, left and right, respectively). In contrast, both chimeric RNAs with full-length NS5A swaps demonstrated significant impairments in RNA replication. The JFH-1/H5A RNA (H77S NS5A in the JFH-1 background) (Fig. 1B) showed no increase in abundance compared to that of the lethal GND mutant (Fig. 1C, right), while the H77S/J5A RNA demonstrated a >10-fold reduction in RNA abundance compared with that of the parental H77S RNA by 72 h posttransfection (Fig. 1C, left). Consistent with these findings, no core protein expression was observed for cells transfected with either of these full-length NS5A chimeras (Fig. 1D, lanes 2 and 5). These results thus demonstrate that major differences in the sequences and/or structures of the genotype 1a and 2a NS5A proteins preclude them from functioning interchangeably in support of viral RNA replication.
Very different results were obtained with the chimeras in which only domain III of NS5A was exchanged: both replicated in a fashion very similar to that of the related parental RNAs (Fig. 1C). A modest decrease in the accumulation of H77S/J5Ad3 RNA (H77S background with NS5A domain III of JFH-1) (Fig. 1B) relative to that of the parental H77S RNA, evident at 24 h only (Fig. 1C, left), was matched by a small reduction in the level of core protein expression detectable by immunoblotting (Fig. 1D, lane 3 versus lane 1). However, any impact of the domain III swap on viral RNA replication was minimal and was completely undetectable in the JFH-1 background. This indicates that the major incompatibilities that preclude the functional exchange of the full-length NS5A proteins with respect to viral RNA replication reside upstream of domain III and in domain I or II.
Intergenotypic NS5A domain exchanges and infectious virus production.
Infectious virus particles released by RNA-transfected Huh-7.5 cells were quantified by a fluorescent-focus infectivity assay (49, 52). Although no overlay is used in this assay, we have confirmed that there is a linear relationship between the number of focus-forming units (FFU) obtained and the dilution of an HCV inoculum (our unpublished data). This correlation indicates that each focus is initiated by a single infectious particle and validates the assay. Both parental RNAs (H77S and JFH-1) produced infectious virus that was detectable in culture supernatant fluids by 3 days posttransfection, although about 10-fold more infectious virus was secreted into the medium of cells transfected with JFH-1 versus H77S RNA (Fig. 2A), as described previously (49). Nonetheless, the titer of the infectious H77S virus produced in these experiments was sufficient to reliably determine the impact of NS5A domain exchanges on infectious virus production. No infectious virus was released from cells transfected with the H77S/J5A or JFH-1/H5A chimeric RNAs even when monitored up to 5 days after transfection. This is consistent with the impairments found in viral RNA replication described above (Fig. 2A). In contrast, the intergenotypic exchange of domain III only (H77S/J5Ad3 and JFH-1/H5Ad3) resulted in RNAs that could produce infectious virus despite the low level of amino acid sequence identity shared by these domains (46%) (Fig. 1A and 2A).
Fig. 2.
(A) Virus yields (extracellular culture fluids) from RNA-transfected cells after intergenotypic (genotype 1a→2a and genotype 2a→1a) exchanges of NS5A or NS5A domain III. Culture supernatants obtained at day 3 after transfection were assayed for infectious virus. Shown are the mean yields from each chimera in triplicate transfections ± standard deviations (SD). (B) ELISA for the HCV core protein present within supernatant culture fluids of RNA-transfected cells. Data shown represent mean values ± SE determined from duplicate experiments.
Interestingly, when domain III of H77S NS5A was placed within the JFH-1 background, there was a significant increase (variable but averaging almost 10-fold in replicate experiments) in the infectious virus yield (Fig. 2A). As discussed above, this occurred without any significant change in intracellular RNA replication (Fig. 1C, right) or intracellular core abundance (Fig. 1D, lanes 4 and 6). The reverse exchange (domain III of JFH-1 into the H77S background) resulted in a ∼60% decrease in the infectious virus yield at 3 days (Fig. 2A). While there was no difference in the intracellular abundances of the H77S and H77S/J5Ad3 RNAs at 48 to 72 h (Fig. 1C, left), the intracellular core abundance was somewhat reduced in H77S/J5Ad3-transfected cells (Fig. 1D, lane 1 versus lane 3). Collectively, these results are consistent with a major role for domain III of NS5A in the assembly of infectious particles, as described previously (2, 16, 28, 39). These results also demonstrate that domain III from genotype 1a NS5A and domain III from genotype 2a NS5A function interchangeably with respect to virus assembly and release, albeit with various efficiencies, and that the substantially higher yield of infectious virus obtained with JFH-1 than that obtained with H77S is not due to a better functionality in this domain of NS5A. In fact, somewhat surprisingly, the H77S NS5A domain functions more efficiently in virus assembly and release than does the native JFH-1 NS5A domain.
The amount of core protein released into supernatant cell culture fluids has been used as a surrogate measure of the production of infectious JFH-1 virus (29) but does not correlate well with the release of infectious H77S virus from cells (49). As shown in Fig. 2B, the core protein was detected by ELISA in medium from cultures transfected with each of the chimeric RNAs that were capable of producing infectious virus but not with the two RNAs with complete NS5A swaps and impaired viral RNA replication (Fig. 2B). However, while the reduction in amount of infectious H77S/J5Ad3 virus compared to H77S was matched by a comparable reduction in the amount of core protein released into the supernatant fluids, this was not the case with the JFH-1/H5Ad3 chimera. The reproducible increase in virus yield observed with JFH-1/H5Ad3 versus JFH-1 RNA (Fig. 2A) was not matched by an increase in the extracellular core abundance (Fig. 2B). We conclude from these results that the measurement of extracellular core abundance by ELISA is not always a reliable indicator of cell culture-infectious HCV yield despite its use as such in the past.
NS5A expression and infectious virus yields.
While the level of intracellular core expression was greater in cells transfected with JFH-1 RNA versus H77S RNA (and their related domain III chimeras), immunoblot analyses suggested much greater differences in NS5A expression (Fig. 1D). A much stronger NS5A signal, appearing as two distinct bands (presumably hypo- and hyperphosphorylated isoforms), was present in immunoblots of lysates of the genotype 2a JFH-1-transfected cells than in lysates from the genotype 1a H77S-transfected cells. Interestingly, there was a rough correlation between the abundance of NS5A and the production of infectious virus by the 4 different RNAs. The NS5A expression levels were highest with JFH-1/H5Ad3, somewhat lower in cells transfected with JFH-1 RNA, and markedly reduced, evident only as a single band and only with the prolonged exposure of the blots, with H77S (Fig. 1D, compare lanes 6, 4, and 1). An even fainter, single band was at the extreme limits of detection in lysates of cells transfected with H77S/J5Ad3 (Fig. 1D, lane 3). Despite the substantial difference in the amino acid sequences of the H77S and JFH-1 proteins, this striking difference in the apparent abundance of NS5A is unlikely to be due to antigenic variation. The monoclonal antibody used in these blots (9E10) was raised against a genotype 1b (Con1 strain) NS5A protein (22), the amino acid sequence of which is 80% identical to that of H77S NS5A but only 61% identical to that of JFH-1. In addition, similar results were observed with a rabbit polyclonal antibody to genotype 1b NS5A (data not shown). These results are thus more likely to reflect significant differences in absolute NS5A abundances.
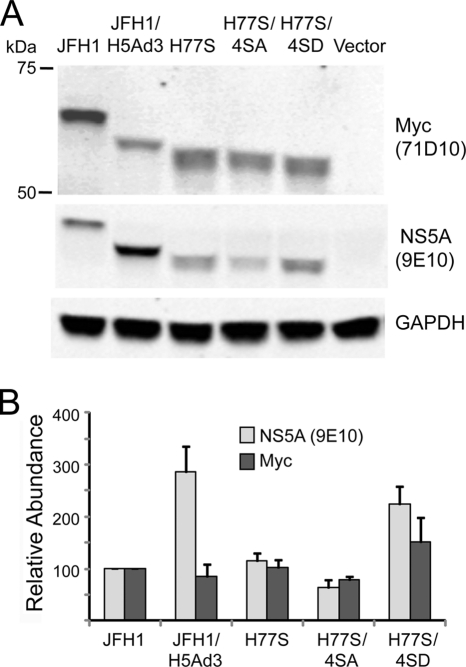
Nonetheless, to confirm that the 9E10 antibody efficiently detects H77S NS5A and to ascertain whether there is any bias in its recognition of genotype 1a versus genotype 2a NS5A, we ectopically expressed these proteins with an N-terminal Myc tag in Huh-7.5 cells. We then probed immunoblots of cell lysates with 9E10 and an anti-Myc antibody in parallel (Fig. 3A), quantifying the signal obtained with each antibody. These results revealed that the JFH-1/H5Ad3 chimeric protein bound significantly more 9E10 than anti-Myc antibody (explaining the more intense labeling of this protein in Fig. 1D) but demonstrated that there is no significant difference in the recognition of H77S versus JFH-1 NS5A by 9E10 (Fig. 3B). This confirms that the remarkable differences observed for immunoblots of NS5A in lysates of cells transfected with H77S versus JFH-1 RNA (Fig. 1D) reflect absolute differences in protein abundance. In contrast to what was observed for cells transfected with replication-competent viral RNAs (Fig. 1D), ectopically expressed JFH-1 NS5A migrated as a single protein band (Fig. 3A). A direct comparison indicated that this was the hypophosphorylated species, while ectopically expressed H77S NS5A appeared to be hyperphosphorylated (data not shown).
Fig. 3.
Ectopically expressed NS5A proteins. (A) Expression vectors encoding various NS5A molecules, each with an N-terminal Myc tag, were transfected into Huh-7.5 cells along with a second vector expressing Gaussia luciferase as a transfection control. Cell lysates were prepared 72 h later and subjected to SDS-PAGE followed by immunoblotting with the 9E10 monoclonal antibody or an anti-Myc antibody. The results shown are representative of data from replicate experiments. (B) 9E10 and anti-Myc antibodies bound by NS5A proteins in parallel immunoblots of cell lysates were detected with an infrared fluorescent probe and quantified by using an Odyssey fluorescent scanner. Results for each antibody were normalized to the amount bound by the JFH-1 protein. The results shown represent the means ± SE from 3 separate transfection experiments. The JFH-1/H5Ad3 protein demonstrated a greater binding of 9E10 than of anti-Myc, but there are no significant differences in the recognitions of JFH-1 and H77S NS5A by these antibodies. The equivalent abundance when the NS5A proteins were probed by anti-Myc antibody suggests that there are no significant differences in the intrinsic stabilities of these proteins. Differences in transfection efficiency, monitored by assessing luciferase activity, were minimal.
Collectively, these results suggest that the infectious virus yield might be limited by the abundance of NS5A more than viral RNA replication. Since lesser differences were observed for the expression levels of the core protein (and core and NS5A are processed from the same polyprotein), the large differences observed in NS5A abundance suggest that the H77S NS5A protein may be less stable than JFH-1 NS5A. Immunoblots showed no consistent difference in the abundances of H77S NS5A and JFH-1 NS5A when these proteins were expressed ectopically in Huh-7.5 cells (Fig. 3), indicating that there is little intrinsic difference in the stabilities of these proteins. While it remains possible that the stabilities of the phosphorylated proteins differ within infected cells, the low level of NS5A expressed by replicating H77S RNA precluded a formal comparison of its stability with the JFH-1 protein.
It would not be surprising if differences in the phosphorylation status of NS5A also impacted the function of the proteins expressed by genotype 1a and 2a RNAs. Differences in the predicted molecular masses of the genotype 1a, 2a, and chimeric NS5A proteins (Fig. 1D) confound a direct comparison of their phosphorylation statuses by simple gel analysis. Nonetheless, there are clear differences evident in the immunoblot shown in Fig. 1D. JFH-1 NS5A was expressed as two isoforms of approximately equal abundance, with a modest increase in the hypophosphorylated form in cells transfected with the JFH-1/H5Ad3 RNA relative to the hyperphosphorylated isoform (Fig. 1D, compare lanes 4 and 6). In some but not all experiments, we also noted a small amount of a third JFH-1/H5Ad3 band migrating with an apparent molecular mass higher than that of the hyperphosphorylated form (Fig. 1D, lane 6, “s”). In contrast, as noted above, only a single NS5A band was evident in cells transfected with H77S and H77S/J5Ad3 RNAs. While the phosphorylation status of the single H77S NS5A band was difficult to assess with these experiments, it is interesting that it migrated more rapidly in SDS-PAGE gels than did the hypophosphorylated JFH-1/H5Ad3 isoform despite having a higher predicted molecular mass in the absence of posttranslational modifications (49.1 versus 47.9 kDa) (Fig. 1D, compare lanes 1 and 6). We were unable to detect H77S NS5A in these cell lysates following treatment with λ-PPase (data not shown). The apparent molecular mass of the H77S protein in SDS-PAGE gels is addressed in additional detail below.
Domain III exchange promotes intracellular assembly of JFH-1 virus.
The enhanced release of infectious virus by cells transfected with the chimeric JFH-1/H5Ad3 RNA compared to that of its JFH-1 parent could be due to either an enhanced assembly of intracellular particles or a facilitation of the release of such particles from the cell. To address this question, we determined the amounts of infectious virus present in intracellular and extracellular fractions of RNA-transfected cells. Multiple freeze-thaw cycles were used to prepare intracellular lysates 3 days after transfection, and the titer of infectious virus present in these lysates and the related culture supernatant fluids was determined by the infectious focus assay. While a somewhat lesser (∼3-fold) increase in the extracellular infectious virus yield was observed for this series of experiments than for those described above when cells were transfected with the JFH-1/H5Ad3 chimera versus JFH-1 RNA, the abundance of intracellular infectious particles was increased ∼30-fold (Fig. 4). Thus, it is likely that the primary impact of the domain III swap is on enhanced viral assembly within the cell and that the observed increase in the release of infectious virus is secondary. Of all infectious virus present in these cultures at 3 days posttransfection, 88% of the JFH-1 virus was present in the extracellular fluids, compared to only 49% of the JFH-1/H5Ad3 virus.
Fig. 4.

Infectious virus titer of cell culture supernatant fluids and intracellular lysates of JFH-1 and JFH-1/H5Ad3 RNA-transfected cells on day 3 after transfection. Lysates were prepared by multiple freeze-thaw cycles. Shown are the means ± SE from duplicate transfections. Percentages are the proportions of the total virus yield released into supernatant (Sup) culture fluids (i.e., virus in supernatant/virus in supernatant + virus in lysate).
Since it is possible that the increase in the amounts of intracellular infectious particles observed with the JFH-1/H5Ad3 RNA could reflect an enhanced interaction between NS5A and the core protein coating lipid droplets (28, 29), we compared the intracellular localizations of core and NS5A in cells transfected with JFH-1/H5Ad3 and JFH-1 RNAs using laser scanning confocal microscopy. These studies demonstrated no detectable differences in the cellular localizations or in the degrees of colocalization of these two viral proteins (data not shown).
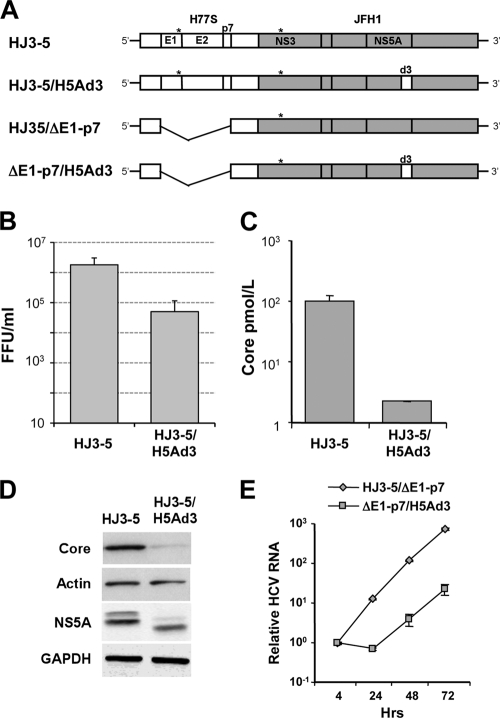
As an alternative approach to assessing the role of the structural proteins in the enhanced production of infectious virus by JFH-1/H5Ad3, we created a similar domain III swap in the background of the HJ3-5 virus, an intergenotypic chimera containing the entire structural protein region (core through NS2) from the genotype 1a H77S virus in the background of the genotype 2a JFH-1 RNA (27, 47) (Fig. 5A). The JFH-1/H5Ad3 construct thus places H77S domain III back in context with the H77S core protein. In contrast to the increase in the infectious virus yield that we observed when domain III from H77S was placed into the JFH-1 background (Fig. 2A), swapping the H77S domain III into HJ3-5 caused nearly a 10-fold reduction in the infectious virus yield (Fig. 5B) and a 30-fold decrease in core protein secretion (Fig. 5C). This was associated with a marked decrease in core protein expression levels as well as a lower abundance of NS5A with the loss of most of the hyperphosphorylated isoform (Fig. 5D).
Fig. 5.
Replication and infectious virus yields from the structural protein chimera HJ3-5 with or without exchange of domain III of NS5A from H77S virus. (A) Schematic showing the organization of the HJ3-5 intergenotypic chimeric RNA and the related NS5A domain III (d3) swap with the H77S sequence. The H77S sequence (core-NS2) is shown as open boxes, while the JFH-1-derived sequence (NS3-NS5B) is shaded; noncoding RNA segments are from JFH-1. The asterisks indicate the location of compensatory mutations (E1 and NS3) that promote yields of infectious virus from the chimera (27). In HJ3-5/ΔE1-p7 and ΔE1-p7/H5Ad3, the E1-p7 sequence has been deleted. (B) Infectious virus production. Shown are the means ± ranges of yields of infectious virus from chimeric RNAs, calculated from duplicate transfections. (C) ELISA for core protein secreted by cells transfected with HJ3-5 and HJ3-5/H5Ad3 RNAs. Means ± SE were calculated from duplicate experiments. (D) Immunoblots for the HCV core protein in lysates of HJ3-5 and HJ3-5/H5Ad3 RNA-transfected cells. Actin served as a loading control. Also shown are immunoblots for NS5A in lysates of cells transfected with the related ΔE1-p7 mutant RNAs; GAPDH was the loading control. (E) Real-time RT-PCR measurements of HCV RNA replication. The relative HCV RNA copy number represents the copy number for each construct relative to a replication-lethal GND mutant, normalized to the value present 4 h after transfection. Results shown represent the means ± SE calculated from triplicate RT-PCR assays.
The NS5A proteins encoded by the JFH-1 and HJ3-5 viruses are identical, and the proteins expressed by these two RNAs each show a slight dominance of the faster migrating of the two major isoforms of NS5A (compare Fig. 1D and 5D). JFH-1/H5Ad3 and HJ3-5/H5Ad3 (which share the same chimeric NS5A sequences in the background of JFH-1 nonstructural proteins) were very different in this regard. JFH-1/H5Ad3 was much like JFH-1, with only a slight dominance of the fast-migrating species, while HJ3-5/H5Ad3 expressed mostly the hypophosphorylated isoform, with little of the more slowly migrating species present (compare again Fig. 1D and 5D). To ascertain whether the domain III swap influenced RNA replication in the context of the HJ3-5 chimera, we compared RNA replication kinetics in cells transfected with modified genomes in which the envelope-coding regions had been deleted (to prevent any confounding influence from infectious virus production and virus spread in the transfected cell culture). These results revealed that the NS5A domain III swap had a substantial, negative effect on RNA replication in the context of the genotype 1a/2a HJ3-5 chimera, resulting in a delay in RNA amplification and a reduction in intracellular viral RNA accumulation of approximately 10-fold (Fig. 5E). This is different from the absence of any such effect when the H77S domain III sequence was placed into the JFH-1 virus (Fig. 1C, right) and explains the reduced core abundance in cells transfected with HJ3-5/H5Ad3 RNA. However, the NS5A abundance was well preserved relative to the amount of core protein present in these cells (Fig. 5D). Collectively, these data provide genetic evidence for interactions between NS5A and the upstream structural proteins that influence the NS5A phosphorylation status and the efficiency of genome amplification. While most previous studies indicated that mutations in domain III have little impact on the replication of viral RNA, recent data from Hughes et al. (16) are also consistent with domain III playing a role in RNA replication.
Virus production mediated by domain III of H77S NS5A is sensitive to a CK II inhibitor.
RNA interference (RNAi)-mediated gene silencing and chemical inhibitor studies reported previously by Tellinghuisen et al. (39) suggested that host CK II phosphorylates Ser-457 in domain III of the JFH-1 NS5A protein and that this is essential for the production of infectious virus. To determine whether the assembly function of domain III in H77S is equivalently sensitive to CK II inhibition, we assessed the effect of the CK II inhibitor 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidaozle (DMAT) on the production of infectious virus by JFH-1/H5Ad3 RNA, which contains H77S domain III in the JFH-1 background. Cells transfected with JFH-1 RNA were treated in parallel and demonstrated a modest decrease in infectious virus yields at 1 μM and a marked decrease at a 10 μM concentration (Fig. 6A, left). While the specificity of a kinase inhibitor is always cause for concern, the data confirm that JFH-1 virus production is inhibited by DMAT (39). However, under the conditions that we used, virus production appeared to be less sensitive to inhibition by the compound than reported previously. Importantly, JFH-1/H5Ad3 RNA was equally sensitive to the inhibition of infectious virus production by DMAT (Fig. 6A, right). The abundances of NS5A and NS2 were moderately reduced at 10 μM, while the abundance of NS3 did not seem to be affected (Fig. 6B). Higher concentrations of DMAT were not tested, as 10 μM was at the cusp of significant cellular toxicity under the conditions used, as determined by WST-1 cellular proliferation assays (Fig. 6C). These results are consistent with the H77S and JFH-1 domain III assembly functions being equivalently sensitive to DMAT inhibition, although the effects of the compound, both on virus production and on cellular toxicity, were less than those previously described (39).
Fig. 6.
Inhibition of virus production by DMAT, an inhibitor of CK II. (A) Following transfection of the indicated RNA, cells were treated with the indicated concentration of DMAT for 48 h. The medium was then replaced with fresh medium (no drug), followed 24 h later by the harvesting of supernatant fluids for virus titration. Means ± SE were calculated from duplicate experiments. (B) Immunoblots for NS5A, NS2, NS3, and GAPDH from cell lysates prepared 72 h after transfection. (C) Cytotoxic effects of DMAT assessed by a WST-1 cellular proliferation assay. Means ± SE were calculated from triplicate experiments.
A conserved cluster of Ser residues in domain III of NS5A regulates production of infectious H77S virus.
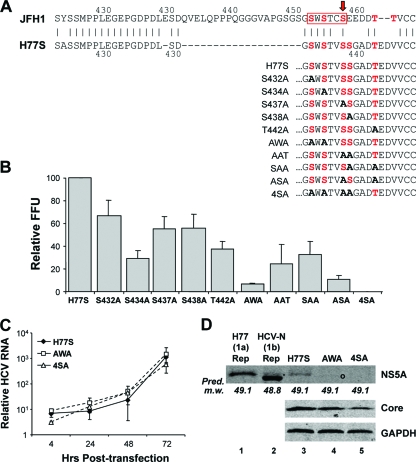
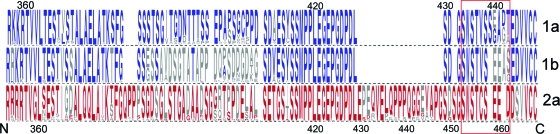
There is only a limited understanding of which of the many potential phosphoacceptor sites in the NS5A sequence are actually phosphorylated during virus replication, the functional role of such phosphorylation, or which cellular kinases are responsible for it. Ser-349, near the C terminus of domain II, was suggested previously to be a major site of phosphorylation in the wild-type H77S NS5A protein (34), while mutational analyses suggested that phosphoacceptor sites in domain III are important for basal phosphorylation (1). While Ala replacements of these residues generally do not affect RNA replication, Tellinghuisen et al. (39) suggested previously that the CK II-mediated phosphorylation of Ser-457 of the JFH-1 NS5A protein (domain III) is essential for intracellular virus assembly. Ser-457 was predicted to be a site of CK II phosphorylation by the Web-based NetPhosK server (5). It is located within an ideal sequence context for CK II phosphorylation, just upstream of several acidic residues (“SEED,” where S is Ser-457) (39). Although the amino acid sequences of domain III of the JFH-1 and H77S proteins have only 46% identity, there are two short regions of exact identity: H77S residues 414 to 428 (also residues 414 to 428 in JFH-1) and residues 431 to 435 (residues 451 to 455 in JFH-1) (Fig. 7A). Interestingly, the latter of these two regions of identity is immediately upstream of Ser-457 in the JFH-1 sequence (Fig. 7A). This sequence also overlaps a domain identified previously by Masaki et al. (28) (residues 452 to 457) as being essential for the recruitment of NS5A to the core protein decorating lipid droplets, a necessary early step in JFH-1 virus assembly (29). While the results of Tellinghuisen et al. (39) and those of Masaki et al. (28) differ significantly in some respects, in aggregate they suggest that 3 potential Ser phosphoacceptor sites within this region (Ser-452, Ser-454, and Ser-457) are important for JFH-1 assembly. All three of these Ser residues are conserved in the H77S sequence (Ser-432, Ser-434, and Ser-437) (Fig. 7A), and their presence in the H77S protein could thus account for the ability of H77S domain III to function in the assembly of the JFH-1/H5Ad3 virus. However, the H77S sequence lacks the CK II “SEED” motif identified previously by Tellinghuisen et al. (39), and the NetPhos 1.0 server predicts instead (with very low probability) the CK II phosphorylation of Ser-438 and Thr-442 (Fig. 7A).
Fig. 7.
Impact of Ala substitutions at potential Ser/Thr phosphoacceptor site residues in domain III of H77S on viral RNA replication and infectious virus production. (A) Possible Ser/Thr phosphoacceptor sites in the C-terminal region of domain III of the NS5A proteins of JFH-1 and H77S viruses. At the top, the H77S and JFH-1 sequences are aligned: Ser residues previously found to be important for the NS5A-core interaction and the assembly and release of infectious JFH-1 virus by Masaki et al. (28) (red box) and Ser-457, identified by Tellinghuisen et al. (39) as a site of CK II phosphorylation (red arrow), are highlighted. Within the related H77S sequence, Ser-438 and Thr-442 are possible sites of CK II phosphorylation predicted by the NetPhos 1.0 server. Below are the C-terminal NS5A sequences of the single- and multiple-Ala-substitution mutants studied. Potential Ser/Thr phosphoacceptor sites studied are shown in red, while Ala substitutions ablating these sites in the mutants are shown in boldface type. (B) Potential phosphorylation sites near the C terminus of domain III of H77S NS5A were mutated to Ala to assess their role(s) in infectious virus production. Shown are the mean virus yields following the transfection of the mutant RNAs, normalized to that of the H77S virus (100% = mean of 160 FFU/ml) ± SE from two independent transfections. (C) HCV RNA replication compared to the replication-lethal GND mutant measured by a real-time RT-PCR assay. The means ± SE were calculated from triplicate RT-PCR assays. (D) Immunoblots for HCV NS5A, core, and GAPDH in lysates of RNA-transfected cells. Pred. m.w., predicted molecular weight (in thousands).
To ascertain the role of these H77S residues in the assembly and release of the H77S virus, we created a series of 5 H77S RNA mutants in which Ser-432, Ser-434, Ser-437, Ser-438, and Thr-442 were individually replaced with Ala and assessed their abilities to produce infectious virus following transfection into Huh-7.5 cells. The results of these experiments, repeated with each construct in at least two replicate experiments with independently transcribed RNAs, are shown in Fig. 7B. Each of these single-site mutations had an adverse impact on infectious virus production, with the most profound effect being associated with S434A (∼70% reduction in the infectious virus yield) and T442A (∼60%). Alanine substitutions at Ser-437 and Ser-438 had only a modest impact (∼45% decrease), while the S432A mutant was least affected (∼35%). Because Masaki et al. (28) found cumulative effects of multiple Ala substitutions in this region of the JFH-1 protein, we also evaluated several H77S mutants in which two of the five potential Ser/Thr phosphoacceptor sites were replaced with Ala. Two of these mutants demonstrated substantially greater defects in virus production: mutant AWA, containing both S432A and S434A substitutions (∼92% reduction), and mutant ASA, containing both S437A and T442A substitutions (∼88% reduction) (Fig. 7B). While none of the double-Ala substitutions that we evaluated completely abolished infectious virus production, no detectable infectious virus was released from cells transfected with a mutant in which each of the 4 suspect Ser residues was replaced with Ala (mutant 4SA) (Fig. 7B). Importantly, the mutants demonstrating the greatest defects in infectious virus production were capable of nearly wild-type RNA replication (Fig. 7C), and cells transfected with these RNAs expressed core at an abundance equal to or only slightly reduced from that of the parental H77S RNA (Fig. 7D, compare lane 3 with lanes 4 and 5). The results of this mutational analysis thus indicate that the assembly and release of infectious genotype 1a H77S virus, like genotype 2a JFH-1 virus (28), are likely to be dependent upon the phosphorylation of several members of a cluster of conserved Ser residues near the extreme C terminus of domain III. While a role for Thr phosphorylation in NS5A has not been identified previously for HCV assembly, Thr-442 appears to be necessary for the efficient production of the H77S virus but is not by itself sufficient in the absence of two or more of the upstream Ser residues.
To assess the impact of these mutations on the phosphorylation status of NS5A, we subjected lysates of RNA-transfected cells to immunoblot analysis. Lysates of cells containing genotype 1a (H77) and genotype 1b (HCV-N) replicons (17, 46) were studied in parallel, as this comparison provided some additional information on the phosphorylation status of NS5A expressed by H77S RNA. Lysates of HCV-N replicon cells demonstrated two distinct NS5A bands, the lower of which accounted for most of the protein present and the upper of which, while of low abundance, comigrated in SDS-PAGE gels with the single NS5A band present in either the H77S replicon cells or cells transfected with H77S RNA (Fig. 7D, compare lane 2 with lanes 1 and 3). Since the genotype 1a H77S and genotype 1b HCV-N proteins have relatively similar predicted molecular masses in the absence of posttranslational modifications (49.1 and 48.8 kDa, respectively), this suggests that the single H77S band that we observed is likely to represent the hyperphosphorylated form of the protein. Importantly, little NS5A protein was detected in lysates of the AWA and 4SA mutant-transfected cells (Fig. 7D, lanes 4 and 5). A very faint band migrating slightly faster than that of H77S NS5A was evident in the AWA lysates with increased exposure times (lane 4, marked by “o”), while no NS5A was detected in 4SA mutant-transfected cell lysates despite the presence of appreciable amounts of the core protein.
Rescue of virus production with Ala-to-Asp substitutions in AWA and 4SA mutants.
To more completely document the loss of infectious virus production accompanying the 4SA mutation in the H77S virus (Fig. 7B), we engineered these mutations into pH77S.3, a modified version of pH77S that produces ∼5-fold more infectious virus from RNA-transfected cells (our unpublished data). In replicate experiments, H77S.3 produced over 700 FFU/ml by 3 days posttransfection, while no virus (<10 FFU/ml) was produced by H77S.3/4SA (Fig. 8A), confirming almost a 100-fold reduction in infectious virus production with Ala substitutions at these 4 residues near the C terminus of NS5A. To determine whether the replacement of these Ala substitutions with a residue containing a phosphomimetic side chain, Asp, would rescue the production of infectious virus, we constructed an H77S.3/4SD mutant as well as H77S.3/DWD in which the Ala residues at positions 432 and 434 (Fig. 7B) were replaced with Asp. In sharp contrast to the AWA mutant, H77S.3/DWD produced almost as much infectious virus as H77S.3, while the 4SD mutant regained the capacity to produce infectious virus, albeit at a level substantially lower than that of H77S.3 (Fig. 8B). Remarkably, NS5A expression, which was much reduced with the AWA and 4SA mutants despite the absence of any significant reduction in viral RNA synthesis, was largely restored with the DWD and 4SD mutants (compare Fig. 7D with 8B, bottom). Taken collectively, the data shown in Fig. 7 and 8 are strongly suggestive of the need for phosphorylation at 2 or more of the 4 Ser residues located at positions 432, 434, 437, and 438 of NS5A for the production of infectious H77S virus. The changes noted for the apparent abundance of NS5A in the mutants (Fig. 7D and 8A) may be due in part to the decreased stability of the mutant proteins. When ectopically expressed as Myc-tagged proteins in Huh-7.5 cells, the abundance of H77S/4SA ranged from 55 to 71% of that of the wild-type H77S protein (depending on whether it was detected in immunoblots with the 9E10 monoclonal antibody or anti-Myc), while H77S/4SD was 1.5- to 1.9-fold more abundant (Fig. 3B).
Fig. 8.
Phosphomimetic Ala-to-Asp substitutions restore production of infectious virus by AWA and 4SA mutant RNAs. Shown are infectious virus yields from cells transfected with H77S.3 and H77S.3/4SA RNAs (A) and H77S.3, H77S.3/DWD, and H77S.3/4SD mutants (B). Results shown are the means ± SE from two transfections with independent RNA transcripts. At the bottom are immunoblots of NS5A and core protein expression with actin (A) and GAPDH loading controls (B).
DISCUSSION
A long-standing limitation of hepatitis C virus research has been a lack of virus strains that are capable of replicating well both in cell culture and in chimpanzees, still the only available animal model of chronic hepatitis C virus. While self-amplifying RNA replicons have been successfully established by using cloned sequences from multiple HCV strains, very few viral RNAs can be coaxed to produce detectable quantities of infectious virus in cell culture (33, 44, 49). This may be due in part to conflicting effects of cell culture-adaptive mutations on RNA replication and infectious particle production (7, 33), but many gaps remain in our understanding of the process of virus assembly. Multiple lines of evidence support a role for NS5A, and in particular domain III of NS5A, in the assembly of infectious HCV particles (2, 16, 28, 39). We studied the role of this domain in the genotype 1a H77S virus by creating chimeric viruses in which the entire NS5A sequence or only domain III was swapped between it and the JFH-1 virus (genotype 2a). Both of these HCV RNAs are competent for infectious virus production when transfected as RNA into permissive cells, but they generate quantitatively different yields of virus. Cells transfected with H77S RNA release between 102 and 103 FFU/ml into cell culture supernatant fluids between 48 and 72 h after transfection under the conditions used in this study, while JFH-1 RNA produces approximately 10-fold more at these early time points (Fig. 2A). While the exchange of the entire NS5A sequence resulted in RNAs that were impaired in their abilities to replicate in transfected cells, the domain III chimeric RNAs replicated well and produced infectious progeny albeit with altered efficiencies. Although the domain III sequences of these two viruses share less than 50% amino acid identity, these experiments demonstrate the existence of a conserved assembly function in domain III that can function in the context of either a genotype 1a or 2a virus background.
A rather surprising result to emerge from these studies was the increase that we observed for the production of infectious JFH-1 virus when domain III from the H77S virus, which produces relatively low virus yields, replaced the related sequence in JFH-1 (Fig. 2A). This result shows clearly that the contrasting capacities of the JFH-1 and H77S genomes to produce infectious particles result from differences in the genomes outside domain III of NS5A. Nonetheless, we observed a good correlation between the apparent abundance of the NS5A protein and the efficiency of infectious virus production: across both chimeras and the 2 parental genomes, the abundance of NS5A and the production of infectious virus were similarly ordered (Fig. 1D). Although matched to some extent by differences in core protein expression levels, the differences in NS5A abundances could not be explained entirely by an altered RNA replication efficiency, as the domain III swaps had a minimal impact on this (Fig. 1C). The contrasts that we observed for the abundances of NS5A in JFH-1 versus H77S RNA-transfected cells were not mirrored by similar differences in the abundances of NS5A when these proteins were expressed ectopically in Huh-7.5 cells (Fig. 3). While this suggests that the stabilities of JFH-1 NS5A and H77S NS5A are not intrinsically different, JFH-1 NS5A was mostly hypophosphorylated when expressed as a solitary protein, and it remains possible that there are significant differences in the stabilities of NS5A expressed from replication-competent H77S and JFH-1 RNAs.
Within the domain III sequences of JFH-1 and H77S, there are 2 regions with striking amino acid identity that reside near the C terminus of NS5A and are separated by a large insertion in the JFH-1 sequence (Fig. 7A). The most C terminal of these regions of identity contains a number of potential phosphoacceptor sites that were strongly implicated in the assembly of the JFH-1 virus by Tellinghuisen et al. (39) and Masaki et al. (28). We observed that Ala substitutions at conserved Ser residues in this region had a strong negative impact on the production of infectious virus from H77S RNA (Fig. 7B), suggesting that the shared domain III assembly functions of the H77S and JFH-1 proteins may be dependent upon these conserved residues. Ala substitutions at these sites also caused a marked reduction in the abundance of NS5A without similar changes in viral RNA replication efficiency or core protein expression (Fig. 7C and D). Although a direct biochemical investigation of NS5A phosphorylation was outside the scope of the experiments that we report in this communication, the latter results, coupled with the positive impact of phosphomimetic substitutions at these residues (Fig. 8B), support the possibility that these are phosphoacceptor sites. Mutations at these residues cause modest changes in the stability of NS5A, as indicated by differences in protein abundance when H77S NS5A was expressed ectopically (Fig. 3B). More pronounced changes in stability could result directly from the altered phosphorylation of NS5A when expressed from a replication-competent RNA or from changes in its ability to interact with other viral proteins, including core (28), or any of its numerous cellular binding partners (43). Unfortunately, NS5A expression from H77S RNA, and particularly from these NS5A mutants, was insufficient for direct pulse-chase measurements of protein stability.
The NetPhosK prediction program (5) suggests that Ser-457 in JFH-1 NS5A may be phosphorylated by CK II. This is consistent with its location upstream of several acidic residues and with the results of Tellinghuisen et al. (39). This residue is also located within the C-terminal region of JFH-1/H77S sequence identity described above (Fig. 7A) and lies within the Ser cluster identified previously by Masaki et al. (28) (residues 452 to 457) as being essential for the recruitment of NS5A to the core protein associated with lipid droplets during early JFH-1 virus assembly (29). The amino acid sequence of this region of JFH-1 NS5A (SWSTCS [residues 452 to 457]) differs from that of H77S by only a single residue (SWSTVS [residues 432 to 437]) (Fig. 7A). However, there are fewer acidic residues downstream of the conserved Ser-437 in H77S (homologous to Ser-457 in JFH-1) and only low-probability predictions of CK II phosphorylation in this region of domain III by the NetPhos I.0 server at Ser-438 and Thr-442. Although infectious virus production by JFH-1 and JFH-1/H5Ad3 RNA was inhibited to a similar extent by DMAT, a CK II inhibitor (Fig. 6), an Ala substitution at Ser-437 in domain III of H77S caused only a modest decrease in infectious virus yield (Fig. 7B). The Ser-437-to-Ala mutation was not as inhibitory to virus production as similar Ala substitutions at Ser-434 or Thr-442. Attempts to assess the impact of the small interfering RNA (siRNA)-mediated knockdown of either or both of the CK II isoforms did not provide a clear answer as to the role of this kinase in the H77S NS5A-mediated production of infectious virus (data not shown). Taken together, however, our results indicate that there is no special role for Ser-437 phosphorylation in the production of infectious H77S virus and suggest instead that virus production is dependent upon multiple Ser residues, and likely Thr-442 as well, acting in a redundant or possibly cooperative fashion. Phosphorylation at Ser-437, or any of the other residues in JFH-1 identified by Masaki et al. (28) or in H77S identified by us (Fig. 7), could modulate phosphorylation elsewhere in NS5A, including at other conserved residues within the C terminus of domain III.
These findings with the H77S virus are thus more consistent with the role of the serine cluster identified previously by Masaki et al. (28) in JFH-1 assembly than with the uniquely important role for Ser-457 phosphorylation suggested previously by Tellinghuisen et al. (39). While there is no clear explanation for the differences observed in these two prior studies of NS5A, it may be important that Masaki et al. (28) studied the assembly of the JFH-1 virus, while Tellinghuisen et al. (39) studied a J6/JFH-1 chimera in which the structural proteins were derived from another genotype 2a virus, J6. While both viruses were entirely genotype 2a in sequence, differences in the structural proteins could well have influenced the requirements for the phosphorylation of residues in domain III of NS5A. However, it is important to recognize in the context of this discussion that no study, including that described here, has yet directly demonstrated the phosphorylation of any residues in domain III of NS5A.
Although the JFH-1 virus has provided a very useful model system that recapitulates the entire HCV life cycle in cultured cells (44, 52), most persons with significant HCV-related liver disease are infected with genotype 1 viruses (51). It is important that findings to emerge from the JFH-1 system are also examined in a genotype 1 background, as we have done here with the role of domain III of NS5A in H77S assembly. An analysis of almost 600 different HCV sequences shows that the NS5A domain III sequence of H77S is broadly representative of all genotype 1a viruses and similar in many respects to the sequence of genotype 1b viruses as well (Fig. 9). Interestingly, this analysis revealed that while the Thr residue present at position 442 is typical of most genotype 1a NS5A sequences, the Thr residue that aligns with it at position 462 of JFH-1 is not found in most genotype 2a viruses. Despite considerable sequence divergence and potential differences in the contributions of specific residues to the assembly of infectious virus among different genotypes, our results point to similar mechanisms for the regulation of HCV assembly by domain III of both genotype 1a and 2a viruses. Still to be explained is why H77S lags so far behind JFH-1 in the production of infectious virus.
Fig. 9.
Sequence logo depiction of amino acid sequence conservation within domain III of NS5A among genotype 1a (224 sequences), genotype 1b (357 sequences), and genotype 2a (18 sequences) strains of HCV from the European HCV Database (euHCVdb) (8). Sequences were aligned with MUSCLE (10), with minor manual modifications, and logos were generated with WebLogo (9). The height of each single-character amino acid code is proportional to the representation of that amino acid at each position. The sequence extending from Ser-432 to Thr-442, which contains potential phosphoacceptor residues in H77S, is boxed in red. To show how representative the H77S sequence is of other genotype 1 viruses, residues identical to those in H77S are shown in blue in the genotype 1a and 1b sequences, while residues that differ from those in H77S are shown in gray. Residues identical to JFH-1 are shown in red in the genotype 2a sequence.
ACKNOWLEDGMENTS
We thank Yinghong Ma, Yuqiong Liang, and Rhykka Connelly for excellent technical assistance and Charles Rice, Tim Tellinghuisen, and Craig Cameron for making cell lines and/or antibodies available.
This study was supported in part by grants U19-AI400035, RO1-AI075090, and R01-DA024565 from the National Institutes of Health.
Footnotes
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Appel N., Herian U., Bartenschlager R. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Appel N., et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blight K. J., Kolykhalov A. A., Rice C. M. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 4. Blight K. J., McKeating J. A., Rice C. M. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649 [DOI] [PubMed] [Google Scholar]
- 6. Boulant S., Targett-Adams P., McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204–2213 [DOI] [PubMed] [Google Scholar]
- 7. Bukh J., et al. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 99:14416–14421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Combet C., et al. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 35:D363–D366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans M. J., Rice C. M., Goff S. P. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:13038–13043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao L., Aizaki H., He J. W., Lai M. M. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gastaminza P., Kapadia S. B., Chisari F. V. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanoulle X., et al. 2009. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 381:634–638 [DOI] [PubMed] [Google Scholar]
- 15. Huang Y., Staschke K., De Francesco R., Tan S. L. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1–9 [DOI] [PubMed] [Google Scholar]
- 16. Hughes M., Griffin S., Harris M. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J. Gen. Virol. 90:1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ikeda M., Yi M., Li K., Lemon S. M. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jirasko V., et al. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546–28562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones C. T., Murray C. L., Eastman D. K., Tassello J., Rice C. M. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaneko T., et al. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320–326 [DOI] [PubMed] [Google Scholar]
- 21. Lemon S. M., Walker C. M., Alter M. J., Yi M. 2007. Hepatitis C virus, p. 1253–1304 In Knipe D. M., et al. (ed.), Fields virology, 5th ed., vol. 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 22. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 23. Lindenbach B. D., et al. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3805–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 25. Love R. A., Brodsky O., Hickey M. J., Wells P. A., Cronin C. N. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83:4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma Y., et al. 2011. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 85:86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma Y., Yates J., Liang Y., Lemon S. M., Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Masaki T., et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyanari Y., et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 30. Moradpour D., et al. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nainan O. V., et al. 2006. Hepatitis C virus genotypes and viral concentrations in participants of a general population survey in the United States. Gastroenterology 131:478–484 [DOI] [PubMed] [Google Scholar]
- 32. Phan T., Beran R. K., Peters C., Lorenz I. C., Lindenbach B. D. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379–8395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pietschmann T., et al. 2009. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reed K. E., Rice C. M. 1999. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J. Biol. Chem. 274:28011–28018 [DOI] [PubMed] [Google Scholar]
- 35. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Shimakami T., et al. 4 November 2010, posting date Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology doi:10.1053/j.gastro.2010.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steinmann E., et al. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tanji Y., Kaneko T., Satoh S., Shimotohno K. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tellinghuisen T. L., Foss K. L., Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tellinghuisen T. L., Foss K. L., Treadaway J. C., Rice C. M. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tellinghuisen T. L., Marcotrigiano J., Gorbalenya A. E., Rice C. M. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 42. Tellinghuisen T. L., Marcotrigiano J., Rice C. M. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tellinghuisen T. L., Rice C. M. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419–427 [DOI] [PubMed] [Google Scholar]
- 44. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wozniak A. L., et al. 2010. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6:e1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yi M., Lemon S. M. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J. Virol. 78:7904–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yi M., Ma Y., Yates J., Lemon S. M. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yi M., Ma Y., Yates J., Lemon S. M. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yi M., Villanueva R. A., Thomas D. L., Wakita T., Lemon S. M. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yount B., Denison M. R., Weiss S. R., Baric R. S. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zein N. N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong J., et al. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]