Abstract
The intrahost evolution of hepatitis C virus (HCV) holds keys to understanding mechanisms responsible for the establishment of chronic infections and to development of a vaccine and therapeutics. In this study, intrahost variants of two variable HCV genomic regions, HVR1 and NS5A, were sequenced from four treatment-naïve chronically infected patients who were followed up from the acute stage of infection for 9 to 18 years. Median-joining network analysis indicated that the majority of the HCV intrahost variants were observed only at certain time points, but some variants were detectable at more than one time point. In all patients, these variants were found organized into communities or subpopulations. We hypothesize that HCV intrahost evolution is defined by two processes: incremental changes within communities through random mutation and alternations between coexisting communities. The HCV population was observed to incrementally evolve within a single community during approximately the first 3 years of infection, followed by dispersion into several subpopulations. Two patients demonstrated this pattern of dispersion for the rest of the observation period, while HCV variants in the other two patients converged into another single subpopulation after ∼9 to 12 years of dispersion. The final subpopulation in these two patients was under purifying selection. Intrahost HCV evolution in all four patients was characterized by a consistent increase in negative selection over time, suggesting the increasing HCV adaptation to the host late in infection. The data suggest specific staging of HCV intrahost evolution.
INTRODUCTION
Hepatitis C virus (HCV) infection is a major cause of liver disease in the world. It is estimated that ∼130 million people are infected with HCV globally (2). HCV is a heterogeneous single-stranded (plus-strand) RNA virus belonging to the Flaviviridae. The HCV genome contains one large open reading frame that encodes a polyprotein which can be processed into 10 mature proteins (34). HCV causes chronic infection in 70 to 85% of infected adults. There is no vaccine against HCV, and current antiviral therapy is effective in only 50 to 70% of chronically infected patients (18).
HCV intrahost evolution is frequently compared to an “arms race,″ implying that the HCV genome constantly changes in order to escape from neutralizing adaptive immunoresponses (53). This concept of constant change is seemingly different from the HIV model, according to which intrahost evolution slows down with CD4+ T-cell depletion and quasispecies diversity decreases with the development of AIDS (48). Our limited understanding of the dynamics of HCV intrahost evolution impedes the development of efficient therapeutic and prophylactic interventions.
Analysis of HCV longitudinal evolution is significantly hindered by difficulties with identifying and with long-term follow-up of patients, starting from the acquisition of infection. In the present study, we explored HCV evolutionary processes during long-term chronic infection in four treatment-naïve patients who were followed up from the acute stage of infection for 9 to 18 years. The results suggested significant adaptation of HCV to the host late in infection and a complex dynamic of intrahost viral subpopulations.
MATERIALS AND METHODS
Samples and nucleic acid isolation.
HVR1 quasispecies analysis using endpoint, limiting dilution PCR (EPLD-PCR) was conducted with 42 serial serum specimens collected from 4 patients who were followed from the acute to the chronic phase of HCV infection over 9 to 18 years. The initial study started with 130 patients who were identified as having non-A, non-B hepatitis (3). All patients were negative for hepatitis B surface antigen and antibodies to HIV. After several screenings and dropouts on the basis of exclusion criteria, 13 were determined to have chronic hepatitis, of which only 4 patients had consistent samples available for a comprehensive molecular analysis starting from the acute phase. The clinical questionnaire during the follow-up of the 4 individuals did not show any other comorbidities or behavioral factors contributing to liver disease (e.g., diabetes, excess alcohol intake, etc.). Acute cases manifested a discrete onset of symptoms consistent with acute viral hepatitis, jaundice, and elevated serum alanine aminotransferase (ALT) levels. Chronically infected persons were anti-HCV positive, and when their ALT level was above the upper limit of normal in two or more follow-up samples, multiple percutaneous liver biopsy specimens were collected during the follow-up period. Histological evaluation showed mild-to-severe chronic liver disease, including chronic active hepatitis, chronic persistent hepatitis, and chronic lobular hepatitis (3). The age, race, gender, risk factors, and follow-up period for each patient are shown in Table 1.
Table 1.
Subjects and specimens
| Patient (genotype) | Age (yr), race, gender, and risk factora | Time of sampling (no. of yrs since acute infection) | No. of variants/no. of clones ofb: |
|
|---|---|---|---|---|
| E1-HVR1 | NS5a | |||
| A (1b) | 80, white, male, BT | 0 | 4/7 | 2/2 |
| 2.3 | 33/45 | 18/47 | ||
| 2.8 | 21/40 | 27/53 | ||
| 7.9 | 34/38 | 53/60 | ||
| 8.8 | 48/50 | 52/61 | ||
| Total | 8.8 | 140/180 | 152/223 | |
| B (1a) | 26, black, female, IDU | 0 | 3/3 | 2/2 |
| 2.8 | 26/43 | 47/56 | ||
| 3 | 18/28 | ND | ||
| 3.3 | 27/53 | 28/51 | ||
| 7.9 | 33/49 | 23/40 | ||
| 9 | 33/42 | 43/43 | ||
| 10.1 | 27/38 | 44/51 | ||
| 11.2 | 18/38 | 39/48 | ||
| 12.2 | 19/32 | 23/46 | ||
| 13.4 | 24/34 | 30/48 | ||
| 15.3 | 14/25 | 32/46 | ||
| 16.2 | 15/33 | 22/48 | ||
| 17.2 | 24/38 | 31/38 | ||
| 18.2 | 17/41 | 23/45 | ||
| Total | 18.2 | 298/497 | 387/562 | |
| C (1a) | 30, black, male, unknown | 0 | 23/60 | 16/44 |
| 0.3 | 41/57 | 18/38 | ||
| 7.1 | 47/58 | 45/52 | ||
| 8.1 | 32/59 | 30/42 | ||
| 9 | 22/54 | 24/45 | ||
| 10.5 | 22/60 | 36/55 | ||
| 11.1 | 22/54 | 33/49 | ||
| 14.6 | 29/58 | 30/42 | ||
| 15 | 24/55 | 45/59 | ||
| 16 | 22/59 | 37/54 | ||
| Total | 16 | 284/574 | 314/480 | |
| D (1a) | 21, unknown, female, IDU | 0 | 2/2 | 3/6 |
| 1.5 | 31/40 | 28/47 | ||
| 1.6 | 21/43 | ND | ||
| 2 | 28/41 | ND | ||
| 2.3 | 31/43 | ND | ||
| 2.5 | 29/47 | ND | ||
| 2.7 | 28/41 | 19/35 | ||
| 4.8 | 37/47 | 22/31 | ||
| 11.6 | 19/19 | 38/55 | ||
| 12.1 | 42/54 | 33/59 | ||
| 14 | 38/51 | 29/58 | ||
| 15 | 44/50 | 39/52 | ||
| 16 | 55/62 | 30/47 | ||
| Total | 16 | 405/540 | 241/390 | |
BT, blood transfusion; IDU, injection drug user.
ND, not done due to not having enough sample volume. Variants, unique sequences; clones, total number of amplicons. Fewer clones were obtained from the acute-infection samples from patients A, B, and D due to low viral titers and sample volumes.
Patient A died after 8.8 years of follow-up.
Sera had been tested for anti-HCV IgG using the Abbott HCV enzyme immunoassay (EIA), version 2.0 (Abbott Laboratories), or the Ortho HCV version 3.0 enzyme-linked immunosorbent assay (ELISA) (Ortho-Clinical Diagnostics Inc., Raritan, NJ). Reactive results were confirmed with supplemental testing using HCV Matrix (Abbott Laboratories) or RIBA 3.0 (Chiron Corp). All patients were negative for hepatitis B surface antigen and antibodies to HIV. Total nucleic acids from anti-HCV-seropositive specimens were extracted from serum by the use of the Roche MagNA Pure LC instrument and the MagNA Pure LC total nucleic acid isolation kit (Roche Diagnostics, Mannheim, Germany) and eluted with 50 μl of buffer according to the manufacturer's instructions. RNA was precipitated and reverse transcribed using both random and specific primers.
Ethics statement.
Blood specimens were acquired with the written informed consent of all study patients, and all research involving human participants was approved by the institutional review board (Centers for Disease Control and Prevention IRB no. 1428). All clinical investigations have been conducted according to the principles expressed in the Declaration of Helsinki.
Quantification and genotyping.
RNA was precipitated and reverse transcribed using both random and specific primers. Reverse transcription was carried out for 60 min at 42°C in a total volume of 20 μl of 5× PCR buffer (Roche), 200 pmol of each deoxynucleotide triphosphate, 1 μg/μl of random primers, 25 units of avian myeloblastosis virus (AMV) reverse transcriptase (Roche), and 40 units of RNase inhibitor (Roche), followed by heating at 95°C for 5 min. HCV RNA was quantified by amplification of the 5′ untranslated region by real-time nested PCR using an in-house assay. To determine the HCV genotype, a segment of the HCV NS5b region, encompassing positions 8275 to 616, was amplified using real-time nested PCR.
EPLD-PCR for HVR1 and NS5A quasispecies analysis.
The determination of HCV quasispecies by EPLD-PCR was performed by following the method in our previous work (40). It involves isolation of individual coexisting sequence variants of HVR1 and NS5A of the HCV genome from serum specimens using a limiting dilution protocol. Nested “hot-start” PCRs were performed in a final volume of 20 μl, using the LightCycler 480 SYBR green 1 master kit (Roche Diagnostics, Mannheim, Germany). The primer sequences to amplify the HVR1 segment are as follows: external primers F1-TGG CTT GGG ATA TGA TGA TGA ACT and R1-GCA GTC CTG TTG ATG TGC CA and internal primers F2-GGA TAT GAT GAT GAA CTG GT and R2-ATG TGC CAG CTG CCG TTG GTG T. The primer sequences to amplify the NS5A segment are as follows: external primers F1-TCA TAG AGG CCA ACC TCC TGT G and R1-TCG ACC ATG ACC CGT CGC TGA G and internal primers F2-AGT GGT GAT TCT GGA CTC TTT CG and R2-CAT GGA GGA ATA GGA CTC AGC GTC. About 96 PCRs were routinely conducted to obtain approximately 48 clones (45% to 55% positivity), which were identified using melting curve analysis and then sequenced. Numbers of clones (range, 20 to 60) amplified varied depending on the viral titer.
Sequence analysis.
Sequencing reactions were performed using the BigDye v3.1 chemistry sequencing kit (Applied Biosystems, Foster City, CA), and products were sequenced using an automated sequencer (3130xl genetic analyzer; Applied Biosystems). Preliminary sequence analysis was conducted using the SeqMan and MegAlign programs from the Lasergene DNA and protein analysis software (version 7.0; DNASTAR Inc., Madison, WI).
Genetic analysis.
Multiple-sequence alignment and evolutionary-distance analysis were performed by using programs in Accelrys GCG, version 11.0 (Genetics Computer Group, Accelrys Inc., San Diego, CA). The extent of quasispecies heterogeneity in each sample was examined by unbiased estimates of nucleotide diversity, calculated according to the method of Nei and Li (31) using the program Arlequin (12a).
Positive selection.
HyPhy (24) was used to determine selective pressures on selected sequences using single-likelihood ancestor counting (SLAC) (23). SLAC involves counting the numbers of nonsynonymous changes (dN) and synonymous changes (dS) and testing whether the dN is significantly different from the dS. The algorithm processes an alignment using likelihood-based branch lengths, nucleotide and codon substitution parameters, and ancestral sequence reconstructions.
Phylogenetic analysis. (i) ML trees.
The program ModelTest (35) was used to establish the best model of RNA substitution for our HCV data. The general time reversible (GTR) model was chosen to create maximum-likelihood (ML) trees (13) using the program HyPhy (24). An initial tree was created using the neighbor-joining approach (45); a search in the tree space was performed using the nearest-neighbor interchange branch on the tree until no further likelihood score improvements could be made.
(ii) MJN.
We applied median-joining networks (MJN) to reconstruct the phylogeny using Network 4.0 (4). Using nucleotide sequences, the MJN method begins by computing the minimum spanning trees (a graph that connects all the sequences with the minimum necessary total length of the branches), following which all the constructed graphs are combined within a single (reticulate) network. In addition, we devised a new approach for the classification of sequences by which topological parameters were used to split the network into natural communities of sequences by means of the modularity algorithm of Girvan and Newman (17). (a) The betweenness of each edge is calculated as the number of shortest paths that go through that edge; (b) the edge with the highest betweenness is removed; and (c) the edge betweenness is recalculated. The process continues until all edges in the network have been removed.
Nucleotide sequence accession numbers.
The HVR1 and NS5A sequences produced in this study have been deposited in the National Center for Biotechnology Information GenBank database under accession numbers for HVR1 (HM350553 to HM352314) and NS5A (HM348899 to HM350552).
RESULTS
Varying temporal patterns of sequence heterogeneity.
Intrahost variants of two variable HCV genomic regions, HVR1 and NS5A, were sequenced from four chronically infected patients who were followed up from the acute stage of infection for 9 to 18 years. Approximately 20 to 60 clones of variants per region were sampled at 5 to 14 time points from the serum of each patient (Table 1). HCV variants in both regions (Fig. 1a) rapidly diverged from the founder population (defined in this work as a population found at the onset of acute infection), consistent with previous observations (25, 48). Supporting this finding, genetic distances between variants from different time points correlated with time for both HCV regions (r = 0.36 to 0.89, P < 0.006) in all patients. However, besides this general trend, there was no clear similarity observed between patterns of divergence in all four patients (Fig. 1a). Additionally, no apparent match was detected between the patterns of nucleotide diversity of different patients (Fig. 1b). Only patients A and D demonstrated a steady increase in HCV diversity. However, HCV in patient D showed a decrease in HVR1 diversity during the last 4 years of infection. HCV in patient B had a variable pattern of diversity characterized by transient increases and decreases during the infection. HCV diversity was consistently low in patient C during approximately the last 7 years of the observed infection (Fig. 1b). Variations in HCV diversity and divergence have frequently been observed in longitudinal studies (12, 38). To understand such variations, we explored additional parameters of HCV evolution in these 4 patients.
Fig. 1.
Divergence (a) and diversity (b) of HVR1 and NS5A quasispecies at different time points in 4 patients. Divergence is calculated as the average distance between variants from different time points and the first time point and is expressed as the percentage of nucleotides that differed over the total length of a region, i.e., 291 bp for HVR1 and 343 bp for NS5A. (c) Distribution of dN/dS values for HVR1 and NS5A quasispecies over different time points.
Time order of phylogeny.
Phylogenetic analysis showed that HCV quasispecies in both regions are generally distributed along the phylogenetic trees in the order reflecting their sampling time (Fig. 2a and b). The time order was less discernible for NS5A than for HVR1 (Fig. 2b). HCV variants sampled during approximately the first 3 years of infection were found primarily on a single major branch in phylogenetic trees, while HCV variants sampled at later time points could be distributed among more than one major branch across phylogenetic trees, e.g., HCV HVR1 and NS5A variants sampled at ∼8.8 years from patient A, ∼9 years from patient B, ∼8.1 years from patient C, and ∼14 years from patient D (Fig. 2a and b). HCV variants sampled during acute infection were found intermixed with variants sampled within approximately the first 3 years in patients A, B, and C. However, the acute HCV HVR1 variants in patient D were phylogenetically distant from early variants and intermixed with HCV HVR1 variants sampled 11.6 to 16 years later (Fig. 2a); notwithstanding that, the acute HCV NS5A variants were found intermixed with early and late variants in the phylogenetic tree (Fig. 2b). This observation suggests that patient D was infected with a complex mixture of HCV variants, some of which were present at sufficiently high frequencies to be sampled only at certain time points.
Fig. 2.
Phylogenetic analysis of the unique HVR1 (a) and NS5A (b) variants obtained from patients A to D. The color bar scale shows the time line of the follow-up since acute infection (in years). Black triangles beside the color bar mark the exact sampling time points. The blue diamonds identify acute-phase HCV variants.
The time order of phylogeny reflects the host selection pressures forcing HCV to evolve during chronic infection. The most intense selection pressure acting on HCV is the neutralizing immune response (53). Because HVR1 contains an HCV-neutralizing epitope(s) (49), it was expected that the intrahost HVR1 phylogeny would have a time-ordered structure. Although this was true overall, there were time points at which this structure was rather obscured, e.g., with HVR1 variants sampled from all patients at the time points shaded in yellow in Fig. 2a and b and variants detected during the last 7 years of the observed infection in patient C.
Two patterns of quasispecies dispersion.
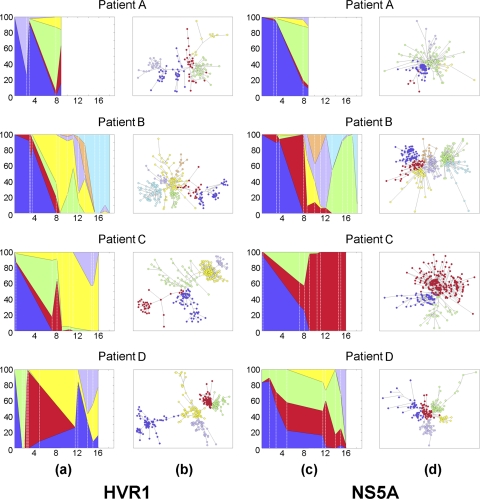
The analysis of viral molecular evolution is often carried out with traditional phylogenetic methods that assume bifurcating trees. However, a bifurcating tree cannot always represent gene evolution accurately when (i) genealogies are multifurcated (one sequence gave origin to many others), (ii) descendant sequences coexist with persistent ancestors, and (iii) there are ambiguous relationships due to homoplastic events (a similarity between sequences that is not the result of common history but the occurrence of reverse or parallel mutations due to convergent evolution) (36). All these phenomena are likely very common at the level of HCV populations, which are found as a genetically heterogeneous ensemble of viral variants. Hence, we applied and explored network approaches, which are better suited for analysis of genetic relationships among closely related variants (30). We used median-joining networks (MJN) to visualize the nucleotide differences between HCV variants in each patient for both the HVR1 and NS5A regions. A time-ordered distribution of HCV variants, similar to the one observed with phylogenetic trees, was also observed along the MJN (Fig. 3). Division of the MJN into communities of sequences (Fig. 3) revealed that during approximately the first 3 years of infection, >80% of the HVR1 and NS5A variants were mapped within a single community in patients A, B, and D. In patient C, however, HVR1 variants could be found for only ∼2 months in a single community at the beginning of infection. During this early period, two single communities alternated between each other in patient A, whereas in patient D, three single communities succeeded each other. Later in infection, HCV variants spread among more than one MJN community and maintained this diversification for the rest of the observed infection, as in patients A and D (continuous-dispersion pattern), or converged into a population evolving within a different single MJN community after ∼9 to 12 years of diversification, as in patients B and C (convergent pattern) (Fig. 3).
Fig. 3.
MJN communities in HVR1 and NS5A in each patient. (a and c) Area graphs of MJN communities in HVR1 and NS5A. Each area represents the percentage of quasispecies that belong to a given community at each time point (in years since acute infection [x axes]); vertical dotted lines delimit time points. (b and d) MJN divided into communities (HVR1 and NS5A) using network topological parameters. Each node corresponds to a unique nucleotide sequence. Areas representing communities (a and c) and MJN nodes (b and d) that belong to these communities are shown with the same colors.
In general, HCV HVR1 variants could be found scattered along the MJN after ∼3 years of infection in patients A and D, frequently without direct connection between clusters of variants identified within and between time points (Fig. 3b and 5b), indicating their independent recent origins. A similar dispersed distribution of the HCV HVR1 variants that have no direct connections to each other in the MJN within and between time points was found in patients B and C for a certain period of time before HCV variants converged into a single community (Fig. 3 and 5a). Both patterns of dispersion were also observed in phylogenetic trees (Fig. 2a and b), but they were more evident in the MJN.
Fig. 5.
Temporal distribution of HVR1 quasispecies in an MJN for patients C (a) and D (b). Each node corresponds to a unique nucleotide sequence, and the radius of each circle is proportional to the frequency of the sequence. Sequences found at a given time point are shown in red; all sequences found before and after the given time point are shown in blue and yellow, respectively. The horizontal bar identifies the exact time points (years since acute infection) marked with black triangles along the bar.
Temporal presence of HCV variants.
Although the majority of the HCV intrahost variants were observed only at certain time points, some variants were detectable at more than one time point (Fig. 4). One HVR1 variant was sampled almost during the entire observation period in patient C. Such variant persistence between time points was more readily detectable using NS5A in all patients (Fig. 4). This parallels the observations made using MJN analysis indicating that the majority of the HCV intrahost variants were observed only at certain time points, but some variants were detectable at more than one time point. These observations indicate that despite the immune selection pressure, many, or at least some, HCV variants were capable of long-term persistence at a frequency sufficiently high to ensure their consistent sampling during infection, and some even increase in frequency over time, as in patient C (Fig. 4a).
Fig. 4.
Presence or absence of a given sequence at different time points. Most of the individual sequences are replaced at every time point for both the HVR1 (top panel) and NS5A (bottom panel) regions in patients A to D. More sequences show persistence in time in the NS5A region than in the HVR1 region. The y axis shows the number of unique variants.
Another important observation is that HCV variants from the final MJN community were detectable as early as 0.3 year in patient C (both the HVR1 and NS5A variants) and ∼3 years after acute infection in patient B (NS5A variants) (Fig. 3). In addition to the aforementioned observation of HCV variants simultaneously sampled from distant regions of the MJN for patient D, these findings suggest that patients B, C, and D were originally infected with a complex mixture of HCV variants representing different MJN communities. Patient A was infected through blood transfusion (Table 1), which makes likely a transmission of numerous HCV variants.
Comparison of HVR1 and NS5A intrahost evolutions.
For patients A, B, and C, a significant correlation between HVR1 and NS5A was found for genetic distances (r = 0.6024 to 0.8566, P < 0.0093) between variants from different time points. For patient D, however, no correlation between the genetic distances of the two regions was found.
Selective sweep and background selection.
Major HVR1 sequence variants, defined in this study as representing >30% of the quasispecies population, characteristically had the smallest sum of genetic distances to all other variants sampled at a given time point, which thus suggests that they have a central position in the variant cloud (data not shown). This central, major variant was associated with a star-like phylogeny and a reduction in heterogeneity that is characteristic of a selective sweep (5, 9). Such a selective sweep was detected during the first 3 years in both HCV regions of all 4 patients (Fig. 5) and coincided with an increase in viral titer (data not shown), suggesting improvement in HCV replicative fitness early in infection.
At later stages of infection in patients B and C, HVR1 underwent another round of reduction in heterogeneity (Fig. 5a, panels 5 to 10) probably associated with background selection (5, 9). This reduction resulted in a single dominant HVR1 community under negative selection (Fig. 1c), purifying the HCV population from less-fit variants.
Reduction in selection pressures over time.
The ratio between nonsynonymous and synonymous (dN and dS) nucleotide substitutions is commonly used as a measure of the strength and direction of natural selection acting on protein coding regions (33). The dN/dS values calculated for variants at each time point declined over time for both HVR1 (r = −0.58, P = 0.0001) and NS5A (r = −0.61, P = 0.0001) in all 4 patients, indicating a reduction in the selection pressure on the HCV genome. In general, NS5A was under negative selection (dN/dS < 0.7). HVR1 experienced either a positive selection (dN/dS = 1.1 to 2.0) that then mitigated, as in patients A and D, or, as in patients B and C, an almost neutral selection (dN/dS = 0.7 to 0.9) that became strongly negative (dN/dS < 0.5) at later stages of infection (Fig. 1C). Under strong purifying selection in patient C, HCV achieved stable adaptation to the host that could be sustained for ∼7 years. This observation indicates that, despite HCV-specific immune responses causing HVR1 diversification, the HCV intrahost evolution may significantly slow down to levels reported in immunocompromised patients (7, 26, 41).
DISCUSSION
Analysis of the intrahost evolution of hepatitis C virus (HCV) is important for understanding mechanisms responsible for establishment of chronic infections and development of efficient preventive and therapeutic measures. However, analysis of HCV's longitudinal evolution is significantly hindered by difficulties in identifying patients and in their long-term follow-up, starting from the acquisition of infection. In the current study, intrahost variants of two variable HCV genomic regions, HVR1 and NS5A, were sequenced from four treatment-naïve chronically infected patients who were followed from the acute stage of infection for 9 to 18 years. The HCV quasispecies from each region were sampled at 5 to 14 time points from each patient using the endpoint limiting dilution PCR, and ∼3,400 clones were obtained (an average of 43 clones per time point per region).
Temporal patterns of communities.
Analysis conducted in this study showed that HVR1 variants in these 4 patients are organized into communities or subpopulations rising to dominance at different time points (Fig. 3). The intrahost HCV population was predominantly found evolving as a single community during approximately the first 3 years of infection, followed by dispersion into several communities or subpopulations. As was shown for all 4 patients, the time order of phylogeny was usually obscured during dispersion into communities. Patients A and D demonstrated the pattern of continuous dispersion for the rest of the observation period. HCV variants in patients B and C converged into another single subpopulation after ∼9 to 12 years of dispersion. The final subpopulation in the last two patients was under negative or purifying selection.
Observation of HCV variants detectable at more than one time point, with certain variants even increasing in frequency over time, as in patient C (Fig. 4 and 5a), indicates that at least some HCV variants are capable of long-term persistence during chronic infection. Most of the individual sequences are replaced at every time point for both the HVR1 and NS5A regions (Fig. 4). More sequences show persistence in time at the NS5A region than at the HVR1 region. Interestingly, the sequences with the highest persistence are usually the most frequent too.
HVR1 and NS5A variants from the final MJN community in patient C were sampled only 0.3 year and NS5A variants in patient B for ∼3 years after acute infection (Fig. 3). In patient D, some NS5A variants sampled at the first time point were found in distant regions of the MJN. All these observations suggest that these 3 patients were infected with an assortment of HCV variants belonging to different MJN communities. These communities seemed to vary in their dominance during infection, allowing the sampling of their members only at some time points. HCV variants from the final MJN community in patient C, and most probably in patient B, were present early during infection and were possibly transmitted to these patients from the source. Later in the infection, these variants grew in frequency and were extensively sampled in our experiments. Patient A was infected through blood transfusion (Table 1), which also implies transmission of numerous HCV variants to this patient.
A low risk of HCV persistence was suggested to be related to transmission of a relatively homogeneous HCV population to a new host (37, 39, 44). In the present study, all 4 patients have indications of transmission of HCV variants representing more than one MJN community. The fact that all these patients established chronic infection lends additional support to the importance of the diverse HCV populations at the initiation of infection for escaping clearance.
Varying rate of evolution and complexity of the founder population.
The observed rate of HCV intrahost evolution varied between time points. It ranged from a significant slowdown during late infection, as in patient C (Fig. 5a, panels 5 to 10), to a rapid jump from one region of the MJN to another during early infection, as in patient D (Fig. 5b, panels 1 to 2). This finding implies various degrees of evolutionary changes at different stages of infection in these 4 patients. However, measures of the evolutionary rate based on calculating differences between nucleotide sequences of viral populations at consecutive time points can be inaccurate in defining the extent of evolution and should be interpreted cautiously. The presence of more than one MJN community, with some evolving in the background, as suggested by the aforementioned observations of the temporal appearance of HCV variants, creates an opportunity for variation in dominance for these communities at different stages of infection, thus confounding the estimation of evolutionary rates calculated using small viral samples. As a result, HCV variants from two consecutive time points may have no direct links to each other in the MJN (Fig. 5a, panels 1 to 2, and Fig. 5b, panels 7 to 8), which suggests that the succeeding viral population is not always immediately derived from the preceding population.
If viral communities preexist, from where do they originate? We hypothesize that many, if not all, communities are originally established within the expanding founder population, which experiences a bottleneck evolution upon transmission followed by a rapid increase in population size. In all 4 patients studied here, the HCV titer was in the range of 6 × 103 to 9 × 104 IU/ml at acute-phase time points (data not shown), suggesting that within a very short period of time after transmission, the HCV population reached ∼107 to 108 viral particles in the bloodstream, starting from a limited number of founder particles. Owing to the large number of progeny genomes generated during such a population expansion, the probability of retaining newly arising mutations in progeny increases. Additionally, changes in selection pressures in a new host can lead to adjustment in epistatic connectivity between sites within the viral genome. This process is analogous to the founder-flush speciation model for a population exploring a new ecological niche (51). In this model, new selectively favorable mutations have a higher probability of survival as a result of the flush increase in population size.
Thus, the first molecular events occurring immediately after HCV transmission may result in the generation of a highly heterogeneous viral population despite the bottleneck. As was shown above, the members of more than one MJN community were sampled at early stages of HCV infection in patients B, C, and D, suggesting transmission of more than one HCV variant, with patient A most probably being infected with many HCV variants through blood transfusion. The transmitted HCV population may acquire, in addition to its original heterogeneity, an extensive genetic variability through the founder-flush process during the initial steps of infection. However, the HCV-specific adaptive immunity likely shapes this founder population into a complex pool of closely related communities, each of which may become dominant at different stages of the HCV infection. HCV's intrahost evolution is, therefore, defined by two distinct processes: incremental changes within viral communities through random mutation and alternations between coexisting communities, reflecting a complex intrahost population dynamic. This consideration implies that succeeding dominant subpopulations may not be directly derived from each other but may rather share an ancestor.
The founder-flush process, although being a powerful tool for producing intrahost heterogeneity, should, however, have limits in its capacity to generate viral variants. Thus, seeding with sequence variants representing different viral communities upon transmission seems to ensure the establishment of the extensive sequence diversity and broad community structure of the HCV population in a new host. It is interesting that HIV infection is usually initiated by a single variant (1, 20, 21), thus limiting the extent of the initial intrahost heterogeneity generated through the founder-flush process and suggesting that HCV and HIV have different requirements for the initial genetic heterogeneity needed to establish chronic infections.
Contributions of the HVR1 and NS5A regions to intrahost evolution.
Chronic HCV infection is frequently associated with a significant reduction in T-cell immune responses (10), thus making humoral immunoresponses the dominant force of selection during long-term infection. HCV HVR1 contains a neutralizing antigenic epitope (49) and is a target for such immune pressure, while evolution of the NS5A region, which harbors T-cell epitopes (32), is not directly affected by neutralization (27). Therefore, it is HVR1 that is under the frequently changing selection pressure of sequence-specific neutralizing antibodies elicited at different stages of chronic infection (53). The similarity observed between the NS5A and HVR1 temporal dispersion patterns among the MJN communities (Fig. 3) seemingly suggests that NS5A evolution merely reflects HVR1 evolution. However, analysis shows that NS5A changes may have different characteristics. In patient B, for example, NS5A quasispecies evolved into the final MJN community ∼1 year earlier than HVR1 quasispecies (Fig. 2b). The disparity between HVR1 and NS5A quasispecies evolution was even more pronounced for patient D. A significant correlation of genetic distances among all time points for HVR1 and NS5A genomic variants was observed for patients A, B, and C. However, such similarity was not observed for the distribution of genetic distances among time points for these two genomic regions in patient D. These observations, together with the finding that NS5A evolved into a single community at the end of the observation period in this patient, while HVR1 variants remained distributed among more than one community (Fig. 3), also suggest an independent role for NS5A in HCV evolution in this patient.
It is important to note that sites across the HCV genome are physically and epistatically linked. Thus, selection on one site may affect the intensity and direction of selection on other sites within the genome. We have shown earlier that such a linkage is globally organized into a scale-free network (8). Both subgenomic regions, HVR1 and NS5A, are significantly separated in the HCV genome and were treated here as independent entities. However, evolutionary changes analyzed in these two subgenomic regions may reflect strong selection pressures acting somewhere else in the HCV genome.
Selection pressure.
Identification of the selective sweep immediately before dispersal into the MJN communities in all 4 patients (Fig. 3 and 5) suggests a role for frequency-dependent selection in intrahost evolution. The most frequent HVR1 variant likely elicits a strong neutralizing immune response against itself, which should result in reduction in fitness and suppression of the community containing that variant. HCV may undergo more than one round of such frequency-dependent selection, as exemplified in the early infection of patient D (Fig. 5b, panels 2 to 8). This period of infection associated with the decrease in HCV fitness seems to be a particularly vulnerable phase in the development of chronic infection and provides a valuable opportunity for initiation of antiviral therapeutic intervention.
In general, HCV's intrahost evolution was characterized by a consistent increase in negative selection during chronic infection in the 4 patients. The final subpopulation in patients B and C was under negative or purifying selection. A significant reduction in HVR1 heterogeneity (Fig. 5a, panels 5 to 10) at later stages of infection in these patients most probably resulted from background selection (5, 9). The increase in viral load observed at this stage likely reflects decline in the effectiveness of neutralizing immunoresponses and, therefore, could be associated with inefficient HCV clearance rather than with improvement in viral replicative fitness.
Adaptation to late stages of infection.
An important indication of improved viral adaptation to the host at later stages of chronic infection for all 4 patients is identification of a positive correlation between viral load and length of infection (r = 0.585, P = 0.0001) and a negative correlation between viral load and dN/dS (r = −0.383, P = 0.012). The existence of the negatively selected HVR1 variants in patients B and C suggests a deep adaptation of these populations to their host. The mechanism of this adaptation is not known. HCV may exploit the preexisting host deficiency in controlling this infection. Although immune escape via exploitation of “holes” in HCV-specific immunity has been observed (54), this mechanism of adaptation should require the frequent availability of various immunological deficiencies in the host population that can be exploited by HVR1 via mutations. Alternatively, as with evasion of innate immunity, HCV may actively affect adaptive immunity, making the host environment conducive to the stable coexistence of the virus with the host. The second mechanism of HVR1 adaptation is consistent with the observation of an HVR1 variant that persisted in patient C for almost the entire observation period but became predominant only during approximately the last 7 years (Fig. 4 and 5a).
In contrast to HIV, HCV does not cause systemic immunodeficiency (47). However, the reduction in selection intensity over the course of HCV infection is clearly attributable to the decline in specific immune responses. Although molecular mechanisms responsible for the decline in the immune pressure on HCV HVR1 at late stages of infection are not known, it can be speculated that such an intrahost environment is caused by B-cell dysfunctions, such as those leading to the hyperactivity and exhaustion observed during chronic HIV (29) and HCV (43) infections; may be related to or associated with competition between antigenic sites (55), a paucity of high-affinity immune cell receptor recognition (54), the “original-antigenic-sin” response (11, 22), or the enhanced antibody-dependent uptake of some HCV variants (28); or may be the result of a combination of these conditions.
Irrespective of the mechanisms responsible for reduction of immune pressure on HVR1, the increase of negative selection in all 4 patients during chronic infections suggests that HCV not merely escapes neutralizing immune responses but also promotes intrahost conditions beneficial for stable viral reproduction through temporal cooperation between viral subpopulations at different stages of chronic infection. Such cooperation may assume different forms (15, 52). In the scenario of “original antigenic sin,″ for example, the succeeding HCV populations may experience a reduction in immune pressure through stimulating moderately cross-immunoreactive memory cells raised against previous populations rather than through eliciting new antibody-producing cells. Thus, we suggest a modification of the continuous-escape (53) or diversification-stabilization (48) model for HCV which implies that a viral population at a given stage has a specific effect on the host environment and reduces host selection pressures for the succeeding population until the final population achieves a state of stable coexistence with the host.
HCV as a noncytopathic virus seems to exploit host mechanisms for reducing immune-related liver damage (6) and for controlling excessive immune responses (42). It adversely affects many functions of the host immune system but does not incapacitate it completely, suggesting a long history of HCV adaptation to humans. Additionally, HCV-host coevolution, besides having immunological effects, may involve mutual virus-host cell adaptation, as has been observed in vitro (56).
Stages of HCV intrahost evolution.
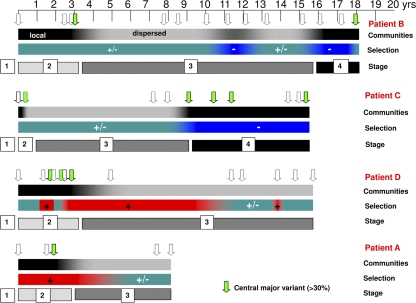
The model presented here suggests that HCV's intrahost evolution starts in the absence of specific immune responses and proceeds toward inefficient neutralizing immune responses. We hypothesize that in order to achieve this goal, HCV evolves through 4 stages (Fig. 6). In stage 1 (implied, but not studied here), HCV establishes itself in a new host before the surge of adaptive immune responses. This stage is the arena for the founder-flush process. Stage 2 covers a period of incremental evolution of viral variants within predominant communities, reflecting the minimal genetic changes actually required for an effective immune escape. Stage 3 is a period of diversification into a set of subpopulations that become prominent following the decline of the previously dominant population. In stage 4, HCV achieves final settlement under strong negative selection. Patients B and C exhibit distinct settlement phases. Patients A and D show neutral selection pressure in the last time points, and probably a few more years of follow-up would likely show settlement under negative selection.
Fig. 6.
Schematic representation of the probable stages of HCV infection over the years in four chronically infected naïve patients. The arrows indicate sampling points from acute infection to 9 to 18 years. The color coding of the arrows corresponds to the percentage of the major variant at the indicated time point of sampling. The green color is for variants whose percentage was >30%, indicating a selective sweep around the third year and purifying selection during the later years of chronic infection. The black and gray bar at the top indicates the dispersion of quasispecies in the MJN, where black indicates single communities (local) and gray is for more than one community (dispersed). The second bar indicates selection pressure, where red indicates positive selection, blue indicates negative selection, and green indicates neutral selection pressure. The last bar indicates the probable stages of HCV infection.
At the final stage, HCV is posited to achieve a stable adaptation to the host, the ultimate goal of intrahost evolution. However, this goal may not always be achieved. As exemplified in patient D, the course of HCV's intrahost evolution seems distressed by strong selection pressure early in infection. Nevertheless, judging from the declining dN/dS values during the last years of infection, HCV evolution in this patient was redirected toward the goal programmed into the genetic composition of the late-stage HCV variants.
Implications.
The duration and intensity of evolutionary processes at different stages likely play an important role in defining clinical outcomes of chronic HCV infections. Staging a chronic HCV infection should assist in determining the optimal timing for the successful application of therapy, since HCV may be differentially sensitive to interventions at particular stages. For example, it is known that patients with acute HCV infection achieve a complete virological response to interferon therapy more frequently than patients with chronic infections (16, 19), suggesting that stages 1 and 2 are most sensitive to therapy. Additionally, the finding that HVR1 in baseline samples of nonresponders and transient responders tends to be negatively selected, whereas HVR1 in samples of sustained responders tends to be positively selected (14), may indicate that HCV at stage 4 is more resistant to combined interferon and ribavirin therapy than at stages 2 and 3.
Our results suggest that the predominant viral subpopulation at the last stage loses its advantages upon transmission, implying that different HCV generations may exhibit variations in transmissibility and, potentially, in virulence. Observation of the reduction in quasispecies diversity associated with severity of liver disease (38, 50) may be interpreted in terms of variation in virulence during stages 3 and 4 of HCV infection. It is also conceivable that sequence variants prevalent at different stages of infection may have somewhat specific immunological properties, which may be explored for the development of hepatitis C prophylactic or therapeutic vaccines.
Although limited to 4 HCV strains because of significant difficulties in obtaining serial specimens from chronically infected treatment-naïve patients over an extended period of time, the findings made in the present work offer a new framework for studying and exploiting HCV's intrahost evolution for clinical and public health interventions.
ACKNOWLEDGMENTS
We acknowledge Omana Nainan (deceased), Harold Margolis, and Ian Williams (Division of Viral Hepatitis, CDC) for prompting this prospective study. We also thank Chong-Gee Teo and John Ward (Division of Viral Hepatitis, CDC) for critical reviews of the manuscript and helpful discussions.
Footnotes
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Abrahams M. R., et al. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alter M. 2007. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 13:2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alter M. J., et al. 1992. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N. Engl. J. Med. 327:1899–1905 [DOI] [PubMed] [Google Scholar]
- 4. Bandelt H., Forster P., Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37–48 [DOI] [PubMed] [Google Scholar]
- 5. Berry A., Ajioka J., Kreitman M. 1991. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics 129:1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Billerbeck E., Bottler T., Thimme R. 2007. Regulatory T cells in viral hepatitis. World J. Gastroenterol. 13:4858–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Booth J., Kumar U., Webster D., Monjardino J., Thomas H. C. 1998. Comparison of the rate of sequence variation in the hypervariable region of E2/Ns1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology 27:27223–27227 [DOI] [PubMed] [Google Scholar]
- 8. Campo D., Dimitrova Z., Mitchell R., Lara J., Khudyakov Y. 2008. Coordinated evolution of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 105:9685–9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charlesworth D., Charlesworth B., Morgan M. 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141:1619–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox A., et al. 2005. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology 42:104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davenport F., Hennesy A., Francis T. 1953. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J. Exp. Med. 98:641–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duffy M., et al. 2002. Comparative rates of nucleotide sequence variation in the hypervariable region of E1/E2 and the NS5b region of hepatitis C virus in patients with a spectrum of liver disease resulting from a common source of infection. Virology 301:354–364 [DOI] [PubMed] [Google Scholar]
- 12a. Excoffier L., Laval G., Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47–50 [PMC free article] [PubMed] [Google Scholar]
- 13. Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368–376 [DOI] [PubMed] [Google Scholar]
- 14. Figlerowicz M., et al. 2010. Hepatitis C virus quasispecies in chronically infected children subjected to interferon-ribavirin therapy. Arch. Virol. 155:1977–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finzi D., Plaeger S., Dieffenbach C. 2006. Defective virus drives human immunodeficiency virus infection, persistence, and pathogenesis. Clin. Vaccine Immunol. 13:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerlach J. T., et al. 2003. Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology 125:80–88 [DOI] [PubMed] [Google Scholar]
- 17. Girvan M., Newman M. 2002. Community structure in social and biological networks. Proc. Natl. Acad. Sci. U. S. A. 99:7821–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heathcote J., Main J. 2005. Treatment of hepatitis C. J. Viral Hepat. 12:223–235 [DOI] [PubMed] [Google Scholar]
- 19. Jaeckel E., et al. 2001. Treatment of acute hepatitis C with interferon alfa-2b. N. Engl. J. Med. 345:1452–1457 [DOI] [PubMed] [Google Scholar]
- 20. Kearney M., et al. 2009. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 83:2715–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klenerman P., Zinkernagel R. 1998. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature 394:482–485 [DOI] [PubMed] [Google Scholar]
- 23. Kosakovsky Pond S. L., Frost S. D. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222 [DOI] [PubMed] [Google Scholar]
- 24. Kosakovsky Pond S. L., Frost S. D. W., Muse S. V. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679 [DOI] [PubMed] [Google Scholar]
- 25. Kumar U., Brown J., Monjardino J., Thomas H. 1993. Sequence variation in the large envelope glycoprotein (E2/NS1) of hepatitis C virus during chronic infection. J. Infect. Dis. 167:726–730 [DOI] [PubMed] [Google Scholar]
- 26. López-Labrador F., et al. 2007. Trends for genetic variation of hepatitis C virus quasispecies in human immunodeficiency virus 1-coinfected patients. Virus Res. 130:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macdonald A., Harris M. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485–2502 [DOI] [PubMed] [Google Scholar]
- 28. Meyer K., Ait-Goughoulte M., Keck Z., Foung S., Ray R. 2008. Antibody-dependent enhancement of hepatitis C virus infection. J. Virol. 82:2140–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moir S., Fauci A. 2009. B cells in HIV infection and disease. Nat. Rev. Immunol. 9:235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morrison D. A. 2010. Using data-display networks for exploratory data analysis in phylogenetic studies. Mol. Biol. Evol. 27:1044–1057 [DOI] [PubMed] [Google Scholar]
- 31. Nei M., Li W. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U. S. A. 76:5269–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neumann-Haefelin C., et al. 2007. Absence of viral escape within a frequently recognized HLA-A26-restricted CD8+ T-cell epitope targeting the functionally constrained hepatitis C virus NS5A/5B cleavage site. J. Gen. Virol. 88:1986–1991 [DOI] [PubMed] [Google Scholar]
- 33. Nielsen R., Yang Z. 2003. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 20:1231–1239 [DOI] [PubMed] [Google Scholar]
- 34. Penin F., Dubuisson J., Rey F., Moradpour D., Pawlotsky J. 2004. Structural biology of hepatitis C virus. Hepatology 39:5–19 [DOI] [PubMed] [Google Scholar]
- 35. Posada D., Crandall K. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 36. Posada D., Crandall K. A. 2001. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 16:37–45 [DOI] [PubMed] [Google Scholar]
- 37. Puro V., et al. 1995. Occupational hepatitis C virus infection in Italian health care workers. Italian Study Group on Occupational Risk of Bloodborne Infections. Am. J. Public Health 85:1272–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qin H., et al. 2005. HCV quasispecies evolution: association with progression to end-stage liver disease in hemophiliacs infected with HCV or HCV/HIV. Blood 105:533–541 [DOI] [PubMed] [Google Scholar]
- 39. Quer J., et al. 2005. Effect of bottlenecking on evolution of the nonstructural protein 3 gene of hepatitis C virus during sexually transmitted acute resolving infection. J. Virol. 79:15131–15141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramachandran S., et al. 2008. End point limiting dilution real time PCR assay for evaluation of HCV quasispecies in serum. J. Virol. Methods 151:217–224 [DOI] [PubMed] [Google Scholar]
- 41. Ramírez S., Pérez-Del-Pulgar S., Forns X. 2008. Virology and pathogenesis of hepatitis C virus recurrence. Liver Transpl. 14:S27–S35 [DOI] [PubMed] [Google Scholar]
- 42. Rehermann B., Shin E. 2005. Private aspects of heterologous immunity. J. Exp. Med. 201:667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosa D., et al. 2005. Activation of naïve B lymphocytes via CD81, a pathogenetic mechanism for hepatitis C virus-associated B lymphocyte disorders. Proc. Natl. Acad. Sci. U. S. A. 102:18544–18549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ross R. S., et al. 2002. Risk of hepatitis C virus transmission from an infected gynecologist to patients: results of a 7-year retrospective investigation. Arch. Intern. Med. 162:805–810 [DOI] [PubMed] [Google Scholar]
- 45. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 46. Reference deleted.
- 47. Semmo N., Klenerman P. 2007. CD4+ T cell responses in hepatitis C virus infection. World J. Gastroenterol. 13:4831–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shankarappa R., et al. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimizu Y., et al. 1996. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology 223:409–412 [DOI] [PubMed] [Google Scholar]
- 50. Sullivan D. G., et al. 2007. Hepatitis C virus dynamics during natural infection are associated with long-term histological outcome of chronic hepatitis C disease. J. Infect. Dis. 196:239–248 [DOI] [PubMed] [Google Scholar]
- 51. Templeton A. 2008. The reality and importance of founder speciation in evolution. Bioessays 30:470–479 [DOI] [PubMed] [Google Scholar]
- 52. Turner P., Chao L. 1999. Prisoner's dilemma in an RNA virus. Nature 398:441–443 [DOI] [PubMed] [Google Scholar]
- 53. von Hahn T., et al. 2007. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 132:667–678 [DOI] [PubMed] [Google Scholar]
- 54. Wölfl M., et al. 2008. Hepatitis C virus immune escape via exploitation of a hole in the T cell repertoire. J. Immunol. 181:6435–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang P., et al. 2007. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc. Natl. Acad. Sci. U. S. A. 104:8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhong J., et al. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]