Abstract
The duck hepatitis B virus (DHBV) reverse transcriptase (P) is translated from the downstream position on a bicistronic mRNA, called the pregenomic RNA, through a poorly characterized ribosomal shunt. Here, the positions of the discontinuous ribosomal transfer during shunting were mapped, and RNA elements important for shunting were identified as a prelude to dissecting the shunting mechanism. Mutations were introduced into the DHBV genome, genomic expression vectors were transfected into cells which support reverse transcription, and P translation efficiency was defined as the ratio of P/mRNA. Five observations were made. First, ribosomes departed from sequences that comprise the RNA stem-loop called ε that is key to viral replication, but the known elements of ε were not needed for shunting. Second, at least two landing sites for ribosomes were found on the mRNA. Third, all sequences upstream of ε, most sequences between the cap and the P AUG, and sequences within the P-coding region were dispensable for shunting. Fourth, elements on the mRNA involved in reverse transcription or predicted to be involved in shunting on the basis of mechanisms documented in other viruses, including short open reading frames near the departure site, were not essential for shunting. Finally, two RNA elements in the 5′ portion of the mRNA were found to assist shunting. These observations are most consistent with shunting being directed by signals that act through an uncharacterized RNA secondary structure. Together, these data indicate that DHBV employs either a novel shunting mechanism or a major variation on one of the characterized mechanisms.
INTRODUCTION
Translation of the large majority of eukaryotic mRNAs is initiated by ribosomal scanning, in which the small ribosomal subunit is recruited to the mRNA by interaction with the 5′ cap structure, followed by linear scanning of the subunit 3′ along the message until it encounters the initiating AUG codon (16, 22). In addition to the standard scanning mechanism, four other mechanisms are known for translation initiation in eukaryotes: (i) leaky scanning, in which the scanning complex passes a potential start codon on the mRNA but then initiates at a subsequent start codon; (ii) reinitiation, in which the ribosome translates one or more short upstream open reading frames (ORFs) and then continues scanning until it initiates translation again at a subsequent start codon; (iii) internal ribosome entry, in which translation initiates from an internal AUG on the mRNA following direct binding of the ribosomal subunit to an internal ribosomal entry sequence on the mRNA; and (iv) ribosomal shunting, in which the ribosome binds to the message at the 5′ cap, scans along the message for a limited distance, and then transfers from a 5′ donor site to a 3′ acceptor site on the mRNA without scanning the intervening region.
Ribosomal shunting has been primarily described in viral messages, including those of cauliflower mosaic virus (8, 10), rice tungro bacilliform virus (9), Sendai paramyxovirus (5, 17), human type C adenovirus (39, 43), human papillomavirus type 18 (27), the prototype foamy virus (31), and duck hepatitis B virus (DHBV) (33). It has also been described for a few cellular mRNAs, including those of HSP70 (43), cIAP2 (19, 34), and β-secretase (15, 28). Ribosomal shunting is believed to be promoted by cis-acting elements within a structured region of the 5′ untranslated region that direct the scanning complex to the acceptor site, where scanning resumes (30). However, no defined sequence or structure has been universally associated with shunting, and the molecular mechanism(s) of ribosomal shunting is poorly understood.
Hepadnaviruses are small DNA-containing viruses that replicate by reverse transcription. Human hepatitis B virus (HBV) is a major cause of liver disease and liver cancer worldwide (32). DHBV is a common model for HBV. The hepadnaviral pregenomic RNA (pgRNA) is the RNA template for reverse transcription, and it is also a bicistronic mRNA encoding the capsid (C) and reverse transcriptase (P) proteins (Fig. 1). In DHBV, the pgRNA has an ∼118-nucleotide (nt)-long 5′ untranslated region (UTR) upstream of the C ORF that contains a stem-loop (ε) which is an essential signal for encapsidation and reverse transcription (11, 14, 23, 37, 38). DHBV strain 3 (DHBV3) is a commonly used DHBV isolate. In DHBV3, the P ORF starts 544 nt downstream of the start site for C and ∼662 nt downstream of the cap. Thirteen AUGs (C2 to C14) are between the C1 AUG and the P1 AUG that initiates P translation. Ten stop codons (S1 to S10) are also between the C1 and P1 AUGs. These stop codons terminate all potential upstream translation products and produce seven small ORFs (sORFs), which have coding potentials of 2 to 29 amino acids.
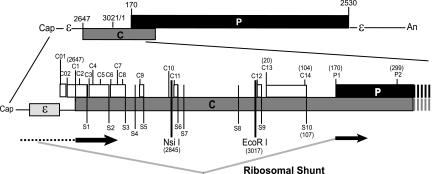
Fig. 1.
Organization of the DHBV3 pgRNA. The pgRNA is 3.3-kb capped and polyadenylated mRNA with a terminal redundancy of approximately 270 nt. The top section shows the relative positions of the major ORFs and functional elements on the pgRNA. The bottom section shows an expanded view of the 5′ end of the RNA, with the location of the features relevant to this study displayed. The genomic positions of the elements are indicated for DHBV strain 3 (nucleotides 3021 and 1 are adjacent to each other within the unique EcoRI site on the circular DNA form of the genome). The ribosomal shunt as it was known at the start of this study is shown at the bottom, with regions that are scanned by ribosomes shown with a dashed line, regions that are translated shown as thick black lines, and the ribosomal shunt shown with gray lines.
We found that DHBV P is synthesized relatively rapidly, despite being located in a very unfavorable position on the pgRNA (40–42) via ribosomal shunting, which allows ∼10% of the ribosomes that bind to the pgRNA to skip over most of the upstream C ORF and initiate P translation at the P1 AUG (33). Translation of the polymerase ORF was also found to initiate at two upstream in-frame AUGs, C13 and C14, in addition to the P1 AUG, and we confirmed that initiation at the C13 AUG occurs through ribosomal shunting (2). C13 and C14 are in frame with P1, and initiation at these start codons leads to production of glycosylated isoforms of P called “pre-P” in the majority of DHBV strains because most DHBV isolates lack the S10 stop codon found downstream of the C14 AUG in DHBV strain 3 (2). In this study, we extended these observations by mapping the shunt donor and acceptor sequences as a prelude to examining the DHBV shunting mechanism.
MATERIALS AND METHODS
Plasmids.
D1.5G is a wild-type overlength DHBV3 (GenBank accession number DQ195079) expression construct containing a 5′ duplication of nucleotides 1658 to 3021 in pBluescript(−) (Stratagene). D1.5G-C1AUG− is a mutant in which the C1 AUG is destroyed by mutating A2647T. D1.5G-POF is a mutant with a frameshift mutation of D1.5G with deletion of nt 435 that causes P translation to terminate after amino acid 88. A series of mutants based on D1.5G were constructed (Table 1). D1.5G was used as the wild-type reference when mutations were introduced upstream of the C1 AUG. D1.5G-C1AUG− was used as the wild-type control when mutations were made within the C ORF to prevent changes in translation of the overlapping C ORF or accumulation of truncated C proteins from interfering with interpretation of the effects on translation of the P ORF. D1.5G-POF was used as the wild-type control when mutations were introduced into the P ORF to avoid confounding effects from possible alterations to the stability or immunogenicity of P.
Table 1.
Plasmids employed
| Plasmid | Description | Background |
|---|---|---|
| D1.5G | Overlength expression vector for wild-type DHBV3 | |
| C1AUG− | Overlength expression vector for DHBV3 in which the C1 AUG was destroyed by mutating A2647T | D1.5G |
| DR1− | Deletion of DR1 | D1.5G |
| DR1-Ran | Replacement of DR1 with random sequences | D1.5G |
| Ran | Replacement of most of sequences between cap and ε with random sequences | D1.5G |
| C012− | Knockout of the two AUGs upstream of core with mutations A2616T and A2630T | D1.5G |
| SL-5′ | Insertion of BamHI-SL at nt 2539 adjacent to the pgRNA start site | D1.5G |
| SL-HindIII | Insertion of BamHI-SL at nt 2559 upstream of ε | D1.5G |
| SL-SalI | Insertion of BamHI-SL at nt 2616 downstream of ε plus an insertion of CTTTGGTA from nt 2616 to nt 2617 | D1.5G |
| SL-2616 | Insertion of the BamHI-SL after nt 2616 downstream of ε | D1.5G |
| SL-2639 | Insertion of BamHI-SL after nt 2639 downstream of ε | D1.5G |
| dl2616-2640 | Deletion of nt 2616 to 2640 | D1.5G |
| dl5 | Deletion of the left stem and bulge of ε | C1AUG− |
| dlBulge | Deletion of the bulge (nt 2571 to 2576) of ε | D1.5G |
| SLM2 | Change of ε bulge sequences nt 2572 to 2576 from TTTAC to CACGT | D1.5G |
| dlU1 | Deletion of the first U of ε (nt 2580) | D1.5G |
| dl2604 | Deletion of an unpaired U in ε at nt 2604 | D1.5G |
| Loop3,4 | Mutation of positions 3 and 4 of the apical loop of ε (G2589A and T2590C) | D1.5G |
| Loop5,6 | Mutation of positions 5 and 6 of the apical loop of ε (T2591C and G2592A) | D1.5G |
| LowerL/R | Complementary mutations in the lower stem of ε by altering nt 2567 to 2569 GTA to CAT and nt 2606 to 2608 TAC to ATG | D1.5G |
| dlUL | Deletion of the upper left stem of ε sequence by removing nt 2577 to 2586 | D1.5G |
| dlAloop | Deletion of the apical loop of ε upper loop by removing nt 2587 to 2593 | D1.5G |
| dlUR | Deletion of the upper right stem of ε by removing nt 2594 to 2603 | D1.5G |
| dlLR | Deletion of the lower right stem of ε by removing nt 2605 to 2616 | D1.5G |
| EcoRI-SL | Insertion of BamHI-SL at the EcoRI site | C1AUG− |
| A-SL | Insertion of BamHI-SL at the A deletion site (nt 2674 to 4) | C1AUG− |
| B-SL | Insertion of BamHI-SL at the B deletion site (nt 2848 to 2844) | C1AUG− |
| C-ESL | Deletion of nt 45 to 94 on the EcoRI-SL background | C1AUG− |
| D-ESL | Deletion of nt 110 to 164 on the EcoRI-SL background | C1AUG− |
| E-ESL | Deletion of nt 5 to 44 on the EcoRI-SL background | C1AUG− |
| POF | Deletion of nt 424T and to produce a stop codon at nt 435 | D1.5G |
| POF-SL-5′ | Insertion of BamHI-SL at nt 2539 adjacent to the pgRNA start site | POF |
| POF-SL-HindIII | Insertion of BamHI-SL at nt 2559 upstream of ε | POF |
| POF-SL-NsiI | Insertion of BamHI-SL at nt 2845 (NsiI site). | POF |
| HBP-571 | Replacement of nt 572 to the P stop codon at nt 2530 with the equivalent region of the HBV P ORF | POF |
| HBP-1171 | Replacement of nt 1172 to the P stop codon at nt 2530 with the equivalent region of the HBV P ORF | POF |
| dl451-1170- | Replacement of nt 451 to 1170 with GTCGAC | POF |
| dl451-760 | Replacement of nt 451 to 760 with GTCGAC | POF |
| dl761-1170 | Replacement of nt 761 to 1170 with GTCGAC | POF |
| dlReII | Replacement of nt 538 to 610 with GTCGAC | POF |
| dl712-738 | Replacement of nt 712 to 738 with GTCGAC | POF |
| dlPhi | Replacement of nt 2514 to 2534 with GTCGAC | POF |
| 8nt-Ran | Mutation of nt 15 to 22 (AGCTTATG) to GTACGCGA | C1AUG− |
| P1loop− | Collapse of the P1 loop and deletion of the C14 and S10 codons by changing nt 96 to 108 from AGAAGCTAATGTA to CATCTCT | C1AUG− |
| P1loop-SalI | Insertion of a SalI site at nt 34 on the P1loop− background | C1AUG− |
| P1loop-SL | Insertion of the BamHI-SL at the SalI site on the P1loop− background | C1AUG− |
RNA fold prediction.
The fold of the 5′ end of the DHBV pgRNA was predicted employing 702 nt from the 5′ end of the pgRNA to 37 nt downstream of the P1 AUG codon using the RNAdraw program (18). The fold of the full-length DHBV pgRNA sequence was predicted using the MFOLD web server (44) to identify possible intragenomic interactions. A chicken (Gallus gallus) 18S rRNA sequence (GenBank accession number AF173612.1) was used to identify sequences complementary to the DBHV “E” region.
Cell culture, transfection, and cell harvesting.
LMH chicken hepatoma cells that produce infectious DHBV when transfected with pgRNA expression vectors such as D1.5G (4) were used for all experiments. The cells were maintained in 1:1 Dulbecco's modified Eagle's medium/Ham F-12 medium with 10% fetal bovine serum. Cells were seeded on 60-mm dishes at a density of 1.2 × 106 cells per plate 18 h prior to transfection. Transfections employed Fugene 6 (Roche) or Lipofectamine (Invitrogen), according to the manufacturers' instructions. Cell lysates were harvested 24 h posttransfection using Tri Reagent (Molecular Research Center), from which protein and RNA were isolated from the same cell lysate, according to the manufacturer's instructions. Previous studies revealed that P levels were proportional to pgRNA levels, as long as the harvesting time posttransfection was not varied (42).
Calculation of translation efficiency.
Whole-cell protein lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to Immobilon-P membranes (Millipore). P was detected with the anti-DHBV P monoclonal antibody (MAb) 11 (epitope amino acids 53 to 61) (40) following incubation with the appropriate IgG-alkaline phosphatase conjugate (Promega). Proteins were visualized by incubation with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Promega). Exposures were carefully monitored to ensure that the Western blot signals were within the linear range of the detection system as we have previously done (42). Where necessary, blots were developed for a range of times to ensure that all samples were analyzed from appropriate exposures. The Western blots were scanned and quantified using ImageQuant software (GE Healthcare). The pgRNA was detected by Northern blotting as described previously (36) using 32P-labeled monomeric DHBV DNA as a probe. RNA signals were detected by phosphorimage analysis and quantified using ImageQuant. Translation efficiency was calculated as the amount of P detected by Western analysis divided by the amount of pgRNA detected by Northern analysis from the same lysate. To permit comparisons between experiments, all data were normalized to translation efficiency of the appropriate wild-type construct that had been transfected in the same experiment and analyzed on the same Western and Northern blots as the test samples. Two constructs produced insufficient pgRNA to permit reliable detection of P in the whole-cell lysates. In these cases, P was concentrated by immunoprecipitation as previously described (2) prior to Western analysis.
RESULTS
Ribosomes depart from sequences that comprise ε, and the region between ε and the C ORF contains an important cis-acting element.
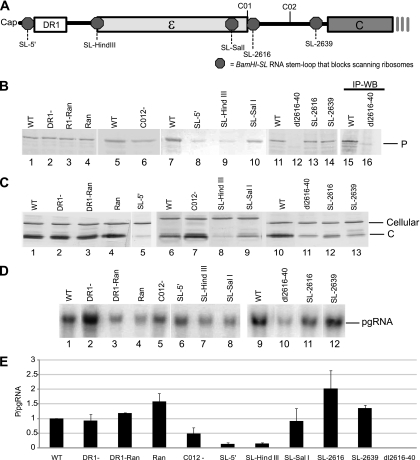
Two elements important for reverse transcription that could also possibly contribute to the shunt donor are found near the 5′ end of the DHBV pgRNA: DR1 and ε (Fig. 2A). DR1 is a 12-nt repeat essential for reverse transcription, and ε is an RNA-stem-loop essential for encapsidation of the pgRNA-P complex and initiation of reverse transcription. To evaluate the role of these elements in shunting, we deleted DR1 (DR1−), replaced DR1 with random nucleotides (DR1-Ran), or replaced the sequences from ∼10 nucleotides downstream of the cap to ε with random sequences (Ran) in the context of the viral genome. These constructs were transfected into LMH cells that support DHBV reverse transcription, and cell lysates were analyzed 1 day posttransfection for P, C, and pgRNA levels by Western and Northern blotting (Fig. 2B to D). To eliminate noise from transfection variation and potential RNA accumulation differences, P translation efficiency was defined as the ratio of P/pgRNA from the same lysate. Northern blots showed that no novel mRNAs were produced (Fig. 2D). None of these alterations near the 5′ end of the genome substantially reduced P synthesis (Fig. 2E).
Fig. 2.
Epsilon sequences contain the donor site, and an element important for shunting is between ε and the C1 AUG. LMH cells were transfected with mutant DHBV genomic expression constructs, and pgRNA and P levels were detected by Northern and Western blotting (WB) of lysates on day 1 posttransfection. P translation efficiency was defined as the amount of P divided by the amount of pgRNA in the same lysate. (A) Positions of relevant elements within the 5′ end of the pgRNA. Octagons represent the positions at which the BamHI-SL was inserted to block scanning ribosomes. (B) Representative Western blots for P. (C) Representative Western blots for C. (D) Representative Northern blots for the pgRNA. (E) P translation efficiency for the mutants with lesions in the 5′ end of the pgRNA. Data are normalized to the activity of the wild-type (WT) construct and shown as the mean ± standard deviation from three to five replicate experiments. IP, immunoprecipitation.
In cauliflower mosaic virus and rice tungro bacilliform virus, a short 5′ proximal ORF is required for efficient shunting (25, 29). In the DHBV pgRNA, there are two sORFs between ε and the C1 AUG which encode peptides of 5 and 14 amino acids. Mutating the AUGs for these sORFs reduced P translation by about 50% (Fig. 2E, C012−). Therefore, the sORFs or the mutated nucleotides appear to contribute to shunting to P ORF, but they are not essential.
To determine if shunting occurred from sequences that comprise ε, we inserted a very stable stem-loop (BamHI-SL; folding stability, −69.2 kcal/mol) that is stable enough to block scanning ribosomes (21) upstream and downstream of ε; we previously confirmed that the BamHI-SL blocks ribosomal scanning (33). Inserting the BamHI-SL upstream of ε (SL-HindIII) blocked P translation as well as it did when ribosomal scanning was blocked by inserting it adjacent to the cap (SL-5′) (Fig. 2E). However, when scanning was blocked downstream of ε at nt 2606 (SL-SalI), 2616 (SL-2616), or 2639 (SL-2639), P translation was 80%, 201%, and 140% that of the wild type, respectively (Fig. 2E). This indicates that most ribosomes depart from sequences that comprise ε.
As expected from previous mutational analyses of the pgRNA (1, 3, 11), pgRNA levels were reduced about 80% when sequences between ε and 6 nt upstream of the C1 ATG (nt 2616 to 2640) were deleted in the construct dl2616-2640 (Fig. 2D, lane 10). P translation was reduced to undetectable levels in this construct (Fig. 2B, lane 12), and increasing the sensitivity of the analysis by immunoprecipitating P rather than detecting it by Western analysis in whole-cell lysates confirmed this result (Fig. 2B, lane 16). The dl2616-2640 pgRNA was a functional mRNA because the C protein is also encoded by this bicistronic message, and C accumulated to easily detectable levels (Fig. 2C, lane 11). Further evidence for lack of P translation from the dl2616-2640 pgRNA was provided by the elevated translation efficiency for C (C/pgRNA), which was roughly twice that of the wild-type pgRNA. This was expected because ∼30 to 50% of the pgRNA in the cytoplasm is unavailable for translation because it is encapsidated into nascent capsid particles, and blocking encapsidation of the pgRNA (which is triggered by binding of P to ε on the pgRNA) increases the amount of the pgRNA available for translation (24). Therefore, not only are nt 2616 to 2640 important for proper accumulation of the pgRNA, but also this region contains an important cis-acting element for shunting. We termed this element “εC” due to its position between ε and the C ORF.
Proper folding of ε and the known functional elements of ε are not needed for shunting.
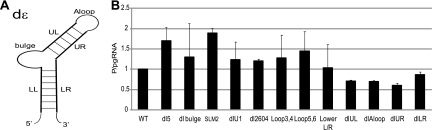
We next analyzed ε (Fig. 3A) in detail, focusing on features known to affect encapsidation and reverse transcription (12, 24). Deleting the lower left stem (dl5), the bulge (dlBulge), altering the sequence of the bulge (SLM2), deleting an unpaired U at nt 2580 (dlU1), deleting an unpaired U at nt 2604 (dl2604), altering sequences in the apical loop (Loop3,4, Loop5,6) or making complementary mutations in the lower stem (LowerL/R) either had no effect on P translation or modestly increased P translation (Fig. 3B). Deleting the upper left stem (dlUL), the apical loop (dlAloop), the upper right stem (dlUR), or the lower right stem (dlLR) all decreased P translation, but the suppression was less than 2-fold. As expected, most mutations to ε that are known to ablate encapsidation (24) led to an increase in P translation, presumably due to availability of a higher proportion of the pgRNA for translation. These data indicate that ribosomes shunt from sequences that comprise ε, but proper folding of ε and the known functional elements of ε are not necessary for shunting.
Fig. 3.
Proper folding of ε and the known functional elements of ε are not needed for shunting. LMH cells were transfected with DHBV genomic expression constructs carrying mutations in ε, and pgRNA and P levels were detected by Northern and Western blotting of lysates on day 1 posttransfection. (A) The structure of DHBV ε showing regions that were deleted or mutated: apical loop (Aloop), upper left stem (UL), Bulge, lower left stem (LL), upper right stem (UR), and lower right stem (LR). (B) P translation efficiency from the mutant constructs shown as the mean ± standard deviation of three to five independent experiments.
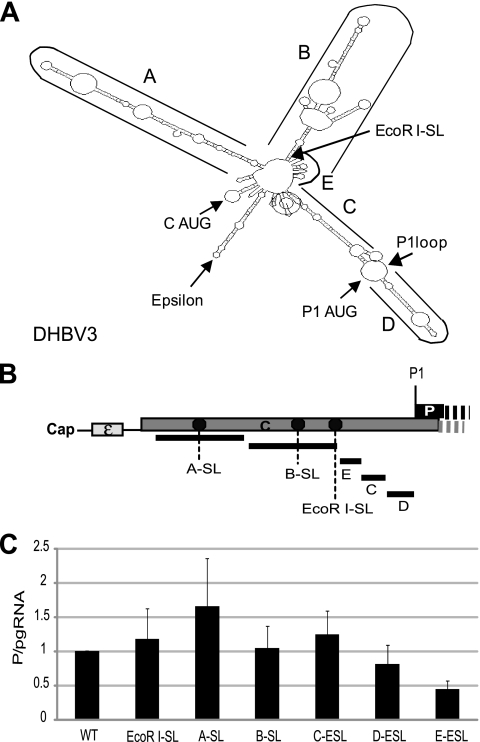
RNA element E may contribute to shunting.
We previously demonstrated that blocking ribosomal scanning at the EcoRI site between the C1 and P1 AUGs (EcoRI-SL; Fig. 4B and C) had minimal effects on P translation (33). Therefore, the acceptor site(s) to which the ribosomes are transferred during shunting must be downstream of the EcoRI site, but sequences upstream from the EcoRI site could still contribute to shunting. To help map sequences that may contribute to shunting, we predicted the fold of the DHBV pgRNA 5′ leader. The DHBV pgRNA 5′ leader was predicted to fold into a 4-armed structure in which prominent features were named A to E (Fig. 4A). This predicted structure was employed only to guide a scanning deletion analysis of the sequences upstream of the P AUG in which we sequentially ablated the predicted elements A to E in the C background (Fig. 4B). To eliminate the possibility that these large deletions may have inadvertently activated ribosomal scanning from upstream of the deletion sites, we then inserted the BamHI-SL into the A and B deletion sites and into the EcoRI site in the C, D, and E mutants. Translation from deletion constructs lacking A, B, C, and D elements either modestly increased P translation or had no effect (Fig. 4C), demonstrating that sequences comprising elements A to D are not needed for shunting. However, P translation from the E deletion construct was reduced to 40% (Fig. 4C, E-ESL), indicating that the E region (nt 5 to 44) may contribute to shunting.
Fig. 4.
Only element E between the C1 and P1 AUGs contributes to shunting. (A) A predicted fold for the 5′ end of the DHBV pgRNA. This predicted fold shows the genomic regions defined as A through E; it was used to guide a scanning deletion analysis of the sequences upstream of the P1 AUG. (B) Relative positions of the deletions and the insertions of the BamHI-SL within the 5′ UTR. (C) P translation efficiency from DHBV genomic expression constructs carrying deletions of the A to E elements. Elements A to E were sequentially deleted. The BamHI-SL was inserted into the A and B deletion sites and into the EcoRI site in the C, D, and E deletion mutants to block fortuitous activation of ribosomal scanning. The mutants were transfected into LMH cells, and P translation efficiency was calculated. The results are shown as the mean ± standard deviations from three independent experiments.
Contribution of sequences within the P ORF to shunting.
To test whether regions of the pgRNA within the P ORF may contribute to shunting, we replaced the P-coding sequences with the LacZ and luciferase genes in the DHBV genomic expression vector D1.5G. Unfortunately, both replacements led to production of novel mRNAs that prevented interpretation of the role of P ORF sequences on shunting.
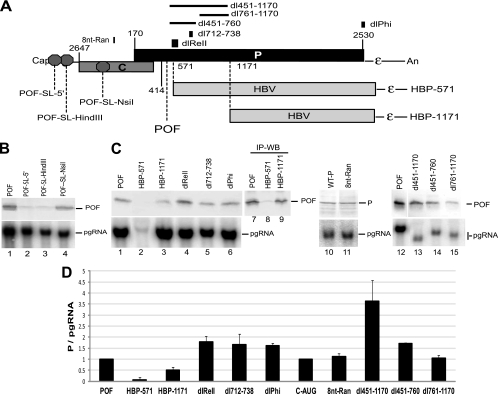
We then made a genomic expression construct in which nt 424T was deleted, producing a stop codon at nt 435 (D1.5G-POF) that truncates P after amino acid 88. Monitoring production of the POF fragment would permit mutational analysis of RNA sequences downstream of nt 435 without risk of artifacts from altering the stability of P if the POF fragment were translated via shunting. Therefore, to evaluate the translation mechanism of the POF protein, we inserted the BamHI-SL within 10 and 30 nucleotides of the cap (POF-SL-5′ and POF-SL-HindIII) and at the NsiI site between the cap and the P1 AUG (POF-SL-NsiI; Fig. 5A). LMH cells were transfected with these genomic expression constructs, and P was detected in whole-cell lysates by Western blotting. Inserting the BamHI-SL near the cap nearly eliminated accumulation of the POF protein (Fig. 5B, lanes 2 and 3), indicating cap-dependent translation. Inserting the BamHI-SL at the NsiI site (Fig. 5B, lane 4) failed to block synthesis of the POF fragment, indicating that ribosomes do not scan linearly from the cap to the P1 AUG along this mutant form of the pgRNA. Northern blots revealed that insertion of the BamHI-SL into the pgRNA did not create a novel mRNA or alter accumulation of the pgRNA (Fig. 5B, lower panel, lanes 1 to 4). Therefore, the POF protein is translated by ribosomal shunting, and hence, it can be used as a marker of translation initiation at P1.
Fig. 5.
Sequences downstream of the P1 AUG are not needed for shunting. Mutations were introduced into genomic expression vectors in the D1.5G-POF or D1.5G background, the mutants were transfected into LMH cells, and POF, P, and pgRNA levels were measured by Western and Northern analysis of the same lysates 1 day posttransfection. (A) Structure of the pgRNA showing the locations of the elements altered in this experiment. The stop codon defining the POF truncated form of P is shown, and octagons represent the insertion sites for the BamHI-SL used to block scanning ribosomes. The regions replaced with HBV sequences are shown. (B) Northern and Western blots showing that the POF fragment is translated by ribosomal shunting. (C) Representative Western and Northern blots. (D) P translation efficiency for the mutants expressed as the mean ± standard deviation from three to five independent experiments.
We then asked whether sequences within the P ORF contribute to shunting. We made two constructs, HBP-571 and HBP-1171, in which sequence from nt 572 or 1172 to the P stop codon at nt 2530 was replaced by the equivalent region of the HBV P ORF (Fig. 5A) in the D1.5G-POF background. Note that there is no nucleotide homology between the DHBV and HBV P ORFs and that HBV P is not translated by shunting (7, 13). P translation was reduced by about 50% in HBP-1171 (Fig. 5C and D). P translation was ablated in HBP-571, but pgRNA levels were also reduced by ∼95% (Fig. 5C, lower panel, lane 2). Increasing the sensitivity of this analysis by immunoprecipitating P revealed that the POF protein was made from the HBP-571 construct but that its translation efficiency (i.e., POF/pgRNA) was only about 10% that of the wild-type POF construct (Fig. 5C, lane 8). We then deleted sequences between nt 451 and 1170 in mutants dl451-1170, dl451-760, and dl761-1170 in the POF background. None of these mutations suppressed P translation, and the dl451-1170 and dl451-760 mutations increased translation, presumably by ablating the second portion of the DHBV encapsidation signal (1, 20). Therefore, the effect of the HBP-571 mutation on P translation was an artifact of the chimeric construct. Together, these data reveal that sequences between nt 451 and 2530 are not essential for shunting.
We made four other mutants in the D1.5G-POF background to test possible roles of specific elements that are known to contribute to DHBV reverse transcription or that were analogous to shunting motifs in other viruses on shunting in DHBV. dlReII deleted nt 538 to 611, which contains the second portion of the DHBV pgRNA encapsidation signal (1, 20). dl712-738 deleted nt 712 to 738, which is complementary to the εC element (nt 2614 to 2640) that we found to be essential for shunting (Fig. 2). dlPhi deleted nt 2514 to 2534, which is complementary to ε and is reported to be important for efficient viral replication (35). Finally, we observed that nt 15 to 22 within the E region that we found contributes to shunting (Fig. 4) was complementary to the chicken 18S rRNA. As mRNA-18S RNA hybridization is an essential component of shunting in the adenoviruses and HSP70 (43), we altered nt 15 to 22 (AGCTTATG to GTACGCGA) in the E element to destroy this homology (8nt-Ran). These four constructs were transfected into LMH cells, and pgRNA and POF or P levels were detected in whole-cell lysates by Western and Northern blotting. None of these mutations reduced pgRNA accumulation (Fig. 5C) or translation from the P1 AUG (Fig. 5C and D). Therefore, these four elements are not needed for shunting.
Ribosomes land at multiple sites.
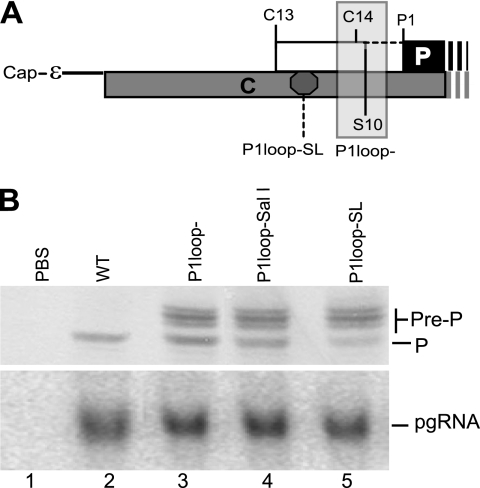
We previously found that translation of the DHBV P ORF initiates at the upstream in-frame C13 and C14 AUGs, in addition to the P1 AUG, and initiation at C13 was demonstrated to occur by ribosomal shunting (2). Translation from the C13 AUG in the absence of the S10 stop codon produces a glycosylated P isoform called pre-P in which the leader sequences appear to be removed following translocation into the endoplasmic reticulum (2). We therefore asked whether ribosomes land at a single acceptor site and then scan to the downstream AUGs or if they land at multiple acceptor sites and initiate near their landing sites. To do this, we employed a derivative of the P1loop deletion (P1loop−) mutant in which the C14 and S10 codons in DHBV strain 3 are deleted, allowing synthesis of the pre-P isoform of P that is initiated at C13 (nt 20). We created a SalI site after nt 34 in the C-P1loop background (P1loop-SalI) and then inserted the BamHI-SL into this site (P1loop-SL) to block scanning from a potential shunt acceptor(s) between C13 and P1 (Fig. 6A). Translation from P1 was efficient in P1loop-SL (Fig. 6B, lane 5), although the amount was 50% less than that of its parental control, P1loop-SalI (Fig. 6B, lane 4). No difference in translation from C13 that produces the larger P isoforms was observed between the P1loop-SL and P1loop-SalI constructs. Because the BamHI-SL is stable enough to block scanning ribosomes but does not impede translating ribosomes (21), these data indicate that ribosomes must land both upstream of the C13 AUG at nt 20 and downstream of the P1loop-SL insertion at nt 34. We previously demonstrated that blocking ribosomal scanning at the NsiI (nt 2845) or EcoRI (nt 3017) sites has little to no effect on P translation (2, 33). Therefore, acceptor regions must lie between nt 3017 and 20 and between nt 35 and the P1 AUG at nt 170. However, because P1 translation was reduced ∼2-fold by inserting the BamHI-SL between C13 AUG and P1 AUG in P1loop-SL, we cannot exclude the possibility that some ribosomes land upstream of nt 20 and then scan downstream to initiate translation at the C13 and P1 AUGs.
Fig. 6.
Ribosomes land at more than one acceptor site. Mutations were introduced into genomic expression vectors in the P1loop− background to permit synthesis of the pre-P protein from DHBV strain 3, the mutants were transfected into LMH cells, and P, pre-P, and pgRNA levels were measured by Western and Northern analysis of the same lysates 1 day posttransfection. (A) Structure of the pgRNA showing the locations of the elements altered in this experiment. (B) Representative Western blot (upper panel) and Northern blot (lower panel) showing accumulation of P, pre-P, and the pgRNA. PBS, pBluescript vector.
DISCUSSION
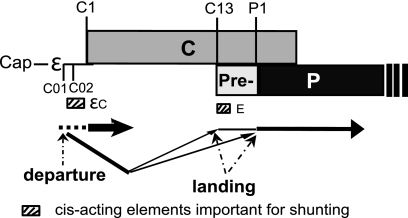
Five observations relevant to ribosomal shunting along the DHBV pgRNA to the P ORF are presented here. First, ribosomes depart from sequences that comprise ε (Fig. 2), but the fold of ε and its known functional elements are not needed for shunting (Fig. 3). Second, ribosomes land at two or more sites on the pgRNA, one between nt 3017 (EcoRI site) and nt 20 (C13) and the other between nt 35 (P1loop-SL) and nt 170 (P1 AUG) (Fig. 6). Third, all sequences upstream of ε, the majority of the sequences between the C1 and P1 AUGs, and sequences between nt 451 and 2530 are dispensable for shunting (Fig. 2, 4, and 5). Fourth, elements on the pgRNA that are known to be involved in reverse transcription or that are predicted to be involved in shunting on the basis of mechanisms documented in other mRNAs are not needed for shunting in DHBV (Fig. 2 and 5). Finally, εC (nt 2616 to 2640) and the E element (nt 5 to 44) contribute to shunting (Fig. 2 and 4), with the contribution of εC possibly involving the C01/C02 sORFs. These observations are summarized in the context of the DHBV pgRNA in Fig. 7.
Fig. 7.
Summary of the DHBV P shunt and the RNA elements directing ribosomal transfer.
The locations at which the ribosomes cease scanning linearly along the pgRNA (the donor site) and where they land (the acceptor sites) were identified by these studies. Sequences that comprise ε were revealed to be the donor site because blocking scanning near the 5′ end of the pgRNA or just 5′ to ε strongly inhibited P translation, whereas blocking translation at three sites just 3′ to ε had little to no effect on shunting (Fig. 2). However, we could not identify sequences within ε that were essential for shunting (Fig. 3). Therefore, although sequences within ε appear to comprise the donor site, their role may be mechanistically passive. We used translation of the P isoform called pre-P that initiates at the C13 AUG (2) to demonstrate that there are at least two acceptor sites where the ribosomes land following shunting. Blocking ribosomal scanning just downstream of C13 (nt 35) had no effect on translation from C13 (Fig. 6), so ribosomes must land upstream of C13 to permit initiation at this codon. They must also land downstream of C13 because the stem-loop that we inserted 3′ to C13 would block ribosomes that scanned past C13, but this mutation reduced translation from the downstream P1 AUG by only 50%. Although these data reveal that ribosomes land between nt 3017 and 20 and between nt 35 and 170, they do not reveal whether the ribosomes land at two or more discrete motifs or if they land relatively randomly within these acceptor regions.
RNA elements other than ε that are candidates for cis-acting shunting sequences because they are important for encapsidation of the pgRNA or reverse transcription or are complementary to a key shunting element include DR1 (nt 2535 to 2546; mutated in constructs DR1− and DR1-Ran), the secondary element of the encapsidation signal (nt 537 to 610; dlReII), phi (nucleotides 2514 to 2534; dlPhi), and sequences complementary to εC (dl712-738). Mutating these sequences revealed that none of them were essential for shunting to the P ORF (Fig. 2 and 5). Therefore, shunting, encapsidation, and reverse transcription are functionally separable processes.
The previously characterized shunting events can be divided into three classes (30). Class I includes cauliflower mosaic virus 35S mRNA (6, 10), rice tungro bacilliform pararetrovirus RNA (25, 26), and the prototype foamy virus RNA (31). Shunting in this group is characterized by scanning along the message, translation of an sORF and release of the product peptide, pausing of the ribosomes at the base of a large stable RNA stem-loop, translocation of the ribosome across the base of the stem-loop to the acceptor site, and reinitiation of translation at the target AUG. Class II includes the adenovirus late mRNAs and the HSP70 mRNA (43). This class is characterized by the presence of multiple strong secondary structures in the 5′ UTR and direct binding between the 5′ UTR and the 18S rRNA. The mechanistic role of the mRNA-rRNA binding is not clear, but the interaction is needed for shunting. Class III includes translation of the Y1 and Y2 proteins from the Sendai virus P/C mRNA. This class is characterized by a lack of a clearly defined donor element, lack of a requirement for an obvious strong secondary structure in the 5′ UTR, an essential short cis-acting element just 3′ of the initiation codon, direct transfer of the ribosomes to the initiation codon, and lack of a requirement for AUG as the start codon (5, 17). Insufficient information exists to place translation of the Sendai virus X protein on the P/C mRNA (5), the β-secretase (28), or the papillomavirus type 18 E1 protein (27) into any of these classes.
Our data regarding shunting on the DHBV pgRNA to the P ORF are not compatible with any of these three shunting mechanisms. The DHBV shunt does not fit into class I because each of the predicted large secondary structures in the pgRNA could be deleted without ablating shunting and because the sORFs near the shunt donor are not essential for P translation (destroying the sORF AUGs in mutant C012 reduced P translation by only 50%, and these sORFs are not encountered by the scanning ribosomes because they are 3′ to the donor site within the ε sequences [Fig. 2]). The DHBV shunting mechanism does not appear to correspond to the mRNA-18S rRNA binding mechanism in class II because ribosomes depart from a discrete donor site (within the ε sequences) and because randomizing the only homology that we have been able to identify between elements important for DHBV shunting and the chicken 18S rRNA had no effect on shunting (Fig. 5D, 8nt-Ran). Therefore, if direct interactions between the pgRNA and the 18S rRNA are needed for shunting, we have not yet identified the complementary sequences in the pgRNA. DHBV shunting is also incompatible with class III because DHBV uses a discrete donor site and multiple acceptor sites (Fig. 2 and 6), and because initiation following shunting is strictly AUG dependent (mutating the P1 AUG leads to initiation at the P2 site, and mutating both the P1 and P2 AUGs leads to initiation at the P3 AUG [33]). Together, these observations indicate that shunting on the DHBV pregenomic RNA employs either a novel mechanism or a substantial variation on one of the characterized mechanisms.
These data identify the donor and acceptor sites on the pgRNA where the ribosomes depart and land during shunting, and they reveal some of the sequences that are important in promoting shunting. However, they do not fully map the sequences that direct shunting. This is partially due to induction of alternative mRNAs from some constructs that prevent interpretation of the data and the inability to examine sequence between P1 (nt 170) and the POF deletion site (nt 424) without possibly altering the half-life of the translation product from P1. However, the ability to identify the donor site as being within the ε sequences but the inability to identify discrete features within ε that are needed for shunting, coupled with the ability of at least two regions ranging from nt 3017 to 170 to act as acceptor sites for the ribosomes, indicates that our failure to fully map the regulatory elements may be primarily due to the nature of the shunting signals themselves. The simplest interpretation is that much of the information required to direct shunting is dependent upon the overall fold of the pgRNA molecules that are serving as P mRNAs, but this hypothesis cannot be evaluated at this time because the conformation(s) that the pgRNA adopts has not yet been fully characterized.
ACKNOWLEDGMENTS
We thank Ann Palmenberg for predicting the structure of the full-length DHBV pgRNA, Xiaohong Cheng for experimental assistance, and Christian Thoma for critical comments on the manuscript.
This work was supported by grant R01 AI38447 from the National Institutes of Health and by Saint Louis University institutional support.
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Calvert J., Summers J. 1994. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J. Virol. 68:2084–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cao F., Scougall C. A., Jilbert A. R., Tavis J. E. 2009. Pre-P is a secreted glycoprotein encoded as an N-terminal extension of the duck hepatitis B virus polymerase gene. J. Virol. 83:1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang C., Hirsch R. C., Ganem D. 1995. Sequences in the preC region of duck hepatitis B virus affect pregenomic RNA accumulation. Virology 207:549–554 [DOI] [PubMed] [Google Scholar]
- 4. Condreay L. D., Aldrich C. E., Coates L., Mason W. S., Wu T. T. 1990. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J. Virol. 64:3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Breyne S., Simonet V., Pelet T., Curran J. 2003. Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins. Nucleic Acids Res. 31:608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dominguez D. I., et al. 1998. Ribosome shunting in cauliflower mosaic virus. Identification of an essential and sufficient structural element. J. Biol. Chem. 273:3669–3678 [DOI] [PubMed] [Google Scholar]
- 7. Fouillot N., Tlouzeau S., Rossignol J. M., Jean-Jean O. 1993. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 67:4886–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Futterer J., Kiss-Laszlo Z., Hohn T. 1993. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73:789–802 [DOI] [PubMed] [Google Scholar]
- 9. Futterer J., et al. 1996. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J. Virol. 70:2999–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hemmings-Mieszczak M., Steger G., Hohn T. 1997. Alternative structures of the cauliflower mosaic virus 35 S RNA leader: implications for viral expression and replication. J. Mol. Biol. 267:1075–1088 [DOI] [PubMed] [Google Scholar]
- 11. Hirsch R. C., Loeb D. D., Pollack J. R., Ganem D. 1991. cis-Acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J. Virol. 65:3309–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu K., Beck J., Nassal M. 2004. SELEX-derived aptamers of the duck hepatitis B virus RNA encapsidation signal distinguish critical and non-critical residues for productive initiation of reverse transcription. Nucleic Acids Res. 32:4377–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hwang W. L., Su T. S. 1998. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol. 79:2181–2189 [DOI] [PubMed] [Google Scholar]
- 14. Junker-Niepmann M., Bartenschlager R., Schaller H. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koh D. C., Mauro V. P. 2009. Reconciling contradictory reports regarding translation of BACE1 mRNA: initiation mechanism is altered by different expression systems. RNA Biol. 6:54–58 [DOI] [PubMed] [Google Scholar]
- 16. Kozak M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867–19870 [PubMed] [Google Scholar]
- 17. Latorre P., Kolakofsky D., Curran J. 1998. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 18:5021–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matzura O., Wennborg A. 1996. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput. Appl. Biosci. 12:247–249 [DOI] [PubMed] [Google Scholar]
- 19. Morley S. J., Coldwell M. J. 2008. A cunning stunt: an alternative mechanism of eukaryotic translation initiation. Sci. Signal. 1:e32. [DOI] [PubMed] [Google Scholar]
- 20. Ostrow K. M., Loeb D. D. 2002. Characterization of the cis-acting contributions to avian hepadnavirus RNA encapsidation. J. Virol. 76:9087–9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pelletier J., Sonenberg N. 1985. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40:515–526 [DOI] [PubMed] [Google Scholar]
- 22. Pestova T. V., et al. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 98:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pollack J. R., Ganem D. 1993. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol. 67:3254–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pollack J. R., Ganem D. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68:5579–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pooggin M. M., Futterer J., Hohn T. 2008. Cross-species functionality of pararetroviral elements driving ribosome shunting. PLoS One 3:e1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pooggin M. M., Ryabova L. A., He X., Futterer J., Hohn T. 2006. Mechanism of ribosome shunting in rice tungro bacilliform pararetrovirus. RNA 12:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Remm M., Remm A., Ustav M. 1999. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 73:3062–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rogers G. W., Jr., Edelman G. M., Mauro V. P. 2004. Differential utilization of upstream AUGs in the beta-secretase mRNA suggests that a shunting mechanism regulates translation. Proc. Natl. Acad. Sci. U. S. A. 101:2794–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryabova L. A., Pooggin M. M., Dominguez D. I., Hohn T. 2000. Continuous and discontinuous ribosome scanning on the cauliflower mosaic virus 35 S RNA leader is controlled by short open reading frames. J. Biol. Chem. 275:37278–37284 [DOI] [PubMed] [Google Scholar]
- 30. Ryabova L. A., Pooggin M. M., Hohn T. 2002. Viral strategies of translation initiation: ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 72:1–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schepetilnikov M., et al. 2009. Molecular dissection of the prototype foamy virus (PFV) RNA 5′-UTR identifies essential elements of a ribosomal shunt. Nucleic Acids Res. 37:5838–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seeger C., Zoulim F., Mason W. S. 2007. Hepadnaviruses, p. 2977–3029 In Knipe D. M., Howley P., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 33. Sen N., Cao F., Tavis J. E. 2004. Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting. J. Virol. 78:11751–11757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sherrill K. W., Lloyd R. E. 2008. Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol. Cell. Biol. 28:2011–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang H., McLachlan A. 2002. A pregenomic RNA sequence adjacent to DR1 and complementary to epsilon influences hepatitis B virus replication efficiency. Virology 303:199–210 [DOI] [PubMed] [Google Scholar]
- 36. Tavis J. E., Ganem D. 1996. Evidence for the activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 70:5741–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tavis J. E., Perri S., Ganem D. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang G. H., Seeger C. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507–6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xi Q., Cuesta R., Schneider R. J. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 18:1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yao E., Gong Y., Chen N., Tavis J. E. 2000. The majority of duck hepatitis B virus reverse transcriptase in cells is nonencapsidated and is bound to a cytoplasmic structure. J. Virol. 74:8648–8657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao E., Schaller H., Tavis J. E. 2003. The duck hepatitis B virus polymerase and core proteins accumulate in different patterns from their common mRNA. Virology 311:81–88 [DOI] [PubMed] [Google Scholar]
- 42. Yao E., Tavis J. E. 2003. Kinetics of synthesis and turnover of the duck hepatitis B virus reverse transcriptase. J. Biol. Chem. 278:1201–1205 [DOI] [PubMed] [Google Scholar]
- 43. Yueh A., Schneider R. J. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 14:414–421 [PMC free article] [PubMed] [Google Scholar]
- 44. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]