Abstract
The incorporation of potentially catalytic groups in DNA is of interest for the in vitro selection of novel deoxyribozymes. A series of 10 C5-modified analogues of 2′-deoxyuridine triphosphate have been synthesised that possess side chains of differing flexibility and bearing a primary amino or imidazole functionality. For each series of nucleotide analogues differing degrees of flexibility of the C5 side chain was achieved through the use of alkynyl, alkenyl and alkyl moieties. The imidazole function was conjugated to these C5-amino-modified nucleotides using either imidazole 4-acetic acid or imidazole 4-acrylic acid (urocanic acid). The substrate properties of the nucleotides (fully replacing dTTP) with Taq polymerase during PCR have been investigated in order to evaluate their potential applications for in vitro selection experiments. 5-(3-Aminopropynyl)dUTP and 5-(E-3-aminopropenyl)dUTP and their imidazole 4-acetic acid- and urocanic acid-modified conjugates were found to be substrates. In contrast, C5-amino-modified dUTPs with alkane or Z-alkene linkers and their corresponding conjugates were not substrates. The incorporation of these analogues during PCR has been confirmed by inhibition of restriction enzyme digestion using XbaI and by mass spectrometry of the PCR products.
INTRODUCTION
In vitro selection is a combinatorial chemical technique in which functional nucleic acids are isolated from random libraries of single-stranded DNA or RNA. The technique has been used extensively to generate high affinity ligands for a range of targets from small organic molecules to proteins. In addition, direct selection for catalysis has yielded nucleic acids that are capable of accelerating the rate of a variety of reactions, including several phosphoryl transfer reactions (1–4), amide bond formation (5–7) and a Diels–Alder reaction (8). These newly selected nucleic acid catalysts lend further weight to the nucleic acid world hypothesis.
The selection of nucleic acids as high affinity ligands and catalysts from random libraries exploits the structural diversity of both single-stranded DNA and RNA molecules, which assume a wide range of structures, sometimes stabilised by metal or ammonium ions. However, in contrast to proteins, the functional group repertoire of nucleic acids is relatively modest. With particular reference to catalysis, the pKa values of the nucleobases, typically considered to be in the region 3.5–4.5 (for protonation of N-1 adenine and N-3 cytosine) or 9–10 (deprotonation of NH-1 guanosine and NH-3 uridine), appear to be unsuited to general acid or general base catalysis, although it is possible in some cases that special local structures may perturb these pKa values (9). Indeed, two naturally occurring ribozymes, the hepatitis δ virus ribozyme (10) and the ribosome (11), seem to use general base catalysis. However, most catalytic nucleic acids appear to use alternative forms of rate acceleration. Thus RNA and DNA appear to lack the acid and base functionalities close to neutrality and the potential for covalent catalysis frequently observed to be present in the active sites of protein enzymes. In addition, nucleic acids are also lacking the potentially positively charged functionality provided in proteins by lysine and arginine residues.
To increase the structural diversity and catalytic potential of nucleic acids there has been recent interest in the potential of modified nucleotides to present these moieties during an in vitro selection experiment. Most selection protocols involve a PCR amplification step allowing multiple rounds of population enrichment and selection to be performed. Libraries are amplified by transcription (RNA) and PCR (DNA) and the resultant nucleic acids must serve as templates for further enzymatic polymerisation following the selection step. Thus, replacing one or more natural nucleoside triphosphates with a modified nucleotide is an attractive route to enhancing functional group diversity provided that base pairing properties are not impaired. Furthermore, the analogue must be a substrate for the relevant polymerase. In addition, modifications to the nucleic acid library could also be used to confer other desirable properties on the resultant functional molecule, for example nuclease resistance or greater stability.
Modified ribonucleoside triphosphates in which the 2′-hydroxyl group has been replaced with a fluorine atom or an amino group are substrates for RNA polymerases and render the resultant RNA nuclease resistant (12). These 2′-modified triphosphates have been used in selection experiments to generate high affinity ligands for human neutrophil elastase (13) and keratin growth factor (14). Eaton and colleagues have described the synthesis of a range of 5-carbonyluridine analogues (15), 5-carboxamidouridine (16) and 8-carboxamidoadenosine derivatives (17), which contain appended alkyl, aryl, hydroxyl and amino functionalities. The triphosphates of 5-benzoyluridine (15), 5-(pyridylmethylcarboxamido)uridine (8,18) and a 5-imidazole-modified uridine (6) have all been reported to be substrates for RNA polymerase and the latter two analogues have completely replaced uridine triphosphate in experiments to isolate RNAs capable of catalysing a Diels–Alder reaction and amide bond formation. McLaughlin and colleagues (19) have recently described synthesis of the triphosphates of 5-(3-aminopropyl)uridine and 5-(2-mercaptoethyl)uridine and their properties as substrates of T7 RNA polymerase have been evaluated. The syntheses of several triphosphates of 2′-deoxy-2′-fluorouridine, 2′-O-methyluridine and 2′-deoxy-2′-fluorocytidine bearing C5 primary amino and imidazole modifications have been described (20). However, the substrate properties were not investigated in this study.
Selection from modified DNA libraries has included pools generated from PCR reactions where dTTP is completely replaced by 5-(1-pentynyl)-2′-deoxyuridine triphosphate (21) and, more recently, by 5-(3-aminopropynyl)-2′-deoxyuridine (22). The modified pools so generated were utilised to evolve DNA molecules with high affinity to human thrombin and ATP, respectively. In addition, Sakthivel and Barbas (23) have described a range of conjugates of 5-(3-aminopropenyl)-2′-deoxyuridine triphosphate which have been demonstrated to be substrates for DNA polymerases typically used in PCR reactions. In subsequent work a urocanic acid conjugate of 5-(3-aminopropenyl)-2′-deoxyuridine triphosphate was used to select a zinc ion-dependent deoxyribozyme (4), although in this case the nucleotide analogue was incorporated into the nucleic acid library using a reverse transcriptase. Hélène and co-workers (24) have demonstrated simultaneous incorporation of 5-(3-aminopropenyl)-2′-deoxyuridine triphosphate (replacing dTTP) and 8-[2-(4-imidazolyl)ethylamino]-2′-deoxyadenosine (replacing dATP) using modified T7 DNA polymerase (Sequenase). However, the analogues were incorporated with reduced efficiency compared to their natural counterparts and no report was made of simultaneous incorporation of both during PCR.
Despite literature precedents that modifications at the 5-position of uridine and 2′-deoxyuridine may be tolerated by polymerases, there has been no systematic study of how the type of linker employed at the 5-position of the nucleobase may change the efficacy of the triphosphate as a substrate during such reactions. In addition, since in vitro selection is essentially an iterative procedure, there is no way of predicting what type of linker would be best for positioning the extra functionality in each selection experiment and thus a range of different linkers would be advantageous. Here we present a systematic study of the substrate properties of 5-modified 2′-deoxyuridine triphosphates bearing free amino or conjugated imidazole groups and a study of their efficiency as substrates for Taq polymerase with respect to flexibility, stereochemistry and length of the C5 linker group.
MATERIALS AND METHODS
Chemicals and reagents
AnalaR glacial acetic acid and tetra-sodium pyrophosphate decahydrate were from BDH. Anhydrous dimethylformamide (DMF), phosphoryl chloride (phosphorus oxychloride), proton sponge® (1,8-bis-dimethylamino naphthalene), tri-n-butylamine (99+%), triethylamine (99%), trimethylphosphate (99+%) and 3 Å molecular sieves were from Aldrich.
The following resins were used: Dowex® 50Wx8 in the H+ form (100–200 mesh); Dowex® AG1X8 from BDH; Sephadex®–DEAE A-25 ion exchange resin (40–120 µm) from Aldrich.
Triethylamine was heated under reflux and distilled from potassium hydroxide pellets before use. Phosphoryl chloride was freshly distilled before use. Trimethylphosphate and tri-n-butylamine were stored over 3 Å molecular sieves before use.
1H NMR (250.134 MHz) and 31P NMR spectra (101.256 MHz) were obtained on a Brüker AC250 and are given in p.p.m. relative to external standards of tetramethyl silane and 85% phosphoric acid, respectively.
Synthesis of 5-modified nucleoside triphosphates
Synthesis of 5-[3-(trifluoroacetamido)propynyl]-2′-deoxyuridine (1). This compound was first synthesised by Hobbs (25). As the experimental details and characterisation are described only briefly by Hobbs (25), the full details of the method are given below. 5-Iodo-2′-deoxyuridine (0.350 g, 1.0 mmol) was dissolved in degassed, anhydrous DMF (7 ml). Copper(I) iodide (0.022 g, 0.20 mmol) was added and the reaction mixture was stirred under an inert atmosphere in the absence of light until the copper(I) iodide had dissolved. Freshly distilled triethylamine (0.3 ml, 2.0 mmol) was added to the reaction mixture, followed by N-propynyltrifluoroacetamide (prepared by analogy with the literature procedure; 26) (0.45 g, 2.97 mmol) and tetrakis(triphenylphosphine)palladium(0) (0.11 g, 0.10 mmol). The reaction mixture was then stirred for 18 h in the absence of light at room temperature. Methanol (20 ml), dichloromethane (20 ml) and excess Dowex AG1X8 (bicarbonate form) were then added to the reaction mixture and the resulting suspension stirred for 30 min, filtered and concentrated by evaporation. The resulting brown oil was then dissolved in methanol, filtered and concentrated and this process was repeated until no further solid precipitated. The residual brown oil was then crystallised from acetonitrile and the brown crystals obtained purified by a further recrystallisation from acetonitrile to give the desired product (0.23 g, 60% yield). Rf (10% MeOH, 90% CH2Cl2) = 0.25.
1H NMR δ (d6-DMSO) 11.60 (1H, br s, NH), 10.10 (1H, br s, NH), 8.21 (1H, s, H6), 6.13 (1H, m, H1′), 5.30 (1H, m, OH), 5.13 (1H, m, OH), 4.32 (3H, s, CH2, H3′), 3.82 (1H, m, H4′), 3.61 (2H, m, H5′, H5′′), 2.10 (2H, m, H2′, H2′′). EI+ MS e/z 377 (7.5% M+).
Synthesis of 5-(E)-[3-(trifluoroacetamido)propenyl]-2′-deoxyuridine (2). This compound was prepared according to the method of Sakthivel and Barbas (23). Rf product (5% MeOH, 95% EtOAc) = 0.23; Rf 5-iodo-2′-deoxyuridine (5% MeOH, 95% EtOAc) = 0.30.
1H NMR δ (d6-DMSO) 11.47 (1H, br s, NH), 9.70 (1H, br s, NH), 8.06 (1H, s, H6), 6.49 (1H, dt, J = 15.9, 6.1 Hz, CH), 6.42 (2H, m, H1′, CH), 5.27 (1H, m, OH), 5.25 (1H, m, OH), 4.24 (1H, m, H3′), 3.98–3.77 (3H, m, H4′, CH2), 3.57 (2H, m, H5′, H5′′), 2.13 (2H, m, H2′, H2′′). EI+ MS e/z 379 (5% M+).
Synthesis of 5-(Z)-[3-(trifluoroacetamido)propenyl]-2′-deoxyuridine (3). This compound was synthesised from 1 using the published procedure and gave identical data to that described (27).
Synthesis of 5-[3-(trifluoroacetamido)propyl]-2′-deoxyuridine (4). The nucleoside 1 (0.200 g, 0.53 mmol) was dissolved in dry methanol (50 ml) and hydrogenated over PtO2. After 1 h the solution was diluted with methanol (50 ml) and the catalyst removed by filtration through Celite, which was washed with a further portion of methanol (100 ml). The solvent was evaporated and the residue adsorbed onto silica gel and then purified by column chromatography (10% MeOH in EtOAc as eluent) to afford a white foam (0.175 g, 0.46 mmol, 88%). Rf product (5% MeOH, 95% EtOAc) = 0.22.
1H NMR (d6-DMSO) 11.30 (1H, s, NH), 9.42 (1H, s, NH), 7.68 (1H, s, H6), 6.15 (1H, J = 6.7 Hz, H1′), 5.24 (1H, m, OH), 5.03 (1H, m, OH), 4.23 (1H, m, H3′), 3.75 (1H, m, H4′), 3.56 (2H, m, H5′, H5′′), 3.16 (2H, m, ArCH2), 2.20–1.97 (4H, m, NHCH2 and H2′, H2′′), 1.63 (2H, m, CH2CH2CH2); FAB+ MS e/z 382 (75%, [M+H]+) calculated for C14H19N3O6F3 382.122595; found 382.126083.
General method for synthesis of the modified triphosphates (5a–8a). This was adapted from the method of Kovacz and Ötvös (28). Modified nucleoside (0.15 g, 0.4 mmol) was dried in vacuo over P2O5 overnight and then dissolved in anhydrous trimethylphosphate (0.6 ml) under an inert atmosphere. Proton sponge (also dried overnight over P2O5) (0.102 g, 0.48 mmol) was added to the solution and dissolved with heating. The reaction mixture was then cooled in an ice bath and freshly distilled POCl3 (50 µl, 0.48 mmol) was then added dropwise via a syringe with stirring. The reaction mixture was stirred on ice for 2 h and the flask then removed. A well-vortexed mixture of 4 equiv bis-tri-n-butylammonium pyrophosphate (3.2 ml of a 0.5 M solution in anhydrous DMF) and 8 equiv tri-n-butylamine (0.8 ml) were then injected in one portion. The reaction mixture was stirred at room temperature for 10 min and then triethylammonium bicarbonate solution (TEAB) (0.1 M, pH 8, 20 ml) was added. The reaction mixture was stirred at room temperature for 1 h and concentrated aqueous ammonia solution (20 ml) was then added. The reaction mixture was then stirred at room temperature overnight, concentrated to remove the ammonia and the resultant mixture diluted with water (30 ml). This crude mixture was purified by anion exchange chromatography on DEAE–Sephadex A-25 using a linear gradient of TEAB (0.05–0.7 M, pH 8, 2 l each). The triphosphates 5a–8a generally eluted between 0.3 and 0.5 M TEAB. Further purification was by preparative reversed phase HPLC on a Zorbax SB-C18 250 × 21.2 mm column (Anachem) using the following gradient: buffer A (0.1 M TEAB, pH 7.5), buffer B (0.1 M TEAB, 30% acetonitrile, pH 7.5), flow rate 8 ml min–1, t = 0, 5% B; t = 5, 5% B; t = 25, 35% B. Retention times were as follows: 5-(3-aminopropynyl)-2′-deoxyuridine triphosphate (5a), 21.7 min; 5-(E)-(3-aminopropenyl)-2′-deoxyuridine triphosphate (6a), 24.7 min; 5-(Z)-(3-aminopropenyl)-2′-deoxyuridine (7a), 25.7 min; 5-(3-aminopropyl)-2′-deoxyuridine (8a), 23.1 min. The desired triphosphates were isolated as their triethylammonium salts in yields as follows: 5a, 800–900 OD280 (60–70%); 6a–8a, 300–400 OD280 (OD268 for 8a) (20–30%).
ES-MS (mobile phase 3:1 acetonitrile:water, negative mode) e/z: 5a (C12H18N3O14P3), 520 (15% [M-H]–); 6a (C12H20N3O14P3), 566 (5% [M-3H+2Na]–), 544 (20% [M-2H+Na]–), 522 (35% [M-H]–); 7a (C12H20N3O14P3), 544 (7.5% [M-2H+Na]–), 522 (15% [M-H]–); 8a (C12H22N3O14P3), 524 (100% [M-H]–).
31P NMR (D2O): 5a, δ = –9.2 (d, γP), –11.1 (d, αP), –22.6 (t, βP); 6a, δ = –6.1 (d, γP), –11.0 (d, αP), –22.1 (t, βP); 7a, δ = –5.8 (d, γP), –10.8 (d, αP), –21.9 (t, βP); 8a, δ = –5.5 (d, γP), –10.6 (d, αP), –21.4 (t, βP).
λmax: 5a, 288 nm; 6a, 282 nm; 7a, 288 nm; 8a, 268 nm.
General synthesis of the modified triphosphate conjugates (5b, 5c, 6b, 6c, 8b and 8c)
Urocanic acid (0.138 g, 1 mmol) or imidazole 4-acetic acid (0.126 g, 1 mmol) was dissolved in anhydrous DMF (10 ml) under argon and the solution cooled in an ice bath. N-hydroxysuccinimide (0.126 g, 1.1 mmol) and dicyclohexyl carbodiimide (0.226 g, 1.1 mmol) were then added sequentially to the stirred solution and the mixture allowed to warm to room temperature over 1 h, then stirred for a further 1 h. The reaction mixture was then refrigerated overnight, filtered and concentrated by evaporation. The desired ester was then dissolved to give a solution of known concentration in DMF (0.5 M for 5b and 5c, 1 M for 6b, 6c, 8b and 8c). All active esters were generated immediately prior to use and yields were assumed to be quantitative.
Nucleoside triphosphates (5a, 6a and 8a) were dissolved in a 1:1 solution of 0.1 M sodium borate buffer, pH 9:DMF (0.5 ml) at ambient temperature. Active ester (3 equiv for synthesis of 5b and 5c; 25 equiv for 6b, 6c, 8b and 8c) was then added as a solution of known concentration in DMF and the pH of the reaction mixture was adjusted with triethylamine to pH 9 if required. The reaction mixture was then stirred overnight at room temperature. This reaction was generally performed on between 30 and 200 OD280 of starting triphosphates 5a and 6a (OD268 for 8a). The reactions were worked up and purified as follows.
For 5b and 5c the reaction mixtures were evaporated and the residue was dissolved in 50 mM aqueous TEAB pH 7.5 (1–2 ml) then filtered and the crude products purified by reversed phase HPLC (conditions described below).
For 6b and 6c the reaction mixtures were evaporated and the residue dissolved in 1:1 water:DMF (made basic with 1 drop of concentrated aqueous ammonia solution) and purified using preparative silica TLC developed with 1:1:1:1 water:ethanol:acetone:concentrated aqueous ammonia. Product-containing bands were excised and extracted with the same eluent and, following evaporation, the product was dissolved in 50 mM aqueous TEAB pH 7.5 (1–2 ml) and purified by reversed phase HPLC (conditions described below).
For 8b and 8c the reaction mixtures were evaporated and the residue was partitioned between 50 mM aqueous TEAB pH 7.5 (20 ml) and dichloromethane (10 ml). The aqueous layer was subjected to purification by anion exchange chromatography on DEAE–Sephadex A-25 eluting initially with TEAB (0.05 M, 0.5 l) followed by a linear gradient of 0.05–0.7 M TEAB (1 l each). Product-containing fractions were combined, evaporated, dissolved in 50 mM aqueous TEAB, pH 7.5, (1–2 ml) and purified by reversed phase HPLC (conditions described below).
HPLC conditions for final purification of the modified triphosphate conjugates were as follows: µpore Bondapak C18 column, 3.9 × 300 mm (Waters), buffer A (0.1 M TEAB, pH 7.5), buffer B (0.1 M TEAB, 30% acetonitrile, pH 7.5), flow rate 1 ml min–1, t = 0, 5% B; t = 5, 5% B; t = 25, 35% B, t = 30, 35% B. Retention times: 5b, 18.5 min; 5c, 27.6 min; 6b, 20.7 min; 6c, 28.1 min; 8b, 20.6 min; 8c, 27.5 min.
Yields after HPLC: 5b, 70 OD from 100 OD of 5a; 5c, 15 OD from 30 OD of 5a; 6b, 30 OD from 100 OD of 6a; 6c, 86 OD from 200 OD of 6a; 8b, 10 OD from 31 OD of 8a; 8c, 15 OD from 50 OD of 8a.
ES-MS (negative mode, mobile phase 3:1 acetonitrile:water): 5b (C17H22N5O15P3), 650 (15% [M-2H+Na]–), 628 (35% [M-H]–); 6b (C17H24N5O15P3), 642 (20% [M-2H+Na]–), 630 (65% [M-H]–); 8b (C17H26N5O15P3), 632 (65% [M-H]–); 5c (C18H22N5O15P3), 662 (20% [M-2H+Na]–), 640 (40% [M-H]–); 6c (C18H24N5O15P3), 664 (10% [M-2H+Na]–), 642 (100% [M-H]–); 8c (C18H26N5O15P3), 666 (5% [M-2H+Na]–), 644 (25% [M-H]–).
31P NMR (D2O): 5b, δ = –7.2 (d, γP), –11.0 (d, αP), –21.9 (t, βP); 5c, δ = –7.3 (d, γP), –11.0 (d, αP), –21.9 (t, βP); 6b, δ = –5.8 (d, γP), –10.8 (d, αP), –21.5 (t, βP); 6c, δ = –5.7 (d, γP), –10.9 (d, αP), –21.8 (t, βP); 8b, δ = –5.9 (d, γP), –10.7 (d, αP), –21.5 (t, βP); 8b, δ = –5.8 (d, γP), –10.7 (d, αP), –21.4 (t, βP).
λmax: 5b, 288 nm; 6b, 289 nm; 8b, 268 nm; 5c, 291 nm; 6c, 287 nm; 8c, 278 nm.
All attempts to perform this reaction with 7a were unsuccessful.
Oligonucleotide synthesis
Primers for PCR reactions were synthesised on an ABI 392 DNA Synthesiser using conventional phosphoramidite protocols. Biotinylated primers were prepared using ‘biotin amidite’ (PE Biosystems) in the final cycle of synthesis. All oligonucleotides were purified by C18 reversed phase HPLC using triethylammonium acetate buffers and a gradient of acetonitrile. Purified oligomers were desalted on a NAP-10 column (Pharmacia).
PCR amplification
A typical PCR reaction contained 100 pg DNA template [pUC19 (Pharmacia) or pLitmus28 (New England Biolabs)], 0.5 µM each of the forward and reverse primers, 1 µl of Taq polymerase (1 U; Promega), 5 µl of 10× magnesium-free polymerase buffer (Promega), 2 mM MgCl2, 100 µM unmodified dNTPs [control reactions dATP, dCTP, dGTP and dTTP (MBI Fermentas); reactions containing modified triphosphates dATP, dCTP and dGTP only] and 500 µM modified dUTP (omitted in control reactions) in a 50 µl reaction. For all reactions a hot start (3 min at 95°C, addition of MgCl2, followed by a further 3 min at 95°C) was used, followed by 30 cycles of amplification (1 min at 95°C, 1 min at 52°C, 2 min at 72°C) and a final incubation for 10 min at 72°C. The primer sequences used were as follows for pUC19 (98 nt) and pLitmus28 (224 nt): forward primer-1, d(GTA AAA CGA CGG CCA GT); reverse primer, d(AAC AGC TAT GAC CAT GA). A shorter PCR product from pUC19 (62 nt) was formed using the above reverse primer and forward primer-2, d(GGG GAT CCT CTA GAG TCG).
The products of PCR reactions were separated by denaturing 12% (100 nt) or 14% (62 nt) PAGE containing 7 M urea. Larger fragments (224 nt) were monitored by 2% agarose gel electrophoresis. The products were visualised following ethidium bromide staining by transillumination and the image recorded with an Ultra-Violet Products CCD camera and the Labworks analysis software.
XbaI digestion of PCR products
PCR products (98 nt) from 4 × 100 µl reactions were precipitated with 1 vol of isopropanol from 0.75 M ammonium acetate solution. Precipitated products were redissolved in 17 µl of water, and 2 µl of Promega 10× XbaI restriction buffer and 1 µl (10 U) of XbaI (Promega) was added. Reaction mixtures were incubated at 37°C and analysed and visualised by gel electrophoresis as described above. The PCR product containing only natural dNTPs was completely digested after 1.5 h, whilst no products of cleavage were observed for any of the PCR products containing analogues after 3.5 h incubation.
Mass spectrometry of PCR products
Shorter PCR products (62–63 nt) were obtained from pUC19 using forward primer-2 and biotinylated reverse primer. The products of 10 × 100 µl PCR reactions (each using 200 pg DNA template and 5 U enzyme) were combined, extracted with phenol/chloroform/ether, desalted on NAP-10 columns (Pharmacia) and then precipitated with 10 vol of ethanol from 0.75 M ammonium acetate solution. DNA precipitates were dissolved in 200 µl of capture buffer (20 mM Tris–HCl pH 7.5, 2 M NaCl, 1 mm EDTA) and captured on magnetic streptavidin beads (Sigma) that had previously been washed three times with the same buffer. The beads were shaken at 37°C for 30 min, washed three times with capture buffer and eluted with 3 × 200 µl of 0.2 M NaOH. Eluates were combined, neutralised by addition of 0.5 vol of 7.5 M ammonium acetate, desalted on a NAP-10 column and precipitated with ethanol. Single-stranded PCR products were dissolved in water (5–10 µl) and used for mass spectrometry.
Mass spectrometry was performed using 1 µl of a solution containing single-stranded PCR product (concentration ∼20 µM) mixed in a 1:1 ratio of matrix (3-hydroxypicolinic acid, 25 mg/ml) on a Brüker Reflex III MALDI-TOF instrument. Dowex 50 W (NH4+) beads were used to desalt the oligomers prior to analysis. A trypsinogen (23982 [M+H]+ and 11991 [M+2H]2+) standard was used to calibrate the instrument.
Unmodified 63 nt pUC19 fragment. d(GGG GAT CCT CTA GAG TCG ACC TGC AGG CAT GCA AGC TTG GCG TAA TCA TGG TCA TAG CTG TTA) [note that Taq polymerase adds an extra dA to the 3′-end (29), which is included here]. Molecular weight calculated 19 494 (63 nt); found 19 535 (difference from calculated molecular weight +41, 0.2%).
Modified 62/63 nt fragment. d(GGG GAT CC CTA GAG TCG ACC U*GC AGG CAU* GCA AGC U*U*G GCG U*AA U*CA U*GG U*CA U*AG CU*G U*U*A). The 63 nt fragment contains the extra dA shown in bold. U* using modified dUTP 5a in PCR reactions: molecular weight calculated 20 118 (63 nt); found 20 223 (difference from calculated mol. wt +105, 0.5%). U* using modified dUTP 5b in PCR reactions: molecular weight calculated 21 535 (62 nt), 21 848 (63 nt); found 21 542 (difference from calculated molecular weight +7, 0.1%, –306, 1.4%)
Mass spectra are available as Supplementary Material.
RESULTS AND DISCUSSION
Design and synthesis of modified triphosphates
Lysine and histidine residues play important roles in the active sites of many protein enzymes, accelerating the rates of reaction by electrostatic, general acid, general base and covalent catalysis. In addition, these residues also play important structural roles in proteins and are involved in electrostatic and hydrogen bonding interactions. In contrast, nucleic acids lack the functional group repertoire normally associated with proteins. To mimic the role of lysine and histidine residues we chose to synthesise 2′-deoxyribonucleoside triphosphates bearing primary amino groups and imidazole residues. In the pyrimidine series C5 was the position chosen to attach the required functionality because of literature precedents (25,30), which suggested that C5-modified pyrimidines should be substrates for DNA polymerases.
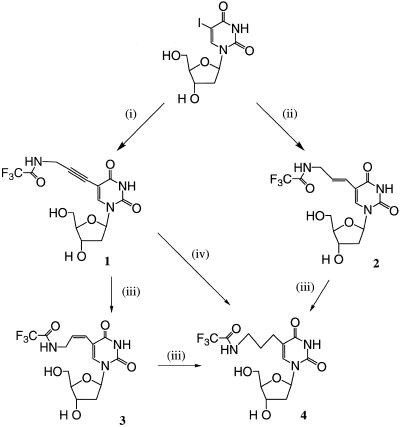
In order to study the role of linker rigidity on the substrate properties of C5-amino-modified 2′-deoxyuridine triphosphates in PCR reactions, we synthesised a series of nucleoside derivatives with variable oxidation state three carbon linkers for the amino functionality. Trifluoroacetyl-protected 5-(3-aminopropynyl)-2′-deoxyuridine (1) was synthesised according to the method of Hobbs (25; Fig. 1). The order of addition of reagents in this palladium-catalysed reaction was found to be critical (see Materials and Methods). Changing the order of addition of reagents resulted in formation of unidentified fluorescent nucleosidic by-products and concomitant decomposition of the catalyst. It is possible that this fluorescent product is the result of a cyclisation reaction mediated by Cu(I), analogous to that described for 5-ethynyl-2′-deoxyuridine (31,32). The corresponding protected alkenes of 5-(E)-(3-aminopropenyl)-2′-deoxyuridine (2) and 5-(Z)-(3-aminopropenyl)-2′-deoxyuridine (3) were prepared by palladium-catalysed coupling of N-propenyltrifluoroacetamide to 5-iodouridine-2′-deoxyuridine (23) or reduction of 1 using NiCl2/NaBH4 (27), respectively. The corresponding protected alkane of 5-(3-aminopropyl)-2′-deoxyuridine (4) was previously synthesised from 1 via reduction with NiCl2/NaBH4 (27). However, we have now found that this compound may be prepared more efficiently by catalytic hydrogenation of 1 using platinum oxide.
Figure 1.
The synthesis of protected C5-amino-modified 2′-deoxyuridines from 5-iodo-2′-deoxyuridine. (i) N-propynyltrifluoroacetamide, Pd(PPh3)4, CuI, Et3N, DMF, 60% yield; (ii) N-propenyltrifluoracetamide, Na2PdCl4, aq NaOAc, pH 5.2, 30–50% yield (23); (iii) NiCl2/NaBH4, MeOH, –78°C, 55% yield (for 1–3) (27); (iv) H2, PtO2 in MeOH, 1 h at room temperature, 88% yield.
Nucleosides 1–4 were converted to the corresponding triphosphates using the method of Kovacz and Ötvös (28). Addition of proton sponge suppresses addition of HCl across the alkyne and alkene functionalities but also accelerates the rate of phosphorylation and was therefore included in the reaction with nucleoside 4. Triphosphates were first deprotected with ammonia and then purified by anion exchange chromatography, followed by reversed phase HPLC. Yields of purified triphosphates were typically 25–50% (best with alkyne derivative 1) and all compounds were characterised by NMR and mass spectrometry.
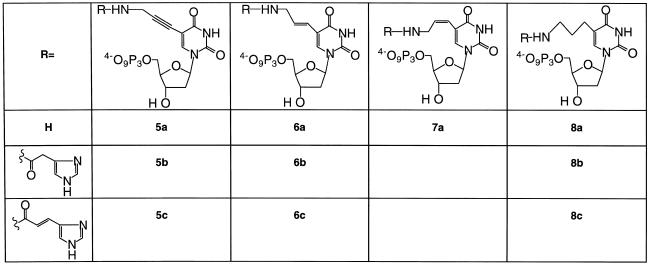
Imidazole conjugates of the C5-amino-modified 2′-deoxyuridine triphosphate derivatives (Fig. 2) were prepared by coupling of the N-hydroxysuccinimide (NHS) esters of either imidazole 4-acetic acid or urocanic acid with the modified triphosphates 5a–8a under basic conditions. The reaction rates of the modified 2′-deoxyuridine triphosphates differed substantially. Thus, whilst reactions of the NHS esters with 5-(3-aminopropynyl)-2′-deoxyuridine 5′-triphosphate (5a) proceeded with 3 equiv of the active ester, reactions of the triphosphates of the trans-alkene (6a) and the alkane (8a) required a much greater excess of the active ester. When large amounts of the active esters were employed it was necessary to remove excess ester and its by-products either by preparative silica TLC or anion exchange chromatography prior to reversed phase HPLC. We have found the former method to be preferable, being simpler and more efficient. Surprisingly, all attempts to make derivatives of the cis-alkene triphosphate (7a) failed. It is possible that this lack of reactivity reflects an inaccessible conformation of 5-(Z)-(3-aminopropenyl)-2′-deoxyuridine 5′-triphosphate (7a) and further studies are currently in progress to investigate this.
Figure 2.
The C5-modified triphosphates employed in this study. The triphosphates were prepared according to the procedures described in Materials and Methods.
PCR reactions
In order to be of use for in vitro selection procedures the modified deoxynucleoside triphosphates are required to be substrates in PCR reactions. Thus, the triphosphates must be good substrates for thermostable DNA polymerases and the modification must not interfere with the fidelity of Watson–Crick base pairing, so that multiple rounds of PCR can take place. Furthermore, in order that the sequence of the final selected DNA can be determined using conventional techniques and to allow faithful copying of the template DNA, the modified triphosphate must completely replace its unmodified counterpart in PCR reactions.
In vitro selection experiments typically involve random DNA libraries of 25–200 nt, which are amplified in successive rounds by PCR. Consequently, we have studied the incorporation of the modified triphosphates during PCR reactions mediated by Taq polymerase using template DNAs varying from 62 to 224 nt. Whilst other thermostable DNA polymerases (Tth from Thermus thermophilus and Vent from Thermococcus litoralis) were investigated, initial studies indicated that the best results for amplification, using both the modified and natural nucleoside triphosphates, were produced using Taq enzyme. In the current study we have therefore concentrated on investigating the substrate properties of the analogues with the cheaper and more widely available Taq enzyme.
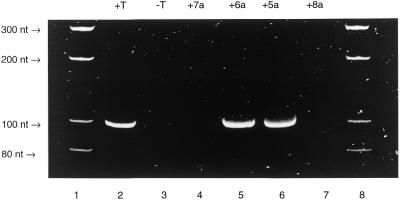
The results of the substrate studies with the modified triphosphates during PCR are summarised in Table 1. Of the four different amine functionalised triphosphates (5a–8a) only the alkyne analogue (5a) and the trans-alkene analogue (6a) were found to be substrates. Both of the analogues 5a and 5b have been prepared previously and were reported not to be substrates for Taq polymerase during PCR (22,23). However, analogue 5a was found to be a substrate for Vent polymerase (22), whilst analogue 6a, prepared previously by Sakthivel and Barbas (23), was found to be incorporated by the thermostable polymerase rTh from T.thermophilus. In contrast to these results, we have found that both 5a and 6a are excellent substrates for Taq polymerase, even with longer DNA templates (224 nt). Sakthivel and Barbas (23) incorporated 6a into a 519 nt PCR fragment using rTh, but were unable to incorporate this analogue using AmpliTaq (PE Biosystems). However, we have found that 6a can be incorporated using Taq polymerase purchased from Promega. The substrate properties of both 5a and 6a with Taq polymerase are comparable with shorter templates (≤98 nt in this study) and PCR reactions using these analogues are comparable in efficiency to that using the four natural triphosphates (Fig. 3). However, the amount of full-length PCR product obtained with the 224 nt template and the analogues (data not shown) was reduced compared to the reaction using the natural dNTPs, with 6a giving a slightly better yield than 5a.
Table 1. Summary of substrate properties of the nucleotide analogues.
| Triphosphate | Template | ||
| |
pUC19 (62 nt) |
pUC19 (98 nt) |
pLitmus28 (224 nt) |
| Amino analogues | |||
| 5a (alkyne) | +++ | +++ | + |
| 6a (trans) | +++ | +++ | ++ |
| 7a (cis) | – | – | – |
| 8a (alkane) | – | – | – |
| Imidazole acetic acid conjugates | |||
| 5b (alkyne) | ++ | ++ | – |
| 6b (trans) | ++ | ++ | – |
| 8b (alkane) | – | – | – |
| Urocanic acid conjugates | |||
| 5c (alkyne) | + | + | – |
| 6c (trans) | ++ | ++ | + |
| 8c (alkane) | – | – | – |
In each case the indicated nucleotide was used as a replacement for dTTP during PCR. Results are shown for the synthesis of PCR proucts of lengths 62–224 nt. Details of conditions are described in Materials and Methods. The absence of PCR product is indicated by –. + indicates that full-length PCR product was obtained (the number of + signs is proportional to the efficiency/yield of PCR product).
Figure 3.
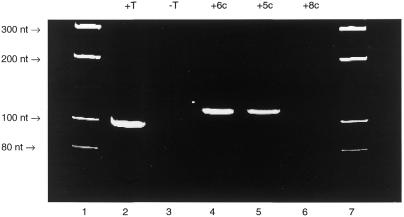
PAGE gel image of the ethidium bromide stained and UV visualised 98 nt PCR fragment derived from pUC19 and the C5-amino-modified triphosphates 5a–8a. The full experimental conditions are given in Materials and Methods. Lane 1, molecular weight markers; lane 2, a PCR reaction containing all four natural triphosphates, dATP, dCTP, dGTP and dTTP; lane 3, a PCR reaction containing dATP, dCTP and dGTP, which does not result in formation of any product; lane 4, a PCR reaction containing dATP, dCTP, dGTP and 7a; lane 5, a PCR reaction containing dATP, dCTP, dGTP and 6a; lane 6, a PCR reaction containing dATP, dCTP, dGTP and 5a; lane 7, a PCR reaction containing dATP, dCTP, dGTP and 8a; lane 8, molecular weight markers. Triphosphates 5a and 6a (Fig. 2) are substrates for Taq polymerase during the PCR reaction but 7a and 8a are not.
It can be concluded that linker rigidity confers favourable substrate properties on the C5-amino-modified triphosphates during PCR with Taq polymerase. Thus, the alkyne (5a) and the trans-alkene (6a) were found to be substrates for the polymerase, but, interestingly, the cis-alkene compound (7a) and the alkane (8a) were not. It is likely that at the pH of the PCR reactions the amino groups of the modified triphosphates are protonated. The cis compound may display this positive charge in such a way that the polymerase cannot bind to the modified triphosphate or is inhibited in catalysing incorporation of this analogue. It is noteworthy that this analogue places the amino functionality closest to the nucleobase. An alternative explanation for the lack of substrate properties of the cis compound during PCR reactions is an inability of DNA containing it to act as a template for further amplification. Further experiments are in progress to clarify this point. Clearly, a greater degree of conformational flexibility is unfavourable, since the alkane triphosphate (8a) is not a substrate in PCR reactions, and in designing C5-modified dUTPs, alkyl linkers should be avoided. The inability of 8a to act as a substrate may have similar origins to the cis compound (7a).
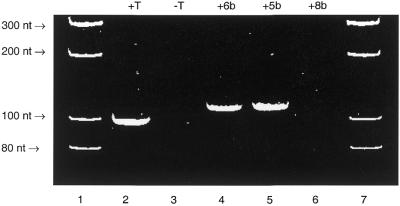
The imidazole triphosphate conjugates (5b, 5c, 6b and 6c) can replace dTTP in PCR reactions using the 98 nt template (Figs 4 and 5). Of the imidazole 4-acetic acid derivatives the alkyne 5b is a comparable substrate to the trans-alkene 6b. However, of the urocanic acid conjugates the alkene 6c is a slightly better substrate, but, furthermore, it is the only conjugated triphosphate which allows synthesis of the longer PCR product (224 nt) (data not shown). Neither 8b nor 8c were found to be substrates for Taq polymerase, analogous to the parent amino alkane triphosphate (8a).
Figure 4.
PAGE gel image of the ethidium bromide stained and UV visualised 98 nt PCR fragment using pUC19 as template and the C5-imidazole-modified triphosphates 5b–8b, derived from imidazole 4-acetic acid. The full experimental conditions are given in Materials and Methods. Lane 1, molecular weight markers; lane 2, a PCR reaction containing all four natural triphosphates, dATP, dCTP, dGTP and dTTP; lane 3, a PCR reaction containing dATP, dCTP and dGTP, which does not result in formation of any product; lane 4, a PCR reaction containing dATP, dCTP, dGTP and 6b; lane 5, a PCR reaction containing dATP, dCTP, dGTP and 5b; lane 6, a PCR reaction containing dATP, dCTP, dGTP and 8b; lane 7, molecular weight markers. Triphosphates 5b and 6b (Fig. 2) are substrates for Taq polymerase during the PCR reaction but 8b is not.
Figure 5.
PAGE gel image of the ethidium bromide stained and UV visualised 98 nt PCR fragment using pUC19 as template and the C5-imidazole-modified triphosphates 5c–8c, derived from urocanic acid. The full experimental conditions are given in Materials and Methods. Lane 1, molecular weight markers; lane 2, a PCR reaction containing all four natural triphosphates, dATP, dCTP, dGTP and dTTP; lane 3, a PCR reaction containing dATP, dCTP and dGTP, which does not result in formation of any product; lane 4, a PCR reaction containing dATP, dCTP, dGTP and 6c; lane 5, a PCR reaction containing dATP, dCTP, dGTP and 5c; lane 6, a PCR reaction containing dATP, dCTP, dGTP and 8c; lane 7, molecular weight markers. Triphosphates 5c and 6c (Fig. 2) are substrates for Taq polymerase during the PCR reaction but 8c is not.
One plausible explanation for the inability of the nucleoside triphosphates of the cis-alkene (7a) and those derived from the alkane (8a–8c) to act as substrates might be derived from the chemical lability of these analogues during PCR. If this were the case, since no PCR products were detected, it is likely that substantial decomposition of the respective triphosphates would occur during the first cycle of PCR. The absence of any PCR product would suggest that such a process would be expected to be complete after the first few cycles of PCR. Thus, solutions of these nucleotides in PCR buffer were analysed by reversed phase HPLC both prior to and after one cycle of PCR. In each case decomposition was minimal (data not shown). Furthermore, we found no evidence that these nucleoside triphosphates were inhibitory to the normal PCR reaction when they were added in equimolar concentration to the four natural nucleotides. These results suggest that the analogues themselves are not substrates during PCR.
From the results obtained here no distinct pattern emerges with the conjugated triphosphates containing trans-alkene or alkyne linkers. Furthermore, the type of pendant group introduced during elaboration of the aminoalkyl side chains does not change the substrate properties in a predictable manner and so we have observed both imidazole acetic acid and urocanic acid conjugates to be substrates. In subsequent work it is likely to be difficult to predict the substrate properties of triphosphates that result from further elaboration of the C5-amino side chain. Thus, preparing both the alkyne and trans-alkene conjugates of any desired functionality would be of value since the substrate properties of these analogues are generally comparable.
Characterisation of modified PCR products
To provide the first direct evidence of incorporation of the modified triphosphates during PCR we attempted to record MALDI-TOF mass spectra of the PCR products. Long PCR products are challenging substrates for mass spectrometry, with difficulties being encountered with desalting prior to analysis and depurination due to the acidic matrix during ionisation (33). In addition, Taq polymerase often adds an additional 2′-deoxyadenylate residue to the 3′-end of PCR products (29), a further complicating factor during mass spectrometry. We chose the shorter 62 nt PCR product as suitable for MALDI analysis. Amplification of the 62 nt template using a biotinylated reverse primer allowed separation of the two strands of the PCR products by capture of the biotinylated strand on magnetic streptavidin beads. After capture, the non-biotinylated PCR product was eluted with sodium hydroxide. Material from 10 PCR reactions was pooled and was sufficient for analysis by MALDI-TOF mass spectrometry.
Mass spectra (Supplementary Material) were obtained for the PCR products resulting from using the four natural dNTPs and those in which dTTP was replaced by modified triphosphates 5a and 5b, respectively. Molecular ions determined for the respective DNA strands were all between 0.2 and 1.5% mass units of the calculated masses (see Materials and Methods). Since the calculated mass differences between the PCR products containing analogues 5a and 5b and that containing the natural nucleotides are 0.6 and 2.4 kDa, respectively, the results demonstrate unambiguously that analogues 5a and 5b have been incorporated into the PCR product.
As has been observed previously (29), mass analysis of the PCR products reveals the addition of an extra dA nucleotide to some of the PCR products. In the case of the natural triphosphates this addition appears to be complete and good resolution of the molecular ion is observed with a mass that is very close to that predicted for the 62 nt product plus an additional 3′-dA. The resolution observed for the PCR products using the modified triphosphates is poorer. It appears likely that when using either 5a or 5b a mixture of products of either 62 or 63 nt is obtained, resulting in a more heterogeneous mixture of molecular ions.
Further evidence for incorporation of the modified triphosphates during PCR analogues, albeit less directly, was obtained by subjecting the respective PCR products to digestion by XbaI (recognition sequence TCTAGA). For the 98 nt PCR products that were obtained using 5a, 5b, 5c, 6a, 6b or 6c in place of dT, no digestion by XbaI was observed following incubation for 3.5 h. In contrast, the unmodified sequence was completely digested under identical conditions within 1 h (data not shown). It is also noteworthy that for the PCR products containing any of the imidazole triphosphate conjugates 5b, 5c, 6b or 6c a slower gel mobility was observed, again suggesting incorporation of the modifications.
CONCLUSIONS
It is clear from this study that incorporation of modifications into DNA during PCR by the use of C5-modified dUTPs can best be achieved, at least when Taq polymerase is used, via C5 linkers comprised of rigid alkynyl and trans-alkenyl moieties. The analogues that we have described allow DNA to be equipped with potential catalytic groups such as primary amino and imidazole functions and thus will be useful for in vitro selection experiments designed to select novel nucleic acid-based catalysts. In our experience chemical syntheses of both C5-alkyne-modified 2′-deoxyuridine and the triphosphate thereof were more efficient and therefore this is the modification of choice if shorter PCR products (≤100 nt) are required. However, for PCR amplification of longer templates (>200 nt) modified dUTPs containing trans-alkenyl linkers are preferable. In contrast, dUTP analogues with C5-alkane or cis-alkenyl linkers should be avoided.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Brian Taylor and Ms Sue Bradshaw for obtaining NMR spectra, and we thank Ms Elaine Frary for her expert technical assistance. We also thank Dr Jon Cummins and Dr Mai Yee Chan (Amersham Pharmacia Biotech) for useful discussions. This work was supported by the Leverhulme Trust, grant no. F/118/AQ (N.M., T.G. and A.S.), the BBSRC, grant no. 50B09442 (S.E.L.), the Wellcome Trust, grant no. 053924 (A.S.), and the EPSRC, grant no. GR/L85749. J.A.G. is a BBSRC Advanced Research Fellow.
References
- 1.Breaker R.R. and Joyce,G.F. (1995) A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem. Biol., 2, 655–660. [DOI] [PubMed] [Google Scholar]
- 2.Cuenoud B. and Szostak,J.W. (1995) A DNA metalloenzyme with DNA-ligase activity. Nature, 375, 611–614. [DOI] [PubMed] [Google Scholar]
- 3.Lorsch J.R. and Szostak,J.W. (1994) In-vitro evolution of new ribozymes with polynucleotide kinase activity. Nature, 371, 31–36. [DOI] [PubMed] [Google Scholar]
- 4.Santoro S.W., Joyce,G.F., Sakthivel,K., Gramatikova,S. and Barbas,C.F. (2000) RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc., 122, 2433–2439. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B.L. and Cech,T.R. (1997) Peptide bond formation by in vitro selected ribozymes. Nature, 390, 96–100. [DOI] [PubMed] [Google Scholar]
- 6.Wiegand T.W., Janssen,R.C. and Eaton,B.E. (1997) Selection of RNA amide synthases. Chem. Biol., 4, 675–683. [DOI] [PubMed] [Google Scholar]
- 7.Lohse P.A. and Szostak,J.W. (1996) Ribozyme-catalysed amino-acid transfer reactions. Nature, 381, 442–444. [DOI] [PubMed] [Google Scholar]
- 8.Tarasow T.M., Tarasow,S.L. and Eaton,B.E. (1997) RNA-catalysed carbon–carbon bond formation. Nature, 389, 54–57. [DOI] [PubMed] [Google Scholar]
- 9.Legault P. and Pardi,A. (1997) Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc., 119, 6621–6628. [Google Scholar]
- 10.Nakano S., Chadalavada,D.M. and Bevilacqua,P.C. (2000) General acid-base catalysis in the mechanism of a hepatitis δ virus ribozyme. Science, 287, 1493–1497. [DOI] [PubMed] [Google Scholar]
- 11.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 12.Aurup H., Williams,D.M. and Eckstein,F. (1992) 2′-Fluoro-2′-deoxynucleoside and 2′-amino-2′-deoxynucleoside 5′-triphosphates as substrates for T7 RNA polymerase. Biochemistry, 31, 9636–9641. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y., Qui,Q., Gill,S.C. and Jayasena,D. (1994) Modified RNA sequence pools for in vitro selection. Nucleic Acids Res., 22, 5229–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagratis N.C., Bell,C., Chang,Y.F., Jennings,S., Fitzwater,T., Jellinek,D. and Dang,C. (1997) Potent 2′-amino- and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol., 15, 68–73. [DOI] [PubMed] [Google Scholar]
- 15.Dewey T.M., Mundt,A.A., Crouch,G.J., Zyzniewski,M.C. and Eaton,B.E. (1995) New uridine derivatives for systematic evolution of RNA ligands by exponential enrichment. J. Am. Chem. Soc., 117, 8474–8475. [Google Scholar]
- 16.Dewey T.M., Zyzniewski,M.C. and Eaton,B.E. (1996) RNA world: functional diversity in a nucleoside by carboxyamidation of uridine. Nucl. Nucl., 15, 1611–1617. [Google Scholar]
- 17.Tu C., Keane,C. and Eaton,B. (1997) Carboxyamidation of purine nucleosides: new secondary 8-carboxamidopurine nucleosides. Nucl. Nucl., 16, 227–237. [Google Scholar]
- 18.Tarasow T.M., Tarasow,S.L., Tu,C., Kellogg,E. and Eaton,B.E. (1999) Characteristics of an RNA Diels–Alder active site. J. Am. Chem. Soc., 121, 3614–3617. [Google Scholar]
- 19.Vaish N.K., Fraley,A.W., Szostak,J.W. and McLaughlin,L.W. (2000) Expanding the structural and functional diversity of RNA: analog uridine triphosphates as candidates for in vitro selection of nucleic acids. Nucleic Acids Res., 18, 3316–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matulic-Adamic J., Daniher,A.T., Karpeisky,A., Haeberli,P., Sweedler,D. and Beigelman,L. (2000) Nucleoside triphosphates for in vitro selection of new catalytic RNAs. Bioorg. Med. Chem. Lett., 10, 1299–1302. [DOI] [PubMed] [Google Scholar]
- 21.Latham J.A., Johnson,R. and Toole,J.J. (1994) The application of a modified nucleotide in aptamer selection—novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine. Nucleic Acids Res., 22, 2817–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battersby T.R., Ang,D.N., Burgstaller,P., Jurczyk,S.C., Bowser,M.T., Buchanan,D.D., Kennedy,R.T. and Benner,S.A. (1999) Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J. Am. Chem. Soc., 121, 9781–9789. [DOI] [PubMed] [Google Scholar]
- 23.Sakthivel K. and Barbas,C.F. (1998) Expanding the potential of DNA for binding and catalysis: highly functionalized dUTP derivatives that are substrates for thermostable DNA polymerases. Angew. Chem. Int. Ed. Engl., 37, 2872–2875. [DOI] [PubMed] [Google Scholar]
- 24.Perrin D.M., Garestier,T. and Hélène,C. (1999) Expanding the catalytic repertoire of nucleic acid catalysts: simultaneous incorporation of two modified deoxyribonucleoside triphosphates bearing ammonium and imidazolyl functionalities. Nucl. Nucl., 18, 377–391. [DOI] [PubMed] [Google Scholar]
- 25.Hobbs F. (1989) Palladium catalysed synthesis of alkynylamino nucleosides. A universal linker for nucleic acids. J. Org. Chem., 54, 3420–3422. [Google Scholar]
- 26.Curphey T. (1979) Trifluoroacetylation of amino acids and peptides by trifluoroacetate. J. Org. Chem., 44, 2805–2807. [Google Scholar]
- 27.Lee S.E., Vyle,J.S., Williams,D.M. and Grasby,J.A. (2000) Novel syntheses of (Z)-alkene and alkane base-modified nucleosides. Tetrahedron Lett., 41, 267–270. [Google Scholar]
- 28.Kovacz T. and Ötvös,L. (1988) Simple synthesis of 5-vinyl- and 5-ethynyl-2′-deoxyuridine-5′-triphosphates. Tetrahedron Lett., 29, 4525–4528. [Google Scholar]
- 29.Clark J.M. (1988) Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res., 16, 9677–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prober J.M., Trainor,R.J., Dam,R.J., Hobbs,F.W., Robertson,C.W., Zagursky,R.J., Cocuzza,A.J., Jensen,M.A. and Baumeister,K. (1987) A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science, 238, 336–341. [DOI] [PubMed] [Google Scholar]
- 31.Woo J., Meyer,R.B. and Gamper,H.B. (1996) G/C-modified oligodeoxynucleotides with selective complementarity: synthesis and hybridization properties. Nucleic Acids Res., 24, 2470–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robins M.J. and Barr,P.J. (1983) Nucleic acid related compounds. 39. Efficient conversion of 5-iodo to 5-alkynyl and derived 5-substituted uracil bases and nucleosides. J. Org. Chem., 48, 1854–1862. [Google Scholar]
- 33.Siegert C.W., Jacob,A. and Köster,H. (1996) Matrix-assisted laser desorption/ionization time of flight mass spectrometry for the detection of polymerase chain reaction products containing 7-deazapurine moieties. Anal. Biochem., 243, 55–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.