Abstract
In Drosophila embryos the protein Naked cuticle (Nkd) limits the effects of the Wnt signal Wingless (Wg) during early segmentation. nkd loss of function results in segment polarity defects and embryonic death, but how nkd affects Wnt signaling is unknown. Using ectopic expression, we find that Nkd affects, in a cell-autonomous manner, a transduction step between the Wnt signaling components Dishevelled (Dsh) and Zeste-white 3 kinase (Zw3). Zw3 is essential for repressing Wg target-gene transcription in the absence of a Wg signal, and the role of Wg is to relieve this inhibition. Our double-mutant analysis shows that, in contrast to Zw3, Nkd acts when the Wg pathway is active to restrain signal transduction. Yeast two hybrid and in vitro experiments indicate that Nkd directly binds to the basic-PDZ region of Dsh. Specially timed Nkd overexpression is capable of abolishing Dsh function in a distinct signaling pathway that controls planar-cell polarity. Our results suggest that Nkd acts directly through Dsh to limit Wg activity and thus determines how efficiently Wnt signals stabilize Armadillo (Arm)/β-catenin and activate downstream genes.
Keywords: Wnt/wingless, naked cuticle, dishevelled, zeste-white 3, segmentation, Drosophila
Secreted Wnt proteins act as potent mitogens and cell-fate regulators in organisms ranging from nematodes to humans. In vertebrates they specify cell fate and control growth in a variety of developmental processes, including brain development, limb formation, axis specification, and gastrulation (for review, see Cadigan and Nusse 1997). In the fruit fly Drosophila, the Wnt protein Wingless (Wg) establishes segment polarity during embryogenesis and is involved in multiple additional patterning events throughout later development (Cadigan and Nusse 1997). wg is first expressed in the developing epidermis in stripes just anterior to cells expressing the engrailed (en) gene and is necessary to maintain en transcription (DiNardo et al. 1988; Martinez Arias et al. 1988). hedgehog (hh) is expressed in the en-expressing cells and positively regulates wg expression in the anterior cells (Ingham et al. 1991; Lee et al. 1992). This positive-feedback loop establishes parasegmental boundaries, the first evidence of the metameric organization of the embryo, between wg- and en/hh-expressing cells. At later stages of embryonic development, a tight balance between Wg and other signaling pathways, such as the Drosophila epidermal growth factor receptor (EGFR), determines whether epidermal cells secrete either naked (smooth) cuticle or hair-like structures called denticles (Dougan and DiNardo 1992; O'Keefe et al. 1997; Szuts et al. 1997). In the absence of wg function, embryos are covered with a lawn of denticles, whereas otherwise wild-type embryos exposed to excess Wg produce a naked cuticle (Martinez Arias et al. 1988; Noordermeer et al. 1992).
Genetic and biochemical studies have lead to the identification of the key components of the Wnt/Wg pathway and have uncovered some of the molecular events that are involved in signal transduction (Fig. 1A). Wg binds 7-pass transmembrane receptors of the frizzled family (Fz or Dfz2), which, in turn, activate the cytoplasmic protein Dishevelled (Dsh; Klingensmith et al. 1994; Theisen et al. 1994; Bhanot et al. 1996). Dsh antagonizes the activity of a large protein complex that, in the absence of Wg signal, results in Armadillo (Arm)/β-catenin phosphorylation and subsequent degradation by the ubiquitin-proteasome pathway (Yost et al. 1996; Aberle et al. 1997; Pai et al. 1997). This multiprotein complex includes Zw3/Glycogen synthase kinase 3β (Gsk3β), Adenomatous Polyposis Coli (APC), Axin, and Arm/β-catenin. Axin constitutes the core of this complex, allowing Zw3/Gsk3β to phosphorylate Arm/β-catenin (Ikeda et al. 1998). In the presence of Wg signal, Zw3 activity is reduced, resulting in stabilized Arm protein, which associates with the dTCF/Pangolin transcription factor to activate context-specific Wg target genes (Brunner et al. 1997; van de Wetering et al. 1997). In addition to transducing Wg signal, some of these components are involved in an Arm-independent pathway that regulates planar cell polarity (Fig. 1A; for review, see Shulman et al. 1998). During morphogenesis, this pathway orients cells in an axis orthogonal to their apical-basal axis by influencing the cytoskeleton (Shulman et al. 1998). It is required for the proper orientation of hairs and bristles on the thorax, abdomen, wing, and leg, as well as for the correct polarity of ommatidia in the eye. Both Fz and Dsh are involved in this pathway, but components downstream from Dsh are distinct from the Wg pathway (Axelrod et al. 1998; Boutros et al. 1998).
Figure 1.
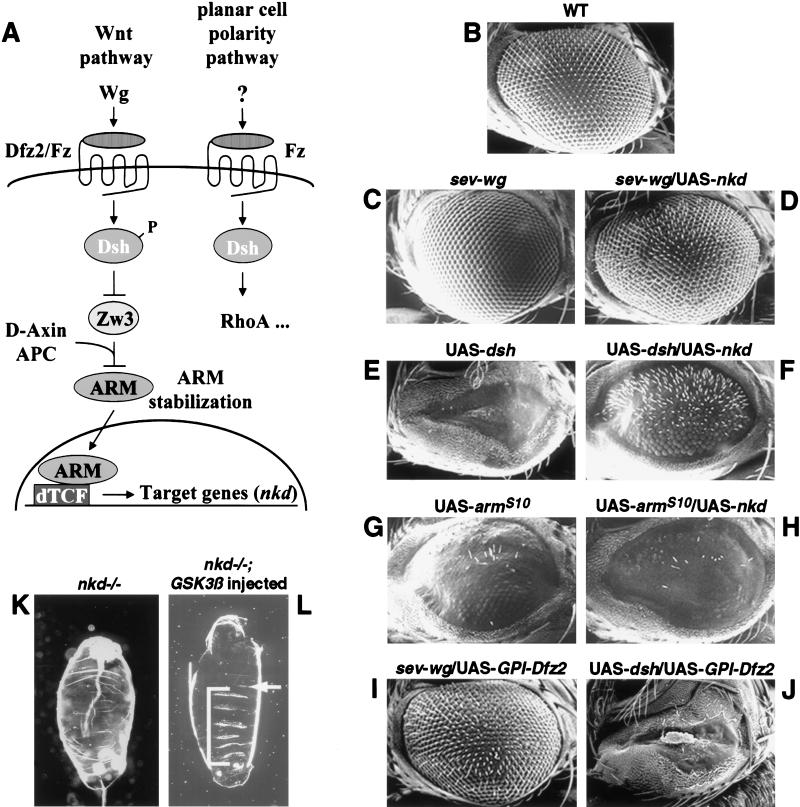
Epistasis study in the eye and embryo. (A) Schematic diagram of the Wg pathway and the planar cell polarity pathway. Arrows and bars show positive and negative actions, respectively. P represents the phosphorylated state of Dsh. See text for details. (B–J) Nkd can suppress Wg and Dsh misexpression eye phenotypes, but not the ArmS10misexpression eye phenotype. Ventral is to the left and anterior up. (B) The wild-type adult eye consists of an array of ommatidia and interommatidial bristles. (C,E) The sev-wg or UAS-dsh eyes lack bristles and/or ommatidia. (G) The UAS-armS10 eye has disrupted ommatidia and loss of bristles (average number of bristles/eye = 11.6; n = 8). (D,F) Co-misexpression of UAS-nkd dramatically suppressed the sev-wg (n = 8) and the UAS-dsh (n = 8) eye phenotypes. (H) Co-misexpression of UAS-nkd did not suppress the UAS-armS10 phenotype (average number of bristles/eye = 5.4; n = 8). UAS-GPI-Dfz2 suppresses the sev-wg loss of bristle phenotype (I) but does not suppress the UAS-dsh phenotype (J). (K,L) Injection of Xenopus GSK3β mRNA into nkd mutant embryos can restore denticles to the nkd embryos. Anterior is up. (K) nkd mutant embryos lack ventral denticle belts. (L) Injection of GSK3β into nkd7H16 embryos restored ventral denticles (brackets). nkd7H16 mutant embryos were identified by a Ubx mutation resulting in the transformation of the first abdominal segment A1 to the third thoracic segment T3 (arrow).
Given the Wnt/Wg pathway's key roles in cell growth and differentiation, it is not surprising to find that perturbations of its activity can lead to tumorigenesis. Integration of the mouse mammary tumor virus into the wnt-1 proto-oncogene locus promotes tumor formation in mice (Nusse and Varmus 1982). In addition, mutations in APC, β-catenin, and Axin that lead to aberrant Wnt activity have been associated with various types of human cancers (for review, see Polakis 2000). Inappropriate activation of Wnt target genes such as c-myc or cyclin D1 may be an important step toward tumor formation (Polakis 2000). It is therefore important to understand how Wnt signals are normally limited during development and cancer progression.
We recently reported the cloning of the naked cuticle (nkd) gene and showed that nkd antagonizes Wg signaling in Drosophila (Zeng et al. 2000). nkd Loss of function leads to embryonic lethality due to segmentation defects (Jürgens et al. 1984). The nkd phenotype is characterized by a loss of ventral denticle belts and resembles the phenotype of embryos with excess Wg signaling, such as embryos overexpressing Wg or those lacking the negative regulators zw3, d-axin, or dAPC2 (Perrimon and Smouse 1989; Noordermeer et al. 1992; Hamada et al. 1999; McCartney et al. 1999). We also showed that the nkd gene itself is regulated by Wg, creating a negativefeedback loop that restricts Wg activity during segmentation (Zeng et al. 2000). Nkd is a novel protein containing a 60–amino acid region that is related to the high affinity Ca2+-binding EF-hand of the recoverin family of myristoyl switch proteins. Here we address how Nkd limits Wg signaling by using a combination of genetic and biochemical approaches. Our results suggest that Nkd can antagonize Wg signaling cell-autonomously through a direct interaction with Dsh.
Results
nkd regulates interommatidial bristle formation
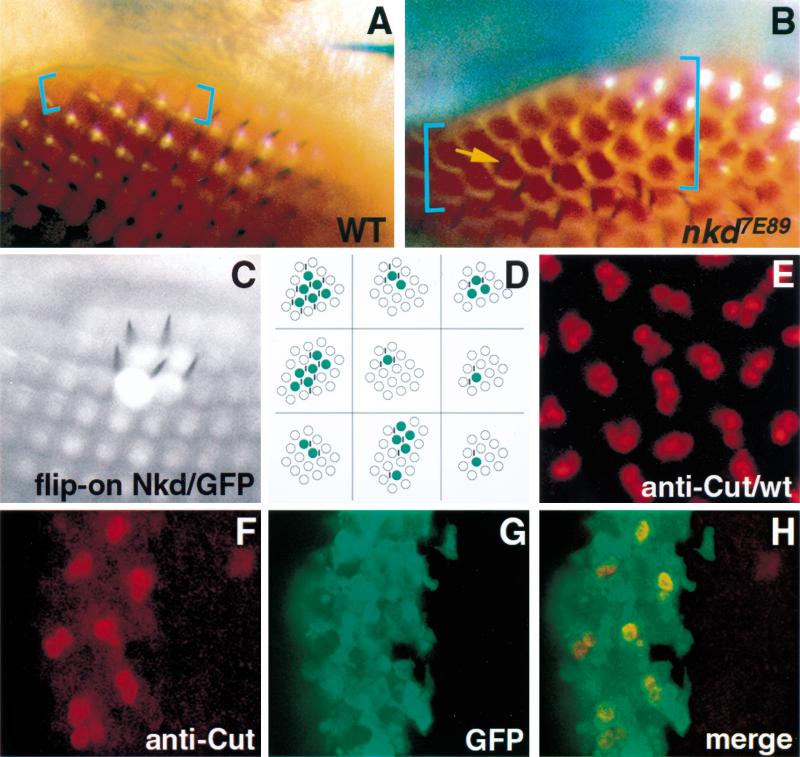
The Drosophila eye is composed of mechanosensory bristles present at vertices of ommatidia (Fig. 1B). Bristle formation is suppressed near the circumferential margin of the eye, and the degree of suppression is least at the extreme dorsum of the head, typically 0–2 ommatidial diameters (Fig. 2A). Previous work showed that Wg signaling, active at the circumference of the developing eye where wg is expressed, is responsible for this suppression of peripheral bristle formation (Cadigan and Nusse 1996; K.M. Cadigan et al., in prep.). To assay the function of nkd in eye bristle formation, we used the EGUF/hid method (Stowers and Schwarz 1999) to make homozygous mutant nkd eyes in nkd/+ animals. In this technique, Flp-mediated recombination between a chromosome mutant for nkd and a chromosome harboring both recessive and dominant cell-lethal mutations is specifically induced in the eye using the eyeless promoter. During eye development, the only cells surviving are those that have lost the cell-lethal chromosome through recombination, producing an eye homozygous mutant for nkd. Examination of eyes mutant for the strong allele nkd7E89 reveals, at the dorsum of the eye, consistent eye bristle suppression 3–5 ommatidial diameters away from the margin, with occasional closer bristles (Fig. 2B). This result suggests that endogenous nkd regulates interommatidial bristle suppression by antagonizing the effects of endogenous Wg in cells farther than one cell diameter away from the Wg source.
Figure 2.
nkd autonomously regulates interommatidial bristle formation. (A) Wild-type eye margin at dorsum of head shows bristle suppression 0–1 ommatidial rows (blue brackets) from eye margin. (B) nkd7E89 mutant eye shows suppression of bristles 3–5 ommatidial rows from eye margin. Occasional bristles are present closer to eye margin (yellow arrow). No bristle phenotype was seen with the weaker nkd9G33 allele, whereas h1 nkd7H16 eyes are small and rough (not shown), possibly due to h/nkd interactions (Zeng et al. 2000) and hence could not be scored for this phenotype. (C,D) Nkd/GFP misexpression clones in sev-wg adult eyes. (C) Bristles are restored only in the region of the clone marked by GFP. (D) Cartoon depiction of nine adult eye clones (green color); vertical black lines represent bristles. (E) Cut nuclear localization in bristle cells of a wild-type pupal eye disc. (F,G) Restoration of bristle cells in a sev-wg pupal eye, revealed by Cut localization (red color), is confined to the region of the Nkd/GFP misexpression clone (green color); the clone begins at the edge of the disc (left) and continues inward (right). (H) Merged image of F and G shows that GFP and Cut colocalize in bristle cell precursor nuclei present in the clone.
Nkd misexpression in the eye blocks Wg activity
To determine how Nkd impinges on the Wg pathway, we tested the ability of Nkd to block the action of the positive regulators Wg, Dsh, and Arm. To do so, we took advantage of a Drosophila eye misexpression system (Cadigan and Nusse 1996). Production of Wg in a subset of photoreceptor cells throughout the eye using a sevenless promoter transgene (P[sev-wg]) prevents formation of interommatidial bristles in a paracrine fashion; otherwise, the eye is normal (Fig. 1C; Cadigan and Nusse 1996). Previous Nkd misexpression experiments did not indicate whether Nkd blocks Wg synthesis, Wg distribution, or cellular responses to received Wg (Zeng et al. 2000). To distinguish between these possibilities, we used the GAL4/UAS binary expression system to evaluate the effect of Nkd (UAS-nkd) on Wg-mediated eye bristle suppression. Misexpression of Nkd alone using multiple repeats of the eye-specific glass (gl) enhancer (GMR) to drive the yeast transcription factor GAL4 (P[GMR-GAL4]) has no visible effect on eye development (data not shown). However, the combination of sev-wg with nkd misexpression results in nearly complete suppression of the P[sev-wg]-induced bristle-loss phenotype (Fig. 1D). Nkd misexpression did not alter the levels or distribution of Wg antigen (data not shown), indicating that Nkd is probably blocking signaling events downstream from Wg.
Nkd misexpression blocks Dsh activity but not Arm activity
The effect of Nkd on the downstream Wg pathway components Dsh and Arm was also tested using the GMR-GAL4 system. Dsh misexpression (UAS-dsh) produces small, bristle-less eyes devoid of ommatidia (Fig. 1E). Nkd strongly suppressed the Dsh misexpression eye phenotype, restoring numerous bristles and ommatidia (Fig. 1F). If the Dsh misexpression eye phenotype is Wg-dependent, its suppression by Nkd could be due to Nkd acting on Wg rather than on Dsh or other downstream components. Previous work suggests that the Dsh misexpression eye phenotype is Wg-independent (Cadigan and Nusse 1996). To confirm the Wg-independence of the Dsh phenotype, a dominant-negative form of Dfz2 (UAS-GPI-Dfz2; Cadigan et al. 1998) was coexpressed with either sev-wg or UAS-dsh. UAS-GPI-Dfz2 effectively suppressed sev-wg-induced bristle loss in the eye (Fig. 1I). Coexpression of UAS-GPI-Dfz2 and UAS-Dsh resulted in some eye necrosis, but it had negligible effects on the UAS-dsh eye phenotype (Fig. 1J). These results confirm that the Dsh misexpression effect in the eye is Wg-independent. Therefore, rescue of the UAS-dsh phenotype by Nkd is not an indirect effect due to suppression of Wg activity.
GMR-driven expression of UAS-armS10, a constitutively activated form of arm, also produces bristle loss and failure of proper ommatidial development (Fig. 1G; Ahmed et al. 1998). Nkd coexpression had no effect on the Arm misexpression phenotype (Fig. 1H). Dsh and Arm misexpression phenotypes were not affected by simultaneous expression of UAS-lacZ, indicating that suppression of the dominant eye phenotypes by Nkd was not due to GAL4 titration (data not shown). The ability of Nkd to block effects of Wg and Dsh but not Arm suggests that Nkd is acting at the level of, or downstream from, Dsh but not downstream of Arm.
Epistasis between nkd and zw3 in the embryo
The relationship between Nkd and Zw3 could not be determined by a similar suppression test because both proteins are negative regulators of Wg. In addition, the subtlety of the nkd phenotype in the eye made this tissue unsuitable for analyzing the epistasis between nkd and zw3. Instead, Zw3/Gsk3β was overproduced in nkd mutant embryos using genetic and mRNA injection methods: We used heat shock promoter (hsp70)-controlled GAL4 to drive Zw3 production, or we used injections with Xenopus gsk3β mRNA. Vertebrate gsk3β genes have sequences very similar to the fly gene and can rescue zw3 mutant embryos (Siegfried et al. 1992). nkd mutants lack ventral denticle belts and are considerably smaller than wild-type embryos (Fig. 1K). Overproduction of Gsk3β or Zw3 in nkd mutants results in partial to almost complete restoration of denticle belts and restoration of more normal embryo size (Fig. 1L; data not shown). Because Zw3 restores denticles to nkd mutants, Zw3 cannot act genetically upstream of the defect in nkd mutants (i.e., by stimulating nkd function) in the linear Wg pathway. Nkd therefore is likely to act upstream of, or in a pathway parallel to, Zw3 and downstream from, or at the level of, Dsh.
Nkd acts cell-autonomously
The eye misexpression results suggest that Nkd antagonizes Wg signaling at the level of, or downstream from, Dsh. Loss-of-function dsh clones revealed that Dsh acts autonomously in Wg-responsive cells (Klingensmith et al. 1994), suggesting that Nkd must also act in Wg-responsive cells. Indeed, previous observations in fly embryos suggest an initial requirement for nkd in cells receiving the Wg signal (Martinez Arias et al. 1988; Dougan and DiNardo 1992; Zeng et al. 2000). Because eye development allows fairly easy production of sharply bounded clones, we chose the eye to assess the cell autonomy of Nkd action. Marked clones of Nkd-misexpressing cells were produced in developing eyes and the range of Nkd action on sev-wg was monitored.
We used the flip-on GAL4 system to make random clones of cells misexpressing both Nkd and a cell-autonomous marker, green fluorescent protein (GFP), in eyes with excess Wg (sev-wg eyes). All clones examined (n = 15) showed suppression of bristle loss, with the suppression consistently within or immediately adjacent to GFP misexpression clones (Fig. 2C,D). No bristles were present outside the clones, indicating a local action of Nkd. To address whether Nkd was acting only in bristle precursor cells, and hence cell-autonomously, we specifically marked those cells with antisera against the Cut nuclear protein in pupal eye discs (Cadigan and Nusse 1996). In the vicinity of Nkd misexpression clones, there was a perfect correlation between GFP and Cut-labeled cells: All Cut-positive bristle precursor cells expressed GFP and hence Nkd; no GFP-negative/Cut-positive cells were found (Fig. 2F-H; n = 8 clones). These results suggest that Nkd was acting within Cut-positive bristle precursor cells to antagonize the inhibitory effects of Wg on bristle cell differentiation.
The dsh; nkd double-mutant resembles dsh mutant embryos
Cuticles derived from embryos lacking wg activity (wg, dsh, or arm) have nearly continuous fields of denticles, whereas HS-wg embryos, or those mutant for the negative regulator zw3, secrete naked cuticle (Martinez Arias et al. 1988; Perrimon and Smouse 1989; Noordermeer et al. 1992). Wg misexpression and double-mutant analyses showed that Wg acts sequentially through Dsh, Zw3, and Arm (Siegfried et al. 1992, 1994; Noordermeer et al. 1994; Peifer et al. 1994). Embryos doubly mutant for wg and zw3 (zw3; wg), as well as zw3 dsh embryos, resemble zw3 embryos, whereas zw3 arm embryos resemble arm embryos, indicating that zw3 acts downstream from dsh and upstream of arm (Siegfried et al. 1992, 1994; Peifer et al. 1994). Mutations in either nkd or zw3 give rise to a naked cuticle phenotype, with posterior expansion of en expression and ectopic wg expression in the developing embryo (Martinez Arias et al. 1988; Perrimon and Smouse 1989). However, in contrast to the naked cuticle phenotype of the zw3; wg embryo, the wg; nkd embryo has a wg-like phenotype (Bejsovec and Wieschaus 1993), indicating a dependence on Wg for the naked cuticle phenotype of nkd mutants (Dougan and DiNardo 1992).
To clarify the relationship between Nkd and other Wg pathway components in the embryo, we made embryos doubly mutant for nkd and dsh or arm, using both genetic means and RNA interference (RNAi). Whereas the nkd gene is strictly zygotic (Zeng et al. 2000), dsh has a maternal contribution that must be removed via germ-line clones to obtain the embryonic dsh phenotype (Perrimon and Mahowald 1987). Females heterozygous for nkd and carrying dsh germ-line clones were crossed to males heterozygous for nkd (see Materials and Methods). Embryos derived from crosses using different combinations of nkd and dsh alleles were counted and grouped according to their cuticle phenotypes (Table 1). The expected Mendelian ratios are 37.5% wild type (3/8), 37.5% dsh (3/8), 12.5% nkd (1/8), and 12.5% dsh; nkd (1/8). Only three phenotypes could be detected—wild type, dsh, and nkd—indicating that the dsh; nkd mutants exhibit one of these phenotypes or die before secreting cuticle. Whereas the observed percentages for the wild-type and nkd categories are very close to the expected percentages (38.2% and 14.3%, respectively), the percentage of dsh embryos is significantly higher (47.5%), suggesting that the dsh; nkd mutant resembles the dsh mutant.
Table 1.
The dsh; nkd double-mutant resembles dsh embryo
| ♀ × ♂
|
wt
|
dsh
|
nkd
|
No. of embryos
|
|---|---|---|---|---|
| dshv26;;nkd7H16× nkd7H16 | 38.7% (144/140) | 47.6% (177/140) | 13.7% (51/146) | 372 |
| dshv26;;nkd7H16× nkd7E89 | 40.2% (181/169) | 45.6% (205/169) | 14.2% (64/56) | 450 |
| dshv26;;nkd7E89× nkd7H16 | 32.2% (75/87) | 50.2% (117/87) | 17.6% (41/29) | 233 |
| dsh477;;nkd7H16× nkd7H16 | 39.2% (98/94) | 48.4% (121/94) | 12.4% (31/31) | 250 |
| Total | 38.2% (498/489) | 47.5% (620/489) | 14.3% (187/163) | 1305 |
Phenotypic distribution of embryos laid by dsh/ovoD1;;nkd/+ females (♀) mated to +/Y;;nkd/TM3 males (♂; see Materials and Methods for details). Four cuticular phenotypes were expected: wild-type (wt), dsh, nkd and unknown corresponding to the dsh; nkd double mutant, with ratios of 3:8 (37.5%), 3:8 (37.5%), 1:8 (12.5%) and 1:8 (12.5%), respectively. Two dsh alleles (dshv26, dsh477) and two nkd alleles (nkd7H16, nkd7E89), all strong alleles, were used in different combinations, as indicated in the first column. Results are presented as percentages followed between parentheses by the observed number of embryos (first number) and the expected number (second number). A total of 1305 embryos were counted and the total percentage for each phenotype is indicated in bold.
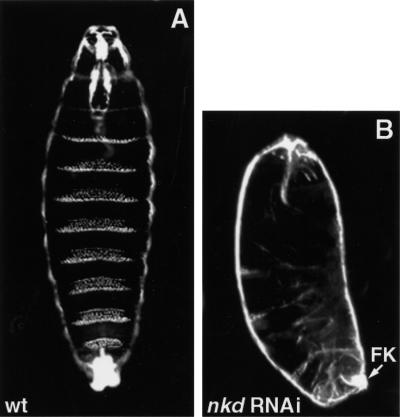
To confirm the dsh; nkd phenotype, we performed RNAi experiments. Injection of nkd double-stranded RNA (dsRNA) into wild-type embryos efficiently mimics nkd loss of function: 76% of the injected embryos (n = 221) develop with greatly reduced denticles compared to wild-type embryos (Fig. 3A,B). The majority of these mutant embryos (69%) show an intermediate to strong nkd cuticle phenotype, the others showing a weak expressivity characterized by a loss of only a few denticles (data not shown). We also tried RNAi with dsh, but, in contrast to nkd, both the penetrance and the expressivity of the dsh phenotype were very weak (data not shown). Increasing the dsh dsRNA concentration had little effect, producing only a fusion between belts A4 and A5 in <5% of the injected embryos and ruling out the utility of nkd and dsh double injections. Instead we injected nkd dsRNA into dsh embryos derived from germ-line clones. Half of the collected embryos are wild type due to rescue by the paternal X-chromosome (see Materials and Methods for the cross). To score only nonrescued dsh mutant embryos, we crossed females carrying germ-line clones to males carrying an X-chromosome GFP balancer and scored GFP-negative embryos after eliminating GFP embryos. Injection of nkd dsRNA into dsh embryos had no effect on the dsh phenotype (n = 66; data not shown), confirming that dsh; nkd double mutants resemble dsh embryos.
Figure 3.
Cuticle phenotypes of embryos injected with nkd dsRNA. (A) Wild-type (wt) embryo shortly before hatching. The ventral side is characterized by denticle belts separated with naked cuticle. (B) nkd RNAi mimics the nkd phenotype. Embryos are shorter and lack denticle belts, resembling nkd−/− embryos (see Fig. 1K). nkd RNAi embryos also show the characteristic defects in the Filzkörper (FK).
arm; nkd double-mutant embryos resemble arm embryos
We used the null allele armYD35 (Peifer and Wieschaus 1990) to generate arm; nkd double-mutant embryos. Embryos homozygous for this allele have a strong arm phenotype, even without making germ-line clones. Male heterozygotes for the strong alleles nkd7H16 or nkd7E89 were crossed to females heterozygous for armYD35 and nkd7H16 (see Materials and Methods). As these crosses generate a majority of wild-type embryos (a ratio of nine wild type to seven mutants), we counted only cuticles from unhatched embryos, which are expected to be mutant for arm (ratio 3:7, 42.9%), nkd (3:7, 42.9%), and arm; nkd (1:7, 14.3%). Like the dsh; nkd embryos, the arm; nkd mutants do not exhibit a distinct phenotype. The results (Table 2) show that the nkd phenotype is found at the expected frequency (42.6%), but the arm phenotype is over-represented (57.4% instead of 42.9%), indicating that this category also contains the arm; nkd embryos. Therefore, the arm; nkd mutant is covered with denticles and resembles arm embryos.
Table 2.
The arm; nkd double-mutant resembles arm embryos
| ♀ × ♂
|
arm
|
nkd
|
No. of unhatched embryos
|
|---|---|---|---|
| armYD35;;nkd7H16 × nkd7H16 | 58.0% (677/500) | 42.0% (490/500) | 1167 |
| armYD35;;nkd7H16× nkd7E89 | 56.6% (571/432) | 43.4% (437/432) | 1008 |
| Total | 57.4% (1248/932) | 42.6% (927/932) | 2175 |
Phenotypic distribution of unhatched embryos laid by armYD35/+;;nkd7H16/+ females (♀) crossed with +/Y;;nkd7H16/TM3 or +/Y;;nkd7E89/TM3 males (♂). Three phenotypes were expected among the unhatched embryos: arm, nkd and arm; nkd, the latter being unknown. The predicted ratios are: 3:7 (42.9%) for arm and nkd, and 1:7 (14.3%) for arm; nkd. Results are presented as in Table 1.
The double-mutant analysis indicates that the nkd phenotype occurs only if wg, dsh, and arm genes are active, confirming the requirement for Wg signaling to generate the nkd phenotype (Dougan and DiNardo 1992; Bejsovec and Wieschaus 1993). Zw3 constitutively represses Wg target-gene transcription, and the role of Wg is to overcome this inhibition (Siegfried et al. 1992). Our results indicate that Nkd, in contrast, is required to oppose Wg signal. Removal of nkd in the absence of wg, dsh, or arm has little effect on cuticle phenotype. Accordingly, increased levels of Nkd do not modify the wg mutant cuticle (Zeng et al. 2000). The negative influence of Nkd could be mediated by inhibition of Dsh activity, stimulation of Zw3 activity, or by interactions with unknown pathway components. To test whether Nkd can directly interact with known Wg signaling components, we used yeast two-hybrid and in vitro binding assays.
Nkd directly interacts with Dsh in yeast and in vitro
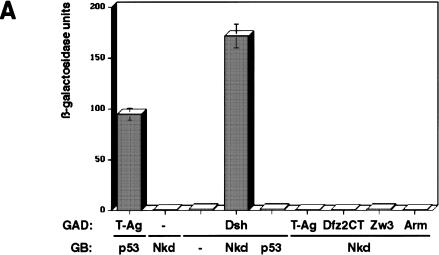
Expression in yeast of full-length Nkd protein fused to the GAL4 DNA-binding domain (GB-Nkd) did not activate transcription by itself (Fig. 4A). When GB-Nkd was coexpressed with Dsh fused to the activation domain of GAL4 (GAD-Dsh), strong β-galactosidase activity was detected, indicating an interaction between Nkd and Dsh (Fig. 4A). The reverse experiment, using GB-Dsh and GAD-Nkd, could not be performed because GB-Dsh activates transcription on its own (data not shown). No interaction between Nkd and Zw3 was detected, nor did Nkd interact with the C-terminal intracellular portion of Dfz2 (Dfz2CT), Arm, or the control protein large T-antigen (T-Ag; Fig. 4A). Nkd also did not interact with D-Axin (data not shown).
Figure 4.
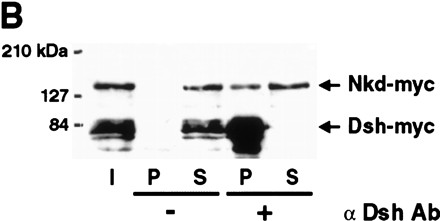
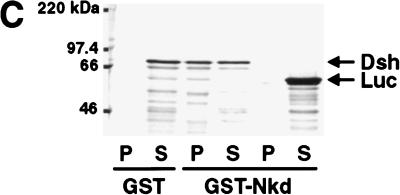
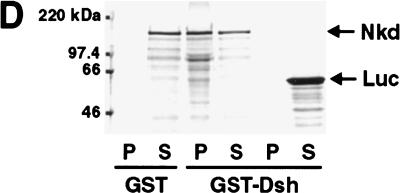
Nkd and Dsh directly interact in the yeast two-hybrid system, coimmunoprecipitation and GST pull-down assays. (A) Interaction between Nkd and Dsh in the yeast two-hybrid system using a liquid culture β-galactosidase assay. Yeast cells were cotransformed with plasmids expressing the GAL4 DNA-binding domain (GB, amino acids [aa] 1–147) either alone (−) or in fusion with Nkd, along with plasmids expressing the GAL4 activation domain (GAD, aa 768–881) alone (−) or in fusion with Dsh. The interaction between GB-Nkd and GAD-Zw3, GAD-Arm, or GAD-Dfz2CT (intracellular portion of Dfz2) was also tested. Murine p53 (aa 72–390) fused to GB and SV40 large T-antigen (T-Ag, aa 87–708) fused to GAD were used as positive controls. For each transformation, results corresponding to the mean of 2-independent yeast colonies are shown. The values are expressed in Miller units. The activation was confirmed using a second reporter gene (ADE2) present in the yeast strain and protein blots of yeast extracts showed that the different fusion proteins accumulated to similar levels (data not shown). (B) Coimmunoprecipitation of Nkd and Dsh in COS-7 cells. Extracts from COS-7 cells expressing Nkd-myc and Dsh-myc were subjected to coimmunoprecipitation with anti-Dsh antibody (α Dsh Ab) (+). As a control, the anti-Dsh antibody was omitted (−). The eluted protein from the beads (P) and one-tenth of the supernatant (S), as well as proteins corresponding to one-tenth of the input (I) were analyzed by Western blot using anti-c-Myc antibody. (C,D) Interaction between Nkd and Dsh in the GST pull-down assay. Bacterially expressed GST-Nkd protein was incubated with [35S]methionine-Dsh, produced and labeled by in vitro translation (C), whereas GST-Dsh was incubated with [35S]methionine-Nkd (D). As controls, GST-Nkd or GST-Dsh proteins were also tested for their interaction with [35S]methionine-Luciferase (Luc). The eluted proteins from the beads (P) and one-tenth of the supernatant (S) were separated by SDS-PAGE and viewed using a phosphorimager (Molecular Dynamics).
The interaction between Dsh and Nkd was confirmed using coimmunoprecipitation and glutathione-S-transferase (GST) pull-down assays. For coimmunoprecipitation tests, COS-7 cells were transfected with vectors expressing Myc-tagged Nkd and Dsh proteins. Immunoprecipitation with anti-Dsh, followed by protein blotting with anti-c-Myc antibody, showed that Nkd and Dsh can be coprecipitated (Fig. 4B). For GST pull-down assays, in vitro translated [35S]methionine Dsh bound to GST-Nkd fusion protein but not to GST or Luciferase protein (Fig. 4C). Similarly, in vitro translated [35S]methionine-labeled Nkd specifically bound GST-Dsh (Fig. 4D). All three protein association assays indicate that Nkd and Dsh can directly interact, in keeping with the epistasis results that suggested a role for Nkd at the level of Dsh or Zw3.
Production of Nkd–GFP fusion protein in larval salivary glands revealed striking colocalization with endogenous Dsh, stained with an anti-Dsh antibody (data not shown), indicating that the two proteins may also interact in vivo. However, we did not detect an association between the two proteins using coimmunoprecipitation experiments with embryo extracts or lysates from Drosophila cell lines (data not shown). The negative results may be due to protein complex dynamics, accessibility to antibodies, low levels of the complex in fly cells, as well as possible modes of regulation of the interaction, which we are currently investigating.
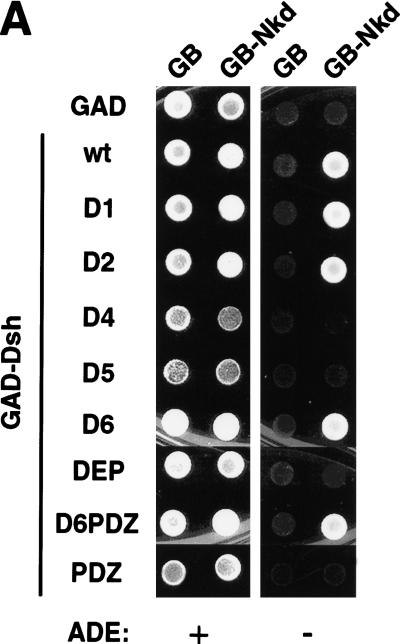
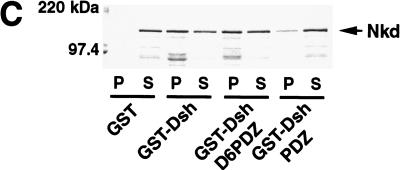
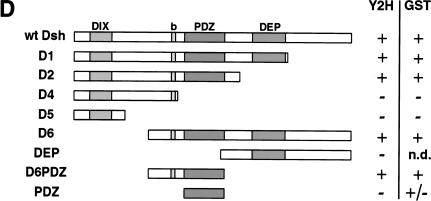
Nkd interacts with the basic/PDZ region of Dsh
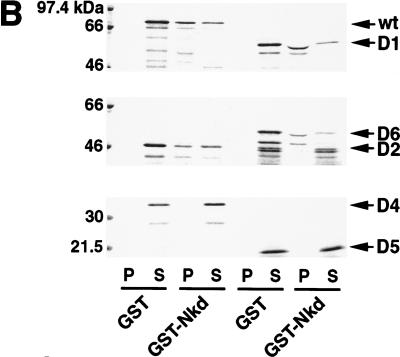
The Dsh protein contains three defined domains: DIX, PDZ, and DEP (Fig. 5D). The DIX (Dishevelled, Axin) domain shares homology with the C-terminal part of D-Axin, the PDZ (PSD-95, Dlg, Zo-1) domain is a modular region involved in protein–protein interactions, and the DEP (Dsh, Egl-10, Pleckstrin) domain is usually found in signaling proteins, although its role remains unclear. In addition, a stretch of basic residues is present between the DIX and PDZ domains. Using both the yeast two-hybrid system and GST pull-down experiments, the region in Dsh that binds Nkd was defined (Fig. 5). Our results indicated that the central region of Dsh containing the basic sequence and the PDZ domain was sufficient for binding Nkd (Fig. 5D). The PDZ domain of Dsh is necessary for the interaction with Nkd but, in contrast to proteins such as casein kinase I or Frat1 (Li et al. 1999; Peters et al. 1999), it cannot efficiently bind Nkd by itself (Fig. 5D).
Figure 5.
Nkd interacts with the basic/PDZ region of Dsh. (A) Interaction of different Dsh deletion mutants, fused to GAD, with GB or GB-Nkd in the yeast two-hybrid system. Yeast growth was evaluated using the ADE reporter gene present in the strain: Yeast colonies expressing the different combinations of fusion proteins, as indicated, were grown on medium containing (+) or lacking (−) adenine. (B,C) Using the GST pull-down assay, [35S]methionine-Dsh D1 to D6 were tested for their capacity to bind GST-Nkd (B), whereas in (C), GST-DshD6PDZ and GST-DshPDZ proteins were incubated with [35S]methionine-Nkd. The curved bands observed with D1 and D6 are due to comigration with a byproduct of GST-Nkd protein (data not shown). (D) Summary of domain mapping results from the yeast two-hybrid system (Y2H) and the GST pull-down assay (GST). (+) Positive interaction, (−) no interaction, (+/−) weak interaction, (n.d.) not determined. The different domains of Dsh, DIX, basic region (b), PDZ, and DEP, are indicated.
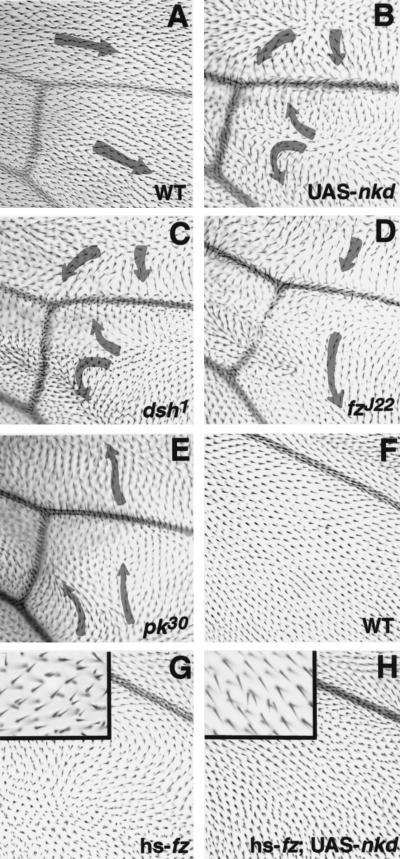
Nkd overexpression specifically affects Dsh function in planar cell polarity
Dsh is a branchpoint connecting two distinct signaling pathways in Drosophila development: the Wg pathway and the planar cell polarity pathway (PCP; Boutros and Mlodzik 1999). nkd mutant clones have normal planar cell polarity (Zeng et al. 2000); so there is no detectable normal role for nkd in the PCP pathway. If Nkd affects Dsh during Wg signaling, as our data suggest, then appropriately timed overexpression of Nkd might be able to specifically alter Dsh function in PCP signaling. Nkd was tested for its ability to interfere with PCP signaling during a time when Fz and Dsh are not appreciably participating in Wg signaling. Timed overexpression of Nkd at 24 h after puparium formation (APF) produces adult flies with wing hair polarity defects that are indistinguishable from those seen in dsh1 mutant adults (Fig. 6A–C). dsh1 is an adult viable allele of dsh that harbors a missense mutation in the C-terminal DEP domain (Axelrod et al. 1998; Boutros et al. 1998). Genetic tests have shown dsh1 to be a null allele for PCP signaling (Perrimon and Mahowald 1987). The Nkd overexpression polarity pattern is reproducible and qualitatively distinct from that produced by complete loss of function of other known PCP mutants, including fz or prickle (pk; Fig. 6D,E; Gubb and Garcia-Bellido 1982). The Nkd overexpression defect is also different from those associated with Fz or Dsh overexpression (data not shown).
Figure 6.
Effects of overproduced Nkd on planar cell polarity. (A) Wild-type (WT) wing pattern in region distal to posterior cross vein; same area shown in B–E. (B) Effect of overproduction of Nkd, which is similar to the phenotype shown in C, but different to the phenotypes shown in D and E. (C) Phenotype of loss of dsh function (dsh1 allele). (D) Phenotype of loss of frizzled function (fzJ22 allele). (E) Phenotype of loss of prickle function (pk30 allele). Arrows indicate hair orientation. (F) Wild-type wing pattern in the area shown in G and H: region posterior to vein 5. (G) Phenotype of heat shock promoter-driven frizzled expression. (H) Phenotype of heat shock promoter–driven fz expression in the presence of UAS-nkd. Almost complete suppression of polarity defects by Nkd overexpression is observed. High magnification shows that fz overexpression also induces double hair cells (G inset) and that their number decreases in the presence of Nkd (H inset).
The PCP phenotype associated with Fz overexpression is sensitive to the dose of dsh (Krasnow and Adler 1994). To determine whether Nkd could similarly titrate Dsh from PCP signaling induced by Fz overexpression, we simultaneously expressed Fz and Nkd. Indeed, overexpressed Nkd suppressed the effects of excess Fz (Fig. 6F–H). Neither excess Nkd nor decreased nkd dosage modified the wing bristle polarity of dsh1 mutant flies (data not shown). The results suggest that Nkd can specifically interfere with Dsh function in planar cell polarity and that this effect requires wild-type Dsh protein.
Discussion
Segment polarity genes encode signaling proteins that establish cell fate within each segment of the Drosophila embryo. A positive feedback loop between the Wg and Hh signaling pathways, active in adjacent cells, specifies parasegmental boundaries during early segmentation. Previous work showed that nkd is necessary to restrict the expression of the Wg target gene en as soon as its expression becomes dependent on Wg (Bejsovec and Wieschaus 1993; van den Heuvel et al. 1993). The inducible antagonist role of Nkd creates a negative feedback loop that limits Wg signaling during early embryogenesis (Zeng et al. 2000). Here we provide experimental evidence that nkd also regulates Wg activity in the eye. Our epistasis study suggests that Nkd acts cell-autonomously to antagonize Wg signaling at the level of, or between, Dsh and Zw3. In agreement with this study, yeast and biochemical experiments reveal that Nkd can bind directly to Dsh. Misexpressed Nkd specifically phenocopies loss of dsh in planar cell polarity, showing that Nkd can specifically interfere with endogenous dsh function.
Our data suggest a model of how Nkd limits early Wg signaling in the embryo. At stage 8–9 of wild-type embryonic development, when early segmental patterning events are taking place, Wg protein maintains the transcription of en/hh in adjacent posterior cells (Klingensmith and Nusse 1994). Cells located farther posterior receive an insufficient amount of Wg for induction of en/hh expression (Vincent and O'Farrell 1992). In nkd mutants at this stage, the more posterior cells ectopically express en/hh in a fashion that is Wg-dependent (Bejsovec and Wieschaus 1993; van den Heuvel et al. 1993). When ectopic en/hh transcription is first observed in nkd mutants at stage 8–9, Wg distribution is apparently normal (Moline et al. 1999; data not shown), indicating that Nkd normally prevents more distant cells from turning on en/hh in response to Wg. Consistent with this requirement, we saw enhanced nkd expression precisely in these more distant cells during stage 9 (Zeng et al. 2000). Our present results suggest that Nkd may act through Dsh in those cells to block Wg activity. Later in development, at stage 10–11, inappropriate Wg distribution may also contribute to the nkd phenotype (Moline et al. 1999). By stage 11, an ectopic stripe of wg expression is induced just posterior to the expanded en/hh domain and is required for the excess naked cuticle seen in nkd mutants (Dougan and DiNardo 1992). This extra wg stripe depends on Wg and Hh activities, as well as on the action of the pair-rule transcription factors Sloppy paired (Bejsovec and Wieschaus 1993; van den Heuvel et al. 1993; Cadigan et al. 1994).
Our experiments further clarify the differences between Nkd and Zw3 in Wg signaling. Embryos lacking both maternal and zygotic zw3 have a naked cuticle phenotype and expanded stripes of en transcription as in nkd mutants (Perrimon and Smouse 1989; Siegfried et al. 1992). Both Zw3 and Nkd are negative regulators of en transcription (Martinez Arias et al. 1988; Siegfried et al. 1992), but en stripe expansion is independent of Wg signaling in zw3 mutants and Wg dependent in nkd mutants (Siegfried et al. 1992; Bejsovec and Wieschaus 1993; van den Heuvel et al. 1993). Cuticles derived from double-mutant embryos illustrate the difference: zw3; wg and zw3 dsh mutants resemble zw3 embryos (Siegfried et al. 1992, 1994; Peifer et al. 1994), whereas embryos doubly mutant for nkd and either wg (Bejsovec and Wieschaus 1993) or dsh (this study) have a wg-like phenotype. In the absence of Wg, Zw3 phosphorylates and causes degradation of Arm, whereas in cells receiving Wg signal, Zw3 activity is inactivated. If Zw3 function is eliminated by mutation, Arm becomes stable and stimulates target-gene transcription, regardless of upstream Wg or Dsh activities. In contrast, the removal of Nkd in wg or dsh embryos does not lead to excess target-gene activity. Therefore, Wg does not act through Nkd—in the sense that it does through Zw3—to regulate en transcription and epidermal patterning. Taken together, our findings show that Nkd acts as a regulatory component that restrains Wg signal transduction.
The phenotypes of double-mutant embryos are consistent with the observation that the nkd gene is itself regulated by Wg signaling (Zeng et al. 2000). The level of nkd mRNA is markedly decreased in wg mutants (Zeng et al. 2000); so complete loss of nkd (i.e., in a nkd mutant) in a wg mutant would not be expected to dramatically alter the wg phenotype. In contrast, zw3 transcripts are ubiquitous during embryonic stages (Bourouis et al. 1990; Siegfried et al. 1990), and no indication of Wg control of zw3 transcription has been found. Various signaling pathways are regulated in a negative feedback loop by components acting in the extracellular space, in the cytoplasm and/or in the nucleus (Perrimon and McMahon 1999; Freeman 2000). Those mechanisms elegantly buffer cellular responses against signal level fluctuations during pattern formation. Our results allow classification of Nkd as a novel intracellular negative feedback antagonist that acts cell-autonomously in the Wg/Wnt pathway in a manner analogous to sprouty in the EGFR pathway, puckered in the JNK pathway, or daughters against dpp in the TGF-β/Dpp pathway (Perrimon and McMahon 1999).
The biochemical and cell biological roles of Dsh in Wg and PCP signaling remain a mystery. Previous work showed that these two pathways employ Dsh in distinct ways, through different domains, to transduce signals (Axelrod et al. 1998; Boutros et al. 1998). Under certain circumstances Wg signaling can titrate Dsh from its function in planar cell polarity (Axelrod et al. 1998). We showed that aberrant Nkd production during times when Dsh is participating in PCP signaling results in polarity defects specific to loss of dsh function, confirming that Nkd can affect the activity of endogenous Dsh. Polarized subcellular localization of PCP components appears to be essential for normal PCP signaling (Tomlinson and Struhl 1999; Usui et al. 1999). Control of Dsh subcellular localization has been implicated as a distinguishing feature in its two distinct roles in signal transduction: In a heterologous system, Fz is capable of recruiting Dsh to the plasma membrane, whereas DFz2, which is not involved in PCP signaling, is not (Axelrod et al. 1998). Recently, Dsh has been shown to regulate convergent extension movements during Xenopus gastrulation by modulating the frequency of filopodial extensions on adjacent faces of individual cells (Wallingford et al. 2000). Upon Wg signal activation, Dsh is phosphorylated, but the biochemical function of this phosphorylation is not known (Yanagawa et al. 1995). By directly interacting with Dsh, Nkd may sequester, degrade, or modify Dsh to block participation in PCP or Wg signaling. Future experiments will seek to elucidate the cellular and biochemical consequences of the interaction between Nkd and Dsh during Wg signaling.
That Nkd can act through Dsh has important implications for the dynamic control of Wg/Wnt signaling. By acting upstream of the β-catenin degradation machinery, Nkd may determine how effectively a given dose of Wnt causes β-catenin accumulation and target-gene activation and thereby influence the sensitivity of a cell to a given amount or type of Wnt ligand. The kinetic and dynamic parameters of the feedback loop involving Wg, Dsh, and Nkd may play key roles in controlling the duration and extent of signaling activity. Tight regulation of this feedback loop is clearly important for normal Drosophila embryonic development, and in various animals it may be subject to spatial and temporal adjustments during evolution or during disease progression. Future experiments will test how the interaction between Nkd and Dsh affects responses to Wnt signals during development and may provide insight into Wnt-associated tumor progression.
Materials and methods
nkd mutant eyes
P[FRT 80B], h1 nkd7H16 or h+ nkd7E89 or h1 nkd9G33/TM3 (Zeng et al. 2000) males were mated to y w; EGUF/EGUF; P[FRT 80B] cl y+ P[GMR-hid]/TM2 females and eyes of nonbalancer progeny were photographed under a Leica M10 stereomicroscope. (EGUF) eyeless-Gal4/UAS-Flp chromosome; (cl) recessive cell lethal (Stowers and Schwarz 1999).
Eye misexpression
All UAS-transgenes were expressed using multiple repeats of the glass (gl) enhancer to drive GAL4 expression P[GMR-GAL4]. The transgenic lines used in the eye epistasis study were P[sev-wg], P[GMR-GAL4], P[UAS-dsh], P[UAS-armS10] (armS10 encodes a constitutively active form of Arm protein with a 54–amino acid deletion in the N-terminal domain; Pai et al. 1997), P[UAS-nkd 3–2], P[UAS-nkd 11], P[UAS-GPI-Dfz2], and P[UAS-lacZ]. All crosses were carried out at 29°C. Adult offspring were prepared for scanning electron microscopy by treatment in a graded series of ethanol, followed by treatment in a graded series of hexamethyldisilazane. Dried samples were mounted on colloidal graphite and a 12-nm gold coat was applied with a Polaron Coating System. Samples were viewed with a Phillips 505 scanning electron microscope and photographed using Kodak 55 Instant Film.
zw3/Gsk3β expression in nkd mutants
A 0–3.5 h collection of P[UAS-zw3]/P[UAS-zw3]; +/+; hs-GAL4, nkd7H16/TM3 embryos was heat-shocked for 15 min at 37°C and allowed to recover overnight at 25°C. The embryos were then dechorionated in 50% bleach. The cuticles were prepared as described by Willert et al. (1999) and were evaluated using phase contrast and darkfield microscopy. For the injection study, pCS2-XGsk3β DNA was linearized with NotI and single-stranded RNA transcripts were synthesized with Sp6 polymerase using Message Machine (Ambion). mRNA transcripts were visualized on a 1% gel and diluted to 1.0, 0.5, and 0.2 μg/mL in ddH2O. Preblastoderm wild-type, nkd7E89, or nkd7H16 embryos were injected posteriorly with mRNA and allowed to complete embryogenesis. Cuticles were evaluated as stated above. Rescue tended to be greatest posteriorly at the site of injection.
nkd misexpression clones
To induce nkd misexpression clones, +/Y ; P[sev-wg], P[UAS-nkd 3–2]; +/+ males were crossed with y, w, P[Actin5C>CD2>GAL4]; P[UAS-GFP]; MKRS, Sb P[hs-FLP]/TM6B, Tb females. Second instar larvae were heat-shocked for 1 h in a 37°C water bath. For evaluation of clones in adult eyes, heads were dissected from the body and the eyes examined for GFP expression and suppression of bristle loss using a Zeiss Axioplan. Images were taken with a Princeton Micromax Digital Camera System. For evaluation of clones in pupal eyes, discs were dissected from pupae 30–36 h after pupal formation and fixed in 4% paraformaldehyde. Mouse monoclonal anti-Cut was used at a concentration of 1:100; rhodamine-conjugated goat anti-mouse antibody (Jackson Immunoresearch Labs) was used at a concentration of 1:200. Clones were evaluated and images taken using a Zeiss Axioskop and the MRC-1000 Laser Scanning Confocal Imaging System.
Crosses for the dsh; nkd and arm; nkd double mutants
The FLP-FRT system was used as described by Chou and Perrimon (1992) to generate the dsh germ-line clones. Females of the genotypes y w dshv26 FRT101/FM7c; +/+; nkd7H16 ru cu ca/TM6B or y w dshv26 FRT101/FM7c; +/+; nkd7E89 ru cu ca/TM6B or y w dsh477 FRT101/FM7c; +/+; nkd7H16 ru cu ca/TM6 were crossed with w ovoD1 v FRT101/Y; FLP38/FLP38; +/+ males. Early pupae were heat-shocked for 3 h in a 37°C water bath. y w dsh FRT101/w ovoD1 v FRT101; FLP38/+; nkd ru cu ca/+ females were selected and crossed with nkd7H16 ru cu ca/TM3 or nkd7E89 ru cu ca/TM3 males. Embryos were collected at 10 or 14 h and incubated at room temperature or 25°C until the first larvae hatched. The embryos were dechorionated in 50% bleach and cuticles were prepared as described by Willert et al. (1999).
For the arm; nkd double mutant, females of the genotype y armYD35/FM7;; +/+ were crossed with +/Y;; nkd7H16 ru cu ca/TM3 males. y armYD35/+;; nkd7H16 ru cu ca/+ females were selected and mated to +/Y;; nkd7H16 ru cu ca/TM3 or +/Y;; nkd7E89 ru cu ca/TM3 males. Embryos were collected for 10 or 14 h and incubated at room temperature for 36 h to let the wild-type larvae crawl away. Cuticles of the unhatched embryos were prepared as indicated above.
RNA interference
The following oligonucleotides containing the T7 promoter were designed as described previously by Kennerdell and Carthew (1998) to amplify a fragment of 797 bp from the nkd cDNA: T7-GAAGAGCCATCACCACCAGTCG (sens) and T7-GTATTGCAGCGTTGGCGTTGC (anti-sens). nkd dsRNA was synthesized from the PCR fragment using the MEGAscript in vitro transcription kit (Ambion) as recommended by the manufacturer. Injections in y w flies and cuticle preparations were made as described by Willert et al. (1999). For injections in dsh embryos, y w ras dsh75 FRT101/FM7; +/+ females were crossed with w ovoD1 v FRT101/Y; FLP38/FLP38 males and heat-shocked as outlined above. y w ras dsh75 FRT101/w ovoD1 v FRT101 females were selected from this cross and mated to Df(1)JA27/FM7c, Kr-GFP10 males. GFP-expressing embryos, which carry the rescue paternal X-chromosome, were eliminated between 19 h and 26 h after injection.
Yeast two-hybrid assay
The full-length nkd coding sequence was inserted into the yeast expression vector pAS2–1 (Clontech). The coding sequences of dsh (wt and mutants), zw3, arm, and the intracellular portion of Dfz2 (Dfz2CT, amino acids 607 to 694) were inserted into the yeast vector pACT2 (Clontech). The Dsh mutants D1 to D6 have been described by Yanagawa et al. (1995). The DEP, D6PDZ, and PDZ mutants correspond to amino acids 334–623, 167–338, and 248–338 of Dsh, respectively. Transformation of yeast strain PJ69–4A was performed using a variation of the lithium acetate method, and β-galactosidase activity (from the LacZ reporter gene) was assayed in a liquid-culture assay using O-nitrophenyl β-D-galactopyranoside as substrate (see Clontech protocols). Yeast-growth assay was performed using the ADE reporter gene and adenine minus medium.
Immunoprecipitation
COS-7 cells were transfected using calcium phosphate precipitation with pcDNA3.1B(−)/Myc-HisB vectors (Invitrogen) expressing Nkd and Dsh proteins tagged with the myc epitope at the C terminus. After 40 h, cells were lysed in TNN75 buffer (25 mM Tris-HCl at pH 8.0, 75 mM NaCl, 0.5% IGEPAL CA-630, 1mM DTT, 1 mM Pefabloc SC, antiprotease cocktail). Immunoprecipitation was carried out in TNN75–10% glycerol using rabbit anti-Dsh antibody (affinity-purified) and protein A Sepharose-4B beads. Immunoprecipitate was washed in TNN75 buffer and proteins were eluted with SDS-loading buffer and then run on SDS protein gels. Western blot was performed using anti-c-Myc antibody and enhanced chemiluminescence (Pierce).
GST pull-down assay
The coding sequence of full-length nkd was inserted into the pGEX-4T-1 vector (Pharmacia Biotech). Plasmid expressing GST-Dsh has been described by Willert et al. (1997). Bacterial lysates containing the GST fusion proteins were prepared as described by Pharmacia Biotech, except MTPBS buffer (150 mM NaCl, 12.5 mM Na2HPO4, 2.5 mM KH2PO4, 1 mM Pefabloc SC, 5 mM DTT, and antiprotease cocktail) was used instead of PBS. The lysates were bound to glutathione-sepharose 4B beads and the beads resuspended in DT80 buffer (20 mM Tris-HCl at pH 8, 80 mM KCl, 0.25% Triton X-100, 1 mM Pefabloc SC, 1 mM DTT). The beads were then incubated with [35S]methionine-labeled Nkd or Dsh (full length or mutant), which were produced using the TNT T7 Coupled Reticulocyte Lysate System (Promega) from pBluescript IIKS (+) plasmids. The beads were washed with DT300 buffer (20 mM Tris-HCl at pH 8, 300 mM KCl, 0.25% Triton X-100) and incubated with SDS-loading buffer to elute the proteins. Samples were run on SDS protein gels.
Planar cell polarity assay
To assay effects of overexpression on planar polarity, white prepupae were selected and placed in plastic vials, aged for 24 h at 25°C, immersed for 2 h in a 37°C water bath and then allowed to develop at 25°C. Wings were mounted in Euparal. The genotypes were UAS-nkd 3–2/+; hs-GAL4/+ (Fig. 6B), hs-fz30.9/+ (Fig. 6G), UAS-nkd 3–2/hs-fz30.9; hs-GAL4, nkd7H16/+ (Fig. 6H); and w, dsh1/Y; UAS-nkd 3–2/+; hs-GAL4, nkd7H16 (data not shown).
Acknowledgments
We thank P. Klein for the pCS2-XGSK3β construct; E. Rulifson for the FLP-out/GAL4 line; K. Willert for anti-Dsh antibody; the Developmental Studies Hybridoma Bank for anti-Cut antibody; S. Stowers and T. Schwarz for EGUF/hid stocks; the Bloomington Stock Center for the ovoD1 fly stock; and the Axelrod, Nusse, and Scott labs for reagents, advice, and encouragement. R.R. was supported by the Association pour la Recherche sur le Cancer and by the Human Frontier Science Program. J.A.M. was supported by an NIH postdoctoral fellowship and the Howard Hughes Medical Institute (HHMI). K.A.W. was supported by a K-08 award from the NIH. J.D.A. was supported in part by DRS 16 of the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation, and by the HHMI. K.C. was supported by a grant from the NIH (RO1 GM59846) and by the HHMI. R.N. and M.P.S. are investigators of the HHMI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL scott@cmgm.stanford.edu; FAX (650) 725-7739.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.869201.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Y, Hayashi S, Levine A, Wieschaus E. Regulation of armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell. 1998;93:1171–1182. doi: 10.1016/s0092-8674(00)81461-0. [DOI] [PubMed] [Google Scholar]
- Axelrod JD, Miller JR, Shulman JM, Moon RT, Perrimon N. Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes & Dev. 1998;12:2610–2622. doi: 10.1101/gad.12.16.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejsovec A, Wieschaus E. Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development. 1993;119:501–517. doi: 10.1242/dev.119.2.501. [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Bourouis M, Moore P, Ruel L, Grau Y, Heitzler P, Simpson P. An early embryonic product of the gene shaggy encodes a serine/threonine protein kinase related to the CDC28/cdc2 + subfamily. EMBO J. 1990;9:2877–2884. doi: 10.1002/j.1460-2075.1990.tb07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Mlodzik M. Dishevelled: At the crossroads of divergent intracellular signaling pathways. Mech Dev. 1999;83:27–37. doi: 10.1016/s0925-4773(99)00046-5. [DOI] [PubMed] [Google Scholar]
- Boutros M, Paricio N, Strutt DI, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. wingless signaling in the Drosophila eye and embryonic epidermis. Development. 1996;122:2801–2812. doi: 10.1242/dev.122.9.2801. [DOI] [PubMed] [Google Scholar]
- ————— Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes & Dev. 1994;8:899–913. doi: 10.1101/gad.8.8.899. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1998;93:767–777. doi: 10.1016/s0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, Sher E, Heemskerk-Jorgens J, Kassis J, O'Farrell P. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan S, DiNardo S. Drosophila wingless generates cell type diversity among engrailed expressing cells. Nature. 1992;360:347–350. doi: 10.1038/360347a0. [DOI] [PubMed] [Google Scholar]
- Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–319. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- Gubb D, Garcia-Bellido A. A genetic analysis of the determination of cuticular polarity during development in Drosophila melanogaster. J Embryol Exp Morphol. 1982;68:37–57. [PubMed] [Google Scholar]
- Hamada F, Tomoyasu Y, Takatsu Y, Nakamura M, Nagai S, Suzuki A, Fujita F, Shibuya H, Toyoshima K, Ueno N, et al. Negative regulation of Wingless signaling by D-axin, a Drosophila homolog of axin. Science. 1999;283:1739–1742. doi: 10.1126/science.283.5408.1739. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Taylor AM, Nakano Y. Role of the Drosophila patched gene in positional signalling. Nature. 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- Jürgens G, Wieschaus E, Nüsslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Wilhelm Roux Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Klingensmith J, Nusse R. Signaling by wingless in Drosophila. Dev Biol. 1994;166:396–414. doi: 10.1006/dbio.1994.1325. [DOI] [PubMed] [Google Scholar]
- Klingensmith J, Nusse R, Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes & Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- Krasnow RE, Adler PN. A single frizzled protein has a dual function in tissue polarity. Development. 1994;120:1883–1893. doi: 10.1242/dev.120.7.1883. [DOI] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H, Weaver CD, Mao J, Farr GH, III, Sussman DJ, Jonkers J, Kimelman D, Wu D. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt- mediated regulation of LEF-1. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A, Baker NE, Ingham PW. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development. 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- McCartney BM, Dierick HA, Kirkpatrick C, Moline MM, Baas A, Peifer M, Bejsovec A. Drosophila APC2 is a cytoskeletally-associated protein that regulates wingless signaling in the embryonic epidermis. J Cell Biol. 1999;146:1303–1318. doi: 10.1083/jcb.146.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moline MM, Southern C, Bejsovec A. Directionality of Wingless protein transport influences epidermal patterning in the Drosophila embryo. Development. 1999;126:4375–4384. doi: 10.1242/dev.126.19.4375. [DOI] [PubMed] [Google Scholar]
- Noordermeer J, Johnston P, Rijsewijk F, Nusse R, Lawrence PA. The consequences of ubiquitous expression of the wingless gene in the Drosophila embryo. Development. 1992;116:711–719. doi: 10.1242/dev.116.3.711. [DOI] [PubMed] [Google Scholar]
- Noordermeer J, Klingensmith J, Perrimon N, Nusse R. dishevelled and armadillo act in the wingless signalling pathway in Drosophila. Nature. 1994;367:80–83. doi: 10.1038/367080a0. [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- O'Keefe L, Dougan ST, Gabay L, Raz E, Shilo BZ, DiNardo S. Spitz and Wingless, emanating from distinct borders, cooperate to establish cell fate across the Engrailed domain in the Drosophila epidermis. Development. 1997;124:4837–4845. doi: 10.1242/dev.124.23.4837. [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development. 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell. 1990;63:1167–1176. doi: 10.1016/0092-8674(90)90413-9. [DOI] [PubMed] [Google Scholar]
- Peifer M, Sweeton D, Casey M, Wieschaus E. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Mahowald AP. Multiple functions of segment polarity genes in Drosophila. Dev Biol. 1987;119:587–600. doi: 10.1016/0012-1606(87)90061-3. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Smouse D. Multiple functions of a Drosophila homeotic gene, zeste-white 3, during segmentation and neurogenesis. Dev Biol. 1989;135:287–305. doi: 10.1016/0012-1606(89)90180-2. [DOI] [PubMed] [Google Scholar]
- Perrimon N, McMahon AP. Negative feedback mechanisms and their roles during pattern formation. Cell. 1999;97:13–16. doi: 10.1016/s0092-8674(00)80710-2. [DOI] [PubMed] [Google Scholar]
- Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature. 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes & Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Shulman JM, Perrimon N, Axelrod JD. Frizzled signaling and the developmental control of cell polarity. Trends Genet. 1998;14:452–458. doi: 10.1016/s0168-9525(98)01584-4. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Perkins LA, Capaci TM, Perrimon N. Putative protein kinase product of the Drosophila segment-polarity gene zeste-white3. Nature. 1990;345:825–829. doi: 10.1038/345825a0. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Chou TB, Perrimon N. wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Wilder EL, Perrimon N. Components of wingless signalling in Drosophila. Nature. 1994;367:76–80. doi: 10.1038/367076a0. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuts D, Freeman M, Bienz M. Antagonism between EGFR and Wingless signalling in the larval cuticle of Drosophila. Development. 1997;124:3209–3219. doi: 10.1242/dev.124.16.3209. [DOI] [PubMed] [Google Scholar]
- Theisen H, Purcell J, Bennett M, Kansagara D, Syed A, Marsh JL. dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development. 1994;120:347–360. doi: 10.1242/dev.120.2.347. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Struhl G. Decoding vectorial information from a gradient: Sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development. 1999;126:5725–5738. doi: 10.1242/dev.126.24.5725. [DOI] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Van den Heuvel, M., Klingensmith, J., Perrimon, N., and Nusse, R. 1993. Cell patterning in the Drosophila segment: Engrailed and wingless antigen distributions in segment polarity mutant embryos. Dev. Suppl. 105–114. [PubMed]
- Vincent JP, O'Farrell PH. The state of engrailed expression is not clonally transmitted during early Drosophila development. Cell. 1992;68:923–931. doi: 10.1016/0092-8674(92)90035-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Rowning BA, Vogeli KM, Rothbacher U, Fraser SE, Harland RM. Dishevelled controls cell polarity during Xenopus gastrulation. Nature. 2000;405:81–85. doi: 10.1038/35011077. [DOI] [PubMed] [Google Scholar]
- Willert K, Brink M, Wodarz A, Varmus H, Nusse R. Casein kinase 2 associates with and phosphorylates dishevelled. EMBO J. 1997;16:3089–3096. doi: 10.1093/emboj/16.11.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert K, Logan CY, Arora A, Fish M, Nusse R. A Drosophila Axin homolog, Daxin, inhibits Wnt signaling. Development. 1999;126:4165–4173. doi: 10.1242/dev.126.18.4165. [DOI] [PubMed] [Google Scholar]
- Yanagawa S, van Leeuwen F, Wodarz A, Klingensmith J, Nusse R. The dishevelled protein is modified by wingless signaling in Drosophila. Genes & Dev. 1995;9:1087–1097. doi: 10.1101/gad.9.9.1087. [DOI] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes & Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- Zeng W, Wharton KA, Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]