Abstract
We investigated the roles of Caenorhabditis elegans MRE-11 in multiple cellular processes required to maintain genome integrity. Although yeast Mre11 is known to promote genome stability through several diverse pathways, inviability of vertebrate cells that lack Mre11 has hindered elucidation of the in vivo roles of this conserved protein in metazoan biology. Worms homozygous for an mre-11 null mutation are viable, allowing us to demonstrate in vivo requirements for MRE-11 in meiotic recombination and DNA repair. In mre-11 mutants, meiotic crossovers are not detected, and oocyte chromosomes lack chiasmata but appear otherwise intact. γ-irradiation of mre-11 mutant germ cells during meiotic prophase eliminates progeny survivorship and induces chromosome fragmentation and other cytologically visible abnormalities, indicating a defect in repair of radiation-induced chromosome damage. Whereas mre-11 mutant germ cells are repair-deficient, they retain function of the meiotic G2 DNA damage checkpoint that triggers germ cell apoptosis in response to ionizing radiation. Although mre-11/mre-11 animals derived from heterozygous parents are viable and produce many embryos, there is a marked drop both in the number and survivorship of embryos produced by succeeding generations. This progressive loss of fecundity and viability indicates that MRE-11 performs a function essential for maintaining reproductive capacity in the species.
Keywords: Meiosis, recombination, DNA repair, checkpoint, C. elegans, Mre11, mre-11
Living organisms expend considerable energy in preserving the integrity of their genomes. Multiple mechanisms have evolved to ensure the fidelity of genome duplication and to guarantee faithful partitioning of chromosomes at cell division. Cells also have the capacity to recognize and repair DNA damage acquired through exposure to genotoxic environmental stresses and to prevent cell cycle progression until repair has been completed (Weinert 1998; Haber 2000). It has become increasingly clear that the mechanisms involved in accomplishing these tasks are intimately related. The interconnectedness of mechanisms responsible for genome maintenance is particularly evident during sexual reproduction when cells undergoing meiosis reduce their diploid chromosome complement by half. Crossover recombination events between the DNA molecules of homologous chromosomes collaborate with sister chromatid cohesion to establish temporary connections between homologs (chiasmata) that allow them to orient toward opposite poles of the meiosis I spindle (Hawley 1988; Moore and Orr-Weaver 1998).
Recently, studies of meiotic recombination, double-strand break (DSB) repair, cell cycle checkpoints, and tumor suppressors have converged to focus attention on complexes containing two well-conserved proteins, Rad50 and Mre11 (Haber 1998; Dasika et al. 1999; Petrini 2000). In vitro studies have demonstrated multiple intrinsic nuclease activities for human and/or yeast Mre11, including 3′ to 5′ ds and ssDNA exonuclease, ssDNA endonuclease, and hairpin opening (Furuse et al. 1998; Paull and Gellert 1998, 1999; Trujillo et al. 1998; Usui et al. 1998; Moreau et al. 1999). These activities are modified in efficiency and specificity by interactions with other components of the respective Mre11 complexes (Paull and Gellert 1998, 1999; Trujillo et al. 1998).
Most of what we know about the in vivo roles of Mre11/Rad50 protein complexes comes from studies in Saccharomyces cerevisiae where Mre11 and Rad50 are known to function in diverse cellular processes required to maintain genome stability. They participate in both homologous recombination and nonhomologous end-joining pathways for repair of DSBs in vegetative cells (Moore et al. 1993; Ivanov et al. 1994, 1996; Moore and Haber 1996; Boulton and Jackson 1998; Tsubouchi and Ogawa 1998; Bressan et al. 1999; Lewis et al. 1999). During meiosis, they play a dual role in both formation and processing (5′ to 3′ resection) of the regulated DSBs that initiate meiotic recombination (Alani et al. 1990; Cao et al. 1990; Johzuka and Ogawa 1995; Nairz and Klein 1997). Additional roles include a requirement for these proteins in maintenance of telomere length (Kironmai and Muniyappa 1997; Boulton and Jackson 1998; Nugent et al. 1998). An intact Mre11p nuclease domain is required for some but not all of these functions (Bressan et al. 1998, 1999; Furuse et al. 1998; Tsubouchi and Ogawa 1998; Usui et al. 1998; Moreau et al. 1999).
Vertebrate Mre11/Rad50 complexes have been implicated in cell cycle checkpoint responses to DNA damage. Evidence for a role in an S-phase DNA damage checkpoint came initially from the finding that hMre11 and hRad50 copurify in a complex with Nbs1, the protein encoded by the gene mutated in the Nijmegen breakage syndrome (NBS) (Carney et al. 1998; Varon et al. 1998). NBS is an inherited disorder associated with an increased sensitivity to ionizing radiation (IR) and a predisposition to cancer (Shiloh 1997); cells from NBS patients fail to suppress DNA synthesis after exposure to IR (the radioresistant DNA synthesis, or RDS, phenotype). More recently, patients with an ataxia-telangiectasia-like disorder with cellular features similar to NBS, including the RDS phenotype, were found to have hypomorphic mutations in hMRE11 (Stewart et al. 1999). Independent evidence that Mre11 and Rad50 function in the response to DNA damage in mammalian cells came from the observation that IR induces localization of Mre11 to sites of DNA damage (Nelms et al. 1998). Nbs1 and Rad50 colocalize with Mre11 in IR-induced foci along with the tumor suppressor gene product Brca1, which physically associates with Rad50 (Maser et al. 1997; Carney et al. 1998; Zhong et al. 1999). These proteins are all constituents of a huge (>20 MD) protein complex that also includes the checkpoint protein kinase Atm (Wang et al. 2000), and phosphorylation of Nbs1 by Atm appears to be important for conferral of radioprotection and for IR-induced inhibition of DNA synthesis in cultured cells (Gatei et al. 2000; Lim et al. 2000; Zhao et al. 2000).
Although these studies have implicated the mammalian orthologs of Mre11 and Rad50 in the cellular response to DNA damage, it has been difficult to discern whether they function in repair per se or in damage detection, transduction of a damage signal, regulation of checkpoint responses, or some combination of these functions. Mre11 has been implicated in maintaining chromosome integrity in chicken DT40 cells through a role in recombination-based repair (Yamaguchi-Iwai et al. 1999), but the hyperrecombinogenic character of the DT40 cell line makes it difficult to draw conclusions about the importance of this role in normal cellular physiology. Efforts to further elucidate the in vivo biological roles of Mre11 and Rad50 in metazoan systems, particularly at the organismal level, have been hampered by the fact that vertebrate cells that lack Mre11 or Rad50, as well as mouse embryos that lack Rad50, are inviable (Xiao and Weaver 1997; Luo et al. 1999; Yamaguchi-Iwai et al. 1999).
We report here our analysis of Mre11 function in the nematode Caenorhabditis elegans, a simple metazoan. Worms homozygous for an mre-11 null mutation are viable, providing an opportunity to investigate the in vivo roles of C. elegans MRE-11 in multiple cellular processes required to maintain genome integrity. This work establishes requirements for CeMRE-11 in meiotic recombination and in repair of IR-induced chromosome damage and demonstrates that CeMRE-11 is dispensable for function of a meiotic G2 DNA damage checkpoint. Furthermore, we find that although CeMRE-11 is not required for viability in the short term, it is crucial in the long term for maintenance of reproductive capacity and thus for perpetuation of the species.
Results
C. elegans mutants defective in mre-11
We isolated our initial mre-11 mutant allele in a genetic screen for C. elegans mutants defective in meiotic chromosome segregation (Kelly et al. 2000). Because males (which possess a single X chromosome) arise among the self-progeny of XX hermaphrodites as a consequence of chromosome missegregation (Hodgkin et al. 1979), meiotic mutants were identified by screening for hermaphrodites producing a high incidence of male embryos (or “Him” phenotype). Most Him mutants identified in this manner also suffer autosomal missegregation and consequently produce broods comprised mainly of dead aneuploid embryos, with a few euploid adult survivors. Because recombination between homologous chromosomes is required to ensure faithful segregation at meiosis I, mutants defective in this prerequisite event are readily recovered using this approach.
One mutation identified in this manner, me41, was mapped to a 0.95-cM region containing ZC302.1 (now designated mre-11), which encodes the C. elegans ortholog of the yeast meiotic and mitotic DNA repair protein Mre11p. Depletion of mre-11 transcripts by RNA interference elicited a robust phenocopy of the me41 mutant. Moreover, sequencing of the mre-11 gene in the me41 mutant revealed a G → A transition that substitutes a lysine for a glutamate residue that is conserved in all Mre11 orthologs; this conserved glutamate is located at a position immediately adjacent to the second block of homology shared between the Mre11 proteins and the Escherichia coli SbcD nuclease (Fig. 1; Sharples and Leach 1995).
Figure 1.
C. elegans MRE-11 predicted protein sequence aligned with orthologs from human, mouse, Drosophila, fission yeast, and budding yeast. The solid underline corresponds to the conserved phosphoesterase domain, with the thicker segments indicating motifs conserved between Mre11 and E. coli SbcD nuclease. The asterisk marks the alternative initiator methionine predicted by cDNAs obtained by 5′ RACE. The position of the glutamate to lysine change caused by the mre-11(me41) missense mutation is indicated above the sequence alignment. The in-frame deletion in mre-11(ok179) removes the segment of sequence encoding the amino acids indicated by the dashed line below the alignment.
A second mre-11 mutant allele, ok179, was generated by the C. elegans Deletion Consortium using a PCR-based strategy to isolate deletions in mre-11. This screen allows identification of mutations in the heterozygous state, therefore there is no bias against recovery of mutations that confer lethality when homozygous. The ok179 allele contains an in-frame deletion that removes 70% of the conserved coding sequence, including nearly half of the highly conserved phosphoesterase domain required for nuclease activities in yeast Mre11p (Fig. 1; Furuse et al. 1998; Usui et al. 1998; Moreau et al. 1999). On the basis of the size and structure of this deletion, the ok179 mutation is expected to eliminate mre-11 function. Because me41 and ok179 fail to complement and confer very similar phenotypes in multiple assays (see below), it is likely that the me41 allele also severely reduces gene function.
For both mre-11 mutant alleles, viable homozygous mutant worms comprised the expected one-quarter of the self-progeny from heterozygous hermaphrodites (105 of 416 for ok179 and 101 of 399 for me41). The fact that homozygous mre-11 mutants are apparently fully viable when derived from a heterozygous parent allowed us to investigate the in vivo roles of CeMRE-11 in multiple processes required to maintain genome integrity.
Sequencing of cDNAs obtained from the Kohara EST library confirmed all intron/exon boundaries predicted by Genefinder for ZC302.1, and several cDNAs included part of the first predicted exon, making it likely that the MRE-11 protein sequence predicted by Genefinder (GI:3881390) is correct. We also generated cDNAs by 5′ RACE that did not contain the first predicted exon but instead contained the trans-spliced leader SL1 spliced to predicted exon 2, providing evidence for a variant protein form that initiates at a methionine corresponding to amino acid 45 of the Genefinder-predicted protein. The two predicted forms of MRE-11 are 772 and 728 amino acids in length, and both contain nonconserved amino-terminal regions (103 or 59 amino acids) upstream of the conserved remainder of the protein.
C. elegans mre-11 is required for meiotic crossing over and chiasma formation
Both mre-11 alleles confer an array of phenotypes diagnostic of a defect in meiotic reciprocal recombination (Dernburg et al. 1998; Zalevsky et al. 1999; Kelly et al. 2000). Homozygous mutant hermaphrodites are morphologically normal and display a Him phenotype indicative of a severe defect in meiotic chromosome segregation (Table 1). They produce a large number of embryos, most of which are inviable, and a large fraction of the progeny that survive to adulthood are male. Cytological analysis of the germ lines of these males revealed that chromosome segregation is also defective during male meiosis.
Table 1.
mre-11 mutants exhibit a Him phenotype indicative of a defect in meiotic chromosome segregation
| Genotype
|
Average no. of eggs laid (no. of broods)
|
Percent inviable embryosa (total no. of embryos)
|
Percent males (total no. of adults)
|
|---|---|---|---|
| Wild type | 281 ± 53 (5) | 0 (1403) | 0 (1403) |
| mre-11(me41)b | 191 ± 36 (9) | 97.9 (1717) | 24 (332) |
| mre-11(ok179)b | 190 ± 38 (10) | 96.9 (1898) | 35 (530) |
99% of the inviable embryos died before hatching; 1% hatched and died as arrested L1–L2 larvae.
The hermaphrodites whose progeny were scored in these experiments were homozygous mre-11 mutants derived from mre-11/+/+ mothers.
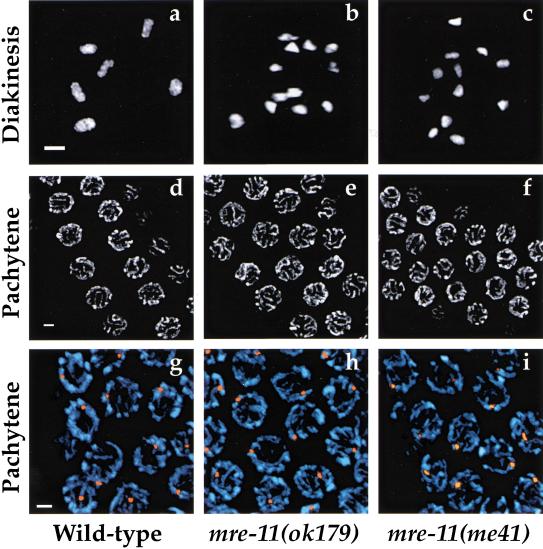
Cytological analysis of oocyte nuclei in late meiotic prophase (diakinesis) demonstrated an absence of chiasmata in mre-11 mutants that readily accounts for the meiotic segregation defect. At this stage, homologous chromosomes have lost the side-by-side alignment that is the hallmark of an earlier stage of prophase (pachytene), but remain attached by chiasmata, temporary physical links established as a result of reciprocal recombination events completed earlier in prophase. Although six DAPI-stained bodies (corresponding to the six pairs of homologs attached by chiasmata) are detectable in ooctyes of wild-type worms, 12 individual DAPI-stained bodies (corresponding to unattached univalent chromosomes) are observed in the majority of mre-11 mutant oocytes (Fig. 2a–c). An average of 11.9 DAPI-stained bodies were resolved in 91 oocyte nuclei from 12 mre-11(ok179) worms, and an average of 11.7 DAPI-stained bodies were resolved in 140 oocyte nuclei from 18 mre-11(me41) worms.
Figure 2.
Chiasmata are absent in mre-11 mutants but pachytene morphology and homolog pairing are normal. (a–c) Single DAPI-stained oocyte nuclei at diakinesis, the final stage of meiotic prophase. Each panel shows a projection of a three-dimensional data stack through an entire nucleus. (a) Wild-type nucleus with six DAPI-stained bodies, corresponding to six pairs of homologous chromosomes attached by chiasmata. (b,c) mre-11 mutant oocytes with 12 univalents, indicating an absence of chiasmata. (d–f) Fields of nuclei at the pachytene stage earlier in meiotic prophase. Images shown are projections halfway through the nuclei to highlight nuclear organization. (d) Pachytene nuclei in wild-type. Discrete tracks of DAPI-stained chromatin corresponding to pairs of side-by-side aligned and synapsed chromosomes are arranged at the periphery of each nucleus. (e,f) Pachytene nuclei in the mre-11 mutants. DAPI-stained chromatin tracks are similar in thickness, number, and arrangement to those seen in wild-type pachytene nuclei. (g–i) Homolog pairing assayed by FISH in wild-type (g) and mre-11 mutant (h,i) nuclei in the pachytene region of the germ-line. DAPI-stained chromosomes are shown in blue, and a probe from the left end of chromosome I is shown in orange. A single hybridization signal indicative of intimate pairing is visible in each nucleus for all three genotypes. Images shown are projections of three-dimensional data stacks through entire nuclei. Scale bars, 2 μm.
Measurement of crossover frequency in the mre-11(ok179) mutant indicates that the absence of chiasmata reflects an underlying defect in crossover formation. We assessed crossing over in a 38-cM interval corresponding to 80% of the X chromosome and found no recombinants among 487 progeny of mre-11(ok179) hermaphrodites (Table 2). A recombination frequency <1% of the wild-type level indicates that crossing over is severely reduced or eliminated in mre-11(ok179) mutants. Because this assay detects recombination in both male and female germ lines of the hermaphrodite, this extreme reduction in crossover frequency indicates that recombination is severely compromised in both spermatocyte and oocyte meiosis.
Table 2.
Severe reduction in crossover frequency in the mre-11 (ok179) mutant
| Genotype of parent
|
No. of recombinants
|
No. of progeny scored
|
Map distance (cM)
|
|---|---|---|---|
| +/(mre-11 or +); dpy-3 unc-3/+ | 602 | 1956 hermaphrodites | 38 |
| mre-11/mre-11; dpy-3 unc-3/+ | 0 | 354 hermaphrodite | <0.2 |
| 133 male |
Normal pairing and alignment of homologs in mre-11 mutant germ lines
A severe deficit in crossover formation could reflect a defect in recombination per se or might result from an inability to establish or maintain pairing and alignment of homologous chromosomes. We ruled out a defect in homolog pairing by cytological analysis of whole-mount germ-line preparations, in which nuclei at all stages of meiotic prophase are represented simultaneously in a temporal/spatial gradient. The morphology of meiotic prophase chromosomes in three-dimensionally preserved mre-11 mutant germ lines appeared normal until the diakinesis stage, when the absence of chiasmata became evident. Figure 2d–f shows the similar appearance of wild-type and mre-11 nuclei at the earlier pachytene stage of meiotic prophase, when homologs are fully paired and aligned along their lengths. Discrete DAPI-stained tracks indicative of paired and synapsed homologs are apparent in both wild-type and mre-11 mutant nuclei. Moreover, fluorescence in situ hybridization (FISH) experiments verified that homologous chromosomal regions are intimately associated in mre-11 mutants (Fig. 2g–i). For probes from three different chromosomes, hybridization signals from the two homologs were either completely coincident or closely juxtaposed in nuclei from the pachytene regions of both wild-type and mre-11 mutant gonads.
mre-11 is required for repair of IR-induced chromosome breaks generated during meiotic prophase
Meiotic recombination is normally initiated by programmed induction of enzymatically generated double-strand DNA breaks (Paques and Haber 1999). Dernburg et al. (1998) demonstrated that artificial induction of DNA breaks by γ-irradiation can bypass the requirement for the putative recombination-initiating enzyme CeSPO-11, producing both crossovers and chiasmata in spo-11 mutants (which normally lack recombination). Kelly et al. (2000) further demonstrated that the response to IR-induced breaks can be used to distinguish between mutants specifically defective in initiation of recombination and mutants defective in subsequent steps in the recombination pathway. They showed that γ-irradiation does not bypass the requirement for msh-5, which acts at a later step to promote the crossover outcome of initiated recombination events. We used this γ-irradiation treatment to further investigate the roles of mre-11.
Exposure of mre-11 mutant germ lines to 5 krad of γ-irradiation revealed a profound defect in the response of germ cells to DNA damage (Table 3; Fig. 3). Whereas wild-type hermaphrodites exhibited only a 6% decrease in progeny survivorship after this treatment, progeny survivorship was reduced by at least two orders of magnitude, and thereby effectively abolished, for both mre-11(me41) and mre-11(ok179) hermaphrodites. In contrast, previous analysis showed that this treatment actually increases the production of viable progeny by 10- to 20-fold in the spo-11 mutant (presumably reflecting improved chromosome segregation as a consequence of chiasma formation) and causes only about a 50% decrease in progeny survivorship for msh-5 mutants (Dernburg et al. 1998; Kelly et al. 2000).
Table 3.
Progeny survivorship is abolished in mre-11 mutants after germ-line γ-irradiation
| Genotype
|
Total embryos
|
Viable adult progeny (observed/expecteda)
|
% Survivorship (relative to unirradiated)
|
|---|---|---|---|
| Wild type | 1535 | 1445/1535 | 94.1 |
| mre-11(me41) | 13,385b | 0/174 | <0.6 |
| mre-11(ok179) | 5580b | 0/67 | <1 |
Germ lines of late-stage L4 larvae were exposed to 5 krad of γ-irradiation from a 137Cs source, and survivorship of their progeny was assessed.
The expected number of viable adult progeny was calculated by multiplying the total number of embryos by the fraction of embryos that survived to adulthood in the broods of concurrently- scored unirradiated control hermaphrodites of the same genotype (0.013 for me41 and 0.012 for ok179).
These numbers are estimates based on multiplying the average number of embryos per brood (determined by counting several complete broods) by the total number of broods examined.
Figure 3.
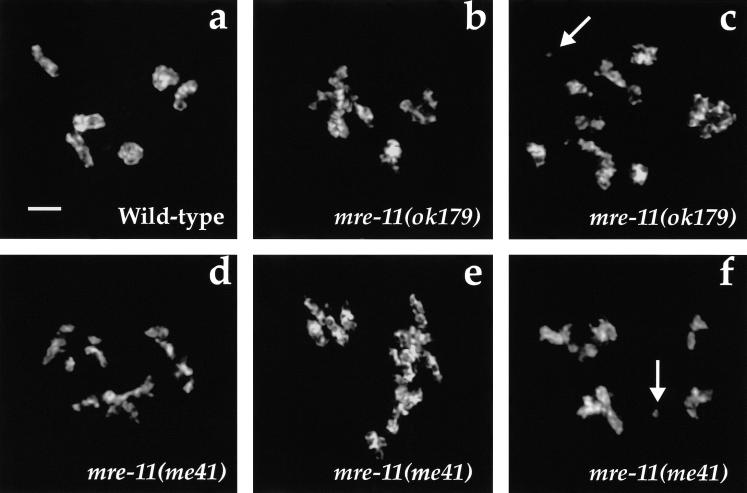
IR-induced chromosome defects in mre-11 mutant oocytes. Wild-type and mre-11 worms at the late L4 larval stage were exposed to 5 krad of γ-irradiation, and their germ lines were fixed with formaldehyde 18 h later and stained with DAPI. Oocyte nuclei examined at the diakinesis stage at this time were likely at the pachytene stage at the time of irradiation. (a) Wild-type oocyte nucleus, which contains six pairs of homologs attached by chiasmata; the chromosomes appear morphologically normal and intact. (b–f) mre-11 mutant oocytes exhibiting multiple chromosomal abnormalities. Chromosomes tend to be clumped or aggregated with a frayed appearance. Chromosome fragments are indicated by arrows in c and f. Images are projections of three-dimensional data stacks through entire nuclei. Scale bars, 2 μm.
Cytological examination of oocyte chromosomes 18 h after irradiation revealed profound defects in the ability of mre-11 mutant germ cells to repair chromosomal damage induced by γ-irradiation (Fig. 3). Whereas diakinesis-stage chromosomes in wild-type oocytes appeared similar to unirradiated controls, the chromatin in oocytes from irradiated mre-11 animals exhibited multiple abnormalities. Chromosomes had a frayed appearance and tended to be clumped together in large masses that made it impossible to resolve them as separate entities. We also observed several clear examples of chromosome fragments (Fig. 3c,f), indicating a failure to properly repair chromosome breaks. This appearance of gross chromosomal abnormalities after germ-line irradiation in mre-11 mutants contrasts sharply with results obtained for spo-11 and msh-5 mutants, in which chromosomes emerged from the treatment appearing morphologically intact and, in the case of spo-11, connected by chiasmata (Dernburg et al. 1998; Kelly et al. 2000).
The meiotic G2 DNA damage checkpoint is functional in mre-11 mutants
The above analysis indicates that at least some germ-line nuclei in irradiated mre-11 mutants can proceed to the diakinesis stage without successfully repairing damaged chromosomes. This likely reflects a defect in the repair process, but might also reflect a defect in a checkpoint mechanism that would normally eliminate germ cells that have sustained substantial damage. Gartner et al. (2000) recently reported that exposure of female C. elegans germ cells to γ-irradiation can induce them to undergo apoptosis at the end of the pachytene stage of meiotic prophase (an extended G2 phase). Induction of apoptosis by irradiation is dependent on a conserved DNA damage checkpoint requiring mrt-2, the C. elegans ortholog of checkpoint genes Schizosaccaromyces pombe rad1+ and S. cerevisiae RAD17. This checkpoint can also be triggered in unirradiated wild-type germ lines by depletion of RAD-51 by RNAi, which presumably results in persistence of unresolved recombination intermediates. We examined whether this meiotic G2 DNA damage checkpoint is operative in mre-11 germ lines.
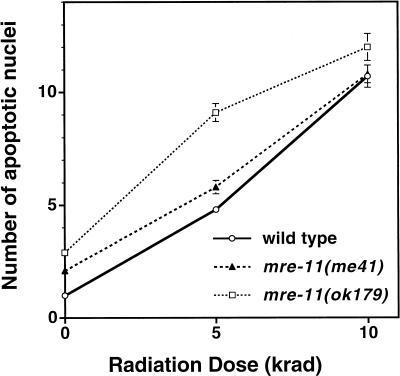
Our analysis of radiation-induced apoptosis indicates that the meiotic G2 DNA damage checkpoint is functional in mre-11 mutant germ lines (Fig. 4). L4 worms were exposed to 0, 5, or 10 krad of γ-irradiation, and their germ lines were scored 24 to 28 h later for the presence of apoptotic germ cells. Both mre-11 mutants and wild-type controls exhibited similar dose-dependent increases in the frequency of apoptotic germ cell corpses detected after irradiation. Thus, mre-11 function is required neither for sensing the presence of γ-irradiation-induced DNA damage nor for triggering apoptosis.
Figure 4.
A functional meiotic G2 DNA damage checkpoint in mre-11 mutants. Late-stage L4 hermaphrodites were exposed to 0, 5, or 10 krad of γ-irradiation, and induction of germ cell apoptosis (a readout of the meiotic G2 DNA damage checkpoint) was assessed 24–28 h later using Nomarski microscopy. The y-axis indicates the number of apoptotic nuclei observed per gonad arm. Fifty-two to seventy gonad arms were scored for each data point; error bars indicate standard error of the mean.
mre-11 is required for maintenance of reproductive capacity
Although mre-11 homozygotes derived from mre-11/+ parents (m+z−) are fully viable and produce large numbers of embryos, there is a marked decrease both in the number and viability of embryos produced by succeeding generations (Table 4). A substantial fraction of mre-11 homozygotes from mre-11/mre-11 parents (m−z−) fail to lay any eggs at all (26 of 112 for ok179 and 51 of 96 for me41). In contrast, failure to lay eggs is unusual among m+z− hermaphrodites (0 of 105 for ok179 and 4 of 101 for me41). For those m−z− worms that do lay eggs, the average number of viable progeny produced per brood is ≤5% of that produced by their m+z− parents (Table 4), making it impossible to propagate either mre-11 mutant as a homozygous strain.
Table 4.
Progressive loss of reproductive capacity in mre-11 mutants
| Genotype
|
Average no. of eggs laid (no. of broods)a
|
% Viable progeny (total no. of embryos)
|
Average no. of viable progeny/brooda
|
|---|---|---|---|
| mre-11(me41) (m+ z−)b | 191 ± 36 (9) | 2.1 (1717) | 4.0 |
| mre-11(me41) (m− z−)c | 61 ± 44 (19) | 0.2 (1151) | 0.1 |
| mre-11(ok179) (m+ z−) | 190 ± 38 (10) | 3.1 (1898) | 5.9 |
| mre-11(ok179) (m− z−) | 54 ± 42 (16) | 0.6 (868) | 0.3 |
Worms that laid no eggs were not included in these averages.
(m+ z−) indicates that the hermaphrodites assessed for progeny production were homozygous mre-11 mutants derived from self-fertilization of mre-11/+ mothers, i.e., mre-11(+) was present maternally (m+) but absent zygotically (z−).
(m− z−) indicates that the hermaphrodites assessed for progeny production were homozygous mre-11 mutants derived from homozygous mre-11 mutant mothers. mre-11(+) was present neither maternally nor zygotically.
Progressive loss of fecundity and viability sets mre-11 mutants apart from many other meiotic recombination-defective mutants that we have analyzed, most of which can be propagated indefinitely as homozygous self-fertilizing strains (Zalevsky et al. 1999; Kelly et al. 2000; K. Hillers, A. MacQueen, K. Reddy, and A.M. Villeneuve, unpubl.). This contrast was illustrated by a direct side-by-side comparison between mre-11 and msh-5(me23), a crossover-defective mutant that produces viable progeny at the same rate as m+z− mre-11 (Kelly et al. 2000). m+z− mre-11 and msh-5 hermaphrodites were plated individually, and the plates were examined 4 wk (approximately eight generations) later. Six of the nine msh-5 worms plated had produced more than 500 viable descendants, two had produced 100–200 descendants, and one had produced no viable hermaphrodite progeny in the first generation. All of the plates with viable progeny contained live animals at multiple developmental stages. In contrast, none of the eight mre-11(me41) and nine mre-11(ok179) worms had produced more than a handful of descendants, and it is unlikely that any had produced progeny beyond the second generation. Of the 17 mre-11 plates, 15 had a few decaying carcasses but no live worms, and the remaining two each contained several carcasses plus a single live worm that was well beyond reproductive age.
To explore the basis of this progressive loss of reproductive capacity, we examined DAPI-stained germ lines of m−z− mutant hermaphrodites. This analysis revealed a variety of abnormalities not seen in m+z− mre-11 mutant or wild-type hermaphrodites. Frayed aggregated chromosomes reminiscent of the chromosome morphology observed after γ-irradiation of m+z− animals were seen in 3 of 31 mre-11(ok179) diakinesis nuclei examined. Likewise, 2 of 17 mre-11(me41) diakinesis nuclei scored possessed frayed aggregated chromosomes. Several mutant worms had disorganized gonads, and in a few cases, no diakinesis-stage nuclei were evident. Furthermore, a severe deficit or absence of sperm was found in several worms that had laid very few or no embryos. In addition to examining hermaphrodites of egg-laying age, we also examined DAPI-stained young adult hermaphrodites at the time of onset of oogenesis, before sperm had been depleted. Large numbers of sperm (>75) were present in both gonad arms of 13 m+z− mre-11 mutant worms. In contrast, 6 of 28 gonad arms in 14 m−z− mre-11 mutant worms possessed <30 sperm nuclei, and an additional 2 of 28 gonads possessed between 30 and 50 sperm nuclei. Consistent with the interpretation that reduced sperm counts are at least partially responsible for reduced embryo production, introduction of sperm by mating with wild-type males substantially increases the number of embryos produced by m−z− mre-11 hermaphrodites.
We also examined several rare surviving progeny arising from m−z− mre-11(ok179) mutant parents. Four of five worms laid no eggs, and five of the eight gonad arms from these four worms had <10 sperm. Two of the 10 diakinesis nuclei examined displayed the frayed aggregated appearance of the chromosomes found in the previous m−z− generation and in irradiated mre-11 diakinesis nuclei. In these two nuclei, we also observed chromosome fragments, which constitute clear evidence of spontaneous chromosome breakage.
Discussion
Roles of mre-11 in meiotic recombination and repair
We have shown that meiotic recombination in C. elegans is profoundly impaired and perhaps eliminated in the absence of mre-11 function, based on an inability to detect chiasmata cytologically or crossover recombination events genetically. This indicates that mre-11 plays a crucial role in meiotic recombination in this metazoan organism. We have also shown that C. elegans mre-11 is required for at least one pathway for repairing chromosome damage induced by IR. C. elegans germ-line nuclei clearly have the capacity to regenerate intact chromosomes after IR exposure, as both progeny survivorship and the cytological appearance of oocyte chromosomes are largely unaffected after irradiation of wild-type germ lines. This capacity is severely compromised in mre-11 mutants, for which the same treatment abolishes progeny survivorship and results in a highly abnormal cytological appearance of oocyte chromosomes. IR-induced cytological abnormalities include chromosome fragments, which are definitive indicators of unrepaired DSBs, and large chromosome aggregates, which may be the result of a misrepair process that comes into play when the repair pathways that require mre-11 are unavailable.
The response of mre-11 mutants to germ-line γ-irradiation contrasts with those of several other meiotic recombination-defective mutants, allowing us to make additional inferences regarding the role of MRE-11 in the recombination pathway. First, we can conclude that mre-11 is not required solely for the initiation step of meiotic recombination since a γ-irradiation treatment that is capable of bypassing the requirement for the recombination-initiating enzyme SPO-11 clearly does not bypass the requirement for mre-11. Rather, the inability of mre-11 mutant germ cells to regenerate intact chromosomes after IR treatment suggests that mre-11 may be required for a process downstream of initiation that is crucial to all meiotic recombination events, both crossover and noncrossover. This role contrasts with that of msh-5, which functions to promote the crossover outcome of initiated recombination events but is apparently not required to regenerate intact chromosomes after irradiation. By analogy to the roles of Mre11p in yeast meiotic recombination, we suggest that a potential requirement for worm MRE-11 downstream of the initiation step could reflect a role in promoting 5′ to 3′ resection of DSBs to generate a substrate for a subsequent strand invasion step (Nairz and Klein 1997; Tsubouchi and Ogawa 1998; Usui et al. 1998; Moreau et al. 1999). However, it is almost certainly an oversimplification to interpret the inability to restore intact chromosomes in mre-11 mutant germ cells as a defect strictly in the meiotic recombination pathway. Although it is clear that wild-type, spo-11, and msh-5 germ cells are capable of repairing IR-induced chromosome damage, we do not know what fraction of damage is being repaired by the homologous recombination pathway that is activated in meiosis and what fraction of damage might be repaired by an alternative pathway, such as nonhomologous end-joining, that also requires mre-11.
Whereas we have excluded the possibility that C. elegans mre-11 functions solely in initiation of meiotic recombination, our data support the possibility that, like yeast MRE11, it may play dual roles in meiotic recombination (Johzuka and Ogawa 1995; Nairz and Klein 1997). That is, it may function not only in a DSB processing/repair step, but also in generating DSBs at the initiation step. Despite their lack of chiasmata, diakinesis-stage chromosomes in mre-11 mutant oocytes appear otherwise morphologically normal unless they are exposed to IR. The fact that chromosomes emerge from meiotic prophase apparently intact in unirradiated mre-11 mutants, coupled with the fact that these mutants exhibit defects in repair of IR-induced DSBs, suggests the possibility that meiotic DSBs are never made in mre-11 mutants. This conclusion must remain tentative at present as we do not know how the number and type of breaks (or other lesions) induced by γ-irradiation compare with the enzymatically generated breaks that normally initiate meiotic recombination.
G2 DNA damage checkpoint function in the absence of MRE-11
Ionizing radiation induces a dose-dependent increase in germ cell apoptosis in mre-11 mutants, indicating a functional meiotic G2 DNA damage checkpoint. Although mre-11 mutant germ lines are repair defective, they are apparently still competent to identify and eliminate (by apoptosis) at least some of the damage caused by IR. This ability to trigger the DNA damage checkpoint in the absence of mre-11 function indicates that MRE-11 cannot be an essential sensor of DNA damage.
Although the G2 DNA damage checkpoint can function to eliminate some damaged germ cells in mre-11 mutants, other germ cells apparently escape death and proceed to diakinesis with obviously abnormal chromosomes. Furthermore, if mre-11 mutants were strictly repair defective but fully checkpoint proficient, one might have anticipated seeing a higher incidence of apoptosis in the mre-11 mutants at the 5-krad dose, because an increased number of lesions capable of triggering the checkpoint presumably would not have been removed. Several possible scenarios could account for these observations. One possibility is that when the MRE-11-dependent repair pathway(s) are unavailable, alternative repair pathways might improperly repair the IR-induced chromosomal damage. This “misrepair” might succeed in eliminating checkpoint-triggering lesions without restoring morphologically normal chromosomes. A second possibility is that different types of DNA lesions might yield different levels of some DNA damage signal, and the subset of lesions normally repaired using MRE-11 may not contribute significantly to the signal triggering checkpoint-induced apoptosis.
Finally, MRE-11 might modulate the signal emanating from damaged DNA. MRE-11 could be involved in processing lesions into forms that can serve as a checkpoint-triggering signal. Evidence from other systems has suggested that ssDNA serves as a potent signal for triggering DNA damage checkpoints (Huang et al. 1996; Abramova et al. 1997; Lee et al. 1998). In S. cerevisiae, Mre11p plays a role in vivo in promoting 5′ to 3′ resection at DSB ends to reveal 3′ ss ends Nairz and Klein 1997; Tsubouchi and Ogawa 1998). Moreover, Lee et al. (1998) showed that whereas deletion of S. cerevisiae MRE11 does not impair a temporary cell cycle arrest elicited by one or two defined DSBs, it does suppress the permanent arrest elicited by two DSBs in wild type or by a single DSB in a mutant with accelerated 5′ to 3′ resection. Their data support a model in which the retardation of 5′ to 3′ resection demonstrated in the yeast mre11 mutant suppresses permanent arrest (under conditions of limited DNA damage) by reducing the amount of ssDNA available to serve as the DNA damage signal. Similarly, loss of C. elegans MRE-11 might reduce the amount of effective DNA damage signal by reducing the 5′ to 3′ resection of DNA ends, thereby dampening the checkpoint response. However, none of these possibilities detract from the primary conclusion that the G2 DNA damage checkpoint can function in the absence of MRE-11.
An essential function for mre-11 in maintaining reproductive capacity
Not only are homozygous mre-11 mutant hermaphrodites from heterozygous parents (m+z−) fully viable, but they also produce viable adult offspring (m−z−) at a frequency typical of many mutants whose sole apparent defect is a lack of crossing over during meiosis (Zalevsky et al. 1999; Kelly et al. 2000). This finding was initially surprising given that vertebrate cells lacking Mre11 or Rad50, as well as mouse embryos lacking Rad50, are inviable (Xiao and Weaver 1997; Luo et al. 1999; Yamaguchi-Iwai et al. 1999). The requirement for Mre11 even in cells that have not been exposed to any exogenous genotoxic insult has implied that Mre11 plays a role in an essential cellular process. Petrini (2000) has proposed that the essential role of the Mre11 complex might be to facilitate repair of DSBs that arise as by-products of DNA replication. On the basis of this hypothesis, it seemed reasonable to anticipate that mre-11 would be an essential gene in C. elegans as well.
Our finding that mre-11 is required for maintenance of reproductive capacity in the nematode provides a framework for reconciling what initially appeared to be a significant difference between vertebrates and worms in the physiological requirement for Mre11. We suggest that progressive loss of reproductive capacity in worm mre-11 mutants and inviability of Mre11−/− vertebrate cells may be, at least in part, different manifestations of the same underlying cellular defects. If we examine the hypothesis that lack of Mre11 impairs the ability to repair spontaneous DSBs (e.g., those that arise as collateral damage of the replication process), we find that the relative timing of onset of problems not attributable to the meiotic recombination defect in C. elegans mre-11 mutants can be considered quite comparable to the timing of appearance of spontaneous chromosome breaks and loss of viability in chicken DT40 cells after depletion of GdMre11 (Yamaguchi-Iwai et al. 1999). DT40 cells undergo ∼9 to 10 cell generations in the absence of Mre11 protein before they cease cycling and start to die, at which time 0.5 chromosome breaks/cell can be detected cytologically. Moreover, DT40 cells are hyperrecombinogenic (Buerstedde and Takeda 1991) and may well have an elevated frequency of spontaneous breaks compared to other cell types. C. elegans has only roughly 10 cell generations in the soma and 10 in the germ line during its entire lifetime, and its genome is one-tenth the size of the chick genome. Given these parameters and the fact that worm embryos apparently receive a maternal endowment of mre-11 mRNA (Reinke et al. 2000), it is not surprising that the (nonmeiotic) lethal consequences of mre-11 loss might first become apparent in the germ lines of m−z− hermaphrodites or in their progeny.
If the reduction in fecundity and progeny viability in m−z− animals is a consequence of persistence or misrepair of spontaneous DSBs, one might have anticipated seeing more cytological evidence of broken chromosomes in their germ lines. However, not all DSBs would be expected to be visible as chromosome breaks. Sister cohesion and other features of higher order chromosome structure may serve to hold together chromosomes with broken DNA molecules, masking their presence. Furthermore, germ-line nuclei that suffered replication-induced breaks during either mitotic or premeiotic S-phase may not enter the meiotic program and, thus, would not be detected by our assays. In addition, nuclei with certain overt types of breaks may be preferentially eliminated by apoptosis, whereas misrepaired chromosomes might persist and confer embryonic lethality. Finally, some lethal lesions may arise later, during subsequent embryonic cell cycles.
We have just made a case that the same underlying DNA metabolism defects that lead to lethality in Mre11−/− vertebrate cells likely contribute to the progressive loss of reproductive capacity in C. elegans mre-11 mutants. However, the reduction in number of embryos produced by m−z− hermaphrodites can be attributed, at least in part, to reduced sperm counts. At first glance, diminished sperm production is not readily explained as a readout of an underlying defect in DNA metabolism. However, recent findings show that regulation of germ cell proliferation and regulation of the sperm/oocyte switch are intimately related (e.g., Puoti and Kimble 1999). Thus, it is plausible that a defect in DNA metabolism could affect the relative timing of germ cell proliferation and that this, in turn, could alter the timing of the sperm/oocyte switch, ultimately resulting in both a reduction and a high variance in the number of zygotes produced.
Because yeast MRE11 has been implicated in telomere length maintenance (Kironmai and Muniyappa 1997; Boulton and Jackson 1998; Nugent et al. 1998), we also considered the possibility that the progressive loss of reproductive capacity of mre-11 mutants might result from telomere loss. Analysis of mrt-2 mutants demonstrated that telomere loss leads to germ line mortality in C. elegans (Ahmed and Hodgkin 2000). However, the rate of telomere shortening in mrt-2 strains is only ∼12 bp/cell division, and it takes at least 8 and usually many more worm generations after homozygosity for mrt-2 to reach functional infertility. Given that mre-11 mutants experience a comparable drop in reproductive capacity within a single worm generation after they become homozygous, we do not favor telomere loss as the major cause of the strain's demise.
C. elegans as a system for investigating function of essential repair proteins
Several DNA repair functions have been identified through research using radiation-sensitive mutant cell lines derived from either human patients or hamster CHO cells (Thompson 1996). However, studies of this type have systematically failed to identify several proteins discovered in fungal systems to be central to double-strand break repair pathways (including Rad51, Rad50, and Mre11), as vertebrate cells that lack these proteins are inviable (Xiao and Weaver 1997; Sonoda et al. 1998; Luo et al. 1999; Yamaguchi-Iwai et al. 1999). The short-term viability of C. elegans mutants that lack MRE-11 has allowed us to investigate the in vivo biological roles of this essential repair protein in a metazoan experimental system. Our preliminary analysis indicates that loss of C. elegans rad-50 function has phenotypic consequences similar to those resulting from loss of mre-11 (K. Hillers and A.M. Villeneuve, unpubl.). This suggests that short-term viability may be a general characteristic of C. elegans mutants that lack essential repair functions and that the C. elegans system may provide a unique opportunity to examine their in vivo roles in metazoan biology at the organismal level. Moreover, it implies that genetic screens for C. elegans mutants defective in meiotic recombination may have the capacity to identify genes encoding previously unknown essential DNA repair proteins.
Materials and methods
Genetics
The following mutations and chromosome rearrangements (strain background Bristol N2) were used (Riddle et al. 1997): LGV: mre-11(me41), mre-11(ok179), nT1 [unc-?(n754) let-?(m435)](IV,V), dpy-11(e224) unc-42(e270); LGX: dpy-3(e27) unc-3(e151). mre-11 alleles were maintained in a heterozygous state using the balancer nT1; heterozygous (Unc) worms were picked periodically to maintain the strain, and mre-11 homozygotes were identified based on absence of the nT1 Unc marker.
me41 was mapped to the left of bP1 (V, 3.15) using the RW7000 strain and STS markers therein as in Williams et al. (1992). me41 was localized very near or to the right of unc-42 (V, 2.20) by a three-factor cross: 20 of 20 Dpy non-Unc and 0 of 10 Unc non-Dpy progeny from dpy-11 unc-42/me41 heterozygotes carried the me41 allele.
Recombination frequencies between dpy-3 and unc-3 were measured as in Kelly et al. (2000).
cDNA analysis
Complete sequences were determined for cDNA clones yk133b9, yk323f11, and yk422g7. The longest clone, yk133b9, begins at coordinate 99 of the Genefinder predicted coding sequence for ZC302.1. We performed 5′ RACE (GIBCO BRL) using gene-specific primer ACTCCTGAATGTTGCTGTGC to prime first strand cDNA synthesis, and the primer CUACUACU ACUAGGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG in conjuction with the nested primer CCGCAATGAATAT CAGTGGC to amplify the cDNA. In the resulting cDNA, the SL1 transplice leader immediately preceded the second predicted exon (beginning at coordinate 129 of the predicted ZC302.1 coding sequence).
Detection of achiasmate chromosomes in oocyte nuclei
Oocyte chromosomes were fixed with Carnoy's fixative and stained with DAPI as in Villeneuve (1994). Because individual univalents or bivalents in some nuclei lie too close to each other to be resolved unambiguously, this method underestimates the frequency of achiasmate chromosomes.
Imaging of meiotic chromosome morphology
Preparation of gonads for cytological analysis was carried out as in Dernburg et al. (1998), with modifications. Dissected gonads were fixed by addition of an equal volume of 7.4% formaldehyde in 1× egg buffer. Tissue was sandwiched between a positively charged glass slide and a siliconized coverslip. The slide was then frozen in liquid N2, the coverslip was quickly removed with a razor, and the sample was transferred to 95% ethanol at −20°C. Slides were postfixed for 10 min at room temperature in M9 containing 3.7% formaldehyde, stained with 0.5 μg/mL DAPI in M9, washed 5× for 5 min in M9 and once for 5 min in 1 M Tris-HCl at pH 8 and mounted for microscopy in 90% glycerol containing 3.6% N-propylgallate, buffered to pH 8 with Tris base. Imaging of DAPI-stained chromosomes was performed using a DeltaVision deconvolution microscopy system as described in Dernburg et al. (1998).
In situ hybridization
FISH was performed using a probe targeting the 5S rDNA locus (chromosome V) and probes derived from YAC clones Y13H5 (left end of I) and Y51E2 (left end of X). Probes were gifts from A. MacQueen and M. Colaiacovo (Stanford University School of Medicine, CA) and were prepared as described in Dernburg et al. (1998) and Zalevsky et al. (1999). Hybridization conditions were as described by Dernburg and Sedat (1998), with modifications. Dissected gonads prepared as above were rehydrated through a series of washes: 3× in 2× SSCT for 2 min, 1× in 25% formamide/2× SSCT for 1 min, 2× in 50% formamide/2× SSCT for 1 min (the second at 37°C), and 1× in 50% formamide/2× SSCT for 3 h. Probes were applied to the sample in hybridization solution, and slides were heated on a 95°C heat block for 2.5 min. After overnight incubation at 37°C, samples were washed 3× in 50% formamide/2× SSCT at 37°C for 10 min, 1× in 25% formamide/2× SSCT at room temperature, and 4× in SSCT for 3 min. Slides were blocked with 1% BSA for 1 h before anti-digoxigenin antibody (Boehringer Mannheim) was applied at 1:100 dilution for 1 h at room temperature and overnight at 4°C. Slides were stained with DAPI as above, rinsed five times in 2× SSCT for 5 min and once in 1 M Tris-HCl at pH 8 for 5 min and mounted as described above.
Acknowledgments
We thank A. Gartner for training in scoring germ-line apoptosis, A. Dernburg and A. MacQueen for assistance with cytology and imaging, and members of the Villeneuve laboratory for helpful discussions and critical reading of the manuscript. We thank the C. elegans Deletion Consortium, the Caenorhabditis Genetics Center, and the National Institute of Genetics (Japan) for sending strains and clones. This work was supported by grants from the National Institutes of Health (GM-53804) and the Searle Scholars Program to A.M.V., and a Junior Faculty Scholar Award from the Howard Hughes Medical Institute to A.M.V.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL villen@cmgm.stanford.edu; FAX (650) 725-7739.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.864101.
References
- Abramova NA, Russell J, Botchan M, Li R. Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc Natl Acad Sci. 1997;94:7186–7191. doi: 10.1073/pnas.94.14.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Baxter BK, Petrini JH. The Mre11–Rad50–Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, III, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW. Mapping three-dimensional chromosome architecture in situ. Methods Cell Biol. 1998;53:187–233. doi: 10.1016/s0091-679x(08)60880-8. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- Haber JE. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- ————— Partners and pathways: Repairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- Hawley RS. Exchange and chromosome segregation in eukaryotes. In: Kucherlapati R, Smith GR, editors. Genetic recombination. Washington, DC.: American Society for Microbiology; 1988. pp. 497–528. [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LC, Clarkin KC, Wahl GM. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lewis LK, Westmoreland JW, Resnick MA. Repair of endonuclease-induced double-strand breaks in Saccharomyces cerevisiae: Essential role for genes associated with nonhomologous end-joining. Genetics. 1999;152:1513–1529. doi: 10.1093/genetics/152.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DP, Orr-Weaver TL. Chromosome segregation during meiosis: Building an unambivalent bivalent. Curr Top Dev Biol. 1998;37:263–299. doi: 10.1016/s0070-2153(08)60177-5. [DOI] [PubMed] [Google Scholar]
- Moore PD, Simon JR, Wallace LJ, Chow TY. In-vitro recombination in rad and rnc mutants of Saccharomyces cerevisiae. Curr Genet. 1993;23:1–8. doi: 10.1007/BF00336741. [DOI] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz K, Klein F. mre11S—a yeast mutation that blocks double-strand break processing and permits nonhomologous synapsis in meiosis. Genes & Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- ————— Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes & Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH. The Mre11 complex and ATM: Collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Puoti A, Kimble J. The Caenorhabditis elegans sex determination gene mog-1 encodes a member of the DEAH-Box protein family. Mol Cell Biol. 1999;19:2189–2197. doi: 10.1128/mcb.19.3.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: Related disorders but genes apart. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Thompson LH. Identifying genes and proteins involved in human DNA repair processes using somatic cell and molecular genetics. Prog Clin Biol Res. 1996;395:175–199. [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Villeneuve AM. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics. 1994;136:887–902. doi: 10.1093/genetics/136.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Weinert T. DNA damage and checkpoint pathways: Molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]