Abstract
Endogenous retroviral sequences are present in high copy numbers in the genomes of all species and may be expressed as RNAs; however, the majority are defective for virus production. Although virus has been isolated from various Old World monkey and New World monkey species, there has been no report of endogenous retroviruses produced from African green monkey (AGM) tissues or cell lines. We have recently developed a stepwise approach for evaluating the presence of latent viruses by chemical induction (Khan et al., Biologicals 37:196–201, 2009). Based upon this strategy, optimum conditions were determined for investigating the presence of inducible, endogenous retroviruses in the AGM-derived Vero cell line. Low-level reverse transcriptase activity was produced with 5-azacytidine (AzaC) and with 5′-iodo-2′-deoxyuridine (IUdR); none was detected with sodium butyrate. Nucleotide sequence analysis of PCR-amplified fragments from the gag, pol, and env regions of RNAs, prepared from ultracentrifuged pellets of filtered supernatants, indicated that endogenous retrovirus particles related to simian endogenous type D betaretrovirus (SERV) sequences and baboon endogenous virus type C gammaretrovirus (BaEV) sequences were induced by AzaC, whereas SERV sequences were also induced by IUdR. Additionally, sequence heterogeneity was seen in the RNAs of SERV- and BaEV-related particles. Infectivity analysis of drug-treated AGM Vero cells showed no virus replication in cell lines known to be susceptible to type D simian retroviruses (SRVs) and to BaEV. The results indicated that multiple, inducible endogenous retrovirus loci are present in the AGM genome that can encode noninfectious, viruslike particles.
INTRODUCTION
Endogenous retroviral sequences are stably integrated, genetically inherited, and present in multiple copies in the genomes of all species. The majority of these sequences are defective; however, some may produce infectious retroviruses (9, 11, 16, 71, 78). In general, newly acquired endogenous retroviral sequences are more likely to be associated with an infectious virus, whereas ancient sequences may be transcriptionally active but defective for virus production (86) or produce noninfectious particles (38). In rodents, endogenous retroviruses can become activated in animals, as a consequence of age (4), or in cell lines, either spontaneously by long-term culture passage (2, 49, 63) or by treatment with a variety of inducers, including biological, immunological, and chemical agents (1, 13, 16, 25, 39, 47, 50). In humans or nonhuman primates (NHPs), spontaneous release of endogenous retroviruses has been reported from tumor tissues and cell lines, as well as from normal placenta (9, 12, 14, 15, 19, 20, 27, 28, 34, 43, 44, 51, 52, 58, 62, 64–66, 69, 72, 73). Endogenous retroviruses have also been isolated from NHP cells by extended cultivation (6 to 8 months) of normal primary cell cultures and cell lines (67, 75, 76, 79). The number of endogenous primate retroviruses isolated thus far is limited, and the virus isolates or the tissues from which they were recovered are not readily available for characterization by current state-of-the-art methods or for the development of reagents for further investigations.
Endogenous retroviruses have been reported from a variety of NHP species, including Old World primates and New World primates, but there has been no evidence of endogenous retroviral particles produced from African green monkey (AGM) tissues or from cell lines derived from this species. The Vero cell line, derived from the kidney of a normal, adult AGM (Chlorocebus species, formerly called Cercopithecus aethiops) (87), is used broadly in research and virus diagnostics as well as in vaccine development due to its broad susceptibility to infection by different viruses. This cell line has been shown to be nontumorigenic at low passage levels (7, 18, 22, 32, 48, 54, 74, 84) and negative for viruses by extensive testing using a variety of assays, including PCR assays and standard chemical induction (29, 74). We have recently developed a stepwise strategy for using chemical inducers to optimize induction conditions for investigating the presence of latent viruses (37). We have used this strategy to evaluate activation of endogenous retroviral sequences in Vero cells, which, although known to contain numerous copies of endogenous retroviral sequences due to their AGM species of origin (5, 8, 10, 26, 31, 36, 42, 57, 68, 81), were not expected to contain an inducible virus, based upon the extensive testing history and broad use of the cell line (29, 74). Here, we report that treatment of Vero cells with 5-azacytidine (AzaC) and with 5′-iodo-2′-deoxyuridine (IUdR) induced endogenous retroviral particles related to ancient simian endogenous type D betaretrovirus (SERV) sequences that are present in all Old World monkeys (83) and distinct from pathogenic, type D simian retroviruses (SRVs) (46). Additionally, particles containing baboon endogenous virus (BaEV)-related type C gammaretrovirus sequences were also induced from AzaC-treated Vero cells. Infectivity analysis of drug-treated Vero cells indicated the absence of a replicating virus using various target cells known to be susceptible to SRVs and BaEV. The results demonstrate the use of optimized chemical induction conditions for investigating infectious, endogenous retrovirus loci in the genomes of primates and other species.
MATERIALS AND METHODS
Cell lines and chemicals.
Cell lines were obtained from American Type Culture Collection (ATCC; Manassas, VA; http://www.atcc.org/). The Vero cell line (AGM kidney cells; ATCC catalog no. CCL-81, lot no. 3645301; passage 120) was grown in Eagle minimum essential medium (EMEM) (modified), with Earle's salts and without l-glutamine (Mediatech, Manassas, VA; catalog no. 15-010-CV), supplemented with 5% fetal bovine serum (FBS), per ATCC instructions (heat inactivated at 56°C for 30 min; HyClone, Logan, UT; catalog no. SH30071.03), 2 mM l-glutamine, 250 U of penicillin per ml, 250 μg of streptomycin per ml, 1× nonessential amino acids (MEM-NEAA 100×; Quality Biological Inc., Gaithersburg, MD, catalog no. 116-078-061), and 1 mM sodium pyruvate (Quality Biological, Inc.), designated complete medium. To maximize reproducibility of results in the induction assays, a cell bank was established at passage 123. A new vial was used in each experiment to maintain similar passage numbers in the induction studies.
For virus-induction studies, Vero cells were treated with different concentrations of IUdR (stock solution, 75 mg per ml in 1 N NH4OH; Sigma, St. Louis, MO; catalog no. 17125), AzaC (1 mg/ml in complete Vero cell medium; Sigma catalog no. A1287), and sodium butyrate (NaBut; 0.9 M in sterile H2O; Sigma catalog no. B5887).
Target cell lines used in infectivity studies were A-204 (human rhabdomyosarcoma; ATCC catalog no. HTB-82), Raji (human B cell lymphoma; ATCC catalog no. CCL-86), and Cf2Th (dog thymus; ATCC catalog no. CRL-1430). Cf2Th cells were grown in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA; catalog no. 119955) supplemented with 20% FBS; A-204 and Raji cells were grown in RPMI 1640 (Quality Biological catalog no. 112-024-101) supplemented with 10% FBS and 1× MEM-NEAA. Additionally, both media contained 2 mM l-glutamine, 250 U of penicillin per ml, and 250 μg of streptomycin per ml.
Growth curve and population doubling time (PDT).
Cells were counted using an automated Guava PCA flow cytometer according to the manufacturer's protocol (Guava ViaCount assay; Hayward, CA). Cells were diluted, and the numbers of viable cells, dead cells, and apoptotic cells were counted in triplicate. The average count of the viable cell numbers was used in the experiments. For cell cycle analysis, 0.5 × 106 cells were processed and stained according to the manufacturer's protocol (Guava cell cycle assay).
To determine the optimum number of cells for obtaining a sigmoidal growth curve, Vero cells (0.5 × 106, 0.75 × 106, and 1.0 × 106) were planted in 5 ml complete medium in 25-cm2 flasks, and viable cells were counted at various times using a Guava PCA cytometer. Cell cycle analysis was done to determine the cell phases. PDT was calculated as 1/k = T (where T = PDT) from the linear curve in the log phase from the formula N = N02kt (where N = total viable cell number at end time t; N0 = total viable cell number at initial time t0, t = hours from N0 to N, and k = regression constant) (60). Results were confirmed in three independent assays.
Drug dose evaluation.
Vero cells (1 × 106; passages 125 to 131) were planted for 16 h in 25-cm2 flasks before replacing medium with fresh medium containing different concentrations of AzaC (0.3125 to 40 μg/ml), IUdR (50 to 3,200 μg/ml), or NaBut (1 to 6 mM). After 48 h of drug treatment, cells were washed with medium three times (designated day 0), trypsinized, and counted by using a Guava PCA cytometer. Another set of flasks were further incubated after the medium change, and the cells were trypsinized and counted at day 3. Untreated cells, at confluence, were used as a control to evaluate cell toxicity and cell recovery based upon the cell-confluence ratio at day 0 and at day 3, which was calculated by dividing the number of viable cells in the drug-treated flask by the number of viable cells at confluence in the untreated control flask (2.3 × 106 cells) and expressed as a percentage. Furthermore, in the case of IUdR-treated cells, additional controls were included to evaluate toxicity due to NH4OH used for dissolving the drug. In case the number of cells in the NH4OH-treated flask was less than that of the untreated control flask, the number of viable cells in the IUdR-treated flask was divided by the number of viable cells in the NH4OH-treated flask before determining the cell-confluence ratio.
Chemical treatment and evaluation for induced retroviruses by the STF-PERT assay.
Vero cells were drug treated under optimized induction conditions: cells (1 × 106) were planted for 16 h and then treated with drug for 48 h (AzaC, 1.25 μg/ml; IUdR, 200 μg/ml; and NaBut, 3 mM); untreated cells were included as a control. For kinetics of virus induction, medium was replaced daily and filtered supernatant collected for detection of reverse transcriptase (RT) activity by the single-tube fluorescent PCR-enhanced reverse transcriptase (STF-PERT) assay (53). Supernatants were collected and filtered (Costar Spin-X centrifuge tube filters, 0.45-μm-pore-size CA membrane; Corning, Corning, NY, catalog no. 8162) on the day of drug removal (day 0), prior to washing the cells, and then daily, at each medium change. Filtered supernatants were stored at −80°C in single-use, 10-μl aliquots for STF-PERT analysis and in 0.5-ml aliquots for additional use. Each sample was tested at a 1:10 dilution (per the assay protocol), and results were obtained from triplicate samples. The PERT assays for testing supernatant from drug-treated cells met the acceptability criteria (53): IUdR, slope = −3.96, y intercept = 48.06, r2 = 0.999; AzaC, slope = −3.14, y intercept = 42.59, r2 = 0.996; NaBut, slope = −3.97, y intercept = 48.05, r2 = 0.996. Negative controls were cells without drug (or with NH4OH in the case of IUdR cell toxicity studies) and were set up in parallel.
RT-PCR.
Total cellular RNAs were extracted by using the RNeasy Plus minikit (Qiagen, Valencia, CA; catalog no. 74134) in combination with the RNase-free DNase set (Qiagen; catalog no. 79254) according to the manufacturer's instructions. Concentration and purity were determined by using UV absorbance.
A low-concentrated (10×) supernatant sample was prepared from normal and drug-treated cells by ultracentrifugation of filtered supernatant (1.5 ml) at 45,000 rpm (Beckman TLA 45 rotor) for 90 min at 4°C. RNA was prepared from the pellet by resuspending it in 130 μl of Promega DNase buffer and adding 10 μl DNase (1 U per μl; RNase-free DNase; Promega, Madison, WI, catalog no. M6101) for incubation at 37°C for 30 min. RNA was extracted from the entire sample by using the QIAamp viral RNA minikit (Qiagen catalog no. 52904).
A high-concentrated (1,000×) supernatant sample was prepared from normal and from AzaC-treated Vero cells (1.25 μg/ml for 48 h) on day 4 after drug treatment (medium was changed on day 1) by ultracentrifugation of pooled (180 ml), filtered supernatant (tube top vacuum filters, 0.45-μm-pore-size CA membrane, Corning catalog no 430314) on a 20% sucrose cushion (25,000 rpm in a Beckman SW-28 rotor for 4 h at 4°C). Pellets were pooled, resuspended in 4 ml phosphate-buffered saline (PBS) (pH 7.4), and ultracentrifuged immediately at 35,000 rpm (Beckman SW-41 rotor) for 90 min at 4°C. The pellet was resuspended in 180 μl in PBS (pH 7.4) and stored in aliquots at −80°C to minimize freeze-thaw of test samples. RNA was extracted from 50 μl using the QIAamp viral RNA minikit after DNase I digestion (1 U per μl), as described above.
One-half of the RNA sample was used for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA; catalog no. 170-8890) according to the manufacturer's instructions. The other half of the RNA was used for control without RT. Additionally, PCR amplification using human β-actin primers was performed to demonstrate absence of cellular DNA according to the manufacturer's protocol (Clontech, Mountain View, CA; catalog no. 639008).
Consensus PCR primers (SRV/SERV) were designed based upon GenBank sequences of SRV-1 type D retrovirus (M11841), SRV-2 complete genome (AF126467), simian Mason-Pfizer D-type retrovirus or SRV-3 (M12349), and simian type D virus 1, complete proviral genome (U85505; designated SERVbab in this paper). The location of the SRV/SERV primers is given in Table 1: a long terminal repeat (LTR) gag fragment (553 bp) was amplified using forward primer F04, 5′-CTGTCTTGTCTCCATTTCT-3′, and reverse primer R10, 5′-ACSGCAGCCATKACTTGYGG-3′; a pol fragment (610 bp) was amplified using forward primer F41, 5′-TACAAGAYCCMTAYACCTA-3′, and reverse primer R46, 5′-TTDGGTGGRTAATGGTTRTC-3′; and an env fragment (548 bp) was amplified using forward primer F65, 5′-CAYATNTCYGATGGAGGAGG-3′, and reverse primer R70, 5′-CCYGTCCARTTTGTRGGTA-3′. PCR conditions were 95°C for 3 min, followed by 35 amplification cycles of 95°C for 30 s, 56°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min.
Table 1.
Comparative nucleotide sequence analysis of cloned SERVagm-Vero DNAa
| Cloned DNA regionb | Position (nt)c | Nucleotide sequence identity (%)d |
|||
|---|---|---|---|---|---|
| SRV-1 | SRV-2 | MPMV | SERVbab | ||
| LTR gag (4) | 395–947 | 77–78 | 81 | 78–79 | 95–96 |
| pol (6) | 4047–4656 | 74–75 | 76 | 73 | 95–96 |
| env (4) | 6501–7048 | 60–61 | 64–65 | 61–62 | 94–95 |
DNA was obtained by RT-PCR from 1,000× supernatant of AzaC-treated Vero cells.
Regions of PCR amplification; number of cloned DNAs analyzed are indicated in parentheses.
Nucleotide positions according to the published SERV sequence of van der Kuyl et al. (83).
Range of values indicate results obtained from analysis of individual clones. SRV-1, accession no. M11841, simian SRV-1 type D retrovirus (L47.1) (61); SRV-2, accession no. M16605, simian retrovirus 2 (55); MPMV, accession no. M12349, simian Mason-Pfizer D-type retrovirus (MPMV/6A) (70); and SERVbab, accession no. U85505, simian type D virus 1 (83).
Primers for amplification of BaEV sequences and PCR cycle conditions were as described previously (82): RT1 and RT2 in the pol region, and ENV1/ENV4 with nested primers ENV2/ENV3 in the env region. Additional primers were made for PCR amplification in the gag region: outer primer pairs were GAG1 (5′-GAGTGGCCCACCCTTCATGT-3′) and GAG2 (5′-CAGTACTGGATCGTGCGGTT-3′), at nucleotide positions 1108 to 1127 and 1697 to 1678, respectively, and inner primer pairs were GAG3 (5′-CCCCGGGACGGAACTTTTGA-3′) and GAG4 (5′-GATGAGGTAGAGGGTCTTGGAAG-3′) at nucleotide positions 1135 to 1154 and 1420 to 1398, respectively. The nucleotide positions are based upon the sequence of the BaEV by Kato et al. (35).
PCRs were done in 25 μl using 2 μl cDNA template, 10× PCR buffer containing 15 mM MgCl2, and 1.5 U Taq DNA polymerase (Roche Molecular Biochemicals, Indianapolis, IN; catalog no. 11647687001). The final concentration of deoxynucleotide triphosphates were 200 μM each, and primers were 1 μM each.
Nucleotide sequence analysis.
PCR-amplified DNA fragments were isolated from agarose gels by using the Zymoclean gel DNA recovery kit (Zymo Research Corporation, Orange, CA; catalog no. D4001) and cloned into the pGEM-T Easy vector (Promega catalog no. A1360). Nucleotide sequences were determined with T7 and SP6 primers by using an ABI 3130xl genetic analyzer according to the manufacturer's standard protocol (Applied Biosystems, Foster City, CA). Sequence analysis and alignment of the sequences were done using nucleotide BLAST (National Center for Biotechnology Information, National Library of Medicine, NIH, Bethesda, MD).
TEM.
Supernatant (160 ml) was collected from Vero cells without drug treatment and from Vero cells on day 4 after drug treatment for 48 h with IUdR (200 μg/ml) and AzaC (1.25 μg/ml) for 48 h. Ultracentrifuged pellets were obtained through a 20% sucrose cushion by using the procedure described above for preparation of the 1,000× supernatant sample. The pellets were fixed overnight in McDowell and Trumps fixative and sent to Charles River Pathology Associates (Durham, NC) for evaluation of viruslike particles. The volume of the pellet was measured by comparison to known standards and processed for transmission electron microscopy (TEM) analysis. Thin sections were cut, stained with methanolic uranyl acetate and Reynolds lead citrate, and examined by TEM. Ten grid spaces were examined and evaluated for numbers of particles with retrovirus-like morphology. The number of viruslike particles in the entire pellet was calculated by multiplying the number of particles tabulated in the examined section by the number of potential sections in the pellet (calculated by dividing the volume of the entire pellet by the volume of the section examined). The limit of sensitivity of the assay was calculated as the smallest detectable amount of virus in the samples or one particle in the section examined by TEM. Thus, to obtain the limit of sensitivity, the number of potential sections in the pellet is multiplied by 1.
Cell pellets (2 × 106 to 4 × 106) for TEM analysis were prepared by trypsinizing cells from normal or AzaC-treated Vero cells, as previously described (41).
Infectivity analysis.
Combined infectivity and coculture studies were set up with cells and supernatant from drug-treated cells that were prepared by planting Vero cells (1 × 106; passage 131) in 25-cm2 flasks for 16 h and then treating the cells with drug for 48 h (1.25 μg/ml AzaC or 200 μg/ml IUdR). Cells were washed three times with plain medium to remove the drug, and fresh complete medium was added (day 0). Medium was replaced the next day (day 1); on day 4, unfiltered supernatant and cells from eight 25-cm2 flasks were pooled and used for infection/coculture with target cells at predetermined cell ratios for equivalent growth of test and target cells. Target control cells were set up without coculture, and control cocultures were set up with target cells and uninduced Vero cells.
In the case of the A-204 and Cf2Th adherent target cells, 2.7 × 106 and 1.5 × 106 cells, respectively, were set up in 10 ml medium for preincubation with 5 ml unfiltered supernatant from AzaC-treated and from IUdR-treated Vero cells (passage 131) at 37°C for 45 min in 75-cm2 flasks, after which trypsinized, AzaC- or IUdR-treated Vero cells (about 3.0 × 106 cells per flask) were added into the corresponding flasks. The coculture ratio of Vero cells to target cells was 1:1 for A-204 cells and 2:1 for Cf2Th cells. In the case of the Raji suspension target cells, 8 × 106 cells in 10 ml per flask were incubated at 37°C for 45 min with unfiltered supernatant from AzaC- or IUdR-treated Vero cells (5 ml), and then all of the cells and supernatant (total volume of 15 ml) were added by replacing the medium in flasks containing drug-treated Vero cells (3 × 106), which had been preincubated for 45 min in 75-cm2 flasks. The target cells were demonstrated to be susceptible to SRV (A-204 and Raji) and to squirrel monkey retrovirus (SMRV) (Cf2Th). Medium was replaced with 13 ml of target cell medium following overnight incubation. In the case of Raji cells, the supernatant was collected and spun at 1,200 rpm (GS-6KR centrifuge with a GH-3.8 rotor; Beckman Instruments, Columbia, MD) for 10 min at 4°C, and the Raji cells were resuspended in 10 ml medium and then added back to the flask containing Vero cells in 10 ml fresh medium. Upon reaching 95% confluence, all of the cells were passaged the next day (day 2) into 162-cm2 flasks. Cultures were propagated in 162-cm2 flasks with passage every 2 to 3 days until termination on day 32.
Extended cell culture was done on AzaC-induced Vero cells (passage 135; 1.0 × 106 cells in 25-cm2 flasks; 1.25 μg/ml in 5 ml). For extended culture, cells were washed 48 h after drug treatment (day 0, the day of drug removal) and then cultured in fresh complete medium. Upon reaching confluence, cells were passaged into 75-cm2 and then into 162-cm2 flasks at the same time as the cocultures and continued in 162-cm2 flasks until termination on day 34. Uninduced Vero cells were included as a control.
The cultures were regularly monitored for cytopathic effect (CPE). Filtered supernatants were collected starting at the first passage at day 4 after coculture or after drug treatment, until termination, and stored at −80°C for the STF-PERT assay.
RESULTS
Determination of cell growth characteristics.
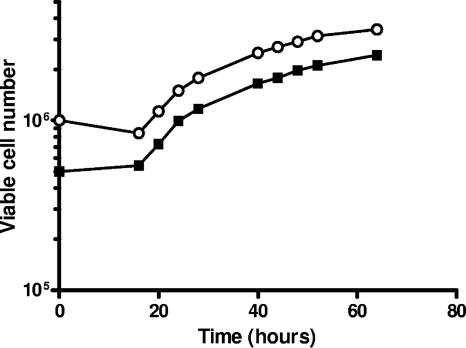
Growth curves for Vero cells were determined using different cell concentrations (0.5 × 106 and 1.0 × 106 cells) (Fig. 1). Similar kinetics were seen in both cases (as well as with 0.75 × 106 cells [data not shown]) with the log phase starting at 16 h after seeding the cells. PDT was determined from 20 h to 40 h in the log phase of the growth curve and calculated as described in Materials and Methods to be 16.9 h and 17.5 h for starting cell concentrations of 0.5 × 106 and 1.0 × 106 cells, respectively. This indicated that any of the tested cell concentrations could be used to obtain a sigmoidal growth curve for determining the beginning and end of the log phase for induction studies. An independent experiment was set up to evaluate the growth curve with cell cycle analysis. The results indicated a high percentage of cells in S phase (28.8%) at 16 h, which corresponded to the beginning of the log phase in the growth curve (data not shown).
Fig. 1.
Vero cell growth curves. Cells were planted in 25-cm2 flasks, and the total numbers of viable cells were counted at various times after planting: 16 h, 20 h, 24 h, 28 h, 40 h, 44 h, 48 h, 52 h, and 64 h after planting. The initial numbers of cells planted are indicated at 0 h: ■, 0.5 × 106 cells; ○, 1 × 106 cells.
Optimization of induction conditions.
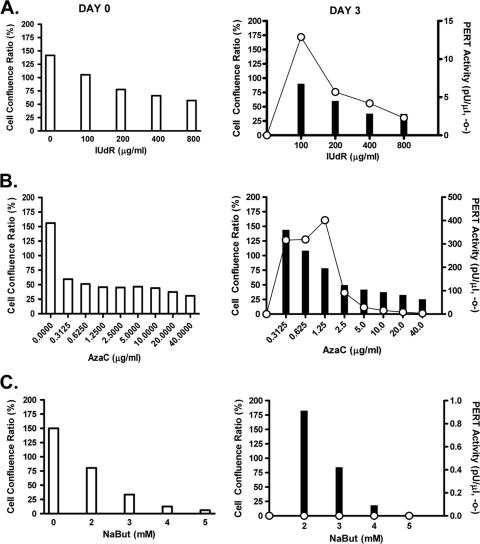
The drug dose range was determined by evaluating cell toxicity and cell recovery based upon the highest drug concentrations that resulted in good culture recovery at 3 to 4 days after drug removal, since the cells without drug treatment reached confluence in 2 to 3 days (37). Cells were planted for 16 h before adding drug, since this corresponded to the beginning of the log phase and a high percentage of cells in S phase. Cells were treated for 48 h (equivalent to 2.7 PDTs [37]) with different drug concentrations (IUdR, 50 to 3,200 μg/ml; AzaC, 0.3125 to 40 μg/ml; and NaBut, 1 to 6 mM). Cell viability was determined on day 0 and on day 3, and the cell-confluence ratio was calculated. Comparison of the results at day 0 and day 3 indicated the drug concentrations (dose range) at which the cells could recover from toxicity (Fig. 2).
Fig. 2.
Drug dose evaluation and PERT activity. Drug dose range for IUdR (A), AzaC (B), and NaBut (C) was determined by evaluating the Vero cell viability after drug treatment with various concentrations for 48 h. Cell toxicity was determined at day 0 (day of drug removal) (open bars), and cell recovery was determined on day 3 (closed bars) and expressed as the percent cell confluence ratio (percentage of the ratio of the total number of viable cells in the drug-treated flask and the total number of viable cells in a confluent untreated 25-cm2 flask). Cells (1 × 106) were planted for 16 h before the drug was added: IUdR, 100 to 800 μg/ml (shown) (50 to 3,000 μg/ml tested); AzaC, 0 to 40 μg/ml; and NaBut, 0 to 5 mM (6 mM, not shown). High cell toxicity with the 800-μg/ml dose of IUdR was due to 0.01 N NH4OH present in the drug. Filtered supernatant from day 3 was analyzed by STF-PERT assay, and results are indicated (○). No RT activity was detected in untreated cells.
To determine the relationship between RT activity and drug dose, filtered supernatants were collected at day 3 from drug-treated cells and analyzed in the STF-PERT assay: increased RT activity corresponded to samples with >50% cell recovery from drug toxicity (Fig. 2). Differences in cell toxicity were seen with different drugs: cells were fairly resistant to IUdR and relatively sensitive to AzaC and NaBut. This was in contrast to the results obtained in previous induction studies with K-BALB mouse cells, where cells were sensitive to IUdR and more resistant to AzaC. In those studies, 30 μg/ml IUdR and 2 μg/ml AzaC (39, 40) or 5 μg/ml AzaC (37; Y. Ma and A. S. Khan, unpublished data) were used with 24 h of drug treatment for virus induction.
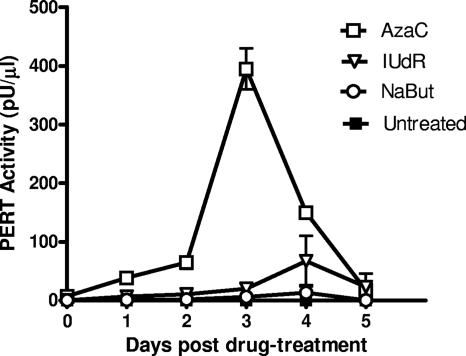
To evaluate the kinetics of RT production with drug treatment, cell-free supernatant was collected daily and tested by STF-PERT assay. Medium for testing was collected from Vero cells treated with different drug concentrations for 48 h, from day 0 (the day the drug was removed) up to day 5, with daily medium change. The results of the highest RT activities obtained with each drug are shown in Fig. 3. A low-level PERT activity was induced with 1.25 μg/ml AzaC, which peaked at day 3, whereas the peak PERT activity induced with 200 μg/ml IUdR was seen at day 4 (Fig. 3). It was noted that a comparable peak of RT activity was seen with 100 μg/ml IUdR at day 3 (Fig. 2). The RT activity with IUdR was lower than that with AzaC at all tested drug concentrations (data not shown). No reproducible PERT activity (>10 pU per μl) was seen in the case of NaBut at any drug concentration (1 to 6 mM) at 24-h or 48-h drug exposure times (Fig. 2C and 3 and data not shown). Additionally, in a separate experiment, if cells were treated with different drug concentrations for only 24 h (equivalent to about 1.5 PDT), the PERT activity was lower and seen only at higher drug concentrations than that of cells with 48 h of drug treatment (data not shown).
Fig. 3.
Evaluation of virus induction from drug-treated Vero cells. Vero cells were treated under optimized cell culture conditions and drug concentrations and evaluated for latent virus induction. Cells (1.0 × 106) were planted in 25-cm2 flasks for 16 h, at which time drug was added (IUdR, 200 μg/ml; AzaC, 1.25 μg/ml; and NaBut, 3 mM). Forty-eight hours later, filtered supernatant was collected (day 0) before removing the drug, and cells were washed and refed with fresh medium. Supernatants were collected daily with medium change and filtered for STF-PERT assay. Control cell cultures were grown in medium without drug.
Evaluation of retroviral sequences in the RT activity produced from drug-treated Vero cells.
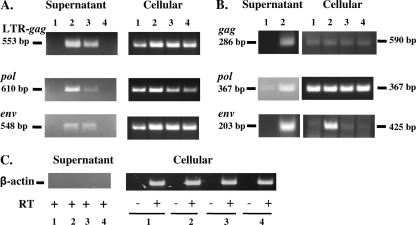
To determine whether the drug-induced PERT activity was associated with endogenous retrovirus particles, cell-free supernatants containing peak RT activity from drug-treated cells (Fig. 3) were ultracentrifuged, and the pelleted material (10× or 1,000×) was analyzed by RT-PCR assays. Consensus SERV/SRV primers amplified fragments from the LTR gag, pol, and env regions in 10× pelleted material of supernatant from AzaC- and IUdR-treated Vero cells (Fig. 4A, lanes 2 and 3, respectively), whereas no fragments were amplified from untreated cells or from NaBut-treated cells (lanes 1 and 4, respectively). The results of the first round of PCR amplification are shown in Fig. 4A, which indicated greater virus induction with AzaC than with IUdR and directly correlated with the PERT results (Fig. 3). Fragments of expected size were PCR amplified from cellular RNAs of untreated and drug-treated cells; increased band intensities were seen in the gag and env regions with all three drugs compared with band intensities of untreated cells. The SERV-related sequences amplified from Vero cells were designated SERVagm-Vero.
Fig. 4.
RT-PCR analysis of drug-treated Vero cells. RNAs were prepared from cells and from filtered supernatant of AzaC-treated cells for RT-PCR using SERV/SRV consensus primers (A), BaEV primers (B), and β-actin primers (C). RNAs from untreated cells were collected on day 2 after planting the cells (lanes 1) and from drug-treated cells on day 4 (medium was changed on day 1 after drug removal) using 1.25 μg/ml AzaC, 200 μg/ml IUdR, and 3 mM NaBut (lanes 2 to 4, respectively). (A) Results using SERV/SRV primers from the LTR gag, pol, and env regions are shown after the first round of PCR with cellular RNAs and with 10× filtered supernatant RNAs. (B) Results using BaEV primers from the gag, pol, and env regions are shown after the first round of PCR with cellular RNAs; in the case of 1,000× filtered supernatant RNAs, results are shown from the first round of PCR using pol primers and from the second round of PCR using gag and env primers. Virus identity was determined by nucleotide sequence analysis. The absence of contaminating cellular DNA in the cellular RNA preparation was demonstrated by RT-PCR with β-actin primers without adding RT. No fragment was amplified with β-actin primers from 10× supernatant RNAs (C) or from 1,000× supernatant preparation (not shown).
BaEV primers amplified fragments from the gag and env regions by nested RT-PCR of 1,000× pelleted supernatant from AzaC-treated Vero cells (Fig. 4B, supernatant column, lane 2). A pol fragment was seen after the first round of PCR amplification in the pelleted supernatant from AzaC-treated Vero cells, as well as weakly in the pellet prepared from supernatant of untreated cells (Fig. 4B, supernatant column, lanes 2 and 1, respectively). PCR-amplified fragments were seen at about the same intensities in cellular RNAs of untreated and drug-treated cells after the first round of PCR amplification with gag and pol primers and strongly in AzaC-treated cells with env primers (Fig. 4B, cellular column, lane 2). The size of the gag and pol PCR-amplified fragments corresponded to the expected size based upon the primers used, whereas the env fragment had 4 nucleotides missing (discussed below). The BaEV-related sequences amplified from Vero cells were designated BaEVagm-Vero.
No fragments were amplified using β-actin primers from 10× supernatant, indicating the absence of contamination with cellular sequences (Fig. 4C), or from 1,000× supernatant (data not shown). To demonstrate the absence of contaminating DNA in the cellular RNA preparations, PCR amplification was done with β-actin primers in the absence of RT: no fragments were seen, whereas fragments of expected size were seen in the presence of RT.
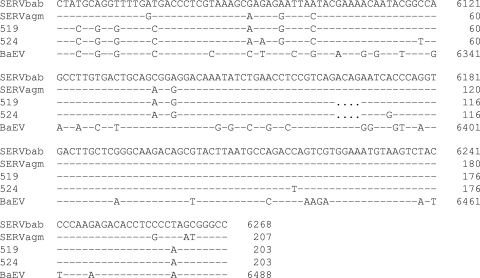
Nucleotide sequence analysis of SERVagm-Vero DNAs cloned from supernatant pellets of AzaC-treated Vero cells indicated a high degree of homology (94 to 96%) in the gag, pol, and env regions to SERVbab sequences compared to those of SRV-1, SRV-2, and SRV-3/MPMV sequences (60 to 81%) (Table 1). Interestingly, nucleotide sequence comparison of SERVagm-Vero and a simian retrovirus 1 isolate SRV_Vero, a recently described endogenous simian retrovirus sequence assembled from DNA fragments originating from Vero cells, indicated less homology (90 to 92%; GenBank accession number HM43845) (85) than that seen with SERVbab. Nucleotide sequence analysis of BaEVagm-Vero DNAs cloned from pelleted supernatant of AzaC-treated cells indicated a high degree of identity to BaEV (95 to 97%) in the pol region, whereas the env sequences were highly related (92 to 94%) to those present in SERVbab (83) (Table 2). There was 83 to 86% sequence homology in the gag region with BaEV, and 83 to 90% homology was seen in the gag and pol regions with BaEV recombinant virus, PcEV. Additionally, there was 69 to 87% nucleotide sequence homology in the gag, pol, and env regions to RD114, an endogenous retrovirus in cats that contains BaEV env (80). Further analysis of env was done by determining nucleotide sequences of 5 BaEVagm-Vero cloned DNAs. The results indicated 4 identical clones (represented by clone 519); clone 524 had 4 different nucleotides. Alignment of BaEVagm-Vero env sequences with the analogous region in SERV and BaEV is shown in Fig. 5. The results indicated that the BaEVagm-Vero env sequences were more closely related to those of SERV than to those of BaEV, except in the regions of the PCR primers; however, they were distinct from SERV env sequences due to the absence of four nucleotides, as well as from that of previously described clones 23.1 and 25.5 that were obtained from baboon DNA by using the same BaEV env primers (82). It should be noted that the four-nucleotide deletion in BaEVagm-Vero sequences would result in the absence of env expression, and therefore particles encoded from these sequences should be noninfectious. Similar analysis of cloned DNAs from the gag, pol, and env regions indicated genetic heterogeneity in the particles produced from SERVagm and BaEVagm sequences in Vero DNA.
Table 2.
Comparative nucleotide sequence analysis of cloned BaEVagm-Vero DNAa
| Cloned DNA regionb | Position (nt)c | Nucleotide sequence identity (%)d |
|||
|---|---|---|---|---|---|
| SERVbab | PcEV | RD114 | BaEV | ||
| gag (4) | 1135–1420 | 0 | 83–86 | 73–74 | 83–86 |
| pol (3) | 3505–3871 | 0 | 88–90 | 85–87 | 95–97 |
| env (5) | 6281–6488 | 92–94 | 0 | 69–71 | 79–81 |
DNA was obtained by RT-PCR from 1,000× supernatant of AzaC-treated Vero cells.
Regions of PCR amplification, with number of cloned DNAs that were analyzed indicated in parentheses.
Nucleotide positions according to the published BaEV sequence of Kato et al. (35).
Fig. 5.
Nucleotide sequence analysis of BaEVagm-Vero env. BaEV env-related clones 519 and 524, isolated from Vero cells in this study, were aligned with SERVbab and BaEV, as well as with SERVagm-Vero, which was also obtained in this study by using consensus SERV/SRV primers. Different nucleotides are shown. Dashes indicate identity; dots indicate missing bases. Nucleotide positions are indicated: SERVbab, based upon simian type D virus 1 (accession no. U85505); BaEV, according to the published BaEV sequence of Kato et al. (35); 519, 524, and SERVagm-Vero, based upon start of the DNA sequences shown.
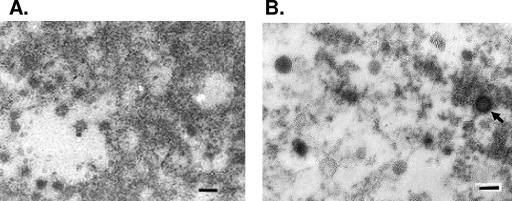
To further demonstrate that the PERT activity produced from drug-treated Vero cells was particle associated, pellets from concentrated, cell-free supernatants of normal and drug-treated cells were analyzed by TEM for viruses and other microbial agents (Fig. 6). Only retrovirus-like particles were reported, albeit few. There were 1.7 × 106 viruslike particles calculated in the AzaC-induced virus pellet prepared from 160 ml supernatant, which is near the limit of detection by TEM. No viruslike particle could be seen in the pellet prepared from the control sample and the IUdR-induced supernatant, which was expected since this had a much lower RT activity than the AzaC sample. Additionally, no particles were seen in cell pellets.
Fig. 6.
TEM analysis. Ultrastructural evaluation of filtered, ultracentrifuged supernatant from normal (without drug treatment) Vero cells (A) and AzaC-treated Vero cells (B). Arrow indicates retrovirus-like particle. Bar, 100 nm.
Infectivity analysis.
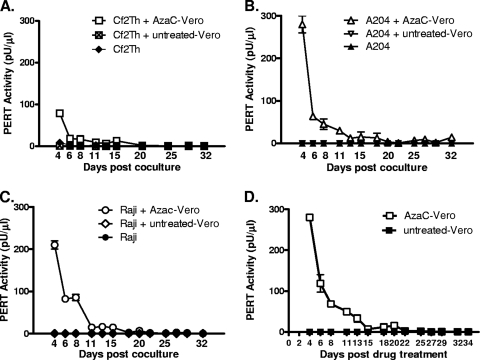
To evaluate whether the RT-containing particles induced from Vero cells were replication competent, AzaC- and IUdR-induced supernatant and cells were used to infect/coculture with cell lines known to be susceptible to replication of different type D simian retroviruses, such as Raji and A-204 for SRV and Cf2Th for SMRV. Additionally, A-204 and Cf2Th are susceptible to replication of BaEV (45), which contains an SERV env via recombination with SERVbab (83) and Raji to other SERV recombinant viruses (24, 59). To enhance the detection of infectious virus, supernatant and cells were used directly upon collection, without freeze and thaw. Additionally, drug-treated Vero cells were cultured for an extended time to allow for amplification of any potential virus with an ecotropic host range. The results using the highly sensitive STF-PERT assay, which can detect 1 to 10 retrovirus particles (53), demonstrated detection of only RT activity in the starting material, which was gradually reduced to background or undetectable levels, but without amplification, even upon long-term culture. These results indicated the absence of an infectious virus that could replicate in Vero cells or that was similar in its host range properties to various known infectious type D simian retroviruses (Fig. 7).
Fig. 7.
Infectivity analysis of drug-induced retrovirus from Vero cells. Vero cells (1 × 106) were planted in 25-cm2 flasks for 16 h and treated with 1.25 μg/ml AzaC or 200 μg/ml IUdR for 48 h. In the case of one flask, medium was replaced daily with fresh medium, and filtered supernatant was collected for STF-PERT assay (D). In the case of the other flasks, which were used in infectivity/coculture studies, medium was changed on day 0 and day 1; on day 4, cells and supernatant were collected, pooled, and used for coculture with target cells at predetermined ratios for equivalent growth of test and target cells (see Materials and Methods): Cf2Th cells (A), A-204 cells (B), Raji cells (C). Filtered supernatant was collected from the cocultured cells starting at the first cell passage on day 4 after coculture until its termination at day 32. Samples were assayed in the STF-PERT assay. Results from the AzaC-treated cells and cocultures are shown. Similar results were seen in the case of IUdR-treated cells, where the induced peak PERT activity was 20-fold less.
DISCUSSION
About 8 to 10% of the genome of NHPs contains endogenous retrovirus-related sequences (23). Although the majority of these sequences are defective, full-length virus genomes may exist that potentially can encode infectious viruses (71). Endogenous retroviruses have been isolated from Old World monkeys, such as the baboon, langur, colobus monkey, and macaques, and from New World monkeys, such as the woolly monkey, squirrel monkey, and owl monkey, as well as from the gibbon ape and a prosimian tree shrew (9, 21, 33, 62, 67, 76–79). However, there has been no report of endogenous retrovirus particles of AGM origin, even though endogenous retroviral sequences related to type C gammaretrovirus murine leukemia virus (MLV) and type D betaretrovirus SRV are present in the genome of African green monkeys and in cell lines derived from this species (56, 83, 85), and MLV-related viral DNAs cloned from AGM tissue were shown to contain functional long terminal repeats (LTRs) (36). Furthermore, African green monkey cell lines, such as Vero and CV-1 (30), have been widely used in research and diagnostics without any report of spontaneous release of virus particles. Additionally, Vero cells have been extensively tested for the presence of viruses, including virus-induction assays for the detection of endogenous retroviruses, and were found to be negative (29, 74).
We recently developed a stepwise approach to evaluate the presence of inducible virus sequences by chemical induction that outlines a strategy for determining optimum induction conditions based upon evaluation of cell growth properties, drug treatment conditions, and use of sensitive assays for known and novel virus detection (37). The study indicated that maximum virus induction was obtained when the drug was added to the cells in the early log phase, when there was a high percentage of cells in the S phase, and when the cells were treated for greater than one PDT before nearing the end of the log phase. Additionally, the optimum drug dose was found to be the highest dose that still had good cell recovery. We used this strategy to investigate whether endogenous retroviral sequences present in AGMs were inducible for virus particles using the well-characterized Vero cell line. Inducers known to activate endogenous retroviruses, such as IUdR, AzaC, and NaBut (16), were used in combination with sensitive broad-detection assays for retroviruses, such as STF-PERT assay (53). In contrast to previous studies (74) and to our initial experiments using induction conditions that had been optimized for K-BALB mouse cells (data not shown), a low level of PERT activity was detected in cell-free supernatants of IUdR- and AzaC-treated Vero cells by following the stepwise virus induction strategy (37). Further characterization of the induced RT activity showed that it could be pelleted by ultracentrifugation and was therefore likely to be particle associated. RT-PCR and nucleotide sequence analysis identified SERV- and BaEV-related sequences in the pellets of RT-containing supernatant from Vero cells; the presence of gag, pol, and env sequences further confirmed that the RT activity was associated with retrovirus particles. TEM analysis demonstrated induced particles in AzaC-treated Vero cells.
A low level of replication-defective retroviral particles was produced from Vero cells under optimized conditions of drug treatment. Efforts to increase virus production under various cell culture and drug conditions known to induce viruses were not successful (e.g., cell synchronization by using serum-free medium for 1 and 2 days, dexamethasone plus IUdR [200 μg/ml] or AzaC [1.25 μg/ml] [3, 16], and IUdR [200 μg/ml] plus AzaC [1.25 μg/ml] [40]). It should be noted that a high dose of IUdR (200 μg/ml) has also been used to activate endogenous retrovirus from culture of the prosimian tree shrew (21). These results indicate a stringent control of virus gene expression in primate cells. It should be noted that the results of chemical virus induction were different in cellular RNA expression and in supernatant RNAs that measured virus production. For example, SERV gag and env RNAs were induced in cells treated with AzaC, IUdR, and NaBut, whereas particle-associated RNAs were induced in the supernatant with only AzaC and IUdR. In the case of BaEV-related RNAs, induction of gag and pol RNAs was not noticed in the cell but only upon analysis of the supernatant. This discordancy in the results emphasizes the need to evaluate cell-free, particle-associated RNAs for investigating the potential of endogenous retrovirus sequences to encode virions.
Analysis of the induced virus particles from Vero cells in infection/coculture studies using cell lines susceptible to SRV, BaEV, and recombinant SERVs (24, 59) followed by detection with the highly sensitive STF-PERT assay demonstrated absence of replication-competent virus. Additionally, long-term culture of the drug-treated Vero cells alone did not result in increased PERT activity. Chemical induction studies indicated the presence of endogenous retrovirus sequences in Vero cells that have the potential to encode noninfectious viruslike particles containing RNAs. Although copackaging of defective retroviral RNAs could result in recombination to generate an infectious virus genome, based upon our data and the extensive testing of Vero cells used for biologicals, there has been no evidence of emergence of an infectious retrovirus. Studies are ongoing to physically and genetically characterize the retroviral particles induced from Vero cells for further evaluation of the potential to generate novel recombinants.
SERV- and BaEV-related sequences have previously been reported in the genomes of baboons and other NHP species, including AGMs (83, 85). It should be noted that the SERV sequences in baboon have been misnamed as SRV-1 sequences in the database (GenBank accession number U85505) (83); furthermore, the recently reported assembled, endogenous retrovirus sequences originating from Vero cells, which are related to SERV in baboons, have also been designated simian retrovirus 1 isolate SRV_Vero (GenBank accession number HM143845) (85). It is important to recognize that SERVs are distinct from SRVs, which are exogenously transmitted and pathogenic in macaques. Interestingly, the SERVagm-Vero sequence associated with induced virus particles had greater nucleotide sequence homology to SERVbab than to SRV_Vero, indicating that it originated from an ERV family in the AGM Vero genome that is distinct from SRV_Vero. The presence of multiple related but distinct families in the AGM genome has been previously described (56). Furthermore, analysis of SERVagm and BaEVagm cloned DNAs in this study demonstrated sequence heterogeneity in the particles induced from Vero cells.
This study demonstrates the presence of inducible retroviral sequences in the AGM genome by use of an algorithm for determining chemical induction conditions optimized for Vero cells. Moreover, this stepwise strategy may be used with emerging broad virus detection technologies to identify novel endogenous retroviruses that can be produced from other primate cells as well as from the cells of other species. This approach may also enhance the sensitivities of the currently available virus detection methods for evaluating new cell substrates used for production of biological products by providing a virus amplification step prior to detection.
ACKNOWLEDGMENTS
We thank Robin Levis, Andrew Lewis, Keith Peden, Hana Golding, Laraine Henchal, Konstantin Chumakov, and Marion Gruber for review of the manuscript.
The work was funded by DMID/NIAID/NIH Interagency Agreement no. Y1-A1-4893-02.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any agency determination or policy.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Aaronson S. A., Anderson G. R., Dunn C. Y., Robbins K. C. 1974. Induction of type-C RNA virus by cycloheximide: increased expression of virus-specific RNA. Proc. Natl. Acad. Sci. U. S. A. 71:3941–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aaronson S. A., Dunn C. Y. 1974. Endogenous C-type viruses of BALB-c cells: frequencies of spontaneous and chemical induction. J. Virol. 13:181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmed M., Schidlovsky G., Harewood K. R., Manousos M., Mayyasi S. A. 1977. Expression of Mason-Pfizer and simian type C viruses in the presence of 5-iododeoxyuridine and dexamethasone. J. Natl. Cancer Inst. 58:1515–1518 [DOI] [PubMed] [Google Scholar]
- 4. Anderson G. W., Plagemann P. G. 1995. Expression of ecotropic murine leukemia virus in the brains of C58/M, DBA2/J, and in utero-infected CE/J mice. J. Virol. 69:8089–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderssen S., Sjottem E., Svineng G., Johansen T. 1997. Comparative analyses of LTRs of the ERV-H family of primate-specific retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology 234:14–30 [DOI] [PubMed] [Google Scholar]
- 6. Reference deleted.
- 7. Bather R., Becker B. C., Contreras G., Furesz J. 1985. Heterotransplantation studies with tissue culture cell lines in various animal and in vitro host systems. J. Biol. Stand. 13:13–22 [DOI] [PubMed] [Google Scholar]
- 8. Benveniste R. E., Heinemann R., Wilson G. L., Callahan R., Todaro G. J. 1974. Detection of baboon type C viral sequences in various primate tissues by molecular hybridization. J. Virol. 14:56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benveniste R. E., et al. 1974. Infectious C-type virus isolated from a baboon placenta. Nature 248:17–20 [DOI] [PubMed] [Google Scholar]
- 10. Benveniste R. E., Todaro G. J. 1974. Evolution of type C viral genes. I. Nucleic acid from baboon type C virus as a measure of divergence among primate species. Proc. Natl. Acad. Sci. U. S. A. 71:4513–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boeke J. D., Stoye J. P. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343–435 In Coffin J. M., Hughes S. H., Varmus H. (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 12. Boller K., et al. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89:567–572 [DOI] [PubMed] [Google Scholar]
- 13. Boyd A. L., Derge J. G., Hampar B. 1978. Activation of endogenous type C virus in BALB/c mouse cells by herpesvirus DNA. Proc. Natl. Acad. Sci. U. S. A. 75:4558–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christensen T., et al. 2000. Molecular characterization of HERV-H variants associated with multiple sclerosis. Acta Neurol. Scand. 101:229–238 [DOI] [PubMed] [Google Scholar]
- 15. Christensen T., Pedersen L., Sorensen P. D., Moller-Larsen A. 2002. A transmissible human endogenous retrovirus. AIDS Res. Hum. Retroviruses 18:861–866 [DOI] [PubMed] [Google Scholar]
- 16. Coffin J. M. 1984. Endogenous retroviruses, p. 1109–1203 In Weiss R., Teich N., Varmus H., Coffin J. (ed.), RNA tumor viruses: molecular biology of tumor viruses, 2nd ed., vol. 1 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 17. Reference deleted.
- 18. Contreras G., Bather R., Furesz J., Becker B. C. 1985. Activation of metastatic potential in African green monkey kidney cell lines by prolonged in vitro culture. In Vitro Cell. Dev. Biol. 21:649–652 [DOI] [PubMed] [Google Scholar]
- 19. Eichberg J. W., et al. 1981. In vivo activation of the baboon endogenous virus. Proc. Soc. Exp. Biol. Med. 166:271–276 [DOI] [PubMed] [Google Scholar]
- 20. Faff O., et al. 1992. Retrovirus-like particles from the human T47D cell line are related to mouse mammary tumour virus and are of human endogenous origin. J. Gen. Virol. 73(Pt. 5):1087–1097 [DOI] [PubMed] [Google Scholar]
- 21. Flugel R. M., Zentgraf H., Munk K., Darai G. 1978. Activation of an endogenous retrovirus from Tupaia (tree shrew). Nature 271:543–545 [DOI] [PubMed] [Google Scholar]
- 22. Furesz J., Fanok A., Contreras G., Becker B. 1989. Tumorigenicity testing of various cell substrates for production of biologicals. Dev. Biol. Stand. 70:233–243 [PubMed] [Google Scholar]
- 23. Gibbs R. A., et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234 [DOI] [PubMed] [Google Scholar]
- 24. Grant R. F., et al. 1995. Characterization of infectious type D retrovirus from baboons. Virology 207:292–296 [DOI] [PubMed] [Google Scholar]
- 25. Grolle P. F., Bentvelzen P., van Noord M. J. 1973. Activation of endogenous C-type oncornavirus by 5-bromodeoxyuridine in cultured embryonic rat fibroblasts. Biomedicine 19:148–151 [PubMed] [Google Scholar]
- 26. Haltmeier M., et al. 1995. Identification of S71-related human endogenous retroviral sequences with full-length pol genes. Virology 209:550–560 [DOI] [PubMed] [Google Scholar]
- 27. Heberling R. L., Kalter S. S., Barker S. T., Weislow O. S. 1975. Isolation and biological properties of endogenous baboon (Papio cynocephalus) type C viruses. Bibl. Haematol. 1975:158–160 [DOI] [PubMed] [Google Scholar]
- 28. Hefti E., Ip J., Giddens W. E., Jr., Panem S. 1983. Isolation of a unique retrovirus, MNV-1, from Macaca nemestrina. Virology 127:309–319 [DOI] [PubMed] [Google Scholar]
- 29. Horaud F. 1992. Absence of viral sequences in the WHO-Vero Cell Bank. A collaborative study. Dev. Biol. Stand. 76:43–46 [PubMed] [Google Scholar]
- 30. Hronovsky V., Plaisner V., Benda R. 1978. CV-1 monkey kidney cell line—a highly susceptible substrate for diagnosis and study of arboviruses. Acta Virol. 22:123–129 [PubMed] [Google Scholar]
- 31. Johansen T., Holm T., Bjorklid E. 1989. Members of the RTVL-H family of human endogenous retrovirus-like elements are expressed in placenta. Gene 79:259–267 [DOI] [PubMed] [Google Scholar]
- 32. Johnson J. B., Noguchi P. D., Browne W. C., Petricciani J. C. 1981. Tumorigenicity of continuous monkey cell lines in in vivo and in vitro systems. Dev. Biol. Stand. 50:27–35 [PubMed] [Google Scholar]
- 33. Kalter S. S., Heberling R. L. 1976. Discovery of baboon endogenous type C virus. Cancer Res. 36:4197. [PubMed] [Google Scholar]
- 34. Kalter S. S., Heberling R. L., Hellman A., Todaro G. J., Panigel M. 1975. C-type particles in baboon placenta. Proc. R. Soc. Med. 68:135–140 [PMC free article] [PubMed] [Google Scholar]
- 35. Kato S., Matsuo K., Nishimura N., Takahashi N., Takano T. 1987. The entire nucleotide sequence of baboon endogenous virus DNA: a chimeric genome structure of murine type C and simian type D retroviruses. Jpn. J. Genet. 62:127–137 [Google Scholar]
- 36. Kessel M., Khan A. S. 1985. Nucleotide sequence analysis and enhancer function of long terminal repeats associated with an endogenous African green monkey retroviral DNA. Mol. Cell. Biol. 5:1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khan A. S., et al. 2009. Proposed algorithm to investigate latent and occult viruses in vaccine cell substrates by chemical induction. Biologicals 37:196–201 [DOI] [PubMed] [Google Scholar]
- 38. Khan A. S., et al. 1998. The reverse transcriptase activity in cell-free medium of chicken embryo fibroblast cultures is not associated with a replication-competent retrovirus. J. Clin. Virol. 11:7–18 [DOI] [PubMed] [Google Scholar]
- 39. Khan A. S., Muller J., Sears J. F. 2001. Early detection of endogenous retroviruses in chemically induced mouse cells. Virus Res. 79:39–45 [DOI] [PubMed] [Google Scholar]
- 40. Khan A. S., Sears J. F. 2001. Pert analysis of endogenous retroviruses induced from K-BALB mouse cells treated with 5-iododeoxyuridine: a potential strategy for detection of inducible retroviruses from vaccine cell substrates. Dev. Biol. (Basel) 106:387–393 [PubMed] [Google Scholar]
- 41. Khan A. S., Sears J. F., Muller J., Galvin T. A., Shahabuddin M. 1999. Sensitive assays for isolation and detection of simian foamy retroviruses. J. Clin. Microbiol. 37:2678–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim H. S., Hyun B. H., Takenaka O. 2002. Isolation and phylogeny of endogenous retrovirus HERV-F family in Old World monkeys. Brief report. Arch. Virol. 147:393–400 [DOI] [PubMed] [Google Scholar]
- 43. Komurian-Pradel F., et al. 1999. Molecular cloning and characterization of MSRV-related sequences associated with retrovirus-like particles. Virology 260:1–9 [DOI] [PubMed] [Google Scholar]
- 44. Langat D. K., Wango E. O., Owiti G. O., Omollo E. O., Mwenda J. M. 1998. Characterisation of retroviral-related antigens expressed in normal baboon placental tissues. Afr. J. Health Sci. 5:144–152 [PubMed] [Google Scholar]
- 45. Lavelle G., Foote L., Heberling R. L., Kalter S. S. 1979. Expression of baboon endogenous virus in exogenously infected baboon cells. J. Virol. 30:390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lerche N. W., Osborn K. G. 2003. Simian retrovirus infections: potential confounding variables in primate toxicology studies. Toxicol. Pathol. 31(Suppl.):103–110 [DOI] [PubMed] [Google Scholar]
- 47. Lerner-Tung M. B., Doong S. L., Cheng Y. C., Hsiung G. D. 1995. Characterization of conditions for the activation of endogenous guinea pig retrovirus in cultured cells by 5-bromo-2′-deoxyuridine. Virus Genes 9:201–209 [DOI] [PubMed] [Google Scholar]
- 48. Levenbook I. S., Petricciani J. C., Elisberg B. L. 1984. Tumorigenicity of Vero cells. J. Biol. Stand. 12:391–398 [DOI] [PubMed] [Google Scholar]
- 49. Lieber M. M., Todaro G. J. 1973. Spontaneous and induced production of endogenous type-C RNA virus from a clonal line of spontaneously transformed BALB-3T3. Int. J. Cancer 11:616–627 [DOI] [PubMed] [Google Scholar]
- 50. Long C. W., Suk W. A., Greenawalt C. 1978. Activation of endogenous type C virus by amino acid alcohols. Virology 88:194–196 [DOI] [PubMed] [Google Scholar]
- 51. Lower R., Lower J., Frank H., Harzmann R., Kurth R. 1984. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J. Gen. Virol. 65(Pt. 5):887–898 [DOI] [PubMed] [Google Scholar]
- 52. Lyden T. W., Johnson P. M., Mwenda J. M., Rote N. S. 1994. Ultrastructural characterization of endogenous retroviral particles isolated from normal human placentas. Biol. Reprod. 51:152–157 [DOI] [PubMed] [Google Scholar]
- 53. Ma Y. K., Khan A. S. 2009. Evaluation of different RT enzyme standards for quantitation of retroviruses using the single-tube fluorescent product-enhanced reverse transcriptase assay. J. Virol. Methods 157:133–140 [DOI] [PubMed] [Google Scholar]
- 54. Manohar M., Orrison B., Peden K., Lewis A. M., Jr 2008. Assessing the tumorigenic phenotype of Vero cells in adult and newborn nude mice. Biologicals 36:65–72 [DOI] [PubMed] [Google Scholar]
- 55. Marracci G. H., et al. 1995. Simian AIDS type D serogroup 2 retrovirus: isolation of an infectious molecular clone and sequence analyses of its envelope glycoprotein gene and 3′ long terminal repeat. J. Virol. 69:2621–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martin M. A., Bryan T., McCutchan T. F., Chan H. W. 1981. Detection and cloning of murine leukemia virus-related sequences from African green monkey liver DNA. J. Virol. 39:835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martin M. A., Bryan T., Rasheed S., Khan A. S. 1981. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc. Natl. Acad. Sci. U. S. A. 78:4892–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mayer R. J., Smith R. G., Gallo R. C. 1974. Reverse transcriptase in normal rhesus monkey placenta. Science 185:864–867 [DOI] [PubMed] [Google Scholar]
- 59. Nandi J. S., Van Dooren S., Chhangani A. K., Mohnot S. M. 2006. New simian beta retroviruses from rhesus monkeys (Macaca mulatta) and langurs (Semnopithecus entellus) from Rajasthan, India. Virus Genes 33:107–116 [DOI] [PubMed] [Google Scholar]
- 60. Paul J. 1975. Cell and tissue culture, 5th ed. Churchill Livingstone, Edinburgh, United Kingdom [Google Scholar]
- 61. Power M. D., et al. 1986. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science 231:1567–1572 [DOI] [PubMed] [Google Scholar]
- 62. Rabin H., Benton C. V., Tainsky M. A., Rice N. R., Gilden R. V. 1979. Isolation and characterization of an endogenous type C virus of rhesus monkeys. Science 204:841–842 [DOI] [PubMed] [Google Scholar]
- 63. Rasheed S., et al. 1976. Spontaneous release of endogenous ecotropic type C virus from rat embryo cultures. J. Virol. 18:799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reitz M. S., et al. 1976. Primate type-C virus nucleic acid sequences (woolly monkey and baboon types) in tissues from a patient with acute myelogenous leukemia and in viruses isolated from cultured cells of the same patient. Proc. Natl. Acad. Sci. U. S. A. 73:2113–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seifarth W., et al. 1995. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J. Virol. 69:6408–6416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Serafino A., et al. 2009. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 315:849–862 [DOI] [PubMed] [Google Scholar]
- 67. Sherwin S. A., Todaro G. J. 1979. A new endogenous primate type C virus isolated from the Old World monkey Colobus polykomos. Proc. Natl. Acad. Sci. U. S. A. 76:5041–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shih A., Coutavas E. E., Rush M. G. 1991. Evolutionary implications of primate endogenous retroviruses. Virology 182:495–502 [DOI] [PubMed] [Google Scholar]
- 69. Smith C. A., Moore H. D. 1988. Expression of C-type viral particles at implantation in the marmoset monkey. Hum. Reprod. 3:395–398 [DOI] [PubMed] [Google Scholar]
- 70. Sonigo P., Barker C., Hunter E., Wain-Hobson S. 1986. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell 45:375–385 [DOI] [PubMed] [Google Scholar]
- 71. Stoye J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914–R916 [DOI] [PubMed] [Google Scholar]
- 72. Stromberg K., Benveniste R. 1983. Efficient isolation of endogenous rhesus retrovirus from trophoblast. Virology 128:518–523 [DOI] [PubMed] [Google Scholar]
- 73. Stromberg K., Huot R. I. 1981. Preferential expression of endogenous type C viral antigen in rhesus placenta during ontogenesis. Virology 112:365–369 [DOI] [PubMed] [Google Scholar]
- 74. Swanson S. K., et al. 1988. Characterization of Vero cells. J. Biol. Stand. 16:311–320 [DOI] [PubMed] [Google Scholar]
- 75. Todaro G. J., et al. 1978. Isolation and characterization of a new type D retrovirus from the Asian primate, Presbytis obscurus (spectacled langur). Virology 84:189–194 [DOI] [PubMed] [Google Scholar]
- 76. Todaro G. J., Benveniste R. E., Sherwin S. A., Sherr C. J. 1978. MAC-1, a new genetically transmitted type C virus of primates: “low frequency” activation from stumptail monkey cell cultures. Cell 13:775–782 [DOI] [PubMed] [Google Scholar]
- 77. Todaro G. J., Lieber M. M., Benveniste R. E., Sherr C. J. 1975. Infectious primate type C viruses: three isolates belonging to a new subgroup from the brains of normal gibbons. Virology 67:335–343 [DOI] [PubMed] [Google Scholar]
- 78. Todaro G. J., Sherr C. J., Benveniste R. E. 1976. Baboons and their close relatives are unusual among primates in their ability to release nondefective endogenous type C viruses. Virology 72:278–282 [DOI] [PubMed] [Google Scholar]
- 79. Todaro G. J., et al. 1978. Endogenous New World primate type C viruses isolated from owl monkey (Aotus trivirgatus) kidney cell line. Proc. Natl. Acad. Sci. U. S. A. 75:1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van der Kuyl A. C., Dekker J. T., Goudsmit J. 1999. Discovery of a new endogenous type C retrovirus (FcEV) in cats: evidence for RD-114 being an FcEV(Gag-Pol)/baboon endogenous virus BaEV(Env) recombinant. J. Virol. 73:7994–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van der Kuyl A. C., Dekker J. T., Goudsmit J. 1995. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J. Virol. 69:7877–7887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. van der Kuyl A. C., Dekker J. T., Goudsmit J. 1995. Full-length proviruses of baboon endogenous virus (BaEV) and dispersed BaEV reverse transcriptase retroelements in the genome of baboon species. J. Virol. 69:5917–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. van der Kuyl A. C., Mang R., Dekker J. T., Goudsmit J. 1997. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J. Virol. 71:3666–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. van Steenis G., van Wezel A. L. 1981. Use of the ATG-treated newborn rat for in vivo tumorigenicity testing of cell substrates. Dev. Biol. Stand. 50:37–46 [PubMed] [Google Scholar]
- 85. Victoria J. G., et al. 2010. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J. Virol. 84:6033–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Weiss R. A. 2000. Ancient and modern retroviruses. Acta Microbiol. Immunol. Hung. 47:403–410 [PubMed] [Google Scholar]
- 87. Yasumura Y., Kawakita Y. 1963. Studies on SV40 in tissue culture—preliminary step for cancer research in vitro. Nihon Rinsho 21:1201–1215 [Google Scholar]