Abstract
We show here that the white spot syndrome virus (WSSV) immediate-early protein IE1 interacts with the Penaeus monodon TATA box-binding protein (PmTBP) and that this protein-protein interaction occurs in the absence of any other viral or cellular proteins or nucleic acids, both in vitro and in vivo. Mapping studies using enhanced green fluorescent protein (EGFP) fusion proteins containing truncations of IE1 and PmTBP delimited the interacting regions to amino acids (aa) 81 to 180 in IE1 and, except for aa 171 to 230, to aa 111 to 300 in PmTBP. A WSSV IE1 transactivation assay showed that large quantities (>800 ng) of the GAL4-IE1 plasmid caused “squelching” of the GAL4-IE1 activity and that this squelching effect was alleviated by the overexpression of PmTBP. Gene silencing of WSSV ie1 and PmTBP by pretreatment with double-stranded RNAs (dsRNAs) prior to WSSV challenge showed that the expression of these two target genes was specifically inhibited by their corresponding dsRNAs 72 and 96 h after dsRNA treatment. dsRNA silencing of ie1 and PmTBP expression also significantly reduced WSSV replication and the expression of the viral early gene dnapol (DNA polymerase gene). These results suggest that WSSV IE1 and PmTBP work cooperatively with each other during transcription initiation and, furthermore, that PmTBP is an important target for WSSV IE1's transactivation activity that can enhance viral gene expression and help in virus replication.
INTRODUCTION
White spot syndrome virus (WSSV), the type species of the genus Whispovirus, family Nimaviridae (42), has a wide host range and is a lethal agent infecting penaeid shrimp (35). WSSV has spread globally and has caused huge economic losses to the shrimp farming industry (10, 27, 28, 35). WSSV is a large double-stranded DNA (dsDNA) virus (43), and although the complete sequence of the WSSV genome has been known for several years (7, 40, 45), knowledge of the biological functions of the viral proteins is still quite poor. During infection by large DNA viruses, such as baculoviruses and herpesviruses, gene expression is regulated such that the immediate-early (IE) genes are transcribed first, followed by the expression of the early and late genes (1, 2, 11, 14). To date, 18 WSSV IE genes have been identified (20, 25). Although the functions of most of the corresponding WSSV IE proteins have not yet been studied, many viral IE genes are known to encode multifunctional transcriptional regulators that both positively and negatively modulate viral early and late gene expression (17, 37, 44). Once expressed, the IE gene products may then function as regulatory trans-acting factors and may serve to initiate viral replicative events during infection. In the cascade of viral regulatory events, successive stages of virus replication are dependent on the proper expression of the genes in the preceding stage. Thus, viral IE genes are critically important in the virus infection cycle.
Upon infection, expression of the WSSV genes can be divided into immediate-early, early, and late phases (25, 29). However, the mechanism of the switch between WSSV immediate-early, early, and late gene expression is largely unknown, and only two studies (16, 24) have reported on the promoters and regulatory sequences that might be involved. An analysis of WSSV gene expression profiles in shrimp tissue showed that 37 of the 64 early genes (58%) and 28 of the 58 late genes (48%) contain a consensus TATA box [TATA(A/T)A] 0 to 300 nucleotides (nt) upstream of the translational start codon (29). In silico identification of putative promoter motifs of WSSV also showed that the upstream region of WSSV early genes contains a TATA box and an initiator that is similar to Drosophila RNA polymerase II core promoter sequences, suggesting that the virus uses the cellular transcription machinery to generate early transcripts (7, 25, 30). All of this evidence suggests that TATA box sequences have a functional role in the regulation of the transcription of WSSV genes.
Transcriptional factor IID (TFIID) is a protein complex that is required for the transcription initiation of all three eukaryotic RNA polymerases (8). TFIID consists of TATA box-binding protein (TBP) and TBP-associated factors (TAFs). This protein complex can interact with distant transcription factors and with RNA polymerase and its accessory proteins, all of which must be present before transcription can begin (33, 41). The distant transcription factors bind to regulatory sequences in the promoter region of the gene that is being transcribed, and our previous study (26) suggested that the WSSV IE gene product IE1, which contains conserved regions, including an N-terminal transactivation domain and a C-terminal DNA binding domain, might also act as a distant transcription factor with enhancer activity. This was further supported by an in vitro assay of a fusion protein that consisted of WSSV IE1 linked to the GAL4 DNA binding domain. This assay showed that WSSV IE1 can activate transcription and can transactivate a minimal promoter containing only a TATA homolog (26). If WSSV IE1 acts as a transcription factor, it is therefore reasonable to hypothesize that it becomes functional by attaching to the TATA-dependent assembly of general transcription factors via specific protein-protein interaction. The IE proteins of several other DNA viruses are known to interact with TBP (15, 18, 21, 22, 36), and in the present study, WSSV IE1's ability to bind to Penaeus monodon TBP (PmTBP) was investigated. The PmTBP gene coding region was cloned, and recombinant PmTBP was expressed. Interaction between WSSV IE1 and PmTBP was revealed by a coimmunoprecipitation analysis and glutathione S-transferase (GST) pulldown assay. Coimmunoprecipitation was also used with a series of IE1 and PmTBP truncation mutants to identify the regions in these two proteins that are involved in their interaction. To achieve a better understanding of how the activity of IE1 impacts the transcriptional machinery, the effect of this activator on the recruitment of the PmTBP protein was also analyzed in living cells. Finally, in vivo gene silencing experiments suggested the importance of the roles played by WSSV IE1 and PmTBP in the regulation of WSSV replication and gene expression.
MATERIALS AND METHODS
Cloning and sequencing of P. monodon TBP cDNA.
Degenerate primers Pm-F (5′-AAYGCNGARTAYAAYCCNAA-3′) and Pm-R (5′-NCCNACCATRTTYTGDATYTT-3′) were used to amplify a DNA fragment, using the template cDNA prepared from a P. monodon hepatopancreas, by low-stringency PCR. A 216-bp PCR product was isolated. To obtain the full-length cDNA, this fragment was used to design primers for the 5′ and 3′ rapid amplification of cDNA ends (5′ and 3′ RACE), using a commercial 5′/3′ RACE kit (Roche). The resultant PmTBP-specific primers used for 5′ and 3′ RACE are listed in Table 1. The final amplification products were cloned into pGEM-T Easy vector (Promega) and sequenced. The PmTBP deduced amino acid (aa) sequence was compared with TBP sequences of Drosophila melanogaster, Anopheles gambiae, Spodoptera frugiperda, Mus musculus, and Homo sapiens (GenBank accession no. NP523805, XP309748, P53361, NP038712, and NP003185, respectively). The multiple sequence alignment of the TBPs from these different species was done in CLUSTAL_X (38).
Table 1.
Primers used for 5′ and 3′ RACE
| Gene | Primer sequence (5′–3′) | Usage |
|---|---|---|
| PmTBP-SP1 | GTCTGGAGTCCTGCTCACTC | 5′ RACE |
| PmTBP-SP2 | TGTCCGTGGTTCTCGAATAC | 5′ RACE |
| PmTBP-SP3 | CATGATGACTGCAGCAAATCG | 5′ RACE |
| PmTBP-SP4 | AAAGTATGCGAGAATAGTGC | 3′ RACE |
Coimmunoprecipitation.
PCR cloning was used to insert the full-length WSSV IE1 and PmTBP genes into V5- or FLAG-tagged vectors containing the heat-inducible Drosophila heat shock protein 70 gene promoter (pDHsp-V5-His and pDHsp-FLAG-His [19]), and the plasmids pDHsp-PmTBP-V5-His, pDHsp-PmTBP-FLAG-His, pDHsp-IE1-V5-His, and pDHsp-IE1-FLAG-His were constructed. To express enhanced green fluorescent protein (EGFP) fusion proteins, the EGFP gene DNA fragment was isolated from the commercial vector pEGFP-N1 (Clontech) and cloned into the HindIII/BamHI-digested pDHsp-V5-His plasmid to generate pDHsp-EGFP-V5-His. Next, by using the WSSV genomic DNA or pDHsp-PmTBP-V5-His as a PCR template, various truncated IE1 or PmTBP gene fragments were cloned into pDHsp-EGFP-V5-His to express different truncated forms of EGFP-V5-tagged IE1 and PmTBP. The primer sequences used for these constructions are listed in Table 2. For DNA transfection, Sf9 insect cells were seeded into a six-well plate (8 × 105 cells/well) and grown in Sf-900 II serum-free medium (SFM; Invitrogen) at 27°C. Plasmids containing the appropriate genes (including the empty vector) were transfected into the Sf9 cells for 16 to 18 h and then heat shocked at 42°C in a water bath for 30 min. Six hours after the heat shock, the cells were washed with 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) and lysed in 140 μl of NP-40 lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40) supplemented with a protease inhibitor cocktail tablet (Roche). The lysis procedure was carried out on ice for 10 min with occasional shaking. The lysate was centrifuged at 12,000 × g for 5 min, and an aliquot of the supernatant (20 μl) was reserved for Western blot analysis to confirm the expression of the transfected genes. The remaining supernatant (120 μl) was then incubated with 15 μl of anti-FLAG M2 affinity gel (Sigma) at 4°C overnight with rotation. The gel was then washed five times in 150 μl of NP-40 lysis buffer. Aliquots of the total cell lysates and immunoprecipitated complexes were separated by 15% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. V5-tagged fusion proteins were detected with a rabbit anti-V5 antibody (Sigma) and a goat anti-rabbit IgG–horseradish peroxidase (HRP) conjugate (Sigma). FLAG-tagged proteins were detected with a rabbit anti-FLAG antibody (Sigma) and a goat anti-rabbit IgG–HRP conjugate. Protein band signals were visualized using a chemiluminescence reagent (ECL; Perkin-Elmer). In addition, to ensure that interactions between IE1 and PmTBP were not being mediated by nucleic acids, the cell lysates were also pretreated with nucleases (2 U DNase I [Invitrogen] and 0.5 μg RNase [Sigma]) in reaction buffer (20 mM Tris-HCl, pH 8.4, 2 mM MgCl2, 50 mM KCl) for 15 min at room temperature before being subjected to the immunoprecipitation assay.
Table 2.
Primers used to construct full-length and truncated IE1 and PmTBP expression plasmids
| Plasmid | Primer name (sequence [5′–3′]a) |
|---|---|
| pDHsp-IE1-V5-His | IE1-F (CCCAAGCTTCTCAAGATGGCCTTTAATTTTG) |
| IE1-R (TCCCCGCGGTACAAAGAATCCAGAAATCTCA) | |
| pDHsp-PmTBP-V5-His | PmTBP-F (CGCGGATCCATGGATCACATGCTGCCTTCCCCG) |
| PmTBP-R (TCCCCGCGGTTGTTTTTTGAAGCTTTTTAATAT) | |
| pDHsp-IE1-FLAG-His | IE1-F/IE1-R |
| pDHsp-PmTBP-FLAG-His | PmTBP-F/PmTBP-R |
| pDHsp-EGFP-V5-His | EGFP-F (CCCAAGCTTACCATGGTGAGCAAGGGCGAGGAG) |
| EGFP-R (CGCGGATCCCTTGTACAGCTCGTCCATGCC) | |
| pDHsp-EGFP-IE11–80-V5-His | IE1-1-F (CGCGGATCCATGGCCTTTAATTTTGAAGAC) |
| IE1-80-R (TCCCCGCGGGAGATTCTCCATATCTTCTGC) | |
| pDHsp-EGFP-IE181–224-V5-His | IE1-81-F (CGCGGATCCAACAGTGGTTCCCATGTCAAG)/IE1-R |
| pDHsp-EGFP-IE181–120-V5-His | IE1-81-F/IE1-120-R (TCCCCGCGGGTCCATGTATTGGGGAAAAGT) |
| pDHsp-EGFP-IE1101–140-V5-His | IE1-101-F (CGCGGATCCAACTGGATTGAAGAGACTGAT) |
| IE1-140-R (TCCCCGCGGCCTAGCCATCCATTTCATGGC) | |
| pDHsp-EGFP-IE1121–160-V5-His | IE1-121-F (CGCGGATCCGGTGGGGATGGTTCACGTGGG) |
| IE1-160-R (TCCCCGCGGTATGGATGCCGCATTTTCTAC) | |
| pDHsp-EGFP-IE1141–180-V5-His | IE1-141-F (CGCGGATCCGATGTGACTTTCTTTGTGTTTG) |
| IE1-180-R (TCCCCGCGGATTGTCAACAATCACCTTTAC) | |
| pDHsp-EGFP-IE1161–200-V5-His | IE1-161-F (CGCGGATCCTGGATGTACCAAAAACTACTA) |
| IE1-200-R (TCCCCGCGGCACTGGGCCTGGGTACTTGCA) | |
| pDHsp-EGFP-IE1181–224-V5-His | IE1-181-F (CGCGGATCCGCATCAAACCCAATGTTTTCT)/IE1-R |
| pDHsp-EGFP-PmTBP1–110-V5-His | PmTBP-F/PmTBP-110-R (TCCCCGCGGGGGCCCTCCTATGGGGGTGGAA) |
| pDHsp-EGFP-PmTBP111–300-V5-His | PmTBP-F (CGCGGATCCATGACCCCCATGACCCCACAC)/PmTBP-R |
| pDHsp-EGFP-PmTBP111–170-V5-His | PmTBP-111-F/PmTBP-170-R (TCCCCGCGGCCGTGGTTCTCGAATACGCAT) |
| pDHsp-EGFP-PmTBP141–200-V5-His | PmTBP-141-F (TCCCCGCGGGAGCTGAAGAAAATTGCACTC) |
| PmTBP-200-R (TCCCCGCGGCGCATACTTTCTTGCCGCAAG) | |
| pDHsp-EGFP-PmTBP171–230-V5-His | PmTBP-171-F (TCCCCGCGGACAACAGCCCTCATCTTCTCC) |
| PmTBP-230-R (TCCCCGCGGGATAGGGAACTTAACGTCACA) | |
| pDHsp-EGFP-PmTBP201–260-V5-His | PmTBP-201-F (TCCCCGCGGAGAATAGTGCAAAAGCTTGG) |
| PmTBP-260-R (TCCCCGCGGAGGCTTGACCATTCGGTAGAT) | |
| pDHsp-EGFP-PmTBP231–300-V5-His | PmTBP-231-F (TCCCCGCGGCGATTGGAGGGTCTTGTTCTC)/PmTBP-R |
| pET-28b-PmTBP | PmTBP-BamHI-F (CGCGGATCCAAGCATGGATCACATGCTGCCT) |
| PmTBP-XhoI-R (CCGCTCGAGTTTTTTGAAGCTTTTTAATATA) | |
| pAcMNPVie1Pro-V5-His | AcMNPV ie1-F (GGAATTCCATATGTCGATGTCTTTGTGATGCGCG) |
| AcMNPV ie1-R (CCCAAGCTTAGTCACTTGGTTGTTCACGATC) | |
| pAcMNPVie1Pro-PmTBP-V5-His | PmTBP-F/PmTBP-R |
Restriction enzyme cutting sites are underlined.
In vitro protein synthesis and GST pulldown assay.
A full-length PmTBP expression plasmid, pET-28b-PmTBP, was constructed by using PCR to clone the PmTBP gene coding region into the commercial vector pET-28b(+) (Novagen). Primers used for PCR cloning are listed in Table 2. The resulting plasmid, pET-28b-PmTBP, was transformed into Escherichia coli BL21 Codon Plus cells (Stratagene) and used for protein production. The transformed cells were grown in Luria-Bertani medium supplemented with kanamycin (50 μg/ml), and protein expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The induced bacteria were collected in ice-cold 1× phosphate-buffered saline (PBS) containing 10% glycerol and a protease inhibitor cocktail tablet. After sonication, the bacterial proteins were solubilized in 1× PBS containing 1.5% sodium lauryl sarcosine, and the recombinant PmTBP-His protein was purified by use of nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen). GST and GST-IE1 fusion proteins were also used in these assays, and they were expressed and purified according to the method of Liu et al. (26). The purified PmTBP-His, GST, and GST-IE1 proteins were all pretreated with nucleases as described above. Subsequently, PmTBP-His (1 μg) was incubated with GST-IE1 (10 μg) or GST (10 μg) bound to glutathione-Sepharose beads in 150 μl NETN buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, and a cocktail tablet of protease inhibitors) at 4°C for 3 h. After five 10-min washes with NETN buffer, the proteins that bound to the beads were resolved by 15% SDS-PAGE followed by Western blotting. The presence of PmTBP-His protein was detected using anti-His monoclonal antibody (Sigma) and goat anti-mouse IgG–HRP conjugate secondary antibody (Sigma).
Dual-luciferase reporter assay.
The firefly luciferase reporter plasmid G5p35BAS-Luc contains five copies of the GAL4 DNA binding site upstream of the Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) p35 basal promoter, and it was constructed as described previously (26). Another internal control reporter plasmid, phRL/AcMNPVie1, was used to monitor and normalize transfection efficiency. This plasmid used the AcMNPV ie1 promoter to drive the expression of the Renilla luciferase gene, and it was constructed as described by Liu et al. (24). Two effector plasmids were also constructed from pWSSV-V5-His as described previously (26): pWSSV-GAL4-V5-His encoded the GAL4 DNA binding domain (aa 1 to 147), while pWSSV-GAL4-IE1-V5-His consisted of the GAL4 DNA binding domain upstream of the in-frame WSSV ie1 coding sequence. To construct the PmTBP expression plasmid, the Orgyia pseudotsugata multicapsid nucleopolyhedrovirus ie2 promoter of pIZ/V5-His (Invitrogen) was replaced by the AcMNPV ie1 promoter to produce the plasmid pAcMNPVie1Pro-V5-His. The full-length PmTBP gene was then inserted into the pAcMNPVie1Pro-V5-His vector by PCR cloning to yield the plasmid pAcMNPVie1Pro-PmTBP-V5-His. The relevant primers are listed in Table 2.
For DNA transfection, Sf9 insect cells were seeded into a 24-well plate (2 × 105 cells/well) and grown overnight. The firefly luciferase reporter gene plasmid G5p35BAS-Luc, the internal control plasmid phRL/AcMNPVie1, and the effector plasmids were cotransfected into the Sf9 cells by use of Effectene reagent (Qiagen). The amounts of each plasmid used in the various transfections are indicated in Fig. 7. The total quantity of DNA in each transfection was kept constant by adding the empty vector pWSSV-V5-His or pAcMNPVie1Pro-V5-His as necessary. Cells were collected 48 h after transfection, and cell lysates were prepared according to the instruction manual for a dual-luciferase assay system (Promega). Luciferase activity was measured with a luminometer (Labsystems). Firefly luciferase activity values were then normalized against the activities of the Renilla luciferase to correct for transfection efficiency, and data were expressed as relative luciferase activities. Independent triplicate experiments were performed for each plasmid, and the means and standard deviations (SDs) were calculated.
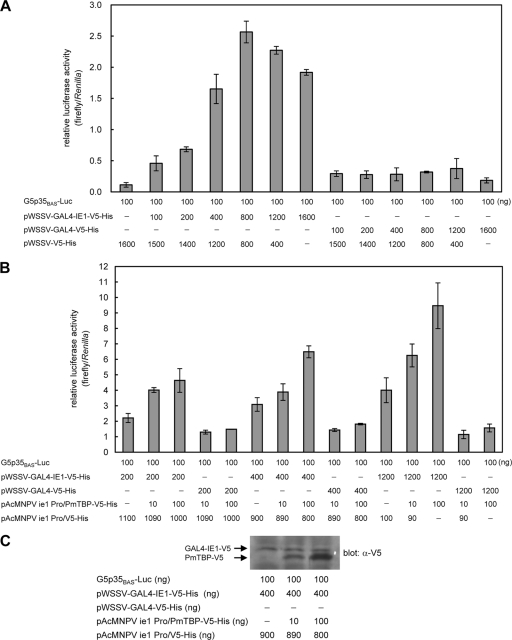
Fig. 7.
Dose-dependent effects of WSSV IE1 and PmTBP. (A) Relative luciferase activities in Sf9 cells that were cotransfected for 48 h with 100 ng of the reporter plasmid G5p35BAS-Luc and the indicated quantity of the pWSSV-GAL4-IE1-V5-His or pWSSV-GAL4-V5-His effector plasmid. The total amount of DNA used for cotransfection was held constant by adding pWSSV-V5-His vector. A Renilla luciferase reporter plasmid (10 ng; phRL/AcMNPVie1) (data not shown) was used to correct for transfection efficiency. Data show the means for three repetitions, and error bars show the SDs. (B) Relative luciferase activities in Sf9 cells cotransfected as described above, with the addition of the indicated quantity of the PmTBP expression plasmid pAcMNPVie1Pro-PmTBP-V5-His. The empty plasmid pAcMNPVie1Pro-V5-His was added to the DNA mix as indicated to hold the total amount of DNA constant. phRL/AcMNPVie1 (10 ng) was used to correct for transfection efficiency. Data show the means for three repetitions, and error bars show the SDs. (C) Western blot analysis to confirm the expression of GAL4-IE1-V5-His and PmTBP-V5-His. Cell lysates were taken from Sf9 cells that were cotransfected as indicated. Protein extracts were separated by SDS-PAGE and examined by Western blot analysis with an anti-V5 antiserum.
WSSV inoculum preparation.
The inoculum used in this study was prepared from the WSSV-TW strain (GenBank accession no. AF440570), which originated from a batch of WSSV-infected P. monodon shrimp collected in Taiwan in 1994 (43). Briefly, 0.5-g samples of the original frozen infected specimens of P. monodon were minced and then homogenized in 4.5 ml of sterile PBS. After centrifugation at 400 × g for 10 min at 4°C, the supernatant was filtered through a 0.45-μm membrane and used to infect adult specific-pathogen-free (SPF) Litopenaeus vannamei shrimp (body weight, 45 g; High Health Aquaculture Inc., HI) by injection as described previously (39). Hemolymph was collected from moribund shrimp. After removal of the hemocytes by centrifugation and filtration, followed by 4× dilution with PBS, the resulting suspension was stored at −80°C and used as a viral stock.
Experimental animals.
For the gene knockdown experiments, the more readily available SPF white shrimp, L. vannamei (∼5 g of body weight), was used instead of P. monodon. The batches of L. vannamei specimens used in this study were purchased from a commercial farm located in Pingtung, Taiwan, and were maintained in a water tank system with a salinity of 30 ppt at 22 to 25°C.
dsRNA synthesis.
Double-stranded RNA (dsRNA) was generated by in vitro transcription. Briefly, DNA templates corresponding to the ie1, PmTBP, and EGFP sequences were amplified by PCRs using gene-specific primers that incorporated the RNA polymerase T7 promoter sequence at the 5′ terminus. Separate sense and antisense RNA strands were then produced using the T7 RiboMAX Express large-scale RNA production system (Promega). Each corresponding single-stranded RNA was mixed together and annealed to generate dsRNA by incubation at 70°C for 10 min followed by gently cooling down to room temperature. The template DNA and any remaining single-stranded RNA were then removed using DNase and RNase following the manufacturer's protocols. After precipitation, dsRNA was verified by agarose gel electrophoresis, quantified by UV spectrophotometry, and stored at −80°C for in vivo gene silencing experiments.
In vivo gene silencing.
Shrimp (L. vannamei; mean weight, ∼5 g) were divided into five groups, with eight shrimp in each group. Each experimental shrimp was treated with dsRNA at a dosage of 1 μg dsRNA/g of shrimp body weight. The dsRNAs were prepared at a concentration of 1 μg dsRNA in 10 μl PBS and delivered into the shrimp by intramuscular injection. Dosages for each group were as follows: 50 μl of PBS (group 1), 2.5 μg EGFP dsRNA plus 2.5 μg ie1 dsRNA (group 2), 2.5 μg EGFP dsRNA plus 2.5 μg PmTBP dsRNA (group 3), 5 μg EGFP dsRNA only (group 4), and 2.5 μg ie1 dsRNA plus 2.5 μg PmTBP dsRNA (group 5). Forty-eight hours after injection, all of these shrimp were challenged with WSSV inoculum (100× dilution of the viral stock) by muscular injection. At 24 and 48 h postchallenge, the gills were collected from four of the shrimp in each group and preserved in liquid nitrogen for the subsequent assays. Total RNA was extracted from the gills with TRIzol reagent (Invitrogen) according to the procedure provided by the supplier. The extracted RNAs were then treated with DNase I to remove any residual DNA. The DNase I-treated total RNAs were primed with an oligo(dT) anchor primer (Roche) and reverse transcribed with SuperScript III reverse transcriptase (RT; Invitrogen) at 50°C for 60 min. Semiquantitative RT-PCR was used to evaluate the knockdown efficiency for ie1 expression. The expression level of a shrimp elongation factor gene, EF-1α, was used as an internal template control. The primer sequences used are listed in Table 3.
Table 3.
Primer sequences used for dsRNA production, RT-PCR, and real-time RT-PCR analysis
| Gene name | Primer name (sequence [5′–3′]) | Usage |
|---|---|---|
| ie1 | IE1-F (GAAGACTCTACAAATCTCTTT) | dsRNA production |
| IE1-R (CTTGCACCTACACGCATTACA) | dsRNA production | |
| IE1-T7-F (GGATCCTAATACGACTCACTATAGGGAAGACTCTACAAATCTCTTT) | dsRNA production | |
| IE1-T7-R (GGATCCTAATACGACTCACTATAGGCTTGCACCTACACGCATTACA) | dsRNA production | |
| PmTBP | PmTBP-F (CTGGAATGTTCGGTTCAGCC) | dsRNA production |
| PmTBP-R (GGCTTCGTAAATCTGGTTTC) | dsRNA production | |
| PmTBP-T7-F (GGATCCTAATACGACTCACTATAGGCTGGAATGTTCGGTTCAGCC) | dsRNA production | |
| PmTBP-T7-R (GGATCCTAATACGACTCACTATAGGGGCTTCGTAAATCTGGTTTC) | dsRNA production | |
| EGFP | EGFP-F (ATGGTGAGCAAGGGCGAGG) | dsRNA production |
| EGFP-R (TTACTTGTACAGCTCGTCCAT) | dsRNA production | |
| EGFP-T7-F (GGATCCTAATACGACTCACTATAGGATGGTGAGCAAGGGCGAGG) | dsRNA production | |
| EGFP-T7-R (GGATCCTAATACGACTCACTATAGGTTACTTGTACAGCTCGTCCAT) | dsRNA production | |
| ie1 | CGCGGATCCATGGCCTTTAATTTTGAAGAC | RT-PCR |
| TCCCCGCGGTACAAAGAATCCAGAAATCTC | RT-PCR | |
| EF-1α | GGAGATGCACCACGAAGCTC | RT-PCR |
| TTGGGTCCGGCTTCCAGTTC | RT-PCR | |
| PmTBP | GCAGCAGGGTCAGCAGATG | Real-time RT-PCR |
| CATTAAGGTGTGTGGCGTGAAC | Real-time RT-PCR | |
| dnapol | TCCAGCTCCATCACCCTCTT | Real-time RT-PCR |
| TGAATTTGGAATTGGTGTCGTAGA | Real-time RT-PCR | |
| EF-1α | TGCTCTGGACAACATCGAGC | Real-time RT-PCR |
| CGGGCACTGTTCCAATACCT | Real-time RT-PCR |
Relative quantitative real-time RT-PCR.
Total RNA was extracted and cDNA was synthesized as described above. The cDNAs were subjected to real-time RT-PCR analysis in a 20-μl reaction mixture containing 10 μl of Power SYBR green PCR master mix (Applied Biosystems), 2 μl of cDNA template, 4 μl of double-distilled H2O, and 2 μl (3 μM) (each) of forward and reverse primers for either the PmTBP or WSSV dnapol (DNA polymerase) gene or for the housekeeping gene control, shrimp EF-1α. The analysis was performed using an ABI Prism 7300 real-time PCR system (Applied Biosystems), and the PCR conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, with one dissociation stage at 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. The dissociation curve of each PCR product was a single peak. The ΔΔCT method was used to monitor the transcription levels of the shrimp PmTBP and WSSV dnapol target genes. The threshold cycle (CT) values of the amplified target genes (CT PmTBP and CT dnapol) and the internal control gene (CT EF-1α) in each sample were computed by the SDS program, using default parameters. ΔCT values were then calculated by subtracting CT EF-1α from CT PmTBP and CT dnapol. To normalize the data, the ΔCT value for each tested sample was subtracted from the mean ΔCT value for the calibrator group (i.e., the group 1 shrimp injected with PBS only) to produce a value called ΔΔCT. The change in transcription level of each sample relative to the mean transcription level of the calibrator group was then expressed as 2−ΔΔCT. WSSV loads in these experimental animals were quantified by the IQ Real WSSV quantitative system (GeneReach Biotechnology Corp.), a commercial real-time PCR system for WSSV load quantification based on the TaqMan assay strategy. Experimental data are presented as means ± standard errors of the means (SEM). In all experiments, statistically significant differences (P < 0.05) were assessed by a Duncan multiple-range test after one-way analysis of variance (ANOVA).
Nucleotide sequence accession number.
The PmTBP cDNA sequence was submitted to GenBank under accession number HQ599317.
RESULTS
Isolation and sequence analysis of PmTBP cDNA.
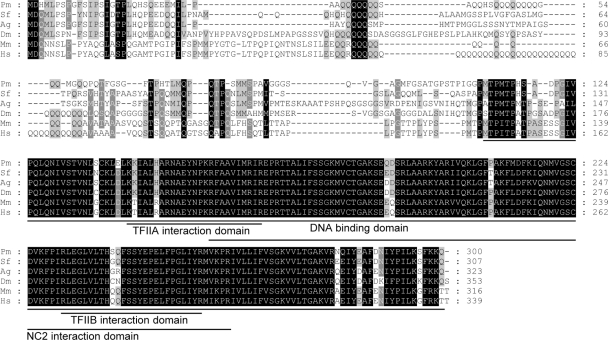
A partial sequence of PmTBP was cloned using degenerate primers based on the highly conserved C-terminal region of TBP (32). After 5′ and 3′ RACE, the full-length, 1,056-nt transcript of PmTBP was obtained, and its complete cDNA sequence was deposited in GenBank under accession no. HQ599317. The full-length PmTBP transcript contains a 5′-untranslated region (5′ UTR) of 84 nt, an open reading frame of 903 nt, and a 3′ UTR of 69 nt (data not shown). A multiple sequence alignment of the TBPs from several different species is shown in Fig. 1. The highly conserved C-terminal region contains several functional domains, including the DNA binding domain and TFIIA, TFIIB, and NC2 (negative cofactor 2) interaction domains, which in PmTBP are located at aa 111 to 300, aa 144 to 168, aa 231 to 256, and aa 158 to 262, respectively.
Fig. 1.
Multiple sequence alignment of deduced amino acid sequence of PmTBP and the sequences of five other ancient eukaryotic TBP family members. Pm, P. monodon; Dm, Drosophila melanogaster; Ag, Anopheles gambiae; Sf, Spodoptera frugiperda; Mm, Mus musculus; Hs, Homo sapiens. Identified conserved domains, including the DNA binding domain, the TFIIA interaction domain, the TFIIB interaction domain, and the NC2 interaction domain, are underlined.
WSSV IE1 interacts with PmTBP.
Interaction between WSSV IE1 and PmTBP was investigated using a coimmunoprecipitation assay in which FLAG-tagged IE1 (IE1-FLAG) and V5-tagged PmTBP (PmTBP-V5) were coexpressed in Sf9 insect cells. As shown in Fig. 2A, the inputs PmTBP-V5 and IE1-FLAG were expressed successfully in Sf9 cells. In the coimmunoprecipitation assay, complexes consisting of PmTBP-V5 plus IE1-FLAG were coimmunoprecipitated by anti-FLAG M2 affinity gel and detected by Western blotting using anti-V5 antibody. A pilot experiment confirmed that the IE1-FLAG protein could be precipitated efficiently by the anti-FLAG antibody (data not shown), and the binding specificity of IE1-FLAG with anti-FLAG M2 affinity gel was reconfirmed by subjecting the IE1-FLAG protein to incubation with anti-hemagglutinin (HA) antibody-conjugated beads (data not shown). A reverse experiment using FLAG-tagged PmTBP (PmTBP-FLAG) and V5-tagged IE1 (IE1-V5) produced the same results (Fig. 2B).
Fig. 2.
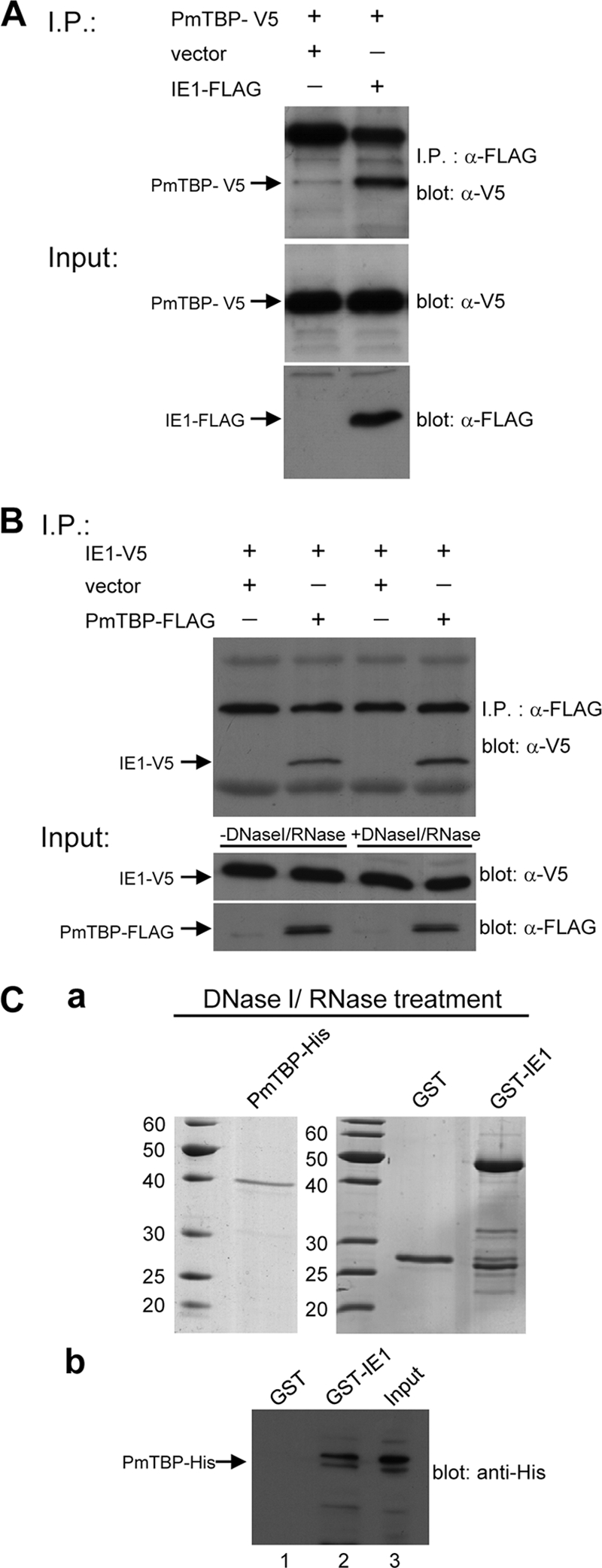
WSSV IE1 interacts with PmTBP. (A) Coimmunoprecipitation of V5-tagged PmTBP (PmTBP-V5) with FLAG-tagged IE1 (IE1-FLAG) from transfected cells. Sf9 cells were transfected with plasmids expressing PmTBP-V5 or IE1-FLAG or with empty plasmid (vector) as indicated. Six hours after heat shock, the cell lysates were harvested and immunoprecipitated with anti-FLAG M2 affinity resins, and then the immunoprecipitated complexes were subjected to Western blot analysis with an anti-V5 antibody probe. In the lower two panels, input expression was confirmed by Western blotting using either anti-V5 antibody or anti-FLAG antibody as a probe. The arrows indicate the expressed PmTBP-V5 and IE1-FLAG. (B) Interaction of WSSV IE1 with PmTBP does not require nucleic acids. After FLAG-tagged PmTBP (PmTBP-FLAG), V5-tagged IE1 (IE1-V5), or both were expressed in Sf9 cells, the lysates were pretreated with DNase I and RNase and subjected to coimmunoprecipitation. Inputs used in the coimmunoprecipitation assays, with or without DNase I and RNase pretreatment, were analyzed by Western blotting using either anti-V5 antibody or anti-FLAG antibody as a probe (lower two panels). Arrows indicate the expressed PmTBP-FLAG and IE1-V5. (C) GST pulldown assays of WSSV IE1 with PmTBP. (a) SDS-PAGE of DNase I- and RNase-pretreated purified histidine-tagged PmTBP (PmTBP-His), GST-IE1, and GST proteins. (b) DNase I/RNase-pretreated PmTBP-His was incubated with GST (lane 1) or GST-IE1 (lane 2), pelleted, washed, and detected by immunoblotting with anti-His antibody. PmTBP-His was also loaded into the gel (lane 3) for Western blot analysis.
WSSV IE1-PmTBP interaction does not require the presence of nucleic acids.
Previous results have shown that WSSV IE1 binds to DNA (26). To examine whether the presence of nucleic acid was required for the complex formation between IE1 and PmTBP, total cell extracts from pDHsp-IE1-V5-His- and pDHsp-PmTBP-FLAG-His-cotransfected Sf9 cells were treated with both DNase I and RNase before the immunoprecipitation assay. The DNase I and RNase degradation of nucleic acids in the cell lysate was confirmed by a PCR using WSSV IE1 primers (data not shown). As shown in Fig. 2B, the DNase I and RNase treatment had no impact on the WSSV IE1 and PmTBP interaction, suggesting that nucleic acids are not required for IE1-PmTBP complex formation. A GST pulldown assay was also performed with purified PmTBP-His, GST, and GST-IE1. After treatment with DNase I and RNase (Fig. 2C, panel a), PmTBP-His and GST-IE1 were still able to associate specifically with each other (Fig. 2C, panel b). This pulldown experiment confirmed the specific interaction between WSSV IE1 and PmTBP shown above (Fig. 2A and B) and also confirmed that this interaction is not mediated by DNA, RNA, or any other protein.
Mapping of WSSV IE1's PmTBP binding domain.
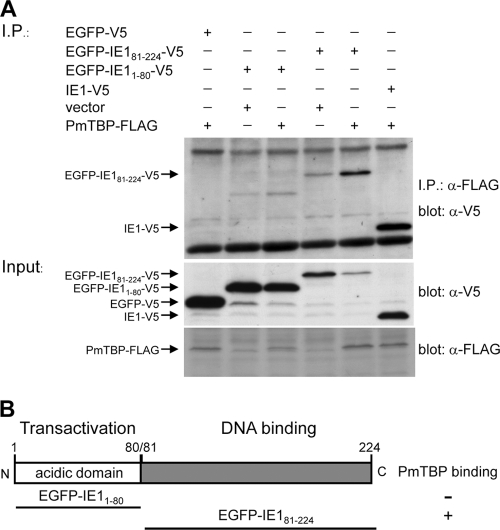
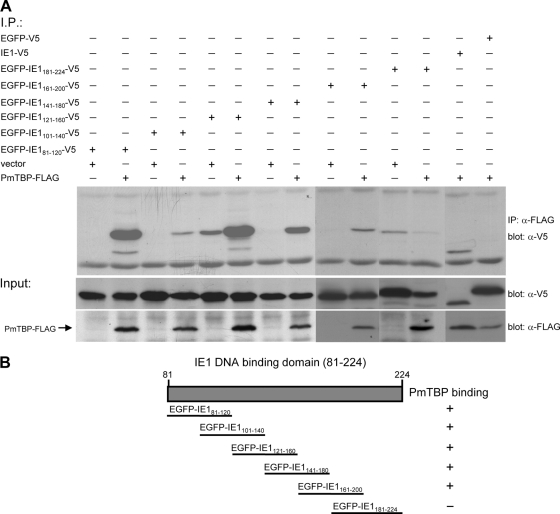
To map the regions of WSSV IE1 which interact with PmTBP, two WSSV IE1 gene fragments were cloned into pDHsp-EGFP-V5-His to construct the plasmids pDHsp-EGFP-IE11–80-V5-His and pDHsp-EGFP-IE181–224-V5-His. These plasmids express the fusion proteins EGFP-IE11–80-V5 and EGFP-IE181–224-V5, which correspond to IE1's transactivation domain and DNA binding domain, respectively. After cotransfection with the plasmid pDHsp-PmTBP-FLAG-His in Sf9 cells, the cell lysates were immunoprecipitated with anti-FLAG antibody-conjugated beads. The immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotting using anti-V5 antiserum. The results showed that PmTBP-FLAG interacted specifically with the WSSV IE1 DNA binding domain (EGFP-IE181–224-V5) but not with its activation domain (EGFP-IE11–80) (Fig. 3). To further identify the subregions within the WSSV IE1 DNA binding domain that confer the ability to bind with PmTBP, a series of overlapping EGFP-V5-fused WSSV IE1 DNA binding domain truncation mutants was constructed to cover the entire length of the IE1 DNA binding domain. Coimmunoprecipitation assays with PmTBP-FLAG showed that almost all of the fragments interacted with PmTBP; only the C-terminal fragment (aa 181 to 224) showed no PmTBP binding activity (Fig. 4).
Fig. 3.
Identification of the WSSV IE1 domain that interacts with PmTBP. (A) Coimmunoprecipitation of PmTBP-FLAG with IE1-V5, EGFP-V5, and two EGFP-V5 fusion proteins, containing the IE1 N-terminal transactivation domain and the IE1 C-terminal DNA binding domain (EGFP-IE11–80-V5 and EGFP-IE181–224-V5, respectively). In the lower two panels, Western blot analysis confirms the expression of the inputs in cell lysates from cotransfected Sf9 cells. (B) Schematic of the modular organization of the WSSV IE1 protein and a summary of the interaction domain analysis results.
Fig. 4.
Mapping the subregions of the WSSV IE1 DNA binding domain that facilitate interaction with PmTBP. (A) Coimmunoprecipitation of PmTBP-FLAG with EGFP-V5 fusion proteins containing different subregions of the WSSV IE1 DNA binding domain. Panels under the coimmunoprecipitation results confirm the expression of the inputs. (B) Schematic representation of WSSV IE1 DNA binding domain and the analyzed subdomains.
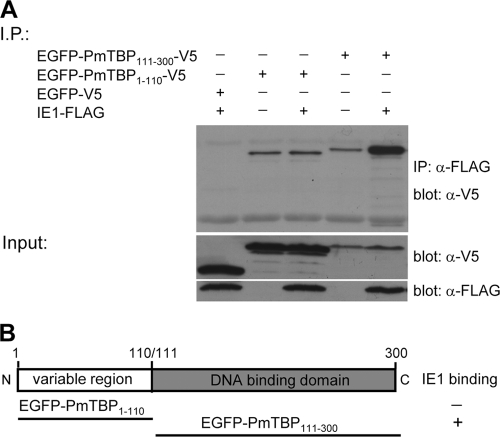
Mapping of PmTBP's WSSV IE1 binding domain.
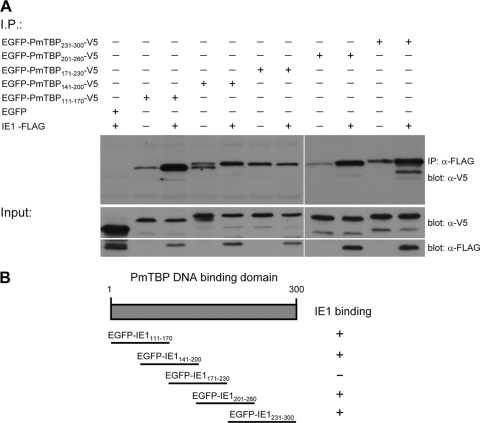
The WSSV IE1 binding regions of PmTBP were identified using a similar strategy to that described above. PmTBP's N-terminal variable region (aa 1 to 110) and C-terminal DNA binding region (aa 111 to 300) were cloned into pDHsp-EGFP-V5-His. The resulting constructs, pDHsp-EGFP-PmTBP1–110-V5-His and pDHsp-EGFP-PmTBP111–300-V5-His, were cotransfected into Sf9 cells with pDHsp-IE1-FLAG-His, and the extracted lysates were subjected to coimmunoprecipitation assays. The results showed that only the PmTBP C-terminal DNA binding domain (EGFP-PmTBP111–300-V5) interacted with WSSV IE1 (Fig. 5). As before, a series of overlapping EGFP-V5-fused PmTBP truncation mutants was constructed. Figure 6 shows that IE1 bound to almost all of the PmTBP C-terminal DNA binding domain fragments but not to the fragment spanning aa 171 to 230. Some of the bands seen in Fig. 6 were the result of nonspecific binding. This coimmunoprecipitation experiment was repeated three times, and nonspecific binding of the negative controls was observed consistently. The two types of bands can be distinguished clearly because the coprecipitated proteins always produced stronger bands than those that were the result of nonspecific binding.
Fig. 5.
Identification of the PmTBP domain that interacts with WSSV IE1. (A) Coimmunoprecipitation of IE1-FLAG with EGFP-V5 and two EGFP-V5 fusion proteins, containing the PmTBP N-terminal variable region and the PmTBP C-terminal DNA binding domain (EGFP-PmTBP1–110-V5 and EGFP-PmTBP111–300-V5, respectively). In the lower panels, Western blot analysis confirms the expression of inputs in cell lysates from cotransfected Sf9 cells. (B) Schematic of the modular organization of the full-length PmTBP protein and a summary of the interaction domain analysis results.
Fig. 6.
Mapping the subregions of the PmTBP DNA binding domain that facilitate interaction with IE1. (A) Coimmunoprecipitation of IE1-FLAG with EGFP-V5 fusion proteins containing different subregions of the PmTBP DNA binding domain. Panels under the coimmunoprecipitation results confirm input expression. (B) Schematic representation of the PmTBP DNA binding domain and the analyzed subdomains.
Effect of PmTBP overexpression on WSSV IE1 transcriptional activation.
Previously reported cotransfection experiments with Sf9 cells showed that a GAL4-IE1 fusion protein activated the transcription of a reporter construct with 5 upstream GAL4 binding sites, namely, G5p35BAS-Luc (26). In this study, cotransfection with different amounts of pWSSV-GAL4-IE1-V5-His showed that the stimulatory effect of GAL4-IE1 was dose dependent and that maximum transactivation activity was reached at 800 ng pWSSV-GAL4-IE1-V5-His (Fig. 7A). However, further increases in the transfection amount (1,200 ng and 1,600 ng) of pWSSV-GAL4-IE1-V5-His resulted in a phenomenon called “squelching”: the reporter activity was not elevated but was weakened. This squelching effect was eliminated by overexpression of PmTBP in the cotransfected Sf9 cells, which produced a further dose-dependent increase in luciferase activity (Fig. 7B). The amount of PmTBP present in the cotransfected cells was monitored by Western blotting (Fig. 7C).
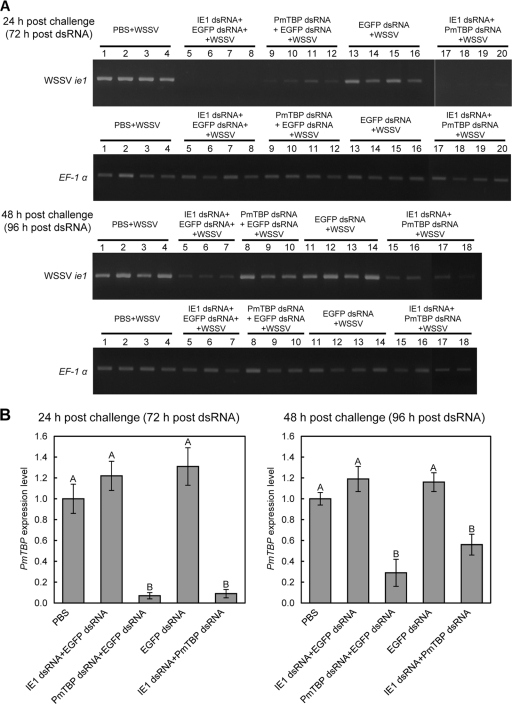
Silencing efficiencies of WSSV ie1 and PmTBP.
To characterize the roles of WSSV IE1 and PmTBP in regulating viral replication and gene expression, ie1 and PmTBP gene knockdown experiments were performed on WSSV-challenged shrimp. Two days prior to WSSV challenge, shrimp were injected with ie1 and PmTBP dsRNAs, and gills were collected at 24 and 48 h post-WSSV challenge. ie1 gene silencing efficiency was monitored using semiquantitative RT-PCR (Fig. 8A, lanes 5 to 8 and 17 to 20 for 24 h post-WSSV challenge and lanes 5 to 7 and 15 to 18 for 48 h post-WSSV challenge). PmTBP silencing efficiency was also monitored, but the expression levels of PmTBP were too low to be analyzed by semiquantitative RT-PCR, so real-time RT-PCR was used instead. Figure 8B shows that PmTBP expression was significantly suppressed both 72 and 96 h after dsRNA injection. These results indicated that injection of gene-specific dsRNAs led to substantial and specific depletion of the cognate mRNAs. We also noted that ie1 expression was noticeably inhibited in the EGFP dsRNA- and PmTBP dsRNA-injected shrimp at 24 h (Fig. 8A, top panel, lanes 9 to 12).
Fig. 8.
dsRNA knockdown of WSSV ie1 and PmTBP expression. (A) Semiquantitative RT-PCR detection of WSSV ie1 expression in gills of L. vannamei shrimp at 24 and 48 h post-WSSV challenge [i.e., 72 and 96 h after pretreatment with the indicated dsRNA(s)]. Shrimp EF-1α was included as a template control. RT-PCR products were analyzed in a 1.5% agarose gel. (B) Real-time RT-PCR quantification of expression levels of PmTBP after dsRNA pretreatment and WSSV challenge. Transcripts were measured in duplicate for every individual shrimp (n = 4 [or 3] for each experimental condition) and normalized against EF-1α. Values are means ± SEM. Statistically significant differences are indicated by different letters (A and B) (P < 0.05).
Effects of WSSV ie1 and PmTBP silencing on WSSV replication and gene transcription.
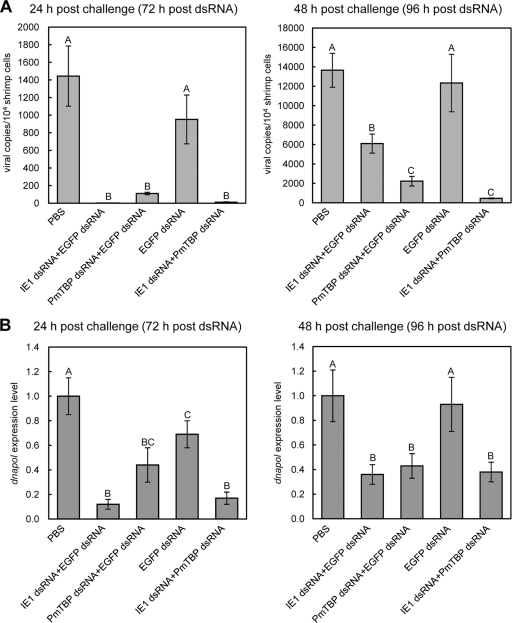
Both 24 h and 48 h after challenge, WSSV replication was effectively inhibited by ie1 dsRNA and by PmTBP dsRNA, both separately and combined (Fig. 9A). Virus loads in these shrimp were much lower than those in the shrimp injected with PBS and EGFP dsRNA. At 48 h postchallenge, there was a slight recovery in WSSV replication in shrimp injected with ie1 dsRNA only. In the second part of this assay, we monitored the transcription profile of a WSSV gene essential for DNA replication, i.e., dnapol. dnapol expression was significantly downregulated at both 24 h and 48 h in all of the ie1 dsRNA- and PmTBP dsRNA-injected shrimp, except for the PmTBP/EGFP dsRNA group at 24 h (Fig. 9B).
Fig. 9.
Real-time RT-PCR quantification of WSSV loads and dnapol expression levels after pretreatment with ie1 and PmTBP dsRNAs. WSSV copy numbers (A) and EF-1α-normalized WSSV dnapol expression levels (B) were determined for dsRNA-pretreated shrimp 24 and 48 h after WSSV challenge. One sample was taken from each experimental shrimp (n = 4 [or 3] for each group) and quantified in duplicate. Values are means ± SEM. Statistically significant differences are indicated by different letters (A, B, and C) (P < 0.05).
DISCUSSION
WSSV IE1 is a potent trans-activator (26), and like all other transcriptional activators, it includes a DNA binding domain and an activation domain (31). Figure 3 shows that WSSV IE1 interacts with PmTBP through its DNA binding domain, not through its activation domain. This experiment was repeated several times, and the same results were always observed. Further confirmation was provided by the following experiment, which showed that almost all of the subcloned WSSV IE1 DNA binding domain fragments were able to bind with PmTBP (Fig. 4). This result was surprising, because for all other IE proteins for which the location of the TBP binding domain is known, it has always been found in the activation domain. Examples include human cytomegalovirus IE2 (6), Epstein-Barr virus (EBV) Zta (21), and herpes simplex virus type 1 (HSV-1) ICP4 (5). Two of these IE proteins (IE2 and Zta) are known to bind specifically to the C terminus of TBP (12, 21), and this is also the case for WSSV IE1 (Fig. 5 and 6). This was expected because the TBP C-terminal DNA binding domain is one of the most evolutionarily conserved protein domains known, whereas the TBP N-terminal region varies greatly between taxa (13) (Fig. 1). We also note that Fig. 1 shows that P. monodon TBP resembles the human, mouse, and Drosophila TBPs in having a glutamine-rich region which is not found in the TBPs of Anopheles gambiae and Spodoptera frugiperda. (The evolutionary significance of this, if any, is not currently known.)
It is possible for proteins with DNA binding activity to associate indirectly with each other through intermediate nucleic acids. However, in the case of WSSV IE1 and PmTBP, Fig. 2B shows that they were able to associate even when no nucleic acids were present. This was further confirmed by in vitro interaction assays with nuclease-treated purified recombinant proteins (Fig. 2C). We therefore concluded that the interaction between IE1 and PmTBP is direct and does not depend on additional bridging factors.
Interaction between WSSV IE1 and PmTBP is critical for promoter activation by IE1 in vitro, and as shown in Fig. 7A, increasing the intracellular concentration of GAL4-IE1 augments transactivation by IE1. However, this is true only to a certain point, and above a certain critical amount (800 ng in Fig. 7A), additional GAL4-IE1 has the effect of inhibiting transactivation instead. This phenomenon has already been reported for many other transcriptional activators and is known as “squelching” (3, 4, 31). The squelching effect is due to a limiting amount of the activator target protein. This implies that if the WSSV IE1 target protein is PmTBP, then overexpression of this protein should relieve squelching. Conversely, if a rate-limiting intermediate molecule is involved, then increasing the amounts of these two proteins should increase squelching, since the intermediary factor would then be titrated out by both the activator and the general transcription factor. We found that transfection with 10 and 100 ng of PmTBP expression vectors resulted in dose-dependent increases in luciferase activity and that this was true even at a “squelching” concentration (1,200 ng) of pWSSV-GAL4-IE1-V5. Based on these observations, we concluded that there is a direct interaction between WSSV IE1 and PmTBP and that the squelching effect was in fact due to the limiting amount of PmTBP.
Like other viral IE proteins, WSSV IE1 is thought to play a critical role in WSSV replication (23). Here we provided more evidence that dsRNA silencing of ie1 leads to reduced WSSV genome replication (Fig. 9A). Presumably, this is because smaller amounts of IE1 result in lower expression levels of its target early and late genes. Although the downstream target genes of IE1 are still unknown, we found here that ie1 silencing caused reduced expression of at least one of the WSSV genes involved in the viral replication cycle, i.e., dnapol (Fig. 9B). dsRNA silencing of PmTBP had a similar effect (Fig. 9B), which suggests that PmTBP binds to the TATA box that is known to be present in many WSSV early and late genes. We note that double silencing with the ie1 dsRNA and the nontarget EGFP dsRNA control resulted in a very low viral copy number in the first 24 h after infection but that the viral copy number increased considerably in the next 24 h (Fig. 9A). In the course of a WSSV infection, the viral copy number usually increases dramatically from 24 to 48 h after infection (9), and since RNA interference in vivo is probably unable to completely suppress expression of the target gene in every cell throughout the whole animal, it is likely that some WSSV IE1 transcripts will escape from some infected cells. With increasing time after challenge, as more IE1 transcripts accumulate, the ie1 dsRNA interference effect will increasingly be reduced, so that by 48 h after infection, it is not surprising that there are many more viral copies. This effect was not seen after double silencing with ie1 dsRNA and PmTBP dsRNA. Presumably, this is because simultaneous silencing of two target genes has a synergistic effect that substantially increases the suppression of IE1 expression.
The data presented here clearly suggest that WSSV IE1 interacts directly with PmTBP. This kind of direct interaction between an IE protein and a component of the cellular transcription machinery may have several functions. For example, the adenovirus IE protein EIA, which binds to TBP and to the sequence-specific transcription factor ATF-2, is thought to stimulate transcription through TBP after recruitment to the promoter by ATF-2 (15, 18, 22). Similarly, EBV Zta binds a DNA motif upstream of the target promoter and appears to stimulate transcription by stabilizing the binding of TBP to DNA (21). Lastly, Sampath and DeLuca (34) have shown that HSV-1 ICP4 is essential for the formation of the TBP and RNA polymerase II transcription complexes that promote expression of the viral early and late genes in virus-infected cells. It seems likely that WSSV IE1 will eventually be shown to fulfill one or more of these functions, but more work will be needed to investigate these various possibilities.
In addition to directly binding to PmTBP in the transcription complex to enhance or repress downstream gene expression, recombinant WSSV IE1 is also known to bind directly to DNA in vitro (26). WSSV IE1 therefore seems to resemble EBV Zta in that it appears to interact both directly and indirectly with its target DNA. However, to confirm this hypothesis, further characterization of WSSV IE1 will be needed.
ACKNOWLEDGMENTS
This investigation was supported financially by the National Science Council (NSC 98-2311-B-002-013-MY2-2).
We are indebted to Paul Barlow for his helpful criticism.
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Blissard G. W. 1996. Baculovirus-insect cell interactions. Cytotechnology 20:73–93 [DOI] [PubMed] [Google Scholar]
- 2. Blissard G. W., Rohrmann G. F. 1990. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 35:127–155 [DOI] [PubMed] [Google Scholar]
- 3. Boyer T. G., Berk A. J. 1993. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 7:1810–1823 [DOI] [PubMed] [Google Scholar]
- 4. Caron C., et al. 1993. Functional and biochemical interaction of the HTLV-1 tax1 transactivator with TBP. EMBO J. 12:4269–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carrozza M. J., Deluca N. A. 1996. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol. Cell. Biol. 16:3085–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caswell R., et al. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691–2698 [DOI] [PubMed] [Google Scholar]
- 7. Chen L.-L., et al. 2002. Transcriptional analysis of the DNA polymerase gene of shrimp white spot syndrome virus. Virology 301:136–147 [DOI] [PubMed] [Google Scholar]
- 8. Cormack B. P., Struhl K. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69:685–696 [DOI] [PubMed] [Google Scholar]
- 9. Du H. H., Li W. F., Xu Z. R., Kil Z. S. 2006. Effect of hyperthermia on the replication of white spot syndrome virus (WSSV) in Procambarus clarkii. Dis. Aquat. Organ. 71:175–178 [DOI] [PubMed] [Google Scholar]
- 10. Escobedo-Bonilla C. M., et al. 2008. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis. 31:1–18 [DOI] [PubMed] [Google Scholar]
- 11. Friesen P. D., Miller L. K. 1986. The regulation of baculovirus gene expression. Curr. Top. Microbiol. Immunol. 131:31–49 [DOI] [PubMed] [Google Scholar]
- 12. Hagemeier C., Walker S. M., Caswell R. C., Kouzarides T., Sinclair J. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hernandez N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291–1308 [DOI] [PubMed] [Google Scholar]
- 14. Honess R. W., Roizman B. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horikoshi N., et al. 1991. Direct interaction between adenovirus EIA protein and the TATA box binding transcription factor IID. Proc. Natl. Acad. Sci. U. S. A. 88:5124–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang X. D., et al. 2010. Shrimp NF-κB binds to the immediate-early gene ie1 promoter of white spot syndrome virus and upregulates its activity. Virology 406:176–180 [DOI] [PubMed] [Google Scholar]
- 17. Kovacs G. R., Choi J., Guarino L. A., Summers M. D. 1992. Functional dissection of the Autographa californica nuclear polyhedrosis virus immediate-early 1 transcriptional regulatory protein. J. Virol. 66:7429–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee W. S., Kao C. C., Bryant G. O., Liu X., Berk A. J. 1991. Adenovirus EIA activation domain binds the basic repeat in the TATA box transcription factor. Cell 67:365–376 [DOI] [PubMed] [Google Scholar]
- 19. Leu J.-H., Kuo Y.-C., Kou G.-H., Lo C.-F. 2008. Molecular cloning and characterization of an inhibitor of apoptosis protein (IAP) from the tiger shrimp, Penaeus monodon. Dev. Comp. Immunol. 32:121–133 [DOI] [PubMed] [Google Scholar]
- 20. Li, et al. 2009. Identification of the immediate-early genes of white spot syndrome virus. Virology 385:267–274 [DOI] [PubMed] [Google Scholar]
- 21. Lieberman P. M., Berk A. J. 1991. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 5:2441–2454 [DOI] [PubMed] [Google Scholar]
- 22. Liu F., Green M. R. 1990. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell 61:1217–1224 [DOI] [PubMed] [Google Scholar]
- 23. Liu H., Soderhall K., Jiravanichpaisal P. 2009. Antiviral immunity in crustaceans. Fish Shellfish Immunol. 27:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu W.-J., Chang Y.-S., Wang A. H.-J., Kou G.-H., Lo C.-F. 2007. White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J. Virol. 81:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu W.-J., Chang Y.-S., Wang C.-H., Kou G.-H., Lo C.-F. 2005. Microarray and RT-PCR screening for white spot syndrome virus immediate-early genes in cycloheximide-treated shrimp. Virology 334:327–341 [DOI] [PubMed] [Google Scholar]
- 26. Liu W.-J., et al. 2008. Transactivation, dimerization, and DNA-binding activity of white spot syndrome virus immediate early protein IE1. J. Virol. 82:11362–11373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lo C.-F., et al. 1996. White spot syndrome baculovirus (WSBV) detected in cultured and captured shrimp, crabs and other arthropods. Dis. Aquat. Organ. 27:215–225 [Google Scholar]
- 28. Lo C.-F., Peng S.-E., Chang Y.-S., Kou G.-H. 2005. White spot syndrome—what we have learned about the virus and the disease, p. 421–433 In Walker P. J., Lester R. G., Bondad-Reantaso M. G. (ed.), Diseases in Asian aquaculture. V. Proceedings of the 5th Symposium on Diseases in Asian Aquaculture Fish Health Section, Asian Fisheries Society, Manila, Philippines [Google Scholar]
- 29. Marks H., Vorst O., van Houwelingen A. M., van Hulten M. C. W., Vlak J. M. 2005. Gene-expression profiling of white spot syndrome virus in vivo. J. Gen. Virol. 86:2081–2100 [DOI] [PubMed] [Google Scholar]
- 30. Marks H., Ren X.-Y., Sandbrink H., van Hulten M. C. W., Vlak J. M. 2006. In silico identification of putative promoter motifs of white spot syndrome virus. BMC Bioinformatics 7:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ptashne M. 1988. How eukaryotic transcriptional activators work. Nature 335:683–689 [DOI] [PubMed] [Google Scholar]
- 32. Rasmussen C., Rohrmann G. F. 1994. Characterization of the Spodoptera frugiperda TATA-binding protein: nucleotide sequence and response to baculovirus infection. Insect Biochem. Mol. Biol. 24:699–708 [DOI] [PubMed] [Google Scholar]
- 33. Roeder R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327–335 [PubMed] [Google Scholar]
- 34. Sampath P., DeLuca N. A. 2008. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 immediate-early, early, and late promoters in virus-infected cells. J. Virol. 82:2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sánchez-Paz A. 2010. White spot syndrome virus: an overview on an emergent concern. Vet. Res. 41:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith C. A., Bates P., Rivera-Gonzalez R., Gu B., Deluca N. A. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA binding protein and TFIIB. J. Virol. 67:4676–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stenberg R. M. 1996. The human cytomegalovirus major immediate-early gene. Intervirology 39:343–349 [DOI] [PubMed] [Google Scholar]
- 38. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsai M.-F., et al. 1999. Long-term presence of white spot syndrome virus (WSSV) in a cultivated shrimp population without disease outbreaks. Dis. Aquat. Organ. 38:107–114 [Google Scholar]
- 40. van Hulten M. C. W., et al. 2001. The white spot syndrome virus DNA genome sequence. Virology 286:7–22 [DOI] [PubMed] [Google Scholar]
- 41. Verrijer C. P., Tjian R. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338–342 [PubMed] [Google Scholar]
- 42. Vlak J. M., et al. 2004. Nimaviridae, p. 187–192 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA [Google Scholar]
- 43. Wang C.-H., et al. 1995. Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Penaeus monodon. Dis. Aquat. Organ. 23:239–242 [Google Scholar]
- 44. West J. T., Wood C. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150–5163 [DOI] [PubMed] [Google Scholar]
- 45. Yang F., et al. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811–11820 [DOI] [PMC free article] [PubMed] [Google Scholar]