Abstract
Coronaviruses are a family of enveloped single-stranded positive-sense RNA viruses causing respiratory, enteric, and neurologic diseases in mammals and fowl. Human coronaviruses are recognized to cause up to a third of common colds and are suspected to be involved in enteric and neurologic diseases. Coronavirus replication involves the generation of nested subgenomic mRNAs (sgmRNAs) with a common capped 5′ leader sequence. The translation of most of the sgmRNAs is thought to be cap dependent and displays a requirement for eukaryotic initiation factor 4F (eIF4F), a heterotrimeric complex needed for the recruitment of 40S ribosomes. We recently reported on an ultrahigh-throughput screen to discover compounds that inhibit eIF4F activity by blocking the interaction of two of its subunits (R. Cencic et al., Proc. Natl. Acad. Sci. U. S. A. 108:1046–1051, 2011). Herein we describe a molecule from this screen that prevents the interaction between eIF4E (the cap-binding protein) and eIF4G (a large scaffolding protein), inhibiting cap-dependent translation. This inhibitor significantly decreased human coronavirus 229E (HCoV-229E) replication, reducing the percentage of infected cells and intra- and extracellular infectious virus titers. Our results support the strategy of targeting the eIF4F complex to block coronavirus infection.
INTRODUCTION
Coronaviruses are ubiquitous enveloped single-stranded positive-sense RNA viruses that contain a 27- to 32-kb genome and cause a variety of respiratory, enteric, and neurologic diseases in mammals and fowl (12). Although they have been known since the mid 1960s, human coronaviruses (HCoV) (23) gained notoriety following the 2002-2003 outbreak of one of their family members, SARS-CoV (severe acute respiratory syndrome coronavirus). By virtue of an RNA-dependent RNA polymerase, a series of 3′-coterminal nested subgenomic mRNAs (sgmRNAs) are generated that contain a common 5′ leader sequence, but the mechanism by which they recruit ribosomes is not known (11).
In eukaryotes, cellular translation is thought to occur by one of two mechanisms: a cap-dependent mechanism involving eukaryotic initiation factor 4F (eIF4F) and a cap-independent mechanism in which ribosomes are recruited internally to an internal ribosome entry site (IRES) via specific mRNA structural motifs present upstream of the initiation codon. eIF4F consists of eIF4E, the cap-binding protein; eIF4A, an RNA helicase; and eIF4G, a large scaffolding protein. There are two isoforms of eIF4G (eIF4GI and eIF4GII) that are 46% identical. All structural features of eIF4GI are present on eIF4GII. eIF4E is the least abundant initiation factor, and it is thought that this renders cap recognition the rate-limiting step in initiation. The availability of eIF4E to assemble into the eIF4F complex is regulated by the mTOR pathway due to sequestration by binding to a negative regulator, 4E-BP (there are three 4E-BPs in a cell, with the best-characterized one being 4E-BP1). Upon activation by upstream signals, mTOR phosphorylates 4E-BP1, which causes the eIF4E–4E-BP1 binary complex to dissociate. Since eIF4G and the 4E-BPs share binding sites on eIF4E, the removal of 4E-BP1 allows eIF4E to enter the eIF4F complex. When mTOR activity is reduced (e.g., amino acid starvation or energy depletion), eIF4F levels are diminished due to the sequestration of eIF4E by 4E-BPs, and cap-dependent translation is squelched. The overexpression of 4E-BP1 in cells impairs coronavirus replication, suggesting that the translation of some viral sgmRNAs or mRNA encoding a cellular factor(s) required for viral replication is cap dependent (5).
We recently reported on an ultrahigh-throughput screen that identified several new inhibitors of eIF4E-eIF4G interaction (7). Herein, we report on the characterization of an inhibitor of eIF4E-eIF4G interaction, called 4E2RCat. We demonstrate that this inhibitor is capable of blocking coronavirus replication as monitored by viral protein expression and the production of infectious virus.
MATERIALS AND METHODS
Ultrahigh-throughput screening for eIF4E-eIF4G inhibitors.
High-throughput screening for eIF4E-eIF4G interaction inhibitors has been described in detail previously (7, 8). In brief, a time-resolved fluorescence resonance energy transfer (TR-FRET)-based high-throughput assay was miniaturized to a 1,536-well format and optimized. It was used to screen a library of 217,341 compounds from the Molecule Library Screening Centers Network, from which 4E2RCat (PubChem no. 2287238) was identified.
In vitro translations.
In vitro transcriptions and translations of bicistronic mRNA reporters were performed as described previously (16). Firefly (FF) and renilla (Ren) luciferase (Luc) activities (RLU) were measured using a Berthold Lumat LB 9507 luminometer. To visualize in vitro-translated proteins, reactions were performed in micrococcal nuclease-treated Krebs extract in the presence of [35S]methionine. Translations were analyzed on a 10% SDS-polyacrylamide gel, followed by staining with Coomassie blue and destaining to ensure equal loading. Gels were further incubated for 1 h in En3Hance (Perkin), dried, and exposed to X-Omat film (Kodak).
GST pulldowns.
For pulldown experiments, either glutathione S-transferase (GST)-eIF4GI517-606 or bovine serum albumin (BSA) was coupled to Affi-Gel-10 (Bio-Rad). To this end, 1 mg of protein was incubated at 2.5 mg/ml with 400 μl of a 50% slurry of Affi-Gel-10 beads in 10 ml AC buffer (20 mM HEPES, pH 7.5, 10% glycerol, 1 mM dithiothreitol [DTT], 1 mM EDTA) end over end for 12 h. After being centrifuged for 10 min at 1,200 × g, the supernatant was removed and beads incubated with AC buffer supplemented with 75 mM NaCl and 80 mM ethanolamine for 1 h end over end at 4°C. After an additional spin to remove the supernatant, beads were blocked for 1 h with 8 ml AC buffer supplemented with 75 mM NaCl and 1 mg/ml BSA. Beads were washed once with 20 ml AC buffer supplemented with 1 M NaCl and three times with AC buffer supplemented with 75 mM NaCl. Beads were stored as 50% slurry in AC buffer containing 75 mM NaCl.
For pulldown experiments involving eIF4E, 20 μl of a 50% slurry of Affi-Gel matrix coupled to either GST-eIF4GI517-606 or BSA was preincubated with 100 μM 4E2RCat. Recombinant eIF4E also was preincubated with 4E2RCat or vehicle (DMSO) for 1 h end over end at room temperature. Recombinant eIF4E and Affi-Gel-bound proteins then were mixed and incubated for another hour end over end at room temperature. Beads were washed three times for 10 min with 10 volumes of binding buffer, resuspended in SDS-PAGE loading dye, and resolved by 10% SDS-PAGE. Western blotting was used to detect the presence of eIF4E (Santa Cruz).
Pulldowns with GST-eIF4GII555-658 and GST-4E-BP1 were performed with 2.5 μg of the GST fusion proteins and 0.25 μg eIF4E. Proteins were individually incubated for 1 h at room temperature in the presence of 25 μM 4E2RCat in binding buffer (20 mM Tris, pH 7.5, 100 mM KCl, 10% glycerol, and 0.1% NP-40). After incubation, eIF4E was added to the GST-tagged partners and incubated for another hour, followed by addition to glutathione beads for another hour at room temperature. Beads were washed three times with 10 volumes of binding buffer and eluted for 1 h using reduced glutathione (10 mM). The eluents were separated on 10% SDS-PAGE followed by transfer to a polyvinylidene difluoride (PVDF) membrane (Millipore) and Western blot analysis. Primary anti-GST and anti-eIF4E antibodies were from Santa Cruz. Secondary antibodies were from Jackson Immuno Research.
In vivo metabolic labeling.
In vivo [35S]methionine labeling was performed by seeding 60,000 cells/well in 24-well plates 24 h prior to treatment. Cells were treated for 4 or 24 h in the presence of increasing concentrations of 4E2RCat. For the last hour, medium was replaced by methionine-free Dulbecco's modified essential medium (DMEM) supplemented with 10% dialyzed serum, and for the last 15 min cells were labeled with [35S]methionine (150 to 225 μCi/ml). Cells were washed with PBS and lysed in RIPA buffer (20 mM Tris7.5, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 20 mM ß-glycerophosphate, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 4 μg/ml aprotinin, 2 μg/ml leupeptin, 2 μg/ml pepstatin). Samples were precipitated with trichloroacetic acid (TCA), and radioactivity was determined by scintillation counting. Protein concentrations in each sample were measured using the Bio-Rad DC protein assay (Bio-Rad Laboratories) and used to standardize the counts obtained after TCA precipitation.
eIF4F pulldown experiments.
For pulldown experiments, a ribosome salt wash (RSW) was incubated in the presence of 1% dimethylsulfoxide (DMSO) or 25 μM 4E2RCat for 1 h at 30°C. Following incubation, 50 μl of 50% m7GTP-Sepharose beads (GE Healthcare) was added to the reaction mixtures. After end-over-end rotation for 2 h at 4°C, beads were washed three times with 500 μl of LCB (20 mM HEPES, pH 7.5, 100 mM KCl, 0.2 mM EDTA) and once with 50 μl of LCB containing 1 mM GTP. Proteins were eluted with 50 μl of LCB containing 1 mM m7GTP for 10 min on ice. Eluents were separated by 10% SDS-PAGE and transferred to a PVDF membrane (Millipore), and Western blot analysis was performed. Primary antibodies used were anti-eIF4E (Santa Cruz), anti-eIF4GI (Bethyl), and anti-eIF4A (9). Secondary antibodies were from Jackson Immuno Research.
Modeling of 4E2RCat to eIF4E.
Computational solvent mapping (4) of the three-dimensional structures of eIF4E has been performed previously (7) and was used to identify the binding site for 4E2RCat. The available X-ray structures of human eIF4E were downloaded from the Protein Data Bank. Mapping was performed after removing all other molecules except for m7GTP, since 4E2RCat does not compete with 5′ mRNA cap structures for binding to eIF4E. The atomic coordinates of 4E2RCat were obtained from PubChem, but the molecule was considered flexible in docking. MGLtools (version 1.5.4) was used to prepare the ligand and receptor for docking and the graphical front end for setting up and running the AutoDock docking software. AutoDock Vina 1.1.0 (with standard settings) was used to perform the docking (24). The most likely binding pose for 4E2RCat was selected using an algorithm that scores bound poses based on the degree of overlap between the compound and the atom densities calculated from the solvent-mapping results, and the most likely binding pose for 4E2RCat was also the lowest energy pose.
Virus and cell lines.
Strain 229E of human coronavirus (HCoV-229E) originally was obtained from the American Type Culture Collection (ATCC) and was grown in the human L132 cell line (ATCC CCL5), which was expanded in alpha minimum essential medium (α-MEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS). L132 cells were infected in triplicate at a multiplicity of infection (MOI) of 0.1 and incubated for 2 h at 33°C in the absence of any compound. After two washes with phosphate-buffered saline (PBS), cells were incubated in α-MEM supplemented with 1% (vol/vol) FBS and containing DMSO (negative control) or compounds at the indicated concentrations at 33°C for up to 48 h.
For poliovirus infections, 3 × 105 HeLa cells were seeded per well in a six-well plate the day before infection. For virus adsorption, cells were washed with PBS and the Mahoney strain of poliovirus type 1 was added at 2 PFU/cell in 200 μl serum-free DMEM. Cells were incubated at room temperature for 30 min with gentle rocking, followed by the removal of the medium and two washes with PBS. At this point, fresh DMEM (containing 10% heat-inactivated FBS) containing either vehicle (1% DMSO) or 50 μM 4E2RCat was added to the cells, and cells were incubated for 4 h at 37°C. For the last 30 min, [35S]methionine (150 μCi/ml) was added to the wells. For harvesting, cells were washed with PBS and lysed in PLB (PBS, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1 mM PMSF). Samples were analyzed on a 10% SDS-polyacrylamide gel, followed by being stained with Coomassie blue and being destained to ensure equal loading. Gels were further incubated for 1 h in En3Hance (Perkin), dried, and exposed to X-Omat film (Kodak).
Quantitation of infectious virus titers by IPA.
The immunoperoxidase assay (IPA) was performed on L132 cells as previously described (13). Briefly, the primary antibody used was monoclonal antibody (MAb) 5-11H.6 directed against the S protein of HCoV-229E. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (KPL). Immune complexes were detected by incubation with 0.025% (wt/vol) 3,3′-diaminobenzidine tetrahydrochloride (Bio-Rad) and 0.01% hydrogen peroxide in PBS, and infectious virus titers were calculated by the Karber method as previously described (13).
Determination of cell viability.
To determine cell viability in the presence of 4E2RCat, 200,000 L132 cells/well were seeded in six-well plates 24 h before treatment. The next day, cells were treated with 12.5 μM 4E2RCat for the indicated times, after which the cells were processed for annexin V/propidium iodide staining. To this end, cell medium was collected. Cells were washed with 1 ml PBS, which was collected as well, and trypsinized in 200 μl 0.05% trypsin-EDTA. Cells were pooled with previously collected supernatants and spun for 2 min at 2,000 rpm and 4°C. The cell pellet was washed with 2 ml cold PBS, followed by another spin. After the second spin, the cell pellet was resuspended in 100 μl annexin V binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) and propidium iodide added to a final concentration of 5 μg/ml. After the addition of 5 μl annexin V-fluorescein isothiocyanate (FITC) (BD Biosciences), samples were incubated for 15 min in the dark at room temperature and diluted with 400 μl annexin V binding buffer. Fluorescence-activated cell sorter (FACS) analyses were performed using a FACScan instrument from BD Biosciences and CELLQUEST software.
Immunofluorescence assay (IFA) and semiquantitative analyses to determine percent S protein-positive cells.
L132 (9 × 104) cells were seeded on coverslips and grown overnight in α-MEM supplemented with 10% (vol/vol) FBS and infected the next day. At specific time points following treatment, cells were fixed with 4% paraformaldehyde (PFA) in PBS for 30 min at room temperature. Fixed cells were permeabilized by incubation with cold methanol for 5 min, followed by a wash in PBS and the detection of viral antigens. The primary antibody was mouse MAb 5-11H.6, and the secondary antibody was Alexa Fluor-488 mouse-specific goat antibody (Invitrogen). After three washes with PBS, fixed cells were incubated for 5 min with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) at 1 μg/ml to stain the DNA in the nucleus.
To determine the percent viral S protein-positive cells, 10 fields of L132 cells (containing a total of 150 to 250 cells; ×200 magnification with a Nikon Eclipse E800 microscope) were counted for each compound tested in three independent experiments. Green cells were scored as HCoV-229E S protein positive out of the total amount of cells (DAPI stained in blue) for each condition.
RESULTS
Characterization of 4E2RCat, an inhibitor of eIF4E-eIF4G interaction.
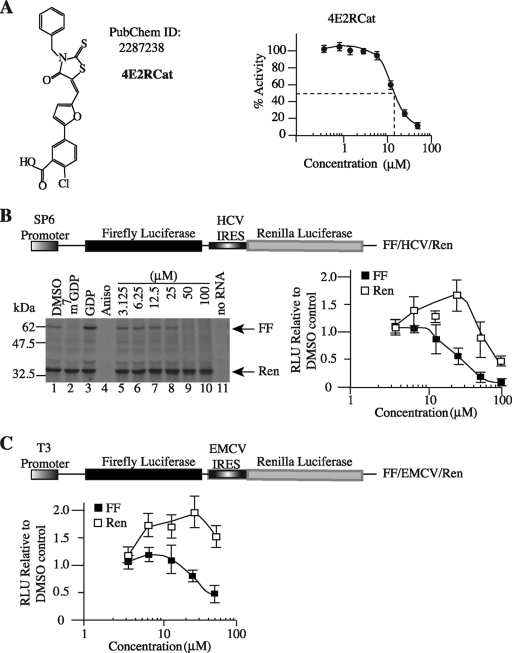
Compound 4E2RCat was identified from an HTS campaign undertaken to identify inhibitors of eIF4E-eIF4G interaction (Fig. 1A) (7). The titration of 4E2RCat in a TR-FRET-based assay monitoring the interaction between eIF4E and eIF4GI revealed a 50% inhibitory concentration (IC50) of 13.5 μM (Fig. 1A). The activity of 4E2RCat was tested on the translation of FF/hepatitis C virus (HCV)/Ren mRNA, a bicistronic mRNA construct that contains a cap-dependent firefly (FF) cistron and a viral IRES-dependent renilla (HCV/Ren) cistron that is not dependent on eIF4F for its initiation (Fig. 1B). The specificity of the bicistronic mRNA was assessed using m7GDP as a specific inhibitor and GDP as a nonspecific inhibitor of cap-dependent in vitro translation. As expected, m7GDP inhibited the translation of FF but not HCV/Ren (Fig. 1B, compare lane 2 to lane 1), whereas GDP had no effect on the translation of either of the two cistrons (Fig. 1B, compare lane 3 to lane 1). The elongation inhibitor anisomycin equally inhibited the translation of the cap-dependent FF and IRES-dependent Ren cistron (compare lane 4 to lane 1). 4E2RCat inhibited cap-dependent translation in a dose-dependent manner (Fig. 1B, compare lanes 5 to 10 to lane 1). Translation from the HCV IRES was not affected by 4E2RCat except at 100 μM, suggesting some off-target effects at this very high concentration (Fig. 1B). We note that an increase in HCV-driven translation is observed upon the inhibition of cap-dependent translation at 25 μM and likely reflects the increased availability of free ribosomes, an effect previously reported with other eIF4F inhibitors (2, 3, 7). Another IRES, the encephalomyocarditis virus (EMCV) IRES, which requires eIF4A and eIF4G but not eIF4E for initiation, also was tested, and we observed that 4E2RCat inhibited cap-dependent FF translation but not EMCV IRES-driven Ren translation (Fig. 1C).
Fig. 1.
Inhibition of cap-dependent translation by 4E2RCat. (A) Schematic diagram illustrating the structure of 4E2RCat. An 8-point dose-response curve of 4E2RCat in a TR-FRET assay is provided to the right. (B) Inhibition of translation by 4E2RCat. Schematic representation of FF/HCV/Ren bicistronic construct used for in vitro translation studies (top). In vitro translations were performed in Krebs extracts programmed with FF/HCV/Ren in the presence of [35S]methionine, and a representative autoradiograph of the products after fractionation on 10% SDS-PAGE is provided (bottom left). Translations contained vehicle (1% DMSO) (lane 1), 500 μM m7GDP (lane 2), 500 μM GDP (lane 3), 50 μM anisomycin (lane 4), the indicated concentrations of 4E2RCat (lanes 5 to 10), or no RNA (lane 11). FF and Ren RLU values (relative to DMSO controls) from two independent experiments are provided with the standard errors of the means (SEM) indicated (bottom right). (C) Schematic representation of FF/EMCV/Ren bicistronic construct used for in vitro translation studies (top). RLU values (relative to those of the DMSO control) from two independent in vitro translations performed in Krebs extract programmed with FF/EMCV/Ren mRNA are provided with the SEM indicated (bottom).
4E2RCat blocks interaction of eIF4E with its binding partners.
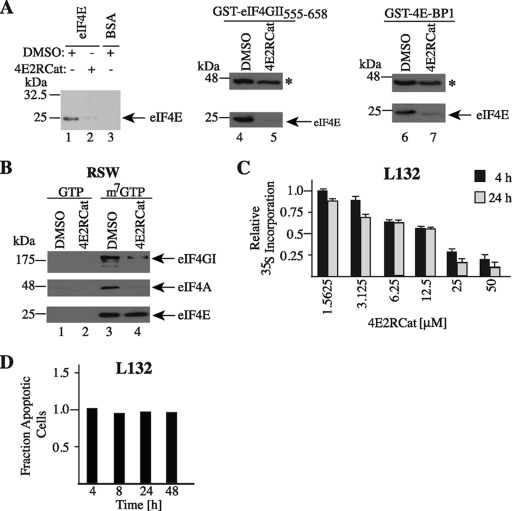
We next investigated the ability of 4E2RCat to inhibit the interaction of eIF4E with its binding partners (Fig. 2). To this end, we performed pulldown assays with eIF4E and either eIF4GI, eIF4GII, or 4E-BP1 (Fig. 2A). We noted that 4E2RCat inhibited interactions between eIF4E and GST-eIF4GI517-606 (compare lane 2 to 1), eIF4E and GST-eIF4GII555-658 (compare lane 5 to 4), and eIF4E and GST-4E-BP1 (compare lane 7 to 6). We also gauged the ability of 4E2RCat to disrupt preformed eIF4F complex. 4E2RCat was added to RSW, a rich source of eIF4F complex, and m7GTP affinity pulldowns were performed to purify eIF4E and its binding partners. The m7GTP eluents demonstrated equivalent recovery of eIF4E from RSW containing either vehicle or 4E2RCat but a significant reduction in the amount of copurifying eIF4GI and eIF4A (Fig. 2B). Compound 4E2RCat inhibited protein synthesis in vivo (Fig. 2C), and this was not a consequence of increased cell death (Fig. 2D).
Fig. 2.
Inhibition of eIF4E-eIF4G and eIF4E-4E-BP1 interaction by 4E2RCat. (A) Assessing the effect of 4E2RCat on the interaction between eIF4E and its binding partners. On the left, eIF4E (lanes 1 and 2) or BSA (lane 3) and Affi-Gel-coupled GST-eIF4GI517-606 were incubated in the presence of vehicle (1% DMSO) or 100 μM 4E2RCat, and the effects on interaction were assessed in pull-down assays as described in Materials and Methods. The gels in the middle and on the right show the consequences of 4E2RCat on eIF4E-GST-eIF4GII555-568 and eIF4E-GST-4E-BP1 interaction. The asterisk denotes the position of the migration of the GST fusion protein. (B) Effect of 4E2RCat on eIF4F assembly. RSW was incubated with vehicle or 25 μM 4E2RCat for 1 h at 30°C, followed by pulldowns using 50 μl of 50% m7GTP-Sepharose beads (GE Healthcare) for 2 h end over end at 4°C. GTP and m7GTP eluents are presented. (C) Inhibition of translation in vivo by 4E2RCat. L132 cells were exposed to the indicated concentrations of 4E2RCat for 4 or 24 h, after which metabolic labeling was performed. Results are the averages from triplicates with the errors of the means shown, and values are standardized against total protein content. (D) 4E2RCat does not induce cell death. Fraction of apoptotic cells following exposure of L132 cells to 12.5 μM 4E2RCat for the indicated time periods. Samples were prepared as described in Materials and Methods, and flow cytometry was performed to determine the fraction of apoptotic cells relative to the value for the DMSO vehicle control, which was set to 1.
SAR and in silico modeling of 4E2RCat.
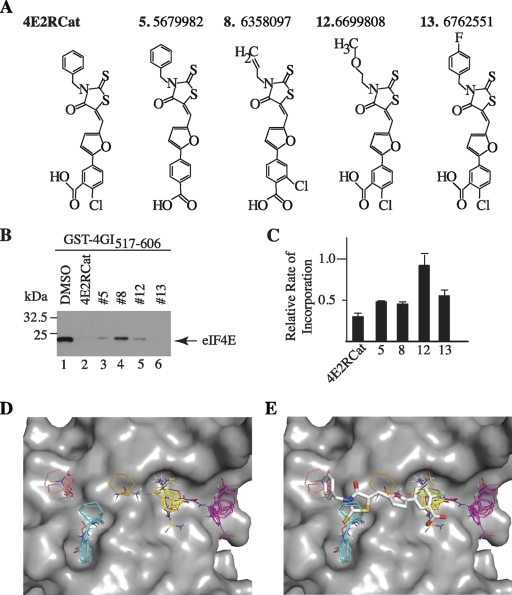
To determine if we could find compounds related to 4E2RCat, we performed structure-activity relationship (SAR) analysis on 4E2RCat using the TR-FRET assay and tested 19 analogs in in vitro translation assays (data not shown). The four more potent compounds (Fig. 3A) were further tested in pulldown assays with eIF4E and eIF4GI517-606 (Fig. 3B) and for effects on cellular protein synthesis (Fig. 3C). None of the congeners tested were as potent as 4E2RCat in preventing eIF4E-eIF4G interaction and in blocking protein synthesis in vivo (Fig. 3B and C).
Fig. 3.
SAR and in silico analysis of 4E2RCat. (A) Chemical structures of 4 of the 19 most potent congeners tested that inhibited cap-dependent translation in vitro. (B) Affi-Gel pulldown experiments with GST-eIF4GI517-606 and eIF4E in the presence of either DMSO (1%) or the indicated compounds at a final concentration of 100 μM. (C) Inhibition of in vivo protein synthesis of analogs of 4E2RCat. MDA-MB-231 cells were treated for 4 h with the indicated compounds at a concentration of 25 μM, after which metabolic labeling was performed. Results are the averages from duplicates with the errors of the mean shown, and values are standardized against total protein content. (D) Location of the largest hot spots of eIF4E. The site (shown in yellow) binds 24 probe clusters and defines the main hot spot. The other large consensus sites are shown in magenta (22 probe clusters), cyan (19 probe clusters), and salmon (10 probe clusters). A small consensus site is shown in ochre (5 probe clusters) and indicates a shallow channel connecting two other consensus sites. (E) The most likely binding pose of 4E2RCat. The predicted hot spots are superimposed for reference.
In silico probing of the surface of eIF4E for potential hot spots for small molecule binding identified five shallow pockets (Fig. 3D). Modeling the binding of 4E2RCat shows the potential to bind into four out of the five pockets (Fig. 3E). This is predicted to clash with eIF4E-eIF4G and eIF4E-4E-BP1 interaction and is consistent with the pull-down experiments described above.
4E2RCat inhibits coronavirus replication.
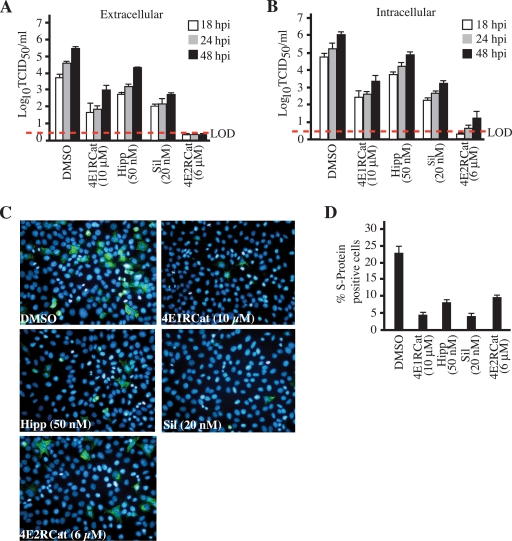
We next sought to test whether 4E2RCat could be used to block the replication of human coronavirus, strain 229E, which previously was shown to be inhibited upon the overexpression of 4E-BP1 (5). The inhibition of translation with the eIF4A inhibitors hippuristanol or silvestrol caused a 10- to 100-fold reduction in infectious virus titers released from infected cells (Fig. 4A). In the presence of 4E1RCat, a previously reported inhibitor of eIF4E-eIF4G interaction (7), viral titers were reduced 100-fold (Fig. 4A). In the presence of 4E2RCat, no extracellular infectious virus could be detected. We also scored for the presence of intracellular infectious viral particles and found similar results: significantly reduced viral progeny upon the inhibition of eIF4A or upon exposure to 4E1RCat or 4E2RCat, although the latter was more potent (Fig. 4B). In two different experiments, a small amount of infectious HCoV-229E was present in infected cells: <10 TCID50/ml at 24 h postinfection (hpi) and <20 TCID50/ml at 48 hpi. This represents a reduction of almost 106-fold compared to levels for control DMSO-treated cells. Furthermore, all compounds reduced the percentage of cells expressing the viral S protein by ∼2- to 4-fold (Fig. 4C and D).
Fig. 4.
Inhibition of coronavirus replication by 4E2RCat. (A) Coronavirus replication is eIF4F dependent. Following infection and exposure to either vehicle (0.2% DMSO) or the indicated concentrations of hippuristanol, silvestrol, 4E1RCat, or 4E2RCat, infectious viral titers released from the cells (extracellular) were determined, and the averages from three independent experiments are presented. Error bars denote standard errors of the means. LOD, limit of detection (denoted by the red dashed line). (B) Effects of eIF4A and eIF4E-eIF4G inhibition on intracellular infectious virus production. (C) Effects of eIF4A and eIF4E-eIF4G inhibition on the production of HCoV-229E S protein. Following infection and exposure to vehicle or compounds for 24 h, infected cells were processed for immunofluorescence using a mouse IgG1 MAb, 5-11H.6, followed by AlexaFluor-488 anti-mouse goat antibody (green). Nuclei are stained with DAPI (blue). (D) Percent S protein-positive cells in HCoV-229E-infected cells.
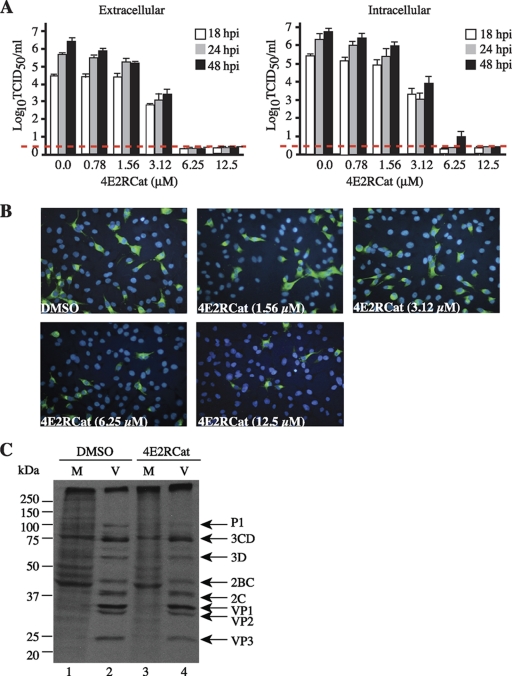
Titration of 4E2RCat onto HCoV-229E-infected L132 cells showed a dose- and time-dependent response in the detection of intracellular and extracellular virus production, with very little extracellular virus at 48 h postinfection at 6.25 μM (Fig. 5A). Accordingly, the decrease in the percentage of cells expressing viral S protein also was dose dependent (Fig. 5B). Unlike findings previously reported for the murine coronavirus MHV-JHM strain (21), HCoV-229E did not induce a host shutoff upon the infection of L132 cells (data not shown). This, and the absence of immunoprecipitating antibodies to detect other coronavirus proteins besides S, prevented us from detecting viral proteins and, consequently, the ongoing protein synthesis of other viral products.
Fig. 5.
Effects of 4E2RCat on coronavirus infection. (A) 4E2RCat inhibits coronavirus replication in a dose- and time-dependent manner. Following the exposure of HCoV-229E-infected L132 cells to the indicated concentrations of 4E2RCat for the indicated periods of time, infectious viral titers released from the cells (extracellular) or from intracellular infectious virus were determined, and the averages from three independent experiments are presented. Error bars denote SEM. The limit of detection is denoted by the red dashed line. (B) Dose-dependent inhibition of HCoV-229E S protein production by 4E2RCat. Following infection and exposure to vehicle or the indicated concentrations of 4E2RCat for 24 h, infected cells were processed for immunofluorescence using mouse IgG1 MAb 5-11H.6 followed by AlexaFluor-488 anti-mouse goat antibody (green). Nuclei are stained with DAPI (blue). (C) Metabolic labeling of HeLa cells infected with type 1 poliovirus (Mahoney) followed by treatment with vehicle (1% DMSO) or 50 μM 4E2RCat for 4 h.
To determine if 4E2RCat had potent off-target effects that would block the replication of any virus, we tested its ability to inhibit poliovirus protein expression. Poliovirus, a member of the picornaviruses, does not require eIF4E for the expression of its proteins, and its translation is cap independent, recruiting ribosomes internally through the use of an IRES (18). At concentrations that significantly reduced host protein synthesis (Fig. 2C), 4E2RCat had no effect on the synthesis of poliovirus proteins (Fig. 5C). These results suggest that the observed inhibition of coronavirus replication by 4E2RCat is not the consequence of nonspecific toxicity at the cellular level.
DISCUSSION
Little is known concerning the mechanism of coronavirus genomic and sgmRNA translation. A genomic species and series of sgmRNA species containing a common 5′ leader and 3′ terminus are generated during the viral life cycle, where presumably the first open reading frame (ORF1) of the transcript dictates the polypeptide synthesized (11). The translation of the coronavirus sgmRNAs is thought to be cap dependent (20). Consistently with this, the overexpression of a nonphosphorylatable form of the translation repressor, 4E-BP1, has been shown to reduce viral replication by ∼90% (5). At ∼8 h postinfection, Akt signaling is activated in SARS-CoV-infected cells, an event that leads to increased mTOR activity and favors eIF4F formation and the increased translation of select mRNAs (see Introduction) (14). Furthermore, murine coronavirus induces p38 mitogen-activated protein kinase (MAPK) activity, which leads to the Mnk-dependent phosphorylation of eIF4E, an event associated with the stimulation of protein synthesis (1).
The genomic RNA and all sgmRNAs contain a common 5′ leader sequence of 65 to 90 nucleotides predicted to fold into a modestly stable secondary structure (ΔG = −14 kcal/mmol) (17). There is a large (24- to 400-fold) difference in expression levels observed among the sgmRNA transcripts (25), and thus it has been proposed that additional cis- and/or trans-acting features of the sgmRNAs are responsible for these differences (22). Hence, we would not necessarily expect all sgmRNAs to be equally inhibited by inhibitors of translation, such as 4E2RCat. Further experiments are required to determine the cap dependency of the individual sgmRNAs and, in particular, if they show a high dependency on eIF4F for ribosome recruitment, a feature that may explain why complete inhibition of coronavirus replication is observed at 6.25 μM (Fig. 5A), a concentration that inhibits host protein synthesis by only ∼40% (Fig. 2C) and reduces the production of the HCoV-229E S protein in infected cells by ∼2.4-fold (Fig. 4D).
Inhibiting the cap-dependent translation of coronaviruses represents a particular vulnerability in the viral replication cycle, since it is one of the steps in which viral replication is dependent on host translation factors. It is known from studies where cap-dependent translation has been targeted in preclinical cancer models that reducing eIF4E levels to inhibit this step using antisense oligonucleotides (10), inhibiting eIF4A with small molecule inhibitors (3, 6), or blocking eIF4E-eIF4G interaction with 4E1RCat (7) is well tolerated in vivo. The efficacy of 4E2RCat in vivo on animal infection models remains to be established.
A third inhibitor of eIF4E-eIF4G interaction, called 4EGI-1, has been reported; it differs from 4E2RCat in that 4EGI-1 increases the interaction between eIF4E and 4E-BP1 (15). One would predict that 4EGI-1 also inhibits coronavirus replication, but clearly that has to be tested. The modeling of 4E2RCat bound to eIF4E predicts that this compound binds to the same region used by eIF4G and 4E-BP1 to interact with eIF4E (Fig. 3). Consistently with this, we found the interaction of both eIF4G and 4E-BP1 with eIF4E being inhibited by 4E2RCat (Fig. 2). The modeling results also suggest a way to improve the specificity and potency of 4E2RCat (Fig. 3). 4E2RCat occupies four of the five “hot spots” present in the eIF4G/4E-BP binding region, and extending 4E2RCat into the fifth unoccupied hot spot (shown in magenta in Fig. 3) could improve the affinity of 4E2RCat for eIF4E.
We note that 4E2RCat has a stronger inhibitory effect on extra- and intracellular coronavirus infectious virus production than the eIF4A inhibitors hippuristanol and silvestrol (Fig. 4A and B). Hippuristanol prevents eIF4A from binding to RNA (2), whereas silvestrol sequesters eIF4A from the eIF4F complex (3, 6). It may be that coronavirus translation is less dependent on eIF4A for initiation but still requires intact eIF4E-eIF4G dimers for ribosome recruitment. Alternatively, other helicases implicated in translation initiation, such as DHX29 (19), might substitute for eIF4A in coronavirus replication. Our results do not exclude the possibility that in addition to inhibiting translation, 4E2RCat could also affect viral gene expression at other steps, although 4E2RCat does not appear to be inducing a nonspecific antiviral state at the cellular level, since high concentrations of 4E2RCat had little effect on poliovirus replication (Fig. 5C). We note that the efficient inhibition of translation from the genomic RNA (giving rise to the pp1a and pp1ab polyproteins) or of only one of the six HCoV-229E sgmRNAs (RNAs 2 to 7) could suffice to curtail a productive infection. This may be the case in our study, where the production of the viral S protein (encoded by sgmRNA 2) appears partially inhibited by 6 μM 4E2RCat, while infectious virus production is almost completely abrogated (Fig. 4A and D).
We previously reported on another inhibitor of eIF4G-eIF4E interaction, called 4E1RCat (7). In our hands, 4E2RCat appears to be a more potent inhibitor than 4E1RCat, even though both compounds target the same interaction. Whether this relates to more potent in vivo inhibitory properties remains to be determined. Although both compounds exhibit similar IC50s in an in vitro TR-FRET assay monitoring eIF4E-eIF4G interaction (7 and this paper), one cannot assume that similar IC50s are to be expected in the viral replication assay described herein, since chemicophysiological differences between each compound could dramatically alter their behavior ex vivo and in vivo.
In summary, we have shown that the inhibition of cap-dependent translation, using small molecule inhibitors of eIF4E-eIF4G interaction or inhibiting eIF4A activity, is an effective strategy to curtail coronavirus replication. In particular, targeting the ribosome recruitment step of protein synthesis blocked viral gene expression in cells and inhibited the formation of infectious viral progeny (Fig. 4 and 5). Our results are consistent with the idea that the translation of at least some coronavirus sgmRNAs is eIF4F dependent and provide a rationale for the development and use of small molecule inhibitors of translation initiation as potential antiviral agents targeting human and animal coronaviruses, including SARS.
ACKNOWLEDGMENTS
We thank Marilyn Carrier and Isabelle Harvey for excellent technical assistance.
Fellowship support was from a CIHR Cancer Consortium Training Grant Award and a Cole Foundation Award to R.C. This work was supported by grants from the National Institutes of Health (1 R01 CA114475 and MH081216-01) and the Canadian Cancer Society Research Institute (17099) to J.P., a grant MT-9203 from the Institute of Infection and Immunity, Canadian Institutes of Health Research, to P.J.T., and NIH PHS 5U54 HG003918 to R.D. and H.F.
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Banerjee S., Narayanan K., Mizutani T., Makino S. 2002. Murine coronavirus replication-induced p38 mitogen-activated protein kinase activation promotes interleukin-6 production and virus replication in cultured cells. J. Virol. 76:5937–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bordeleau M.-E., et al. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213–220 [DOI] [PubMed] [Google Scholar]
- 3. Bordeleau M.-E., et al. 2008. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 118:2651–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brenke R., et al. 2009. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics 25:621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burgui I., Yanguez E., Sonenberg N., Nieto A. 2007. Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 81:12427–12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cencic R., et al. 2009. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One 4:e5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cencic R., et al. 2011. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc. Natl. Acad. Sci. U. S. A. 108:1046–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cencic R., Yan Y., Pelletier J. 2007. Homogenous time resolved fluorescence assay to identify modulators of cap-dependent translation initiation. Comb. Chem. High Throughput Screen. 10:181–188 [DOI] [PubMed] [Google Scholar]
- 9. Edery I., et al. 1983. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J. Biol. Chem. 258:11398–11403 [PubMed] [Google Scholar]
- 10. Graff J. R., et al. 2007. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 117:2638–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lai M. M., Cavanagh D. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lai M. M. C., Perlman S., Anderson L. J. 2007. Coronaviridae: coronavirus replication, p. 1305–1335 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 13. Lambert F., Jacomy H., Marceau G., Talbot P. J. 2008. Titration of Human coronaviruses, HCoV-229E and HCoV-OC43, by an indirect immunoperoxidase assay. In Cavanagh D. (ed.), SARS and other coronaviruses: laboratory protocols. Methods in molecular biology, vol. 454, chapter 8 Humana Press, Totowa, NJ: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. 2004. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology 327:169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moerke N. J., et al. 2007. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128:257–267 [DOI] [PubMed] [Google Scholar]
- 16. Novac O., Guenier A. S., Pelletier J. 2004. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 32:902–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Page K. W., Britton P., Boursnell M. E. 1990. Sequence analysis of the leader RNA of two porcine coronaviruses: transmissible gastroenteritis virus and porcine respiratory coronavirus. Virus Genes. 4:289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pelletier J., Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325 [DOI] [PubMed] [Google Scholar]
- 19. Pisareva V. P., Pisarev A. V., Komar A. A., Hellen C. U., Pestova T. V. 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135:1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siddell S., Snijder E. J. 2008. An introduction to nidoviruses, p. 1–13 In Perlman S., Gallagher T., Snijder E. J. (ed.), The nidoviruses. ASM Press, Washington, DC [Google Scholar]
- 21. Siddell S., Wege H., Barthel A., ter Meulen V. 1981. Coronavirus JHM: intracellular protein synthesis. J. Gen. Virol. 53:145–155 [DOI] [PubMed] [Google Scholar]
- 22. Tahara S. M., et al. 1994. Coronavirus translational regulation: leader affects mRNA efficiency. Virology 202:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Talbot P. J., Jacomy H., Desforges M. 2008. Pathogenesis of human coronaviruses, other than severe acute respiratory syndrome coronavirus, p. 313–324 In Perlman S., Gallagher T., Snijder E. J. (ed.), The nidoviruses. ASM Press, Washington, DC [Google Scholar]
- 24. Trott O., Olson A. J. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y., Hussain S., Wang H., Ke M., Guo D. 2009. Translational control of the subgenomic RNAs of severe acute respiratory syndrome coronavirus. Virus Genes. 39:10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]