Abstract
Pseudorabies virus (PRV), a member of the Alphaherpesvirinae, has a complex multilayered extracellular virion that is structurally conserved among other herpesviruses. PRV virions contain a double-stranded DNA genome within a proteinaceous capsid surrounded by the tegument, a layer of viral and cellular proteins. The envelope layer, which encloses the capsid and tegument, contains viral transmembrane proteins anchored in a phospholipid bilayer. The viral and host proteins contained within virions execute important functions during viral spread and pathogenesis, but a detailed understanding of the composition of PRV virions has been lacking. In this report, we present the first comprehensive proteomic characterization of purified PRV virions by mass spectrometry using two complementary approaches. To exclude proteins present in the extracellular medium that may nonspecifically associate with virions, we also analyzed virions treated with proteinase K and samples prepared from mock-infected cells. Overall, we identified 47 viral proteins associated with PRV virions, 40 of which were previously localized to the capsid, tegument, and envelope layers using traditional biochemical approaches. Additionally, we identified seven viral proteins that were previously undetected in virions, including pUL8, pUL20, pUL32, pUL40 (RR2), pUL42, pUL50 (dUTPase), and Rsp40/ICP22. Furthermore, although we did not enrich for posttranslational modifications, we detected phosphorylation of four virion proteins: pUL26, pUL36, pUL46, and pUL48. Finally, we identified 48 host proteins associated with PRV virions, many of which have known functions in important cellular pathways such as intracellular signaling, mRNA translation and processing, cytoskeletal dynamics, and membrane organization. This analysis extends previous work aimed at determining the composition of herpesvirus virions and provides novel insights critical for understanding the mechanisms underlying PRV entry, assembly, egress, spread, and pathogenesis.
INTRODUCTION
Pseudorabies virus (PRV) is a swine alphaherpesvirus closely related to the human pathogens herpes simplex virus type 1 and 2 (HSV-1 and HSV-2) and varicella-zoster virus (VZV) (96, 113). PRV is a pantropic, neuroinvasive virus with a broad host range and has been used extensively as a powerful tool for studying the architecture of mammalian neuronal circuits (35). The infectious particle of PRV, known as the mature virion, is a multilayered structure that is conserved among all herpesvirus families. At the core of the virion is a linear double-stranded DNA genome that is packaged within a proteinaceous capsid. The capsid is surrounded by a layer of viral and cellular proteins known as the tegument, which is enclosed within a phospholipid bilayer studded with viral transmembrane proteins and is known as the virion envelope (96, 113). To understand the fundamental mechanisms underlying PRV spread and pathogenesis, a comprehensive understanding of the subunits that compose the highly complex virion structure is necessary.
The initiation of infection, beginning at virion attachment and entry, is dependent upon the proteins that are incorporated within mature virions. Attachment of viral particles to heparan sulfate proteoglycans on the cell surface is dependent upon the glycoproteins gC (97, 140) and gB (53, 77), and virion binding to herpesvirus entry mediators (HVEMs) requires the glycoprotein gD (44, 101). Fusion of the virion envelope with the plasma membrane of the cell requires the glycoproteins gB, gH, and gL (22, 51, 52, 125). Following penetration of capsids and tegument proteins into the cell, many of the tegument proteins previously contained within the virion are released into the cytoplasm (46, 85). These proteins serve a variety of functions that are important for the early stages of infection, including takeover of the host protein synthesis machinery, immune evasion, and modulation of viral and host gene expression. For example, the virion host shutoff tegument protein (vhs) is an endonuclease that degrades mRNAs nonspecifically and thus accelerates the turnover of many host and viral mRNAs (130). The function of vhs is important for inhibiting the interferon-mediated antiviral response during infection in vivo (64, 82). Additionally, upon entry, viral capsids and a specific subset of tegument proteins are trafficked toward the nucleus (46, 85). One of these proteins is the large inner tegument protein pUL36, which facilitates interactions with microtubule-dependent molecular motor proteins (86, 114) as well as docking of capsids to the nuclear pore (25).
Viral membrane and tegument proteins are also critical for proper assembly and morphogenesis of viral particles in infected cells. Following nuclear egress, capsids containing newly replicated viral genomes undergo maturation in the cytoplasm, where they acquire tegument proteins and bud into vesicles that give rise to the final envelope (96). This process is driven by an intricate network of protein-protein interactions between proteins within the capsid, tegument, and envelope layers of the virion (36, 78, 95, 134). For example, both of the envelope glycoproteins gM and gE interact with the tegument protein VP22 (pUL49) and are required for its efficient incorporation into virions (39, 99). In turn, VP22 has also been shown to interact with the tegument protein pUL48, which has been shown to interact with gD and gH (20). Overall, while many PRV gene products have been identified as structural components of virions using a variety of approaches, an understanding of which viral proteins constitute a complete PRV virion is lacking. Furthermore, previous studies have shown that host proteins are packaged in PRV virions and also virions of other herpesviruses. However, little is known about which host proteins are included, whether or not they are incorporated specifically, and, importantly, what functions they may serve during infection.
Mass spectrometry-based proteomic analysis is a powerful and sensitive approach that has been used successfully to identify the protein composition of a variety of complex samples from intracellular organelles (4) to extracellular viral particles (91). While HSV-1 extracellular virions have been analyzed using this approach (83), virions of other alphaherpesviruses, including PRV, have not been subjected to a comprehensive proteomic analysis by mass spectrometry. A previous mass spectrometry study of PRV virions by Michael et al. did not identify all of the previously known virion constituents, particularly viral envelope proteins (99).
In this study, we used two complementary proteomic approaches to characterize the protein content of purified PRV virions. We identified a total of 47 viral proteins, seven of which were not previously detected in PRV virions. Additionally, although our proteomic workflow was not tailored toward phosphopeptide enrichment, we identified four viral proteins in virions that are phosphorylated. Finally, we identified 48 host proteins that are virion constituents, many of which are implicated in pathways that modulate cellular signaling, organization, and dynamics.
MATERIALS AND METHODS
Cell culture and virus strains.
Porcine kidney epithelial cells (PK15) were cultured at 37°C in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (HyClone, Logan, UT) in a 5% CO2 environment. PRV Becker, a wild-type laboratory strain, was propagated in PK15 cells in DMEM supplemented with 2% FBS and penicillin-streptomycin.
Virion purification.
Purification of PRV extracellular virions was performed as previously described for herpes simplex virus type 1 (HSV-1) virions (83), with minor modifications (Fig. 1). Three fully confluent 150-cm2 dishes of porcine kidney epithelial cells (PK15) were mock infected or infected with PRV Becker at a multiplicity of infection (MOI) of 10. These infection conditions were chosen to synchronize the infection and produce a high yield of virions. At 6 h postinfection (hpi), the cells were washed twice with serum-free DMEM, and the medium was replaced with serum-free DMEM containing penicillin-streptomycin. The extracellular medium was harvested at 24 hpi and clarified by centrifugation at 600 × g for 10 min at 4°C. The clarified supernatant was treated with 50 μg/ml DNase I (Sigma-Aldrich, St. Louis, MO) for 30 min at 4°C and subsequently filtered through a 0.45-μm-pore-size filter. Virions were pelleted by centrifugation at 20,000 × g for 45 min at 4°C in a Sorvall SA-600 rotor. The viral pellet was resuspended in 1 ml of MNT buffer (30 mM morpholineethanesulfonic acid [MES], 10 mM NaCl, and 20 mM Tris-HCl [pH 7.4]), layered over a 10% Ficoll 400 (Sigma-Aldrich) cushion, and centrifuged at 116,000 × g for 2 h at 4°C in a Beckman SW41 Ti rotor. For protease treatment of virions, virions were resuspended in 1 ml of MNT buffer containing 10 μg/ml proteinase K (Roche, Mannheim, Germany) for 45 min at room temperature and subsequently treated with 2 mM phenylmethylsulfonyl fluoride (PMSF; Roche) prior to density gradient centrifugation (87). The resulting pellet was washed by resuspension in MNT buffer and concentrated by centrifugation at 116,000 × g for 30 min at 4°C. Purified virions were resuspended in MNT buffer and stored at −80°C.
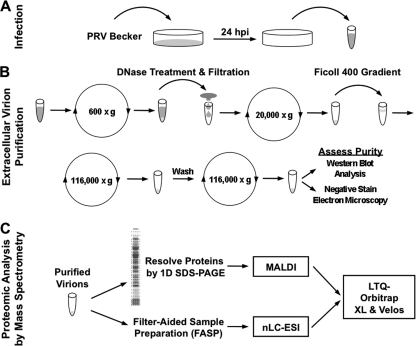
Fig. 1.
Strategy for PRV virion purification and proteomic analysis by mass spectrometry. (A) PK15 cells were infected with PRV Becker at an MOI of 10. At 24 hpi, the extracellular medium was harvested. (B) PRV extracellular virions were enriched as detailed in Materials and Methods. The purity of the preparation was assessed by electron microscopy and Western blot analysis. (C) Enriched extracellular virions were subjected to proteomic analysis by mass spectrometry using two complementary approaches, MALDI or nLC-ESI coupled to an LTQ-Orbitrap XL mass spectrometer.
Microscopy.
Bright-field microscopy images were acquired using a Nikon Ti-Eclipse inverted microscope equipped with a ×20 magnification phase-contrast objective (Nikon Instruments, Tokyo, Japan) and connected to a Cool Snap ES2 camera (Photometrics, Tucson, AZ). For negative-stain electron microscopy (EM), virions in MNT buffer were stained with 1% uranyl acetate and visualized using a Zeiss LEO 912AB transmission electron microscope (Oberkochen, Germany).
SDS-PAGE and Western blot analysis.
Samples were suspended in 4× SDS-PAGE buffer with reducing agent (Invitrogen, Carlsbad, CA), incubated at 70°C for 10 min with agitation, alkylated with 100 mM iodoacetamide at room temperature for 1 h in the dark, and resolved by one-dimensional (1D) gel electrophoresis on 4 to 12% NuPAGE Novex Bis-Tris gels (Invitrogen). For Coomassie staining, the gels were stained with SimplyBlue SafeStain (Invitrogen) according to manufacturer's instructions. For Western blot analysis, gels were transferred to nitrocellulose membranes (Whatman, Keene, NH) using a semidry transfer apparatus. Following transfer, membranes were incubated in blocking solution (5% milk in TBST [50 mM Tris, 200 mM NaCl, 0.1% Tween 20]) for 1 h at room temperature. Membranes were then incubated with blocking solution containing primary antibodies overnight at 4°C. Membranes were washed three times with TBST solution, placed in blocking solution containing horseradish peroxidase (HRP)-conjugated secondary antibodies (1:15,000; KPL, Gaithersburg, MD) for 1 h at room temperature, and then washed as previously described. Proteins were visualized with SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific, San Jose, CA) following the manufacturer's instructions.
Antibodies.
The following antibodies were used in this report for Western blot analysis: rabbit polyclonal anti-UL36-3 at 1:15,000 (gift of T. Mettenleiter) (102), goat anti-UL34 at 1:1,000 (gift of B. Banfield) (23), rabbit anti-UL13 at 1:1,000, mouse monoclonal anti-VP5 IN-13 at 1:2,000 (gift of H. Rziha), rabbit polyclonal anti-gE tail at 1:1,000 (132), mouse monoclonal anti-β-actin AC-74 at 1:5,000 (Sigma Aldrich), mouse monoclonal anti-cytochrome c (Invitrogen), rabbit polyclonal anti-EP0 at 1:1,000 (gift of H. Kida) (136), rabbit polyclonal anti-gI at 1:5,000 (137), rabbit polyclonal anti-gD at 1:5,000 (gift of T. Mettenleiter) (65), mouse monoclonal anti-gC (M1) at 1:10,000 (50), mouse monoclonal anti-gB (M2) at 1:5,000 (50), and mouse monoclonal anti-green fluorescent protein (GFP) at 1:20,000 (Roche).
In-gel and in-solution protein digestion.
For Coomassie-stained gels, the entire gel lane was cut into 1-mm gel slices and destained in 50 mM ammonium bicarbonate containing 50% acetonitrile (ACN). Destained gel slices were dehydrated in 100% ACN and digested in gel with trypsin (Promega, Madison, WI) at 12.5 ng/ml in 50 mM ammonium bicarbonate (10 ml). Peptides were extracted on reverse-phase resin (POROS 20 R2; Applied Biosystems, Foster City, CA) and eluted either in matrix solution (25 mg/ml of 2,5-dihydroxybenzoic acid in 0.1% trifluoroacetic acid–50% methanol–20% ACN) for matrix-assisted laser desorption ionization (MALDI), as previously described (84), or in 0.1% trifluoroacetic acid–70% ACN for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. For in-solution digestion, extracellular virions obtained from the equivalent of one-half of a 150-cm2 plate of infected PK15 cells was solubilized in 0.1 M Tris-HCl (pH 7.6) containing 50 mM dithiothreitol (DTT) and 2% SDS. Extracted proteins were digested in solution with 100 μl of trypsin solution in 100 mM ammonium bicarbonate (1:100, enzyme/protein) by a filter-aided sample preparation method (FASP) as previously described (138). Several filters were tested for the FASP method and Vivacon 500 centrifugal filters (10,000-molecular-weight-cutoff [MWCO]; Sartorius Stedim Biotech, Goettingen, Germany) were found to be the most suitable.
Mass spectrometry.
Peptides deposited on the MALDI target were analyzed on a MALDI linear trap quadrupole (LTQ)-Orbitrap XL (ThermoFisher Scientific), as described previously (84, 104). Briefly, MS spectra from each spot were acquired (resolution [R] of 60,000 at m/z 400). Peptide m/z values from MS spectra were extracted and deisotoped into monoisotopic peak lists using Xtract (Xcalibur, version 2.0.7). Peak lists were searched against the National Center for Biotechnology Information nonredundant protein database, version October 16, 2006, using the XProteo algorithm (http://www.xproteo.com) to obtain a list of putative protein hits for which the quality index, d′, scores were >4 (143). At this threshold, d′ scores greater than 4 correspond to an estimated false-positive rate of 0.05. Putative protein hits were confirmed by at least two unique peptide sequences per protein using MALDI ion trap collision-induced dissociation, as described previously (84). An aliquot of peptides (∼2 μg) generated from in-solution trypsin digestion were analyzed by electrospray ionization (ESI) on an LTQ-Orbitrap XL coupled online to an Ultimate 3000 nanoRSLC system (Dionex Corp., Sunnyvale, CA) running mobile phases: mobile phase A, 0.1% formic acid in water; mobile phase B, 0.1% formic acid in 97% ACN–2.9% water. Peptides were preconcentrated and desalted online with 0.5% trifluoroacetic acid–97.5% water–2% ACN for 5 min at 5 μl/min using a reverse-phase trap column (100 μm by 2.5 cm; particle size, 3 μm) (PepMap C18; Dionex Corp.). Peptides were separated on a nanocapillary reverse-phase column (75 μm by 15 cm; particle size 1.8 μm) (PepMap C18; Dionex Corp.) for 240 min using a linear gradient from 4 to 35% mobile phase B. The mass spectrometer was set to repetitively scan m/z 350 to 1,700 (R = 60,000), followed by data-dependent MS/MS scans on the 10 most abundant ions. For in-gel trypsin digests subjected to ESI, an LTQ-Orbitrap Velos ETD mass spectrometer was employed, as described previously (48). The same instrument parameters described above were used, except that reverse-phase separation was performed by a 90-min linear gradient and data-dependent MS/MS acquisition was performed on the 20 most abundant ions per cycle.
Proteomic data analysis.
MS/MS spectra from LC-MS experiments were extracted using Bioworks (version 3.3.1) or Proteome Discoverer (version 1.2), and peptide spectrum matches (PSMs) were generated by SEQUEST database searching (version 28, revision 12, or version 1.13) against a FASTA protein sequence database comprised of human and pig protein sequences from the SwissProt database (release 2010_04), PRV protein sequences from the TrEMBL database (release 2010_04), and common contaminants, which were all reversed and appended to the forward sequences (44,194 total entries). SEQUEST searches were performed with the following parameters: full enzyme specificity; parent and fragment ion mass tolerances of 10 ppm and 0.5 Da, respectively; fixed modification of carbamidomethyl to cysteine; and variable modifications of oxidation to methionine and phosphorylation to serine, threonine, and tyrosine. PSMs were stored as SRF or MSF files and loaded into Scaffold (version Scaffold_3_00_04; Proteome Software Inc., Portland, OR) using the refinement database search option that employed the X! Tandem algorithm (The GPM, version 2007.01.01.1 [thegpm.org]). XTandem! search parameters were the same as above, except additional variable modifications were used: pyro-Glu of E on the peptide N terminus, S-carbamoylmethylcysteine cyclization of the N terminus, deamidation of asparagine, oxidation of tryptophan, and acetylation of the N terminus. Peptides and protein probabilities were calculated by the PeptideProphet and ProteinProphet algorithms (63). Probability thresholds for peptide and protein identifications were selected empirically to control false-identification rates to less than 1% using protein hits to the reverse database. All proteins were identified by at least two unique peptides. Proteins that were identified by the same peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
RESULTS
Enrichment of PRV virions.
Sample purity is of utmost importance for accurate characterization of virion protein components by mass spectrometry. While no enrichment strategy can yield a perfectly pure sample, the strategy used in this study has several advantages designed to enrich for mature extracellular virions while minimizing contaminants. While many protocols involve separation of mature virions from L particles over a density gradient (commonly using sucrose), the strategy we chose achieves this by pelleting virions through a 10% Ficoll 400 cushion. Sucrose gradients have been reported to damage virions (2), while Ficoll 400 is osmotically inert and, therefore, believed to reduce the stress that fragile membrane-bound structures undergo during enrichment (128). Additionally, as recently shown for HSV-1 virions, top-loading virions over a 10% Ficoll 400 cushion effectively separates less dense L particles from the higher-density mature virions that are pelleted following centrifugation (83). Moreover, since enrichment of PRV virions was performed using a recently established protocol for isolation of HSV-1 virions prior to analysis by mass spectrometry (83), we were able to compare the virion protein content of these two alphaherpesviruses.
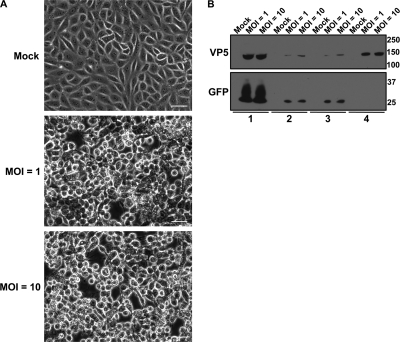
Extracellular virions from the medium of PK15 cells infected with the wild-type laboratory strain PRV Becker (112) were harvested at 24 h postinfection (hpi) (Fig. 1A) and purified (Fig. 1B). To ensure that cellular contaminants due to cell lysis are removed using our virion enrichment strategy, we monitored the purification efficiency of virions isolated from cells infected with PRV 151 (a PRV strain that expresses diffusible GFP) for 24 h. These experiments were performed at MOIs of both 1 and 10 to compare our infection conditions with those previously used for mass spectrometry analysis of virions by Michael et al. (99). While infected PK15 cells at both MOIs displayed the cytopathic effects associated with PRV infection, extensive cell lysis was not observed under either condition (Fig. 2A). As GFP should be retained in intact cells, we monitored the presence of GFP and VP5 in whole-cell lysates, the extracellular medium, and pelleted virions prepared using our purification protocol, excluding the Ficoll 400 cushion and last wash steps. Following 24 h of infection at MOIs of 1 and 10, the vast majority of GFP was retained within the cell bodies of infected cells, thus indicating that cell lysis was minimal and independent of the MOI. Importantly, GFP from the extracellular medium did not cosediment with virions that were pelleted using centrifugation conditions detailed in our enrichment protocol (Fig. 2B). These results suggest that extracellular contaminants are efficiently separated from virions using our purification approach.
Fig. 2.
Confirmation of the virion enrichment strategy. (A) Phase-contrast microscopy images of PK15 cells that were either mock infected or infected at an MOI of 1 or 10 at 24 hpi. Scale bar, 20 μm. (B) PK15 cells were either mock infected or infected with PRV 151 (PRV expressing diffusible GFP) at an MOI of 1 or 10 for 24 h. Virions were purified as illustrated in Fig. 1. The detection of VP5 and GFP was monitored in the following: lane 1, whole-cell lysates; lane 2, the extracellular medium following centrifugation at 600 × g; lane 3, the extracellular medium following centrifugation at 600 × g and filtration through a 0.45-μm-pore-size syringe-driven filter; and lane 4, the crude virion pellet obtained from centrifugation at 20,000 × g.
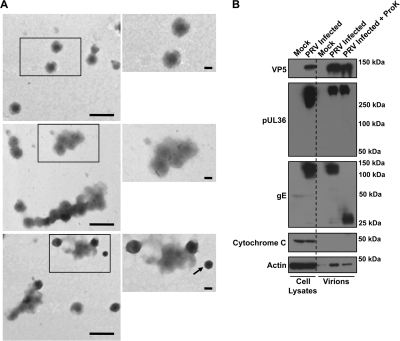
Negative-stain electron microscopy was used to confirm the efficiency of our enrichment strategy for PRV virions. The observed particles were of uniform and expected diameter of approximately 225 nm (49) and spherical and contained an electron-dense DNA core (Fig. 3A). These EM analyses were consistent with the quality and apparent purity of previously reported preparations of herpesvirus virions (56, 60, 61, 83, 128, 133). We rarely observed unenveloped capsids that likely resulted from disruption and loss of the virion envelope layer during the purification procedure or during sample preparation for visualization by electron microscopy. As these capsids are composed of proteins that are also found in mature virions, their presence does not affect the ability to characterize extracellular virions.
Fig. 3.
Assessment of PRV virion purity. (A) Three fields of view containing negatively stained, purified virions visualized by electron microscopy. Scale bars on the lower magnification images represent 500 nm, while those in the magnified regions represent 100 nm. Arrow indicates a capsid without the virion envelope, which was occasionally observed in our preparation. (B) Western blot analysis of viral and host proteins in cell lysates from mock- and PRV Becker-infected PK15 cells and purified virions from mock-treated, untreated, and proteinase K (ProK)-treated samples. Antibodies were used to detect viral proteins, including the major capsid protein VP5, the large inner tegument protein pUL36, and the envelope protein glycoprotein gE. Additionally, samples were probed for actin, a known host virion component, as well as the mitochondrial marker cytochrome c, which is excluded from virions.
We further validated the purity of the virion preparation by Western blot analysis of viral and host proteins known to be present in or absent from virions. To further exclude contaminating cellular proteins present in the extracellular medium, we repeated the virion preparation on PK15 cells that were mock infected. Additionally, to specifically identify proteins contained within virions, we treated extracellular virions with proteinase K. Since PRV virions are enveloped within a phospholipid bilayer, only viral and host proteins contained within the tegument and capsid layers are protected from protease digestion (88, 114).
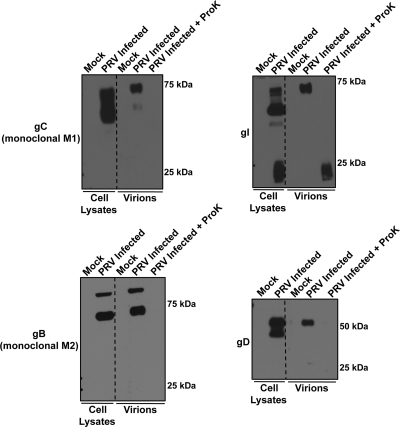
This analysis confirmed the presence of the major capsid protein VP5 and an inner tegument protein, pUL36, in both proteinase K-treated and untreated virions, consistent with our expectation that capsid and inner tegument proteins are mostly protected from protease digestion (Fig. 3B) (114). Additionally, the full-length forms of the PRV type I membrane proteins gC, gD, gI, and gE and both the expected full-length and cleaved forms of gB were identified only in untreated virions, while these proteins were either truncated or undetectable in proteinase K-treated virions (Fig. 3B and 4). These findings are in agreement with digestion of the N-terminal domains of these proteins, which are topologically exposed on the virion surface (54). Further confirming the purity of virion preparation, our findings are consistent with previous work indicating that the mature glycosylated forms of gC, gD, gE, and gI are incorporated into virions (Fig. 3B and 4) (13, 118, 124, 131). Actin, a known component of virions (30, 49, 139), was detected in all three samples (mock, untreated, and proteinase K treated) at different levels, suggesting that actin is present within virions but may also associate nonspecifically on the exterior of the virion envelope. Finally, the mitochondrial marker cytochrome c was undetectable in all virion preparations, confirming that the purified virions are free of large intracellular organelle contaminants such as mitochondria (Fig. 3B). In summary, using a combination of electron microscopy and Western blot analysis, we confirmed that our enrichment approach yielded a highly purified sample of PRV extracellular virions.
Fig. 4.
Sensitivity of viral glycoproteins gC, gB, gI, and gB to proteinase K. Western blot analysis of viral proteins in cell lysates from mock-infected and PRV Becker-infected PK15 cells, and purified virions from mock, untreated, and proteinase K (ProK)-treated samples. gC, gB, gI, and gD are type I transmembrane proteins which have an exposed N-terminal domain on the virion envelope predicted to be sensitive to proteinase K digestion.
Comprehensive characterization of PRV virions by complementary proteomics approaches.
To comprehensively analyze the composition of PRV virions, we took advantage of two complementary mass spectrometry-based strategies (Fig. 1C). First, PRV virion proteins were separated by 1D SDS-PAGE, subjected to in-gel digestion, and analyzed using a MALDI LTQ Orbitrap XL mass spectrometer (84). This approach allows for the rapid detection of proteins and correlation of protein identification with molecular size by 1D SDS-PAGE. To complement this approach, purified virions were analyzed using a nano-LC (nLC)-ESI LTQ Orbitrap XL mass spectrometer following filter-aided sample preparation (FASP), an approach that utilizes in-solution digestion of proteins and therefore avoids sample loss during gel extraction (138). These complementary approaches were performed from independent biological samples, with in-solution digestion of virions representing two technical replicates that were analyzed for each experimental condition (mock, untreated, and proteinase K treated).
Using these approaches, we detected a total of 48 viral proteins (Tables 1 and 2 and Fig. 5B). We readily detected 35 of these proteins using both proteomic approaches. The additional 12 viral proteins were detected using the in-solution digestion approach coupled with nLC-ESI. In part, we attribute this improved sensitivity to increased digestion efficiency and peptide recovery following in-solution digestion. As a result, we chose to further characterize our samples by this method. While positive identification of each protein required a minimum of two unique peptides, 38 of the viral proteins were detected with at least five unique peptides (see Tables S1 and S2 in the supplemental material).
Table 1.
Previously characterized viral virion proteins
| Protein content of PRV virions | Common name | Proposed function(s) | Virion localization | Mass (kDa) | Detection by nLC-ESI/MALDIc | Presence in HSV-1d | Reference(s) |
|---|---|---|---|---|---|---|---|
| EP0a | ICP0 | Gene regulation (transactivator) | Unknown | 44 | +/− | + | 17, 108 |
| IE180 | ICP4 | Gene regulation (transactivator) | Tegument | 150 | +/+ | + | 99 |
| pORF1.2 | Unknown | Unknown | 35 | +/− | NAb | 113 | |
| pUL1a | gL | Viral entry (fusion) | Envelope | 17 | +/+ | + | 68 |
| pUL3.5 | Secondary envelopment and egress | Tegument | 24 | +/+ | − | 38 | |
| pUL6 | Portal protein | Capsid | 70 | +/+ | + | 114 | |
| pUL7 | Interacts with mitochondrial ANT2 | Tegument | 29 | +/− | + | 37 | |
| pUL10 | gM | Secondary envelopment and egress | Envelope | 42 | +/+ | + | 32 |
| pUL11a | Secondary envelopment and egress | Tegument | 7 | +/− | + | 75 | |
| pUL16 | Interacts with UL21 | Tegument | 35 | +/+ | + | 93 | |
| pUL17 | DNA cleavage and encapsidation | Capsid | 64 | +/+ | + | 71 | |
| pUL18 | VP23 | Minor capsid protein | Capsid | 32 | +/+ | + | 114 |
| pUL19 | VP5 | Major capsid protein | Capsid | 146 | +/+ | + | 114 |
| pUL21 | Interacts with UL16 | Tegument | 55 | +/+ | + | 29, 98 | |
| pUL22 | gH | Viral entry (fusion) | Envelope | 72 | +/+ | + | 74 |
| pUL25 | Capsid assembly | Capsid | 57 | +/+ | + | 59 | |
| pUL26 | VP24 | Minor capsid scaffold (protease) | Capsid | 55 | +/+ | + | 114 |
| pUL27 | gB | Viral entry (fusion) | Envelope | 100 | +/+ | + | 28 |
| pUL35 | VP26 | Surface capsid protein | Capsid | 11 | +/+ | + | 123 |
| pUL36 | VP1/2 | Viral entry and egress | Tegument | 324 | +/+ | + | 24 |
| pUL37 | Viral egress | Tegument | 98 | +/+ | + | 73 | |
| pUL38 | Minor capsid protein | Capsid | 40 | +/+ | + | 114 | |
| pUL39 | RR1 | Ribonucleotide reductase | Tegument | 91 | +/+ | − | 122 |
| pUL41 | VHS | Degrades host and viral mRNAs | Tegument | 41 | +/+ | + | 82 |
| pUL43 | Inhibits membrane fusion | Envelope | 38 | +/− | − | 66 | |
| pUL44 | gC | Viral entry (virion attachment) | Envelope | 51 | +/+ | + | 13 |
| pUL46 | VP11/12 | Unknown | Tegument | 76 | +/+ | + | 76 |
| pUL47 | VP13/14 | Secondary envelopment and egress | Tegument | 80 | +/+ | + | 76 |
| pUL48 | VP16, α-TIF | Gene regulation (transactivator) | Tegument | 45 | +/+ | + | 38 |
| pUL49 | VP22 | Major tegument protein | Tegument | 25 | +/+ | + | 39 |
| pUL49.5 | gN | Immune evasion | Envelope | 10 | +/+ | − | 57 |
| pUL51 | Secondary envelopment and egress | Tegument | 25 | +/+ | + | 81 | |
| pUL53 | gK | Secondary envelopment and egress | Envelope | 34 | +/− | − | 67 |
| pUS2 | Tegument protein | Tegument | 28 | +/+ | + | 23 | |
| pUS3 | PK | Protein kinase, inhibits apoptosis | Tegument | 43 | +/+ | + | 47 |
| pUS4 | gG | Immune evasion, secreted | Envelope | 53 | +/− | + | 116 |
| pUS6 | gD | Viral entry | Envelope | 45 | +/+ | + | 13 |
| pUS7 | gI | Cell-cell spread | Envelope | 39 | +/+ | + | 13 |
| pUS8 | gE | Cell-cell spread | Envelope | 62 | +/+ | + | 13 |
| pUS9 | Axonal sorting | Envelope | 11 | +/− | + | 15 | |
| Putative false negatives | |||||||
| pUL13 | VP18.8 | Protein kinase | Tegument | 41 | −/− | + | 27, 106 |
| pUL56 | ORF1 | Unknown | Envelope | 22 | −/− | + | 62 |
Detected in virions not treated with proteinase K.
No known HSV-1 orthologue.
+/+, detected by both nLC-ESI and MALDI-based proteomic workflows; +/−, identified by only nLC-ESI workflow; −/−, absent in both nLC-ESI and MALDI workflows.
As determined by MS (83).
Table 2.
Novel viral virion proteinsa
| Protein | Common name | Proposed function(s) | Mass (kDa) | Detection by nLC-ESI/MALDIb | Presence in HSV-1c |
|---|---|---|---|---|---|
| pUL8 | DNA replication, helicase/primase complex | 71 | +/− | − | |
| pUL20 | Viral egress | 17 | +/+ | + | |
| pUL32 | DNA packaging, capsid precursor | 52 | +/− | − | |
| pUL40 | RR2 | Ribonucleotide reductase | 34 | +/+ | − |
| pUL42 | DNA replication, viral polymerase subunit | 40 | +/− | − | |
| pUL50 | dUTPase | dUTPase | 29 | +/+ | + |
| pUS1 | RSp40/ICP22 | Gene regulation | 40 | +/− | − |
Only proteins detected in virions following proteinase K treatment are shown.
+/+, detected by both nLC-ESI and MALDI-based proteomic workflows; +/−, identified only by nLC-ESI workflow.
As determined by MS (83).
Fig. 5.
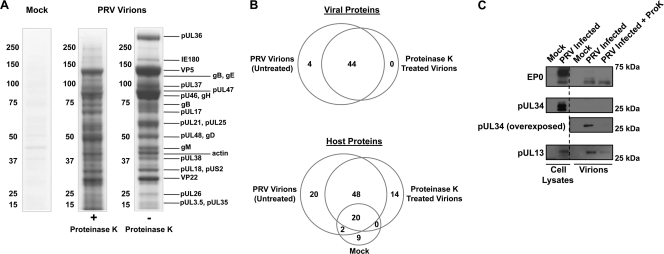
Identification of viral and host virion proteins. (A) Mock, proteinase K-treated, or untreated purified virions prepared from 75-cm2 PK15 cells that were PRV Becker infected or mock infected. Samples were run on a 4 to 12% 1D SDS-PAGE gel and stained with Coomassie blue. In-gel digestion followed by MALDI LTQ Orbitrap XL analysis allowed us to identify the relative position of viral proteins along the length of the gel as indicated. (B) Venn diagrams of viral (top) and host (bottom) proteins detected in untreated, proteinase K-treated, and mock samples by in-solution digestion followed by analysis using a nLC-ESI LTQ Orbitrap XL mass spectrometer. (C) Western blot analysis of viral proteins in cell lysates from mock- and PRV Becker-infected PK15 cells and purified virions from mock, untreated, and proteinase K (ProK)-treated samples. EP0 is a known virion component of PRV virions, while pUL34 is a component of primary virions and should not be present in mature extracellular virions. Using mass spectrometry, EP0 and pUL34 were detected in untreated but not proteinase K-treated virions. The tegument protein pUL13 is a known virion component but was not detectable in virions by mass spectrometry and is therefore a false negative.
Viral protein composition of PRV virions.
These complementary approaches enabled us to perform a comprehensive and unbiased analysis of the protein content of PRV extracellular virions. We identified 40 of the 42 previously identified viral proteins that are constituents of the capsid, tegument, and envelope layers of PRV virions (Table 1). This prior knowledge of virion composition was established, in most cases, by targeted studies on individual viral proteins of interest, and their association within virions was determined using a variety of different experimental approaches. Of direct relevance to our work is the elegant study by Michael et al. (99), which used quantitative mass spectrometry to measure the incorporation levels of a specific subset of viral and cellular proteins present in PRV (Kaplan strain) virions from a limited variety of mutant backgrounds. This previous work has provided a foundation for characterizing protein composition of PRV virions. Our results complement and extend their findings (see comparison of the identified viral proteins in Table S1 in the supplemental material). For example, our analyses have identified envelope proteins that were not previously detected in this mass spectrometry-based study, including gL, gN, gK, gI, gG, pUL43, and pUS9 (see Tables S1 and S3). Of the previously reported viral protein components, only two viral proteins (pUL13 and pUL56) were not detected in any of our samples (Table 1). However, we were able to confirm the presence of pUL13 in proteinase K-treated and untreated virions by Western blot analysis (Fig. 5C). It is possible that these proteins are not abundant virion components and were below the threshold of detection or were not amenable to detection by this particular proteomic workflow. Additionally, we detected 36 PRV orthologues of HSV-1 viral proteins recently identified as constituents of HSV-1 virions by mass spectrometry (83) (Tables 1 and 2). This finding is consistent with previous studies that suggest that these two alphaherpesviruses share a high degree of similarity in virion assembly and packaging.
Identification of viral capsid and tegument components via proteinase K treatment.
To identify proteins specifically contained within the tegument and capsid layers of virions, as opposed to those that are present on the envelope surface, we compared the protein content of untreated and proteinase K-treated virions. Visualization of these samples on Coomassie-stained 1D SDS-PAGE gels revealed differences in the banding pattern between the untreated and proteinase K-treated virion samples (Fig. 5A).
Notably, the high-molecular-mass band present in the gel region in which pUL36 was detected by MALDI MS was reduced following proteinase K treatment. A substantial amount of evidence exists to support the conclusion that pUL36 is an inner tegument protein with key functions during viral entry (10, 11, 18, 24, 41, 46, 69, 79, 86, 100, 103, 117) and, therefore, should be protected from proteinase K digestion. In agreement with this, by Western blot analysis we detected only the full-length pUL36 protein and no evidence of a cleaved product in both untreated and proteinase K-treated samples (Fig. 3B). We therefore hypothesized that the reduced upper band in the Coomassie-stained gel lane containing proteinase K-treated virion proteins could represent other proteins, besides pUL36, that are sensitive to proteinase K digestion. To test this, we analyzed by nLC-MS/MS the upper gel region that contained the pUL36 band. Several host proteins of similar molecular masses to pUL36 were identified that could contribute to the loss of Coomassie staining upon proteinase K treatment. Basement membrane-specific heparan sulfate proteoglycan core protein (∼468 kDa) was identified only in the untreated sample, while filamins A and B (∼280 kDa) showed a 10-fold reduction in number of assigned spectra upon proteinase K treatment, suggesting that their relative abundance was reduced. Consistent with our initial mass spectrometry and Western blot analyses, pUL36 was confidently identified in both samples with similar protein sequence coverage. However, LC-MS/MS analysis revealed that the total number of spectra assigned to pUL36 within the upper gel region was reduced by 2-fold, suggesting that a fraction of pUL36 in the proteinase K-treated virion sample was accessible to proteinase K digestion. This fraction of pUL36 is likely associated with the subset of virions that were damaged during sample isolation, a common drawback of enrichment approaches that are dependent upon density-gradient sedimentation. Since pUL36 is a large protein and contains many proteinase K digestion sites, it may be highly susceptible to protease in virions that are slightly permeabilized or damaged. Nonetheless, our mass spectrometry and Western blot analyses support the conclusion that PRV pUL36 is an inner tegument protein.
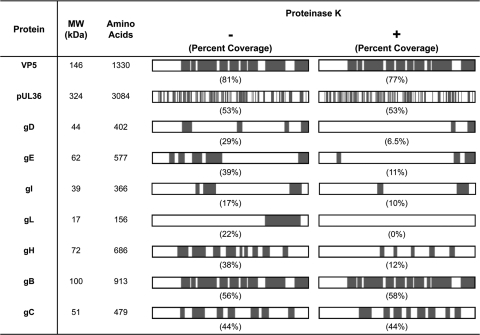
Analysis of the peptide sequence coverage of viral proteins identified by mass spectrometry provided specific information regarding their availability for proteinase K digestion. For VP5, peptides spanning almost the entire length of the protein were identified in both untreated and proteinase K-treated virions (Fig. 6). These results were in agreement with the Western blot analyses that showed the full-length version of this protein in the presence or absence of proteinase K treatment (Fig. 3B).
Fig. 6.
Peptide sequence coverage of viral proteins in untreated and proteinase K-treated virions. Schematic representations of each protein are oriented with the N terminus on the left and the C terminus on the right. Regions of the viral proteins that correspond to the peptides we detected by mass spectrometry are indicated in gray, and the percent sequence coverage (by amino acids) is listed in parentheses. The major capsid protein VP5 and the inner-tegument protein pUL36 are contained within the envelope and are therefore predicted to be resistant to proteinase K treatment. The envelope glycoproteins gD, gE, gI, gL, gH, gB, and gC are all predicted to contain domains that are exposed on the virion surface and potentially available for protease cleavage.
Similar analysis of the viral type I membrane proteins gE, gI, and gD, which have an exposed N-terminal domain on the virion envelope (16), revealed that the majority of the peptides in this domain were not detected following proteinase K treatment. However, we readily detected peptides in the C terminus of these proteins in both proteinase K-treated and untreated virions. This is consistent with localization of this domain inside the virions, where it is protected from proteinase K. For gB and gC, which are also type I membrane proteins with topologies and membrane orientations similar to those of gE, gI, and gD, we unexpectedly detected similar peptide sequence coverage in both the untreated and proteinase K-treated samples (Fig. 6). As a complementary approach, we examined cleavage of these glycoproteins by Western blot analysis, which confirmed that the N termini of gB, gC, gE, gI, and gD are all sensitive to proteinase K digestion (Fig. 3B and 4).
Interestingly, four viral proteins were completely sensitive to proteinase K treatment (i.e., were detected in untreated but not in proteinase K treated virions) (Fig. 5C): pUL34, gL, pUL11, and EP0. This finding can be interpreted considering previous work on their function, localization, and topology. The membrane protein pUL34 is a well-characterized constituent of primary virions that is not incorporated into mature extracellular virions (43). Although identification of pUL34 in untreated virion preparations may indicate the presence of immature virus particles (such as primary virions), we argue that this is a remote possibility for the following three reasons. (i) pUL34 is not detected after proteinase K treatment. Since pUL34 is a type II C-terminally anchored membrane protein, the N-terminal domain of this protein, which accounts for most of the protein length, is topologically found within primary immature virions in the tegument layer. Therefore, if primary virions were present in our preparation, pUL34 would be protected from proteinase K digestion (72, 94, 120). (ii) We did not detect other markers of immature virions, including full-length pUL26, VP21, and pre-VP22a and processed VP22a (83); rather, we identified VP24, the C terminus of pUL26 that is expected to be present in mature virions. (iii) We did not detect pUL31, a well-characterized binding partner of pUL34 that would be expected to reside in immature virions (43). In contrast, gL is a key component of mature extracellular virions and is known to function in mediating fusion of the virion envelope with the plasma membrane prior to entry (70). This protein has no transmembrane domain and is found in a complex with the N terminus of the viral membrane protein gH on the surface of virions and infected cells (3, 22, 68, 70). Therefore, the absence of gL from virions following proteinase K treatment is consistent with its localization on the virion exterior.
For pUL11 and EP0, only two unique peptides were detected by mass spectrometry in untreated virions. This suggests that they may be present in low copy numbers or may be more difficult to detect by mass spectrometry. The latter is likely the case for pUL11, which is a small protein (7 kDa) with an amino acid composition that generates short (<6 amino acids) tryptic peptides not readily detected by mass spectrometry. In contrast, EP0 is a 44-kDa protein that had a low (4.9%) peptide sequence coverage by mass spectrometry. Previous work by Yao and Courtney proposed that the HSV-1 orthologue of EP0 (ICP0) is present in small amounts in the tegument layer of HSV-1 virions (141), suggesting that it also may not be abundant in PRV virions. Therefore, EP0 was likely at the limit of detection of our proteomic approach. Given that previous reports defined PRV EP0 as a virion component (17, 108), we performed Western analysis for EP0, confirming its presence in both untreated and proteinase K-treated virions (Fig. 5C).
Novel viral virion components.
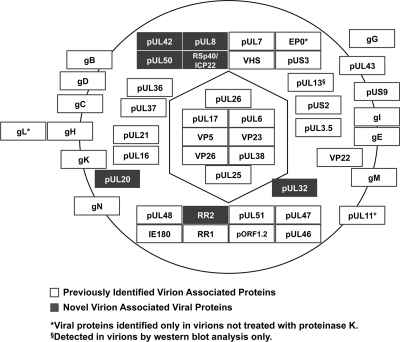
In addition to previously identified PRV virion components, we identified seven viral proteins that may be novel virion constituents. These include pUL8, pUL20, pUL32, pUL40 (small subunit of ribonucleotide reductase), pUL42, pUL50 (dUTPase), and RSp40/ICP22, as illustrated in Table 2. Interestingly, the HSV-1 orthologue of PRV RSp40/ICP22 is important for modulating viral gene expression as well as proper incorporation of virion structural components but was not detected in HSV-1 extracellular virions (83, 110). pUL20 is a putative integral membrane protein with multiple transmembrane domains (89) that functions in viral egress (5, 31, 42) and has been found in HSV-1 virions (135). To our knowledge, this is the first instance in which both of these proteins have been identified in PRV virions.
The remaining five novel virion components have well-characterized enzymatic functions in viral genome replication (pUL8 and pUL42), genome cleavage and encapsidation (pUL32), and ribonucleotide synthesis (pUL40 and pUL50) (40, 45, 58, 119, 121). Previously, virion incorporation of only three of these components has been studied. Specifically, pUL8, pUL42, and pUL50 have been shown not to incorporate into virions, using Western blot analysis of purified virions (58, 110, 113). However, since these proteins were still detected in virions following proteinase K digestion, they are likely virion components. Since these proteins lack predicted membrane-interacting domains and are not known capsid components (96, 114), we have tentatively assigned them to the tegument layer. Further targeted biochemical analysis will be necessary to establish a role for these proteins within the virion.
Phosphorylation of virion components.
Although our biochemical fractionation and proteomic approach (Fig. 1) was primarily designed to identify the protein content of PRV virions, we were also able to detect by mass spectrometry phosphorylation of several virion proteins: pUL26, a minor capsid protein, and three tegument proteins—pUL36, pUL46, and pUL48. For pUL26, a single phosphorylation event was confidently localized to Ser300, while two phosphoserines were identified in pUL36 at Ser2726 and Ser2969 (Table 3; see also Fig. S1 in the supplemental material). While the single phosphorylation events detected on pUL46 and pUL48 could not be unambiguously localized to a particular residue, their localization could be constrained to the N terminus of pUL46 (amino acids 25 to 46) and the C terminus of the alpha-TIF domain of pUL48 (amino acids 315 to 354) (Table 3; see also Fig. S1). Further studies will be required to discern whether these phosphorylations play a biological role in the viral life cycle.
Table 3.
Identified phosphoproteins in mature virions
| Protein | Sequence(s)a | zb | Xcc | Ascored | Mass accuracy (ppm) |
|---|---|---|---|---|---|
| pUL26 | RSPS300PEPRPPAR | 2 | 1.43 | 34 | −0.21 |
| RSPS300PEPRPPAR | 3 | 2.46 | 26 | −0.62 | |
| pUL36 | QQQRPPPPAS2969PLGGLGR | 3 | 2.42 | NA | −0.78 |
| LHIPPPQPVQLEGVVVPLPAS2726PETPAPAQTQPPR | 4 | 5.01 | 48 | −1.00 | |
| pUL46 | TRASCAMLLAAPGEVLTAAVAALR | 3 | 3.77 | 0 | −0.57 |
| pUL48 | DHTYCAMRPPSPVASYGSTAEALLPPPSPSAVLPCAMDPTPPAR | 4 | 3.68 | 0 | 0.99 |
Sequences of tryptic phosphopeptides detected by mass spectrometry. Positions of phosphorylated residues localized by Ascore are indicated by subscripts. Numbers are relative to full-length protein sequences. Modification of cysteine by iodoacetamide is indicated as CAM. Corresponding representative MS/MS spectra are provided in Fig. S1 in the supplemental material.
Observed charge state.
Cross-correlation score (Xc) is the primary metric for the SEQUEST algorithm that correlates with the proportion of positively matched fragment ions within tandem MS spectra.
Primary scoring metric for localization of the phosphorylated residue within the peptide sequence. Scores greater than 19 equal an estimated false positive rate of <1% and were considered confidently localized. NA, not applicable, indicating that the Ascore was not needed to determine localization.
Host protein composition of PRV virions.
Host proteins are well-known constituents of herpesvirus virions (8, 56, 61, 83, 99, 133, 144). Using our approach, we identified 48 cellular proteins that were detected in proteinase K-treated and untreated virions but absent in samples isolated from mock-infected cells (Fig. 5B and Table 4). We identified an additional 20 cellular proteins in all three samples (Fig. 5B; see also Table S5 in the supplemental material). These proteins included actin (139), heat shock proteins (HSP70/HSC70) (99), and annexin A1 and A2 (99), which are previously determined PRV virion components (99), raising the possibility that some proteins in this class exist as both virion and extracellular medium (nonspecific) components.
Table 4.
Cellular proteins detected in PRV virions and comparison to other herpesviruses
| Protein function and namea | PRVb | HSV-1c | HCMVd | EBVe |
|---|---|---|---|---|
| Small GTPases | ||||
| Arf1 | + | + | ||
| Arf3 | + | + | ||
| Rab1A | + | + | ||
| Rab2A | + | + | ||
| Rab6A | + | |||
| Rab7A | + | + | ||
| Rab14 | + | |||
| Metabolism | ||||
| Glutathione S-transferase | + | |||
| Long-chain fatty acid CoA ligase 4 | + | |||
| Thioredoxin | + | + | ||
| Chaperone | ||||
| HSP27 | + | + | + | |
| Membrane trafficking and organization | + | |||
| Annexin A8 | + | |||
| Syntaxin-12 | + | |||
| VAMP3/cellubrevin | + | |||
| Cytoskeleton | ||||
| Alpha-actinin-1 | + | + | + | |
| Cofilin-2 | + | |||
| Destrin | + | |||
| Myosin light chain 3 | + | |||
| Myosin-Ic | + | |||
| Radixin | + | |||
| Transgelin-2 | + | + | ||
| WD repeat-containing protein 1 | + | |||
| RNA bindinge | ||||
| DEAD box helicase 3 | + | + | + | |
| Eukaryotic initiation factor 4A-I | + | + | ||
| Eukaryotic translation initiation factor 4H | + | + | ||
| Eukaryotic translation initiation factor 5A-1 | + | |||
| HNRNPA2B1 | + | |||
| HNRNPF | + | |||
| HNRNPK | + | |||
| Poly(rC)-binding protein 1 | + | |||
| HNRNPH3 | + | |||
| Signaling | ||||
| 14-3-3 protein beta/alpha | + | + | ||
| 14-3-3 protein epsilon | + | + | + | |
| 14-3-3 protein sigma | + | |||
| 14-3-3 protein theta | + | + | ||
| Annexin V | + | + | ||
| Calmodulin | + | |||
| Casein kinase II subunit alpha | + | + | + | |
| Growth factor receptor-bound protein 2 | + | |||
| Guanine nucleotide-binding protein subunit alpha-2 | + | |||
| MAPK1 | + | |||
| Protein phosphatase PP1-alpha catalytic subunit | + | + | ||
| Protein phosphatase PP1-beta catalytic subunit | + | |||
| RhoA | + | |||
| Cell adhesion | ||||
| Integrin beta-1 | + | |||
| Leukocyte surface antigen CD47 | + | |||
| Cell cycle | ||||
| Galectin-3 | + | |||
| Ion channel | ||||
| Chloride intracellular channel protein 1 | + |
Fourteen proteins were exclusively detected in proteinase K-treated virions (see Table S6 in the supplemental material). These proteins likely result from the data-dependent acquisition methods that we employed as the proteinase K treatment altered the sample complexity by removing numerous host proteins. This allowed the identification of additional proteins that were obscured by more abundant or detectable proteins in the untreated sample. In support of this, nearly all the proteins unique to the proteinase K-treated sample were identified by two unique peptides (see Table S6), suggesting that they were at the lower limit of detection. While it is unclear whether this group represents true virion components, some of these proteins have been previously suggested to function during virion assembly and egress. For example, dominant negative disruption of vacuolar sorting protein 4 (Vps4), an important component of the multivesicular body-ESCRT pathway, has been demonstrated to inhibit replication of HSV-1 (19, 26). Vps4 was shown to associate with purified HSV-1 virions by Western blot analysis (111).
Although the host protein content of PRV virions remains poorly characterized, of the 48 host proteins we identified, several are known virion components of PRV, as well as of other herpesviruses (Table 4). For example, we detected mitogen-activated protein (MAP) kinase, which is recruited to virions by the PRV tegument protein pUS2 (88). Eighteen out of the 48 host proteins we detected were found by mass spectrometry in extracellular virions of other herpesviruses, including HSV-1, the betaherpesvirus human cytomegalovirus (HCMV), and the gammaherpesvirus Epstein-Barr virus (EBV) (6, 56, 83, 133) (Table 4). For example, the small GTPases Arf1, Arf3, Rab2A, and Rab7A were also previously detected in HSV-1 virions (83). These proteins may be common virion constituents of the alphaherpesvirus subfamily.
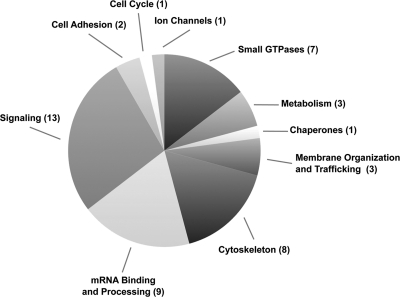
Manual categorization of the identified proteins according to their known molecular functions revealed that many participate in common cellular pathways (Table 4 and Fig. 7). Proteins belonging to the Rab superfamily of small GTPases, as well as SNAREs such as VAMP3/cellubrevin, facilitate organization and trafficking of intracellular membranes. We also identified proteins that modulate key cellular signaling pathways, including 14-3-3 proteins and subunits of the casein kinase II (CKII) and protein phosphatase 1A (PP1A) complexes. Further work is necessary to determine whether these proteins are functionally required for virion entry, assembly, spread, or pathogenesis.
Fig. 7.
Functional classification of host proteins in PRV virions. We identified a total of 48 host proteins that were present in untreated and proteinase K-treated virions but not in samples from mock-infected cells. These proteins were manually classified according to their known cellular functions based on previous studies.
DISCUSSION
Previous mass spectrometry studies have characterized the viral and host protein content of several herpesvirus virions, including HSV-1 (83), Kaposi's sarcoma-associated herpesvirus (KSHV) (8, 144), murine gammaherpesvirus 68 (MHV68) (9), alcelaphine herpesvirus 1 (AlHV-1) (33), EBV (56), murine cytomegalovirus (MCMV) (61), rhesus monkey rhadinovirus (RRV) (107), and HCMV (6, 133). Additionally, Michael et al. utilized stable isotope labeling with amino acids in cell culture (SILAC) in combination with mass spectrometry to quantitatively analyze changes in the incorporation of select virion components in different mutant PRV strains. However, the proteins identified in this study did not represent all of the virion constituents that were identified by additional approaches, especially viral envelope proteins (see Tables S1 and S3 in the supplemental material) (99). Therefore, in this current study, we performed a comprehensive analysis of the protein content of PRV, including both viral and host proteins.
To produce purified virions suitable for the high sensitivity of mass spectrometry, we utilized an isolation approach previously used to purify HSV-1 virions (83). This yielded a purified sample of extracellular virions as confirmed by negative-stain electron microscopy and Western blot analysis (Fig. 1B and 3). We then analyzed the protein content of virions using two complementary mass spectrometric approaches. The first approach involved in-gel digestion and analysis on a LTQ Orbitrap XL mass spectrometer configured with a MALDI ionization source. Luo et al. recently demonstrated that this configuration is suitable for rapid and sensitive detection of proteins present within a complex sample (84). Since sample loss is commonly associated with in-gel digestion strategies, we confirmed and extended our results using a second proteomic approach involving in-solution digestion by FASP and nLC-MS/MS analyses (138). While we reproducibly identified 35 known viral components using both approaches, the in-solution digestion approach enabled the identification of an additional 12 viral proteins associated with PRV virions (Fig. 5B). Therefore, we concluded that this approach is suitable for characterizing samples with the composition and complexity of extracellular virions.
To discriminate between virion components and potential contaminating proteins, we performed our analysis on proteinase K-treated virions as well as samples isolated from mock-infected cells. Protease treatment has been previously utilized to eliminate contaminants present on the surface of KSHV (8, 144) and RRV (107) virions and may be a useful tool for future proteomic studies of other enveloped viral particles or membranous organelles. Analysis of proteinase K-treated PRV virions by mass spectrometry and Western blotting confirmed that capsid and tegument proteins were mostly resistant to digestion. Surprisingly, following proteinase K treatment of virions, we noticed a striking reduction of the high-molecular-weight band in the gel region that contains pUL36 (Fig. 5A). While Zhu and colleagues found evidence that open reading frame 64 (ORF64), the KSHV orthologue of PRV pUL36, is completely sensitive to protease treatment in KSHV virions (144), our data suggest that this is not the case for PRV pUL36. Our Western blot analyses and mass spectrometry data confirm that pUL36 is a tegument protein, a conclusion that is consistent with previous studies on the localization and function of pUL36 in virions (10, 11, 18, 24, 41, 46, 69, 79, 86, 100, 103, 117).
Furthermore, several viral glycoproteins with domains that are exposed on the outer virion envelope were sensitive to protease digestion (e.g., gE, gI, gD, gH, and gL), and the protein sequence coverage obtained by mass spectrometry analysis was in agreement with prior knowledge regarding their domain topology. Unexpectedly, gB and gC, which have topologies and membrane orientations similar to those of gE, gI, gH, and gD, did not display decreased peptide sequence coverage after proteinase K treatment (Fig. 6). We hypothesize that this is because gB and gC are major components of the virion envelope (50, 83), and the sensitivity and sequence-based analysis of the FASP-mass spectrometry approach detected partially digested gB and gC proteins. For readily detected proteins or those with few protease cleavage sites, sequence coverage will not always provide the most discriminating metric for quantitative analysis of protease sensitivity. Indeed, the proteinase K-dependent increase in unique semi-tryptic peptides for gB and gC is supported by our Western blot analysis that confirmed the efficient cleavage of these proteins by proteinase K (Fig. 3B and 4).
Overall, we determined by mass spectrometry that PRV virions contain a total of 47 viral proteins (Table 1 and Fig. 8). This list includes 40 of the 42 previously known viral virion proteins and seven novel viral proteins (Table 2 and Fig. 8). One of the novel virion components, RSp40/ICP22, is an orthologue of HSV-1 ICP22, a protein implicated in multiple aspects of infection. Early in infection, ICP22 induces the formation of discrete nuclear foci containing cellular chaperone proteins known as VICE domains (7). These domains are thought to play an important role in nuclear alterations and protein quality control necessary for viral infection. Additionally, ICP22 has been implicated in modulating viral gene expression (1, 12) and ensuring proper virion morphology (110). However, ICP22 was undetectable in HSV-1 virions (83, 110). Further work is necessary to determine if PRV RSp40/ICP22 is functionally homologous to HSV-1 ICP22. Incorporation of this protein into PRV virions is consistent with a potential role in the early stages of infection. This may be relevant to a successful infection since PRV, unlike HSV-1, does not express RSp40/ICP22 with immediate-early kinetics (113).
Fig. 8.
Schematic summary of previously known and novel viral virion components. Using mass spectrometry, a total of 47 viral proteins were detected in PRV virions including 40 (of the 42) previously identified viral virion proteins (white) and seven novel viral virion proteins (gray). Additionally, while we were unable to detect the viral kinase pUL13, a known component of virions, using mass spectrometry, we were able to confirm that it is a virion component using Western blot analysis (Fig. 5C). Viral proteins associated within the capsid, tegument, and envelope layers are displayed according to their known or putative localizations within virions.
In addition to identifying viral virion components, we detected phosphorylated peptides from pUL26, pUL36, pUL46, and pUL48. Posttranslational modifications of virion proteins may serve a variety of functions during viral infection, such as modulation of viral particle assembly, egress, or entry (14, 34, 55, 80, 105, 126, 127). However, key modifications, such as phosphorylation, can be challenging to detect using mass spectrometry and often require further enrichment (21). Our ability to detect these modifications without enrichment suggests that they are abundant within virions.
Phosphorylation of PRV pUL26, pUL36, pUL46, and pUL48 within virions is, to our knowledge, a novel finding. Phosphorylation of the HSV-1 orthologues of three of these proteins, pUL36, pUL46, and pUL48, has been previously described following virion entry, but the specific sites at which these proteins are modified have not been defined (90, 92, 105, 109, 142). For pUL48, a phosphorylation site has been predicted at Ser375, which is within a consensus casein kinase II (CKII) site near the C terminus. Mutation of this site to alanine disrupts pUL48 transactivation function (109). While the single phosphorylation we detected on PRV pUL48 could not be unambiguously localized to a particular residue, the phosphopeptide we detected contains the same CKII site, which is conserved in PRV and HSV-1 (109). Additionally, the phosphorylation sites within pUL36 (Ser2726 and Ser2969) are within a proline-rich domain near the C terminus. Interestingly, a PRV strain with a large deletion that encompasses this site displays decreased neurovirulence in infected mice (10). Further analyses will determine if this phosphorylation is critical for pUL36 function. For example, this modification may have a role during viral entry or assembly by facilitating nucleocapsid binding (79), nuclear pore docking (25), or recruitment of molecular motor proteins (114). Interestingly, while Morrison et al. have shown that phosphorylation of the HSV-1 orthologues of these proteins occurs only after entry (105), our findings show that these proteins are already phosphorylated in extracellular virions. It is tempting to speculate that phosphorylation may serve to activate viral proteins for specific functions during the early stages of infection.
In addition to viral proteins, herpesvirus virions have been previously shown to contain cellular proteins (8, 56, 61, 83, 99, 133, 144). However, incorporation of host proteins into PRV virions has been poorly characterized. In this study we identified a total of 48 host proteins that were present in both untreated and proteinase K-treated PRV virions but absent in samples from mock-infected cells (Fig. 5B). This number of host proteins was comparable to that previously detected by mass spectrometry in other herpesvirus virions, such as 49 in HSV-1 (83), 71 in HCMV (133), and 43 in EBV (56). Among the host proteins we detected, many have well-characterized functions and participate in a variety of biological pathways, such as cellular signaling, membrane organization and trafficking, RNA processing, and metabolism. Comparison of the host proteins we detected in PRV virions with those detected in other herpesvirus virions by mass spectrometry revealed several interesting similarities. Notably, both PRV and HSV-1 virions share four small GTPase family members (Table 4), while none was detected in HCMV. Additionally, the virions of PRV, HSV-1, and HCMV, but not EBV, contain members of the 14-3-3 family of signaling proteins (56, 83, 99). While these similarities and differences may be due to experimental techniques, the comparison may offer insights into the underlying mechanisms of herpesvirus assembly and the role of host proteins during infection.
In addition to their functions during PRV infection, the mechanisms by which host proteins are localized to virions remain unclear. Several studies have shown that incorporation of certain cellular proteins into virions may be targeted, such as MAP kinase (88). An additional example is the incorporation of cellular actin, which is increased in the absence of the tegument proteins VP22, pUS3, and pUL47 (30, 99). Alternatively, host proteins may be included in virions nonspecifically, and their presence could reflect proteins that are highly abundant within the cell or enriched at sites of viral assembly. Although little is known about the copy numbers of host proteins in mock- and PRV-infected PK15 cells, several studies have investigated the expression of host genes in a variety of PRV- and HSV-1-infected cell types and tissues (115, 129). Interestingly, the mRNA expression levels corresponding to 9 of the 48 cellular proteins we detected were decreased more than 3-fold during PRV infection of rat embryonic fibroblasts (115). In contrast, gene expression of only one of the proteins we detected (thioredoxin) was increased (115). These data do not directly address the relationship between protein abundance and virion incorporation; however, they suggest that the majority of the host proteins were detected in PRV virions despite their unchanged or decreased expression levels during infection. Further targeted studies involving dominant negative proteins or knockdown approaches are necessary to determine the functional roles that these proteins may have during infection.
In summary, we describe the first comprehensive analysis of PRV virion composition. Future analysis of the host and viral protein content of virions will enhance our understanding of the involvement of these proteins in the processes of viral assembly, egress, entry, pathogenesis, and spread.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thomas Mettenleiter (Friedrich Loeffler Institut, Germany), Hiroshi Kida (Hokkaido University, Japan), Hanns-Joachim Rziha (Friedrich Loeffler Institut, Germany), and Bruce Banfield (Queen's University, Canada) for kindly providing antibodies that were used in the study and Margaret Bisher (Princeton University) for technical assistance with the electron microscopy. We also thank Greg Smith (Northwestern University) and members of the Enquist lab for critical reading of the manuscript.
This work was supported by the National Institute on Drug Abuse (DP1 DA026192) to I.M.C., Human Frontier Science Program (RGY0079/2009-C) to I.M.C., National Institutes of Health (R37 NS33506 and P40 RR18604) to L.W.E., Princeton University Start-up funding to I.M.C, and a National Science Foundation Graduate Research Fellowship (DGE-0646086) to T.K.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Advani S. J., Weichselbaum R. R., Roizman B. 2003. Herpes simplex virus 1 activates cdc2 to recruit topoisomerase II alpha for post-DNA synthesis expression of late genes. Proc. Natl. Acad. Sci. U. S. A. 100:4825–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antinone S. E., Zaichick S. V., Smith G. A. 2010. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J. Virol. 84:13019–13030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atanasiu D., et al. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84:3825–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Au C. E., et al. 2007. Organellar proteomics to create the cell map. Curr. Opin. Cell Biol. 19:376–385 [DOI] [PubMed] [Google Scholar]
- 5. Baines J. D., Ward P. L., Campadelli-Fiume G., Roizman B. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baldick C. J., Shenk T. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bastian T. W., Livingston C. M., Weller S. K., Rice S. A. 2010. Herpes simplex virus type 1 immediate-early protein ICP22 is required for VICE domain formation during productive viral infection. J. Virol. 84:2384–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bechtel J. T., Winant R. C., Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bortz E., et al. 2003. Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 77:13425–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Böttcher S., et al. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403–13411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Böttcher S., et al. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 80:9910–9915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowman J. J., Orlando J. S., Davido D. J., Kushnir A. S., Schaffer P. A. 2009. Transient expression of herpes simplex virus type 1 ICP22 represses viral promoter activity and complements the replication of an ICP22 null virus. J. Virol. 83:8733–8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brack A. R., et al. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brideau A. D., Banfield B. W., Enquist L. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brideau A. D., Card J. P., Enquist L. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brideau A. D., Enquist L., Tirabassi R. S. 2000. The role of virion membrane protein endocytosis in the herpesvirus life cycle. J. Clin. Virol. 17:69–82 [DOI] [PubMed] [Google Scholar]
- 17. Brukman A., Enquist L. 2006. Pseudorabies virus EP0 protein counteracts an interferon-induced antiviral state in a species-specific manner. J. Virol. 80:10871–10873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bucks M. A., O'Regan K. J., Murphy M. A., Wills J. W., Courtney R. J. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calistri A., et al. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 81:11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chi J. H. I., Harley C. A., Mukhopadhyay A., Wilson D. W. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253–261 [DOI] [PubMed] [Google Scholar]
- 21. Choudhary C., Mann M. 2010. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11:427–439 [DOI] [PubMed] [Google Scholar]
- 22. Chowdary T. K., et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clase A. C., et al. 2003. The pseudorabies virus Us2 protein, a virion tegument component, is prenylated in infected cells. J. Virol. 77:12285–12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coller K. E., Lee J. I.-H., Ueda A., Smith G. A. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Copeland A., Newcomb W., Brown J. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crump C. M., Yates C., Minson T. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cunningham C., et al. 1992. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J. Gen. Virol. 73:303–311 [DOI] [PubMed] [Google Scholar]
- 28. Curanovic D., Enquist L. 2009. Virion-incorporated glycoprotein b mediates transneuronal spread of pseudorabies virus. J. Virol. 83:7796–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Curanovic D., Lyman M. G., Bou-Abboud C., Card J. P., Enquist L. 2009. Repair of the UL21 locus in pseudorabies virus Bartha enhances the kinetics of retrograde, transneuronal infection in vitro and in vivo. J. Virol. 83:1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. del Rio T., DeCoste C. J., Enquist L. 2005. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J. Virol. 79:8614–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dietz P., et al. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dijkstra J. M., Visser N., Mettenleiter T. C., Klupp B. G. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J. Virol. 70:5684–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dry I., et al. 2008. Proteomic analysis of pathogenic and attenuated alcelaphine herpesvirus 1. J. Virol. 82:5390–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edson C. M. 1993. Phosphorylation of neurotropic alphaherpesvirus envelope glycoproteins: herpes simplex virus type 2 gE2 and pseudorabies virus gI. Virology 195:268–270 [DOI] [PubMed] [Google Scholar]
- 35. Ekstrand M. I., Enquist L., Pomeranz L. E. 2008. The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends Mol. Med. 14:134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fossum E., et al. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathogens 5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fuchs W., et al. 2005. The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress. J. Virol. 79:11291–11299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fuchs W., Granzow H., Klupp B. G., Kopp M., Mettenleiter T. C. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuchs W., et al. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fuchs W., Klupp B. G., Granzow H., Leege T., Mettenleiter T. C. 2009. Characterization of pseudorabies virus (PrV) cleavage-encapsidation proteins and functional complementation of PrV pUL32 by the homologous protein of herpes simplex virus type 1. J. Virol. 83:3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fuchs W., Klupp B. G., Granzow H., Mettenleiter T. C. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fuchs W., Klupp B. G., Granzow H., Mettenleiter T. C. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fuchs W., Klupp B. G., Granzow H., Osterrieder N., Mettenleiter T. C. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Geraghty R. J., Krummenacher C., Cohen G. H., Eisenberg R. J., Spear P. G. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 45. Gottlieb J., Marcy A. I., Coen D. M., Challberg M. D. 1990. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol. 64:5976–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Granzow H., Klupp B. G., Mettenleiter T. C. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Granzow H., Klupp B. G., Mettenleiter T. C. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Greco T. M., Yu F., Guise A. J., Cristea I. M. 2011. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol. Cell Proteomics 10:M110.004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grünewald K., et al. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398 [DOI] [PubMed] [Google Scholar]
- 50. Hampl H., Ben-Porat T., Ehrlicher L., Habermehl K. O., Kaplan A. S. 1984. Characterization of the envelope proteins of pseudorabies virus. J. Virol. 52:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heldwein E. E., Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heldwein E. E., et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 53. Herold B. C., Visalli R. J., Susmarski N., Brandt C. R., Spear P. G. 1994. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J. Gen. Virol. 75:1211–1222 [DOI] [PubMed] [Google Scholar]
- 54. Jacobs L. 1994. Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae. Arch. Virol. 137:209–228 [DOI] [PubMed] [Google Scholar]
- 55. Jakubowicz T., Leader D. P. 1987. A major phosphoprotein of cells infected with pseudorabies virus is phosphorylated by cellular casein kinase II. J. Gen. Virol. 68:1159–1163 [DOI] [PubMed] [Google Scholar]
- 56. Johannsen E., et al. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jöns A., Granzow H., Kuchling R., Mettenleiter T. C. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jöns A., Mettenleiter T. C. 1996. Identification and characterization of pseudorabies virus dUTPase. J. Virol. 70:1242–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kaelin K., Dezélée S., Masse M. J., Bras F., Flamand A. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karger A., Bettin B., Granzow H., Mettenleiter T. C. 1998. Simple and rapid purification of alphaherpesviruses by chromatography on a cation exchange membrane. J. Virol. Methods 70:219–224 [DOI] [PubMed] [Google Scholar]
- 61. Kattenhorn L. M., et al. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187–11197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kehm R., Gelderblom H. R., Darai G. 1998. Identification of the UL56 protein of herpes simplex virus type 1 within the virion by immuno electron microscopy. Virus Genes 17:49–53 [DOI] [PubMed] [Google Scholar]
- 63. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 64. Klopfleisch R., et al. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J. Virol. 80:5571–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Klupp B., Altenschmidt J., Granzow H., Fuchs W., Mettenleiter T. C. 2008. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 82:6299–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Klupp B. G., Altenschmidt J., Granzow H., Fuchs W., Mettenleiter T. C. 2005. Identification and characterization of the pseudorabies virus UL43 protein. Virology 334:224–233 [DOI] [PubMed] [Google Scholar]
- 67. Klupp B. G., Baumeister J., Dietz P., Granzow H., Mettenleiter T. C. 1998. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 72:1949–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klupp B. G., Baumeister J., Karger A., Visser N., Mettenleiter T. C. 1994. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J. Virol. 68:3868–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Klupp B. G., Fuchs W., Granzow H., Nixdorf R., Mettenleiter T. C. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Klupp B. G., Fuchs W., Weiland E., Mettenleiter T. C. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J. Virol. 71:7687–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Klupp B. G., Granzow H., Karger A., Mettenleiter T. C. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J. Virol. 79:13442–13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klupp B. G., Granzow H., Mettenleiter T. C. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Klupp B. G., Granzow H., Mundt E., Mettenleiter T. C. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Klupp B. G., Visser N., Mettenleiter T. C. 1992. Identification and characterization of pseudorabies virus glycoprotein H. J. Virol. 66:3048–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kopp M., et al. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kopp M., Klupp B. G., Granzow H., Fuchs W., Mettenleiter T. C. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Laquerre S., et al. 1998. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J. Virol. 72:6119–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lee J. H., Vittone V., Diefenbach E., Cunningham A. L., Diefenbach R. J. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347–354 [DOI] [PubMed] [Google Scholar]
- 79. Lee J. I.-H., Luxton G. W. G., Smith G. A. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lemaster S., Roizman B. 1980. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J. Virol. 35:798–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lenk M., Visser N., Mettenleiter T. C. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J. Virol. 71:5635–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lin H.-W., Chang Y.-Y., Wong M.-L., Lin J.-W., Chang T.-J. 2004. Functional analysis of virion host shutoff protein of pseudorabies virus. Virology 324:412–418 [DOI] [PubMed] [Google Scholar]
- 83. Loret S., Guay G., Lippé R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Luo Y., Li T., Yu F., Kramer T., Cristea I. M. 2010. Resolving the composition of protein complexes using a MALDI LTQ Orbitrap. J. Am. Soc. Mass Spectrom. 21:34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Luxton G. W. G., et al. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Luxton G. W. G., Lee J. I.-H., Haverlock-Moyns S., Schober J. M., Smith G. A. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lyman M. G., Demmin G. L., Banfield B. W. 2003. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins Us3 and VP22. J. Virol. 77:1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lyman M. G., Randall J. A., Calton C. M., Banfield B. W. 2006. Localization of ERK/MAP kinase is regulated by the alphaherpesvirus tegument protein Us2. J. Virol. 80:7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. MacLean C. A., Efstathiou S., Elliott M. L., Jamieson F. E., McGeoch D. J. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72:897–906 [DOI] [PubMed] [Google Scholar]
- 90. Matsuzaki A., et al. 2005. US3 protein kinase of herpes simplex virus type 2 is required for the stability of the UL46-encoded tegument protein and its association with virus particles. J. Gen. Virol. 86:1979–1985 [DOI] [PubMed] [Google Scholar]
- 91. Maxwell K. L., Frappier L. 2007. Viral proteomics. Microbiol. Mol. Biol. Rev. 71:398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. McNabb D. S., Courtney R. J. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221–232 [DOI] [PubMed] [Google Scholar]
- 93. Meckes D. G., Wills J. W. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 81:13028–13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mettenleiter T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167–180 [DOI] [PubMed] [Google Scholar]
- 95. Mettenleiter T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163–169 [DOI] [PubMed] [Google Scholar]
- 96. Mettenleiter T. C., Klupp B. G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 97. Mettenleiter T. C., et al. 1990. Interaction of glycoprotein gIII with a cellular heparinlike substance mediates adsorption of pseudorabies virus. J. Virol. 64:278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Michael K., Klupp B. G., Karger A., Mettenleiter T. C. 2007. Efficient incorporation of tegument proteins pUL46, pUL49, and pUS3 into pseudorabies virus particles depends on the presence of pUL21. J. Virol. 81:1048–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Michael K., Klupp B. G., Mettenleiter T. C., Karger A. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mijatov B., Cunningham A. L., Diefenbach R. J. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31 [DOI] [PubMed] [Google Scholar]
- 101. Milne R. S., Connolly S. A., Krummenacher C., Eisenberg R. J., Cohen G. H. 2001. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 281:315–328 [DOI] [PubMed] [Google Scholar]
- 102. Möhl B., et al. 2009. Intracellular localization of the pseudorabies virus large tegument protein pUL36. J. Virol. 83:9641–9651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Möhl B. S., et al. 2010. Random transposon-mediated mutagenesis of the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 84:8153–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Moorman N. J., Sharon-Friling R., Shenk T., Cristea I. M. 2010. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol. Cell Proteomics 9:851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Morrison E. E., Wang Y. F., Meredith D. M. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ng T. I., Ogle W. O., Roizman B. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37–48 [DOI] [PubMed] [Google Scholar]
- 107. O'Connor C. M., Kedes D. H. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 80:1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ono E., Watanabe S., Nikami H., Tasaki T., Kida H. 1998. Pseudorabies virus (PRV) early protein 0 activates PRV gene transcription in combination with the immediate-early protein IE180 and enhances the infectivity of PRV genomic DNA. Vet. Microbiol. 63:99–107 [DOI] [PubMed] [Google Scholar]
- 109. O'Reilly D., Hanscombe O., O'Hare P. 1997. A single serine residue at position 375 of VP16 is critical for complex assembly with Oct-1 and HCF and is a target of phosphorylation by casein kinase II. EMBO J. 16:2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Orlando J. S., et al. 2006. ICP22 is required for wild-type composition and infectivity of herpes simplex virus type 1 virions. J. Virol. 80:9381–9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pawliczek T., Crump C. M. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83:11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Platt K. B., Maré C. J., Hinz P. N. 1979. Differentiation of vaccine strains and field isolates of pseudorabies (Aujeszky's disease) virus: thermal sensitivity and rabbit virulence markers. Arch. Virol. 60:13–23 [DOI] [PubMed] [Google Scholar]
- 113. Pomeranz L. E., Reynolds A. E., Hengartner C. J. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69:462–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Radtke K., et al. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathogens 6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ray N., Enquist L. 2004. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol. 78:3489–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Richman D. D., Buckmaster A., Bell S., Hodgman C., Minson A. C. 1986. Identification of a new glycoprotein of herpes simplex virus type 1 and genetic mapping of the gene that codes for it. J. Virol. 57:647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Roberts A. P. E., et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ryan J. P., Whealy M. E., Robbins A. K., Enquist L. 1987. Analysis of pseudorabies virus glycoprotein gIII localization and modification by using novel infectious viral mutants carrying unique EcoRI sites. J. Virol. 61:2962–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sherman G., Gottlieb J., Challberg M. D. 1992. The UL8 subunit of the herpes simplex virus helicase-primase complex is required for efficient primer utilization. J. Virol. 66:4884–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shiba C., et al. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397–2405 [DOI] [PubMed] [Google Scholar]
- 121. Smith C. C. 2005. The herpes simplex virus type 2 protein ICP10PK: a master of versatility. Front. Biosci. 10:2820–2831 [DOI] [PubMed] [Google Scholar]
- 122. Smith C. C., Aurelian L. 1997. The large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is associated with the virion tegument and has PK activity. Virology 234:235–242 [DOI] [PubMed] [Google Scholar]
- 123. Smith G. A., Gross S. P., Enquist L. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. U. S. A. 98:3466–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Solomon K. A., Robbins A. K., Enquist L. 1991. Mutations in the C-terminal hydrophobic domain of pseudorabies virus gIII affect both membrane anchoring and protein export. J. Virol. 65:5952–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Spear P. G., Longnecker R. 2003. Herpesvirus entry: an update. J. Virol. 77:10179–10185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Stevely W. S. 1975. Virus-induced proteins in pseudorabies-infected cells. II. Proteins of the virion and nucleocapsid. J. Virol. 16:944–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Stevely W. S., Katan M., Stirling V., Smith G., Leader D. P. 1985. Protein kinase activities associated with the virions of pseudorabies and herpes simplex virus. J. Gen. Virol. 66:661–673 [DOI] [PubMed] [Google Scholar]
- 128. Szilágyi J. F., Cunningham C. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72:661–668 [DOI] [PubMed] [Google Scholar]
- 129. Szpara M. L., Kobiler O., Enquist L. 2010. A common neuronal response to alphaherpesvirus infection. J. Neuroimmune Pharmacol. 5:418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Taddeo B., Zhang W., Roizman B. 2006. The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. U. S. A. 103:2827–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Tirabassi R. S., Enquist L. 1999. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J. Virol. 73:2717–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tirabassi R. S., Enquist L. 2000. Role of the pseudorabies virus gI cytoplasmic domain in neuroinvasion, virulence, and posttranslational N-linked glycosylation. J. Virol. 74:3505–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vittone V., et al. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ward P. L., Campadelli-Fiume G., Avitabile E., Roizman B. 1994. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J. Virol. 68:7406–7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Watanabe S., Ono E., Shimizu Y., Kida H. 1995. Pseudorabies virus early protein 0 transactivates the viral gene promoters. J. Gen. Virol. 76:2881–2885 [DOI] [PubMed] [Google Scholar]
- 137. Whealy M. E., et al. 1993. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J. Virol. 67:3786–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. 2009. Universal sample preparation method for proteome analysis. Nat. Methods 6:359–362 [DOI] [PubMed] [Google Scholar]
- 139. Wong M. L., Chen C. H. 1998. Evidence for the internal location of actin in the pseudorabies virion. Virus Res. 56:191–197 [DOI] [PubMed] [Google Scholar]
- 140. WuDunn D., Spear P. G. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 63:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yao F., Courtney R. J. 1992. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J. Virol. 66:2709–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zahariadis G., et al. 2008. Cell-type-specific tyrosine phosphorylation of the herpes simplex virus tegument protein VP11/12 encoded by gene UL46. J. Virol. 82:6098–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhang W., Chait B. T. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482–2489 [DOI] [PubMed] [Google Scholar]
- 144. Zhu F. X., Chong J. M., Wu L., Yuan Y. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.