Abstract
Virus-induced alterations in cell morphology play important roles in the viral life cycle. To examine the intracellular events of coxsackievirus B3 (CVB3) infection, green monkey kidney (GMK) cells were either inoculated with the virus or transfected with the viral RNA. Various microscopic and flow cytometric approaches demonstrated the emergence of CVB3 capsid proteins at 8 h posttransfection, followed by morphological transformation of the cells. The morphological changes included formation of membranous protrusions containing viral capsids, together with microtubules and actin. Translocation of viral capsids into these protrusions was sensitive to cytochalasin D, suggesting the importance of actin in the process. Three-dimensional (3D) live-cell imaging demonstrated frequent contacts between cellular protrusions and adjacent cells. Markedly, in spite of an increase in the cellular viral protein content starting 8 h postinfection, no significant decrease in cell viability or increase in the amount of early apoptotic markers was observed by flow cytometry by 28 h postinfection. Comicroinjection of viral RNA and fluorescent dextran in the presence of neutralizing virus antibody suggested that these protrusions mediated the spread of infection from one cell to another prior to virus-induced cell lysis. Altogether, the CVB3-induced cellular protrusions could function as a hitherto-unknown nonlytic mechanism of cell-to-cell transmission exploited by enteroviruses.
INTRODUCTION
Enteroviruses induce fundamental changes in cell morphology. The molecular mechanisms of these changes and the ensuing cellular release of viral progeny are mostly unknown. It is generally assumed that enteroviruses, like many other nonenveloped viruses, require cell lysis for infection spread. For example, the release of coxsackievirus B3 (CVB3) virions from infected cells depends on enhancement of cell membrane permeability caused by viral components (53). However, the lytic escape of enteroviruses can also be complemented by nonlytic virus release, as shown for poliovirus using an autophagosomal pathway (27, 49). Moreover, direct cell-to-cell spread has been suspected to occur with poliovirus in the central nervous system (40). Nonlytic transmission might be particularly important in persistent infection by enteroviruses and various other picornaviruses, such as Theiler's murine encephalomyelitis virus, foot-and-mouth disease virus, and Nora virus, a picorna-like virus (17, 25, 41, 58). Such transmission could be envisioned to provide an important advantage for the virus by helping it to hide inside the cell to avoid immune defense by neutralizing antibodies.
CVB3, a nonenveloped RNA virus, is a member of the genus Enterovirus of the family Picornaviridae. It causes myocarditis and pancreatitis in newborns and may play a role in some chronic diseases, such as dilated cardiomyopathy and type 1 diabetes (4, 5, 26, 30, 42). The icosahedral viral capsids are 29 nm in diameter and consist of the structural proteins VP1, VP2, VP3, and VP4. The 7,400-nucleotide viral RNA genome is polyadenylated, single stranded, and positive sense (39), encoding 11 proteins from a single open reading frame. CVBs invade the intestinal epithelium and bind to the major cellular receptor, a glycoprotein termed the coxsackie-adenovirus receptor (CAR) (12, 15, 16, 44, 61). CVBs enter the cytoplasm by a caveolin-dependent endocytic pathway, with similarities to macropinocytosis (15, 16, 35). CVB3 replicates on the outer surfaces of virus-induced membranous vesicles (8, 12, 20, 61). Viral assembly is a multistep process, in which cleavage of viral protein (VP0) yields mature capsid proteins VP4 and VP2, which, together with VP1 and VP3, form the mature capsid (6). Later in infection CVB3 induces apoptosis through caspase activation in cultured cells (11, 36, 60).
In this study, alterations in the cellular architecture and intracellular distribution of CVB3 proteins were studied in infected cells. The cells were either inoculated with the virus or transfected with the viral RNA by lipofection or cytoplasmic microinjection. Lipofection of viral RNA was used to synchronize viral infection, and comicroinjection of the viral RNA and fluorescent dextran were used to mark the transfected cells (7, 23, 24, 33, 38). Living as well as fixed cells were examined by electron microscopy (EM), wide-field and confocal microscopy, and flow cytometry. CVB3 RNA was shown to induce extensive cellular changes, including extended protrusions connecting adjacent cells. These studies demonstrated for the first time nonlytic intercellular transmission of CVB3.
MATERIALS AND METHODS
Viruses and cells.
CVB3 strain Nancy was obtained from the ATCC (Manassas, VA). Green monkey kidney (GMK) cells were grown in Earle's minimum essential medium (MEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 0.05 mg/ml penicillin, 0.05 mg/ml streptomycin (PAA laboratories GmbH, Linz, Austria), 1 mM l-glutamine (Invitrogen, Carlsbad, CA), and 1% glucose at 37°C in 5% CO2. For infection, the cells were inoculated with CVB3 and kept at 37°C until live-cell studies or fixation was performed (1).
CVB3, coxsackievirus A9 (CAV9), echovirus 11, echovirus 30, parechovirus, rhinovirus, and rotavirus RNAs were isolated from cell culture supernatants with a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The CVB3 RNA concentration in experiments was 120 ng/μl, and for other RNAs it was 100 ng/μl.
Antibodies and chemicals.
CVB3 polyclonal antibody (Ab) was obtained from Pharmacia Fine Chemicals (Uppsala, Sweden). Virus proteins were stained with Ab, followed by Alexa 488-conjugated anti-rabbit IgG. Flow cytometry of cell-associated viral proteins was done with anti-CVB3 monoclonal antibody (MAb) (Millipore, Billerica, MA) conjugated to Atto 488 fluorophore according to the manufacturer's protocol (Innova Biosciences, Cambridge, United Kingdom). Virus neutralization was performed in the presence of virus MAb (2 μg/ml). Microtubules (MTs) were visualized using α-tubulin MAb (Amersham, Buckinghamshire, United Kingdom), followed by Alexa 633-conjugated anti-mouse IgG (Molecular Probes) and actin with tetramethylrhodamine isothiocyanate (TRITC)-phalloidin (Molecular Probes). CAV9, echovirus 11, echovirus 30, parechovirus, rhinovirus, and rotavirus RNA proteins were stained with enterovirus MAb (Dako, Glostrup, Denmark), followed by Alexa 555-conjugated anti-mouse IgG (Molecular Probes). MT- and actin-disintegrating agents, nocodazole and cytochalasin D (Sigma Aldrich, St. Louis, MO), were used to detect CVB3 capsid protein signal. For immuno-EM studies, nanogold-conjugated goat anti-rabbit Ab and HQ-silver enhancement reagents were purchased from Nanoprobes (Yaphank, NY). Epon LX-112 was obtained from Ladd Research Industries (Williston, VT). For microinjection studies, fluorescein isothiocyanate (FITC)-labeled dextrans (40,500 kDa) were obtained from Molecular Probes and TRITC- and FITC-labeled dextrans (150 kDa) from Sigma Aldrich. Cell viability was judged by staining the cells with propidium iodide (PI) (Sigma) and annexin V-Alexa 488 (Molecular Probes). In flow cytometry control studies, apoptosis was induced by staurosporine (STS) or cycloheximide (CHX) obtained from Sigma Aldrich.
Infection.
Viral infection was initiated by either inoculation with virus or microinjection or by lipofection with the viral RNA using the DMRIE-C lipofection reagent (Invitrogen) according to the manufacturer's protocol.
Microinjection into cells was carried out using a semiautomatic system comprising the Transjector 5246 and Micromanipulator 5171 (Eppendorf, Hamburg, Germany) on an inverted microscope. Needles were pulled from glass capillaries (Clark Electromedical Instruments, Reading, United Kingdom) using a P-97 needle puller (Sutter Instruments, Novato, CA). For studies with fixed cells, cultures were grown to 80% confluence on glass coverslips and for live imaging on 21.5-cm2 glass bottom culture dishes (MatTek Cultureware, Ashland, MA). Purified CVB3 RNA was microinjected into the cytoplasm in a microinjection buffer (10 mM Tris-HCl–120 mM KCl, pH 7.4) at a concentration of 120 ng/ml. At various times after microinjection, cells were fixed with 4% paraformaldehyde (PFA) (20 min at room temperature [RT]), followed by labeling with CVB3 Ab and Alexa 488-conjugated anti-rabbit IgG. Three-dimensional (3D) images were obtained with inverted laser scanning confocal microscopes (LSM 510 Axiovert 100 M [Carl Zeiss AG, Jena, Germany] or FV-1000 IX-81 [Olympus, Tokyo, Japan]) using a 63× Plan-Neofluor oil immersion objective with a numerical aperture (NA) of 1.25 or a 60× APO oil immersion objective (NA = 1.35). Stacks of 25 to 35 images with an image size of 800 by 800 pixels were collected. The pixel resolution was adjusted to 110 nm/pixel. Fluorescent dextran (2 mg/ml) was used to identify the microinjected cells. For cell-to-cell spread studies, single cells of confluent monolayers were comicroinjected with RNA and 500 kDa FITC-dextran and incubated in the presence of neutralizing CVB3 Ab. The cells were fixed at 24 h postmicroinjection (p.m.) with 4% PFA and permeabilized with 0.1% Triton X-100. Control studies were performed in the absence of Ab. In microinjection experiments, the cells were stained for virus proteins with CVB3 Ab, followed by Alexa 555-conjugated anti-rabbit IgG.
Flow cytometry.
Double staining with annexin V/propidium iodide (PI) was used to assay cell viability. The cells were inoculated with the virus, and after 8 to 28 h, they were detached by scraping, washed with phosphate-buffered saline (PBS), stained with annexin V-Alexa Fluor 488 conjugate (5 μl) and PI at 2 μg/ml for 15 min, and analyzed with FACSCalibur and CellQuest software (Becton Dickinson, Heidelberg, Germany). The effects of drugs (1 μg/ml STS or 1 μg/ml CHX for 24 h) on cell viability were monitored. Virus proteins were detected with CVB3 monoclonal antibody (1 μg/1 × 106 cells) conjugated to Atto 488. Shortly after, the detached cells were washed with PBS, fixed in fixation buffer (R&D Systems Inc., Minneapolis, MN), and incubated at room temperature for 10 min prior to treatment with permeabilization/wash buffer (R&D Systems Inc.). MAb was added to the cells and incubated for 60 min at 4°C. The cells were washed with permeabilization/wash buffer and resuspended in staining buffer (R&D Systems Inc.) for flow cytometry analysis. The means of one representative experiment were counted from quadruplicate measurements with a total of 4 × 104 cells. Experiments were repeated three times with similar observations.
Live-cell imaging.
Cells were grown to 80% confluence on 21.5-cm2 glass bottom culture dishes (0.16 to 0.19 mm thickness; MatTek Cultureware, Ashland, MA). The images were captured either with a CellObserver HS wide-field microscope (Zeiss) or with a laser scanning confocal microscope (LSM510; Zeiss). Time lapse imaging of virus-induced alterations of cell morphology was performed with a Zeiss CellObserver HS wide-field microscope. The microscope incubator was maintained at 37°C, and the CO2 concentration was adjusted to 5%. The LD Plan-Neofluar 40× (NA = 0.6) objective was used. Images were taken at 5-min intervals. The LSM 510 objective and the sample holder were warmed to 37°C prior to imaging. The 3D stacks of 15 to 30 cross-sectional images were acquired with a Plan-Neofluar 63× oil immersion objective (NA = 1.25). The image size was adjusted to 512 by 512 pixels with an xy resolution of 100 nm/voxel and z of 350 to 500 nm/voxel. A 488-nm argon laser line was used for FITC-dextran excitation, and fluorescence was monitored with a 520- to 560-nm band-pass filter. TRITC-dextran was excited with a 543-nm HeNe laser and monitored with a long-pass filter of 560 nm.
For morphological characterization of uninfected cells, two cell populations individually transfected either with enhanced yellow fluorescent protein (EYFP) or enhanced cyan fluorescent protein (ECFP) were plated onto the same cell culture dish. The cells were imaged with a laser scanning confocal microsope (FV1000; Olympus) using the UPLSAPO 60× (NA = 1.2) water immersion objective. A 514-nm argon laser line was used for EYFP excitation, and fluorescence was monitored using a 530- to 630-nm band-pass filter. ECFP was excited by a 405-nm diode laser and was monitored with a band-pass filter of 460 to 500 nm. The pixel size was 210 nm in x and y and 460 nm in z. Line averaging was set at 2.
Electron microscopy.
For EM, cells on 13-mm glass coverslips were transfected with CVB3 RNA, incubated for 24 h, and fixed with 2.5% glutaraldehyde, 0.1 M Na-cacodylate buffer, pH 7.4. Postfixation with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4) was followed by dehydration and embedding in epoxy resin. Ultrathin sections were cut parallel to the coverslip, poststained with uranyl acetate and lead citrate, and examined with a JEM-100S (JEOL, Tokyo, Japan). For preembedding immunolabeling, the cells were grown on 8.8-cm2 plastic dishes, transfected prior to fixation, and processed as previously described (48). Briefly, the cells were fixed with periodate-lysine-PFA fixative for 2 h at RT and permeabilized with buffer A (0.01% saponin and 0.1% bovine serum albumin [BSA] in 0.1 M phosphate buffer, pH 7.4). Immunolabeling was performed using CVB3 Ab and 1.4-nm gold particle-conjugated goat rabbit IgG. Nanogold was silver enhanced for 9 min with the HQ-silver kit and gold toned with 0.05% gold chloride. The cells were postfixed with reduced 1% osmium tetroxide, dehydrated, and embedded in epoxy resin. Sections parallel to the bottom were cut, stained with uranyl acetate and lead citrate, and examined using a JEM-1200EX electron microscope (JEOL). The center-to-center distances between the virus particles in the protrusions were estimated by 1D Fourier transformation of the line profiles (ImageJ) (2). The most prominent frequency was converted to distance.
Microscopy of fixed cells.
For laser scanning confocal microscopy, cells on glass coverslips were transfected or microinjected with viral RNA (CVB3, CAV9, echovirus 11, echovirus 30, parechovirus, rhinovirus, and rotavirus) or virions or inoculated with virions. In some experiments, the transfected cells were first incubated for 8 h and then for another 8 h with either 60 μM nocodazole or 10 μM cytochalasin D. The cells were fixed at set time intervals after transfection or microinjection with 4% PFA, permeabilized with 0.1% Triton X-100, and stained for virus proteins, MTs, and actin. The cells were embedded with Mowiol-DABCO or ProLong Gold antifade reagent with or without DAPI (4′,6-diamidino-2-phenylindole) (Molecular Probes). In confocal microscopy (Olympus FV-1000), Alexa 488 was excited with a 488-nm argon laser, and fluorescence was collected with a 510- to 540-nm band-pass filter; Alexa 555 was excited with a 543-nm He-Ne laser, and the fluorescence was collected with a 570- to 620-nm band-pass filter; and Alexa 633 was excited with a 633-nm He-Ne laser, and the fluorescence was collected with a 647-nm long-pass filter.
Deconvolution.
To visualize the geometry of the protrusions, deconvolution confocal microscopy was performed with cells comicroinjected with RNA and 500-kDa FITC-dextran. A confocal microsope (Zeiss LSM 510) with a Plan-Neofluar 63× (NA = 1.25; oil) objective was used to acquire 3D stacks. The FITC was excited with a 488-nm laser line, and the fluorescence was collected with a 505- to 550 nm band-pass filter. The image size was adjusted to 1,024 by 1,024 pixels, and 55 slices were imaged with averaging of 4. The voxel size was 95 nm in xy and 400 nm in z. The pinhole was set to 1 Airy unit. The experimental point spread function was collected by imaging with green PS-Speck nanoparticles (Invitrogen). The particles were introduced to cells to simulate the dextran imaging conditions. The cells were washed with PBS prior to the addition of 70 μl of 1:2-diluted green PS-Speck particles. The cells were incubated for 15 min at RT and then fixed with 4% PFA for 20 min at RT. Before imaging of the cells in Dulbecco's modified Eagle's medium (DMEM), the dish was washed with PBS. The deconvolution was performed with Huygens Essential software (SVI, Netherlands). The point spread functions were averaged, and the iterative deconvolution was performed with signal-to-noise set to 10 and the quality threshold set to 0.l.
RESULTS
Infection-induced time-dependent changes in cell morphology and viability.
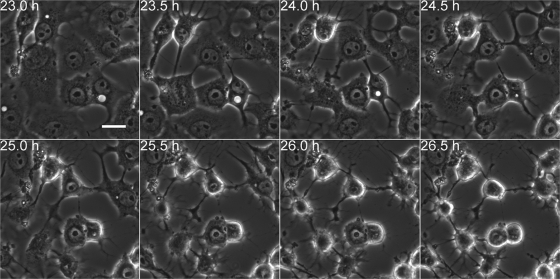
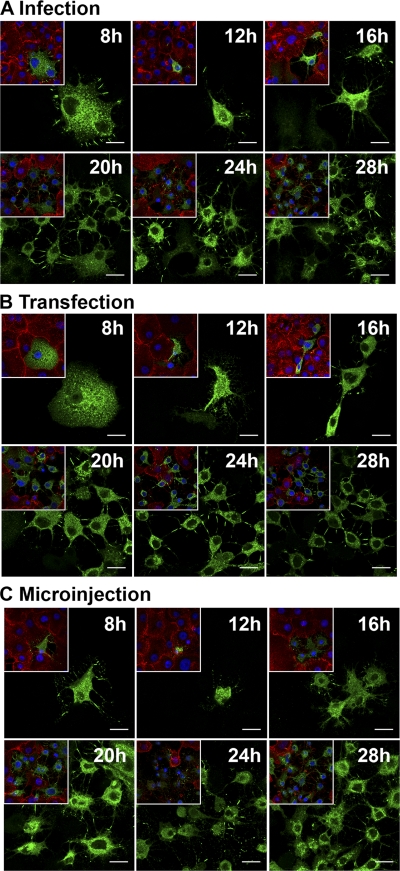
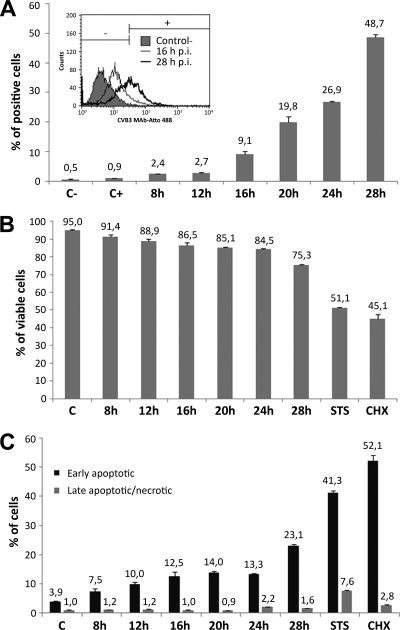
The time-dependent changes in cell morphology were analyzed in viral-RNA-transfected cells by time lapse imaging. Transfection by lipofection was used to synchronize viral infection. Sequential images were taken at 5-min intervals for 8 to 28 h posttransfection by lipofection (p.t.). Imaging of cells from 23 to 26.5 h p.t. revealed time-dependent morphological transformation and emergence of filamentous protrusions. These protrusions had frequent connections with neighboring cells (Fig. 1; see Movie S1 in the supplemental material). In contrast, the untransfected cells displayed the typical cell morphology (see Movie S1 in the supplemental material). Immunofluorescence microscopy verified a time-dependent increase in the number of cells with intracellular viral capsid proteins starting at 8 h postinfection (p.i) or p.t. At 8 h p.i./p.t., viral proteins were distributed homogeneously throughout the cytoplasm of unaltered cells. At 12 h and more clearly at 16, 20, 24, and 28 h p.i./p.t., the presence of viral proteins was accompanied by alterations in the cellular architecture. At 16 h and later in infection/lipofection, viral proteins were enriched in the central regions of the protrusions (Fig. 2A and B). In agreement with these results, flow cytometry of cells at 8, 12, 16, 20, 24, and 28 h p.i. indicated the presence of viral proteins in 2.4, 2.7, 9.1, 19.8, 26.9, and 48.7% of cells, respectively (Fig. 3A). The cells infected by microinjection of viral RNA showed somewhat faster alteration of cell morphology than inoculated or lipofected cells. Already at 8 h p.m., a portion of cells containing viral proteins showed altered morphology, with protrusions (Fig. 2C). At 20 h p.m., 68% ± 4% of the cells (n = 240) expressed viral proteins, all with altered morphology, including protrusions.
Fig. 1.
Viral-RNA-induced time-dependent alterations of cells. Time-lapse imaging with a wide-field microscope was performed to examine the dynamics of morphological transformation of RNA-transfected cells. The images, taken at 23 to 26.5 h p.t., show the gradual emergence of membranous protrusions. Scale bar, 10 μm.
Fig. 2.
Time-related alteration of infected cells. Cells infected by virus inoculation, lipofection, or microinjection of viral RNA were incubated for 8, 12, 16, 20, 24, or 28 h prior to fixation, immunolabeling, and imaging. A general image (inset) and a close-up image of inoculated (A), lipofected (B), and microinjected (C) cells at each time point are shown. Capsid proteins were visualized with CVB3 Ab, followed by Alexa 488-conjugated anti-rabbit IgG (green), actin with TRITC-phalloidin (red), and nuclei with DAPI (blue). Scale bars, 20 μm.
Fig. 3.
Emergence of viral proteins and infection-induced changes in cell viability. Flow cytometry was performed at 8, 12, 16, 20, 24, and 28 h p.i. (A) Percentages of virus protein-expressing cells. The cells were fixed and labeled with CVB3 monoclonal antibody conjugated to Atto 488. Untreated cells (C−) and uninfected immunolabeled cells (C+) were used as controls. (B) The percentage of viable cells was analyzed with annexin V-Alexa 488/propidium iodide double staining. The viable cells were annexin V and PI negative. (C) Percentages of apoptotic cells. Cells in early apoptosis were annexin V positive and PI negative, and cells in late apoptosis or necrosis were both annexin V and PI positive. STS- and CHX-treated cells were included as positive controls. The error bars indicate standard deviations.
The experiments described above indicated that the emergence of viral proteins preceded alteration of the cell morphology. Next, we studied virus infection-induced alterations of cell viability at 8 to 28 h p.i. The number of apoptotic and dead cells was studied by double labeling for annexin V and PI, and flow cytometry analysis. The percentage of viable cells (annexin V-PI double-negative cells) indicated that cell viability was not significantly decreased until 28 h p.i and in the presence of the apoptosis-inducing drugs STS and CHX (Fig. 3B and C). Concomitantly, the cells were analyzed by double staining for the early apoptosis marker annexin V and the late apoptosis/necrotic marker PI to differentiate between early apoptotic and late apoptotic/necrotic cells. Flow cytometry showed that the percentage of apoptotic cells varied from 7% to 13% 8 to 24 h p.i. while the number of early apoptotic cells was significantly increased at 28 p.i. compared with uninfected cells. The percentage of late apoptotic/necrotic cells was only 1.6% at 28 h p.i. In contrast, the drug-treated control cells showed higher percentages of apoptotic and necrotic cells (Fig. 3C). In conclusion, in spite of an increase in the viral protein content of the cells, only a minor decrease in viability or increase in the number of apoptotic cells was observed in infected compared to uninfected cells prior to 28 h p.i.
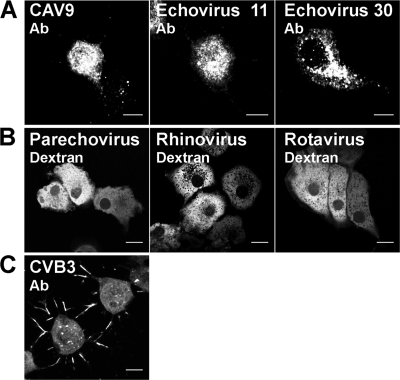
To examine whether it is the viral RNA that explains the alteration of cell morphology, we microinjected positive-sense, single-stranded RNAs ((+)ssRNAs) from CAV9, echovirus 11, echovirus 30, parechovirus, and rhinovirus and double-stranded RNA (dsRNA) from rotavirus into cells. We utilized dextran comicroinjection to mark the RNA-containing cells even in the absence of structural modifications. Enterovirus MAb immunolabeling studies with CAV9, echovirus 11, and echovirus 30 RNAs showed that at 24 h p.m., 20% ± 4% (n = 300), 11% ± 2% (n = 300), and 15% ± 4% (n = 300) of cells expressed viral proteins, respectively. The viral proteins were localized in the cytoplasm, but only a few cells showed alterations in morphology (Fig. 4A). Cells comicroinjected with parechovirus, rhinovirus, and rotavirus RNAs and dextran did not show enterovirus staining or changes in cell morphology (Fig. 4B). CVB3 infection of cells gave rise to a structural conversion similar to that of transfected CVB3 RNA, showing the presence of viral capsid proteins both in the cytoplasm and within the cell surface protrusions (Fig. 4C). Moreover, 94% of cells microinjected with dextran alone (n = 240) survived for 24 h without structural modifications.
Fig. 4.
Cellular effects of RNAs of various viruses. Confocal images of cells comicroinjected with (+)ssRNAs from CAV9, echovirus 11, echovirus 30, parechovirus, and rhinovirus and dsRNA from rotavirus and with fluorescent dextran. At 24 h p.m., the cell were fixed, and viral proteins were visualized with enterovirus MAb followed by Alexa 555-conjugated anti-mouse IgG. (A) Distribution of viral proteins in CAV9, echovirus 11, and echovirus 30 RNA-microinjected cells. (B) Morphology of parechovirus, rhinovirus, and rotavirus RNA-microinjected cells revealed by intracellular fluorescent dextran. (C) Distribution of CVB3 proteins at 24 h postinfection. Capsid proteins visualized with CVB3 Ab followed by Alexa 555-conjugated anti-rabbit IgG. Scale bars, 10 μm.
Viral capsids and capsid proteins localize in protrusions.
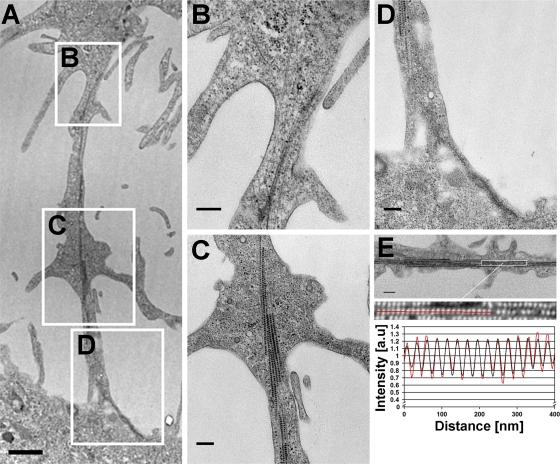
The above-mentioned experiments indicated that infected cells form membranous protrusions. EM images confirmed that these protrusions formed intercellular bridges between adjacent cells (Fig. 5A). High-magnification images showed the presence of cytoskeletal filaments, possibly actin and microtubulin, within the protrusions (Fig. 5B, C, and D). Notably, within the protrusions, viral capsids were seen to align along cytoskeletal filaments at 28 h p.t. (Fig. 5C). The distal parts of the protrusion extended up to the plasma membranes of the adjacent cells (Fig. 5D). The virus- and filament-containing bundles were enriched in the central regions of the protrusions but were absent from distal parts of them. The 1D Fourier transformation analysis showed that the average center-to-center distance between the closely packed particles was 26.9 ± 0.4 nm (Fig. 5E).
Fig. 5.
Virus-containing intercellular bridge between cells. (A) Low-magnification EM image of two neighboring cells and the intercellular bridge at 28 h p.t. Scale bar, 1 μm. (B to D) High-magnification images of the boxed areas in panel A show the protrusion nearest to the infected cell (B), the middle part (C), and the extension to the adjacent cell plasma membrane (D). Scale bars, 250 nm. (E) Fourier transformation analysis of the average distance between the closely packed particles in the protrusion. Scale bar, 250 nm.
The intracellular distribution of viral capsid proteins in viral-RNA-transfected cells was examined by immunoelectron microscopy. Here, viral capsid proteins and capsids were visualized as small, electron-dense grainy spots. At 24 h p.t., viral proteins and capsids were distributed inside cytoplasmic vesicular structures, as well as freely within the cytoplasm. Cells with viral proteins showed altered morphology with fragmented nuclei, chromatin condensation at the nuclear periphery, and protrusions, but with loss of cell surface microvilli. Importantly, viral capsid proteins were observed throughout the protrusions, including the tips, in the close vicinity of the neighboring cells. Untransfected control cells did not show any label (results not shown).
Virus translocation into the protrusions is sensitive to disintegration of the cellular cytoskeleton.
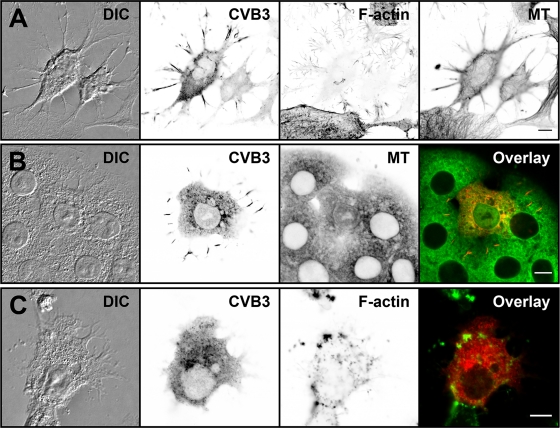
To assess the contribution of cytoskeletal elements to the formation of cellular protrusions and their viral protein content, microtubules and actin were visualized in cells fixed at 24 h p.t. Confocal imaging demonstrated viral capsid proteins, together with microtubules and actin filaments, in the protrusions. Notably, the capsid proteins within the protrusions were not distributed evenly but accumulated into elongated clusters at their central regions. In general, in viral-protein-expressing cells, the F-actin staining was markedly weaker, suggesting that the actin network was specifically affected by the infection (Fig. 6A). To investigate how microtubules contribute to the protrusions and their viral protein content, nocodazole was applied. This microtubule destabilizer affected neither protrusion formation nor the viral protein content (Fig. 6B). On the other hand, treatment with cytochalasin D, an actin-depolymerizing drug, resulted in retention of the viral proteins in the cytoplasm (Fig. 6C). Staining with α-tubulin MAb and phalloidin verified the disintegration of microtubules and actin, respectively, in the drug-treated cells.
Fig. 6.
Cellular distribution of capsid proteins, microtubules, and actin in transfected cells. (A) Confocal single-cross-section image of cells fixed at 16 h p.t. (B and C) Viral protein localization was studied in the presence of a microtubule-destabilizing drug, nocodazole (B), and an actin-depolymerizing drug, cytochalasin D (C), at 16 h p.t. Transfected cells were first incubated for 8 h in the absence and for 8 h in the presence of drugs. The cells were stained for capsids with CVB3 Ab, followed by Alexa 488-conjugated anti-rabbit IgG (red). Actin and microtubules were visualized by TRITC-phalloidin (green) and α-tubulin MAb, respectively, followed by Alexa 633-conjugated anti-mouse IgG (green). Differential interference contrast (DIC) and inverted gray scale images are shown. Scale bars, 10 μm.
To assess the membrane permeability of the protrusions, dextrans of 40 kDa (radius of gyration, 14 nm), 150 kDa (28 nm), and 500 kDa (50 nm) were comicroinjected with viral RNA into cells. Live-cell imaging at 24 h p.m. showed that all dextrans were distributed throughout the cytoplasm and the protrusions. Close examination of living cells revealed that the dextrans extended through the fine and highly branched network of protrusions (Fig. 7). Studies in living cells suggested that a small portion of dextrans might escape from the microinjected cells to the neighboring cells after infection (see Movie S2 in the supplemental material).
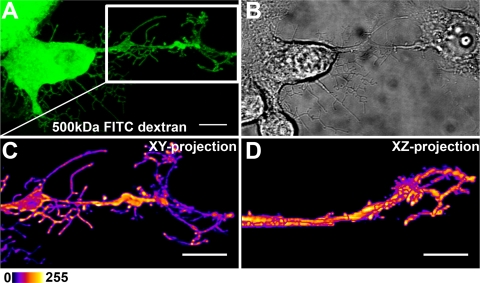
Fig. 7.
3D structure of protrusion. Cell imaging, deconvolution, and 3D space-filling modeling were used to examine protrusion morphology and interactions with neighboring cells. (A) Living cells comicroinjected with RNA and FITC-dextran (green) were imaged at 24 h p.m. (B) DIC image of cells. (C) Magnification of the white box in panel A, showing the protrusion area. (D) The xy and xz renderings of the image reveal the 3D details of the geometry of protrusions located around the periphery of the neighboring cell. Pseudocolor images with intensity increasing from black to white was used to visualize the intracellular distribution of dextran. Scale bars, 10 μm.
CVB3 infection is transmitted from cell-to-cell in the presence of neutralizing viral antibody.
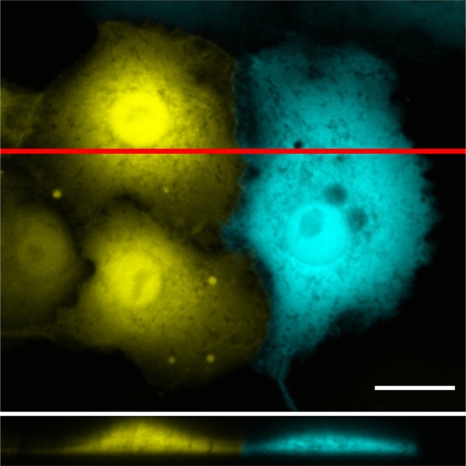
Detailed imaging of infected cells revealed that some of the protrusions exhibited a complex branching pattern. These protrusions frequently extended beyond the cellular edges, making contact with the adjacent cells. Imaging of uninfected cells expressing either EYFP or ECFP in the same monolayer culture showed no significant cell overlapping (Fig. 8). 3D geometry of the cell surface demonstrated that the protrusion tips formed a cage-like structure on the neighboring cell surface (Fig. 7; see Movie S3 in the supplemental material). Images demonstrated dextran accumulation in the enlargements of the branches and even in the tips of the protrusions (Fig. 7A, C, and D). The motility of protrusion tips was analyzed in viral-RNA- and dextran-comicroinjected cells by time-lapse imaging at 24 h p.m. Comicroinjection with dextran was utilized to identify the viral-RNA-containing cells. Imaging of cells for 20 min at 10-s intervals revealed that dextran-containing protrusion tips were mobile and frequently made transient contacts with the plasma membranes of neighboring cells. Notably, the dextran clusters moved inside the protrusions and were occasionally associated with the next cell plasma membrane (see Movie S2 in the supplemental material). At later stages of infection, the movement of protrusions ceased (results not shown).
Fig. 8.
Analysis of cell overlapping in monolayer culture. Shown is confocal live-cell imaging of mixed populations of uninfected cells expressing either EYFP (yellow) or ECFP (cyan). The xz projection reveals the profiles of cells at a location indicated by the red line. Scale bar, 20 μm.
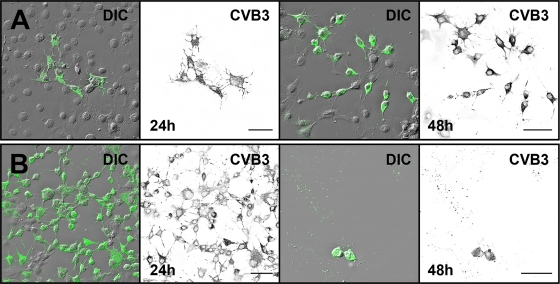
In order to determine the capability of the CVB3-induced protrusions for nonlytic spread of the virus, neutralizing anti-CVB3 MAb was added to the culture medium to eliminate viral spread via the culture medium. Here, for synchronous infection, the cells transfected with viral RNA were incubated with or without the antibody. In the presence of the antibody, the infected cells formed distinct foci surrounded by large areas of uninfected cells (Fig. 9A). In contrast, in the absence of antibody, most cells throughout the culture were infected. This suggested that viruses released from infected cells spread via the culture medium, thus infecting the cells both nearby and at a distance (Fig. 9B). At 48 p.t. in the presence of MAb, the clusters of infected areas were enlarged, whereas in the absence of MAb, the majority of cells were already dead (Fig. 9).
Fig. 9.
Effect of neutralizing antibody on infection spread in RNA-transfected cells. Cells were monitored in the presence (A) and in the absence (B) of neutralizing anti-CVB3 MAb. At 24 h and 48 h p.i., the cells were fixed and immunolabeled with anti-CVB3 Ab, followed by Alexa 488-labeled anti-rabbit IgG (green). DIC and inverted gray scale images are shown. Scale bars, 50 μm.
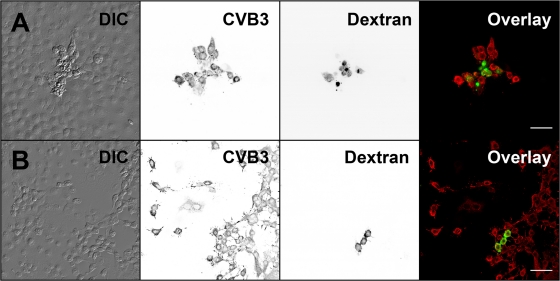
Next, possible cell-to-cell spread of infection was investigated by comicroinjection of RNA and dextran in the presence of neutralizing antibody. Fluorescent dextran was utilized to relocalize the viral-RNA-containing cells. At 12 h p.m., only originally microinjected, i.e., dextran-containing cells expressed viral proteins (results not shown). At 16 h p.m., some of the microinjected cells were surrounded by infected cells juxtaposed with the originally microinjected cells. At 24 h p.m., 23% ± 5% of microinjected cells (n = 311), with viral proteins and modified morphology, were surrounded by a rim of infected cells (Fig. 10A). To assess cell viability, living cells comicroinjected with CVB3 RNA and fluorescent dextran were stained with PI. At 24 h p.m., 31% ± 4% of the microinjected cells (n = 353) were morphologically modified and showed protrusions. A total of 4% ± 2% of the microinjected cells, rounded and without protrusions, were stained with PI (results not shown). Control studies without the neutralizing antibody showed large numbers of infected cells, not only in close proximity to but also far from the microinjection site (Fig. 10B). The overall distribution of infected cells in the absence of antibody pointed to infection spread via the medium after lytic viral release.
Fig. 10.
Intercellular transmission of infection. The spread of infection from the RNA-microinjected cells to neighboring cells was monitored in the presence and absence of neutralizing capsid Ab. Cells were fixed at 24 h after comicroinjection of RNA and dextran. Shown are microinjected and neighboring cells with (A) or without (B) neutralizing Ab. Microinjected cells were identified by FITC-dextran (green), and localization of CVB3 proteins is shown by CVB3 Ab, followed by Alexa 555-conjugated anti-rabbit IgG (red). DIC and inverted gray scale images are shown. Scale bars, 50 μm.
DISCUSSION
Picornaviruses, Theiler's murine encephalomyelitis virus (28, 52) and poliovirus (3, 34, 50) infections elicit profound cellular changes, including nuclear condensation and DNA fragmentation. In this study, we used advanced confocal microscopy and electron microscopy to provide a detailed view of the morphological changes induced by CVB3 RNA in GMK cells. Imaging of infected cells demonstrated that the appearance of viral proteins at 8 h p.i. was followed by extensive morphological alterations, including the emergence of membranous viral-protein- and capsid-containing protrusions at 12 to 24 h p.i. Notably, cell viability was only marginally decreased prior to 28 h p.i.
These CVB3-induced protrusions are different from the previously observed apoptotic membrane blebs generated by actin-myosin cytoskeletal structures (13, 14, 37, 51) yet resemble cellular projections occurring during the late stage of retrovirus (45, 47) and vaccinia (43), measles (19), African swine fever (29), and rice dwarf (54–56) virus infections. Our EM studies revealed that the CVB3-induced protrusions made frequent contacts with the plasma membranes of neighboring cells. Notably, viral capsid-like particles accumulated inside the protrusions. Fourier transformation analysis confirmed that the particles (26.9 ± 0.4 nm) were of a size very similar to that of the icosahedral capsids of CVB3 (29 nm) (39). Immuno-EM, as well as confocal microscopy, confirmed the presence of viral capsid proteins in these protrusions. While the majority of viral capsids were clustered in the central regions of the protrusions, the immuno-EM and confocal studies disclosed viral capsids throughout the protrusion, including parts making contact with the adjacent cells.
Recent advances in live-cell imaging and tracking fluorescent viruses have revealed details of cell-to-cell viral spread via tubular structures (9). With retroviruses, the physical interaction between infected and uninfected cells provides a more efficient way (by 2 to 3 orders of magnitude) to reach their new host cells compared to lytic spread of infection (10, 18, 45). Murine leukemia virus (MLV) uses actin-containing filopodial bridges (45) and lateral actin-myosin-driven movement along the filopodium surface in its cell-to-cell transport. Moreover, human immunodeficiency virus type 1 (HIV-1) exploits membrane nanotubes, in which virus movement is mediated by the underlying actin cytoskeleton, for intercellular spread between T cells (47). Other examples of enveloped viruses utilizing actin-rich cellular projections for intercellular dispersal are herpesviruses (22, 32), African swine fever virus (29), measles virus (19), and vaccinia virus (46, 59). Nonenveloped viruses are usually released into the extracellular space via virus-induced cell lysis. However, some of them exploit other means of cell-to-cell transmission. Prelytic transmission could be of particular use in persistent infection. Direct cell-to-cell spread has been suspected in the transmission of poliovirus from peripheral tissues to the central nervous system (40) and has also been described in CVB-infected myocytes (31), as well as in cells infected by another picornavirus, Ljungan virus (21). Rice dwarf virus, a member of the Reoviridae, provides yet another example of a nonenveloped virus exploiting virus-induced actin-based protrusions for cell-to-cell transfer (54–56). Moreover, this virus requires microtubules for release from insect cells (57). Our confocal microscopy examination showed that the CVB3-induced protrusions contained viral proteins, together with microtubules and actin filaments. Disintegration of actin resulted in cytoplasmic accumulation of viral capsid proteins, whereas depolymerization of microtubules did not inhibit viral translocation into the protrusions. This strongly suggests an important role for actin in the transport of viral particles into the protrusions. Furthermore, EM studies demonstrated the alignment of capsid-like particles along cytoskeletal filaments in the protrusions. These findings indicated that, in addition to the capsids freely diffusing into the protrusions, a significant number of capsids interact actively with the cell cytoskeleton.
To estimate the potential of the protrusions in cell-to-cell transmission, it is crucial to show that they make contacts between adjacent cells. 3D geometry demonstrated that the tips of the protrusions formed a cage-like structure on the neighboring cell surface. Live-cell studies indicated that the branched tips of intercellular protrusions were mobile and frequently located around the periphery of neighboring cells. Although a majority of the dextrans remained in the originally microinjected infected cells, a small portion of dextrans escaped from the microinjected cells after infection. This suggests that viruses might induce nonlytic means, utilized by the dextran, to cross the cell membrane. For example, the nonenveloped rotaviruses and polioviruses possess alternative means of release, including autophagosomes, without cell lysis (4, 27, 49).
To study whether the CVB3-induced protrusions can spread the viral infection into the neighboring cells, we comicroinjected virus RNA and dextran into the cell cytoplasm. This approach in the presence of neutralizing CVB3-specific antibodies can directly demonstrate lateral transmission of infection to the nonmicroinjected cells. A key observation was a cluster of infected cells in close contact with the originally microinjected living cells. This indicates that CVB3 is capable of lateral cell-to-cell transmission of infection.
Based on our results, we suggest a novel mechanism for nonlytic spread of CVB3 infection via cellular protrusions. Protrusion-mediated egress of virus between cells could provide a means of immune evasion that might be particularly important in the establishment of nonlytic persistent infections. However, many fundamental aspects of the molecular mechanisms, as well as the ubiquity, of this viral cell-to-cell transfer remain to be elucidated.
Supplementary Material
ACKNOWLEDGMENTS
First-class experimental support by Kati Vuori, Eija Jokitalo, Mervi Lindman, Eeva Jokela, Eeva Tolvanen, Milla Vuokko, Sami Willman, Pirjo Kauppinen, Raija Vassinen, and Paavo Niutanen is gratefully acknowledged. We are grateful to Klaus Hedman for comments on the manuscript.
The work was supported by the Academy of Finland (contracts 107311 and 129774), the Juvenile Diabetes Research Foundation, and the National Graduate School of Nanoscience.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Abraham G., Colonno R. J. 1984. Many rhinovirus serotypes share the same cellular receptor. J. Virol. 51:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. J. Biophotonics Int. 11:36–42 [Google Scholar]
- 3. Agol V. I., et al. 1998. Two types of death of poliovirus-infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology 252:343–353 doi:10.1006/viro.1998.9438 [DOI] [PubMed] [Google Scholar]
- 4. Andréoletti L., et al. 1997. Experimental CVB3-induced chronic myocarditis in two murine strains: evidence of interrelationships between virus replication and myocardial damage in persistent cardiac infection. J. Med. Virol. 52:206–214 [DOI] [PubMed] [Google Scholar]
- 5. Andréoletti L., et al. 1997. Detection of coxsackie B virus RNA sequences in whole blood samples from adult patients at the onset of type I diabetes mellitus. J. Med. Virol. 52:121–127 [DOI] [PubMed] [Google Scholar]
- 6. Basavappa R., et al. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci. 3:1651–1669 doi:10.1002/pro.5560031005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belsham G. J., Bostock C. J. 1988. Studies on the infectivity of foot-and-mouth disease virus RNA using microinjection. J. Gen. Virol. 69:265–274 [DOI] [PubMed] [Google Scholar]
- 8. Bienz K., Egger D., Pfister T. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. Suppl. 9:147–157 [DOI] [PubMed] [Google Scholar]
- 9. Burckhardt C. J., Greber U. F. 2009. Virus movements on the plasma membrane support infection and transmission between cells. PLoS Pathog. 5:e1000621 doi:10.1371/journal.ppat.1000621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carr J. M., Hocking H., Li P., Burrell C. J. 1999. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology 265:319–329 doi:10.1006/viro.1999.0047 [DOI] [PubMed] [Google Scholar]
- 11. Carthy C. M., et al. 1998. Caspase activation and specific cleavage of substrates after coxsackievirus B3-induced cytopathic effect in HeLa cells. J. Virol. 72:7669–7675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung S. K., et al. 2005. Internalization and trafficking mechanisms of coxsackievirus B3 in HeLa cells. Virology 333:31–40 doi:10.1016/j.virol.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 13. Coleman M. L., et al. 2001. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3:339–345 doi:10.1038/35070009 [DOI] [PubMed] [Google Scholar]
- 14. Cotter T. G., Lennon S. V., Glynn J. M., Green D. R. 1992. Microfilament-disrupting agents prevent the formation of apoptotic bodies in tumor cells undergoing apoptosis. Cancer Res. 52:997–1005 [PubMed] [Google Scholar]
- 15. Coyne C. B., Bergelson J. M. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119–131 doi:10.1016/j.cell.2005.10.035 [DOI] [PubMed] [Google Scholar]
- 16. Coyne C. B., Shen L., Turner J. R., Bergelson J. M. 2007. Coxsackievirus entry across epithelial tight junctions requires occludin and the small GTPases Rab34 and Rab5. Cell. Host Microbe 2:181–192 doi:10.1016/j.chom.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de la Torre J. C., Alarcon B., Martinez-Salas E., Carrasco L., Domingo E. 1987. Ribavirin cures cells of a persistent infection with foot-and-mouth disease virus in vitro. J. Virol. 61:233–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dimitrov D. S., et al. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 67:2182–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duprex W. P., McQuaid S., Hangartner L., Billeter M. A., Rima B. K. 1999. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J. Virol. 73:9568–9575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Egger D., Bienz K. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86:707–718 doi:10.1099/vir.0.80442-0 [DOI] [PubMed] [Google Scholar]
- 21. Ekström J. O., et al. 2007. Replication of Ljungan virus in cell culture: the genomic 5′-end, infectious cDNA clones and host cell response to viral infections. Virus Res. 130:129–139 doi:10.1016/j.virusres.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 22. Favoreel H. W., Van Minnebruggen G., Adriaensen D., Nauwynck H. J. 2005. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc. Natl. Acad. Sci. U. S. A. 102:8990–8995 doi:10.1073/pnas.0409099102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gamarnik A. V., Andino R. 1996. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 15:5988–5998 [PMC free article] [PubMed] [Google Scholar]
- 24. Gamarnik A. V., Boddeker N., Andino R. 2000. Translation and replication of human rhinovirus type 14 and mengovirus in Xenopus oocytes. J. Virol. 74:11983–11987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Habayeb M. S., Ekengren S. K., Hultmark D. 2006. Nora virus, a persistent virus in Drosophila, defines a new picorna-like virus family. J. Gen. Virol. 87:3045–3051 doi:10.1099/vir.0.81997-0 [DOI] [PubMed] [Google Scholar]
- 26. Hyoty H., et al. 1995. A prospective study of the role of coxsackie B and other enterovirus infections in the pathogenesis of IDDM. Childhood Diabetes in Finland (DiMe) Study Group. Diabetes 44:652–657 [DOI] [PubMed] [Google Scholar]
- 27. Jackson W. T., et al. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156 doi:10.1371/journal.pbio.0030156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jelachich M. L., Lipton H. L. 1996. Theiler's murine encephalomyelitis virus kills restrictive but not permissive cells by apoptosis. J. Virol. 70:6856–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jouvenet N., et al. 2006. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell. Microbiol. 8:1803–1811 doi:10.1111/j.1462-5822.2006.00750.x [DOI] [PubMed] [Google Scholar]
- 30. Kandolf R., et al. 1993. Molecular pathogenesis of enterovirus-induced myocarditis: virus persistence and chronic inflammation. Intervirology 35:140–151 [DOI] [PubMed] [Google Scholar]
- 31. Klingel K., et al. 1998. Visualization of enteroviral replication in myocardial tissue by ultrastructural in situ hybridization: identification of target cells and cytopathic effects. Lab. Invest. 78:1227–1237 [PubMed] [Google Scholar]
- 32. La Boissière S., Izeta A., Malcomber S., O'Hare P. 2004. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J. Virol. 78:8002–8014 doi:10.1128/JVI.78.15.8002-8014.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai C. J., Zhao B. T., Hori H., Bray M. 1991. Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc. Natl. Acad. Sci. U. S. A. 88:5139–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. López-Guerrero J. A., Alonso M., Martin-Belmonte F., Carrasco L. 2000. Poliovirus induces apoptosis in the human U937 promonocytic cell line. Virology 272:250–256 doi:10.1006/viro.2000.0405 [DOI] [PubMed] [Google Scholar]
- 35. Marchant D., et al. 2009. ERK MAP kinase-activated Arf6 trafficking directs coxsackievirus type B3 into an unproductive compartment during virus host-cell entry. J. Gen. Virol. 90:854–862 doi:10.1099/vir.0.005868-0 [DOI] [PubMed] [Google Scholar]
- 36. Martin U., et al. 2004. Characterization of coxsackievirus B3-caused apoptosis under in vitro conditions. Med. Microbiol. Immunol. 193:133–139 doi:10.1007/s00430-003-0197-7 [DOI] [PubMed] [Google Scholar]
- 37. Mills J. C., Stone N. L., Erhardt J., Pittman R. N. 1998. Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J. Cell Biol. 140:627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Modalsli K. R., Bukholm G., Mikalsen S. O., Degre M. 1991. Microinjected Coxsackie B1 virus does not replicate in HEp-2 cells. Virology 185:888–890 [DOI] [PubMed] [Google Scholar]
- 39. Muckelbauer J. K., et al. 1995. The structure of coxsackievirus B3 at 3.5 A resolution. Structure 3:653–667 [DOI] [PubMed] [Google Scholar]
- 40. Ponnuraj E. M., John T. J., Levin M. J., Simoes E. A. 1998. Cell-to-cell spread of poliovirus in the spinal cord of bonnet monkeys (Macaca radiata). J. Gen. Virol. 79:2393–2403 [DOI] [PubMed] [Google Scholar]
- 41. Rodriguez M., Oleszak E., Leibowitz J. 1987. Theiler's murine encephalomyelitis: a model of demyelination and persistence of virus. Crit. Rev. Immunol. 7:325–365 [PubMed] [Google Scholar]
- 42. Roivainen M., et al. 1998. Several different enterovirus serotypes can be associated with prediabetic autoimmune episodes and onset of overt IDDM. Childhood Diabetes in Finland (DiMe) Study Group. J. Med. Virol. 56:74–78 [DOI] [PubMed] [Google Scholar]
- 43. Sanderson C. M., Way M., Smith G. L. 1998. Virus-induced cell motility. J. Virol. 72:1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shafren D. R., Williams D. T., Barry R. D. 1997. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J. Virol. 71:9844–9848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sherer N. M., et al. 2007. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 9:310–315 doi:10.1038/ncb1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith G. L., Murphy B. J., Law M. 2003. Vaccinia virus motility. Annu. Rev. Microbiol. 57:323– 342 doi:10.1146/annurev.micro.57.030502.091037 [DOI] [PubMed] [Google Scholar]
- 47. Sowinski S., et al. 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10:211–219 doi:10.1038/ncb1682 [DOI] [PubMed] [Google Scholar]
- 48. Suikkanen S., et al. 2003. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J. Virol. 77:10270–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taylor M. P., Kirkegaard K. 2008. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy 4:286–289 [DOI] [PubMed] [Google Scholar]
- 50. Tolskaya E. A., et al. 1995. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J. Virol. 69:1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Torgerson R. R., McNiven M. A. 1998. The actin-myosin cytoskeleton mediates reversible agonist-induced membrane blebbing. J. Cell Sci. 111:2911–2922 [DOI] [PubMed] [Google Scholar]
- 52. Tsunoda I., Kurtz C. I., Fujinami R. S. 1997. Apoptosis in acute and chronic central nervous system disease induced by Theiler's murine encephalomyelitis virus. Virology 228:388–393 doi:10.1006/viro.1996.8382 [DOI] [PubMed] [Google Scholar]
- 53. van Kuppeveld F. J., et al. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519–3532 doi:10.1093/emboj/16.12.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wei T., Chen H., Ichiki-Uehara T., Hibino H., Omura T. 2007. Entry of Rice dwarf virus into cultured cells of its insect vector involves clathrin-mediated endocytosis. J. Virol. 81:7811–7815 doi:10.1128/JVI.00050-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wei T., et al. 2006. The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J. Virol. 80:8593–8602 doi:10.1128/JVI.00537-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wei T., Shimizu T., Omura T. 2008. Endomembranes and myosin mediate assembly into tubules of Pns10 of Rice dwarf virus and intercellular spreading of the virus in cultured insect vector cells. Virology 372:349–356 doi:10.1016/j.virol.2007.10.034 [DOI] [PubMed] [Google Scholar]
- 57. Wei T., et al. 2009. Association of Rice gall dwarf virus with microtubules is necessary for viral release from cultured insect vector cells. J. Virol. 83:10830–10835 doi:10.1128/JVI.01067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Whitton J. L., Cornell C. T., Feuer R. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3:765–776 doi:10.1038/nrmicro1284 [DOI] [PubMed] [Google Scholar]
- 59. Wolffe E. J., Weisberg A. S., Moss B. 1998. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology 244:20–26 doi:10.1006/viro.1998.9103 [DOI] [PubMed] [Google Scholar]
- 60. Yuan J. P., et al. 2003. Coxsackievirus B3-induced apoptosis and caspase-3. Cell Res. 13:203–209 doi:10.1038/sj.cr.7290165 [DOI] [PubMed] [Google Scholar]
- 61. Zhang Y., Bergelson J. M. 2005. Adenovirus receptors. J. Virol. 79:12125–12131 doi:10.1128/JVI.79.19.12125-12131.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.