Abstract
Major histocompatibility complex (MHC) class II-presented peptides can be derived from both exogenous (extracellular) and endogenous (biosynthesized) sources of antigen. Although several endogenous antigen-processing pathways have been reported, little is known about their relative contributions to global CD4+ T cell responses against complex antigens. Using influenza virus for this purpose, we assessed the role of macroautophagy, a process in which cytosolic proteins are delivered to the lysosome by de novo vesicle formation and membrane fusion. Influenza infection triggered productive macroautophagy, and autophagy-dependent presentation was readily observed with model antigens that naturally traffic to the autophagosome. Furthermore, treatments that enhance or inhibit macroautophagy modulated the level of presentation from these model antigens. However, validated enzyme-linked immunospot (ELISpot) assays of influenza-specific CD4+ T cells from infected mice using a variety of antigen-presenting cells, including primary dendritic cells, revealed no detectable macroautophagy-dependent component. In contrast, the contribution of proteasome-dependent endogenous antigen processing to the global influenza CD4+ response was readily appreciated. The contribution of macroautophagy to the MHC class II-restricted response may vary depending upon the pathogen.
INTRODUCTION
The activation of CD4+ T cells depends upon their recognition of peptides (epitopes) associated with major histocompatibility class (MHC) class II molecules. Conventionally, peptide generation involves the degradation of exogenous (extracellular) antigens in the endosomal network by multiple mechanisms, including unfolding, reduction, and proteolytic degradation. It is now clear that epitopes derived from endogenous antigens (synthesized by the cell) also can be presented on MHC class II molecules (12, 22, 24, 38, 49, 50, 60, 66). Indeed, endogenous antigen expression appears to be an absolute requirement for the presentation of some epitopes. For example, the UV inactivation of A/PR8/34 influenza virus (PR8) or treatment of infected antigen-presenting cells (APCs) with protein synthesis inhibitors prevents the presentation of the NA79 epitope (12). Similar observations have been made for an epitope derived from influenza matrix protein 1 (24) and an epitope derived from the MHC class I H2-Ld molecule (39).
Numerous studies now have demonstrated that endogenous antigens can gain access to MHC class II loading compartments via a variety of intracellular pathways (4, 38, 49, 57, 66, 67). Perhaps the most straightforward route is autophagy, in which cytosolic proteins are delivered to the lysosome via several different mechanisms. Chaperone-mediated autophagy results in the delivery of cytosolic proteins directly to the lysosome based upon the recognition of a KFERQ pentapeptide motif within the target protein, and it was shown to be responsible for the presentation of an epitope derived from glutamate decarboxylase (72). Although 30% of cytosolic proteins contain this motif (70), CMA has not yet been implicated in the class II-restricted response to an infectious agent. Macroautophagy is characterized by the formation of a double-membrane structure that engulfs portions of the cytosol, including proteins, organelles, and/or invading pathogens. The resultant autophagosomes undergo membrane fusion with lysosomes, and the rapid degradation of the contents ensues (18). Macroautophagic vesicles fuse continuously with class II-positive compartments (60), and several epitopes have been shown to depend upon macroautophagy for class II-restricted presentation, including those derived from the C5 protein of the complement pathway (2), neomycin phosphotransferase (45, 53), and the Epstein Barr virus nuclear antigen 1 (EBNA-1) (35, 50). Interestingly, EBNA-1 contains two class II-restricted endogenous epitopes that are not autophagy dependent, suggesting that this protein can be processed via more than one pathway (35). Other studies of macroautophagy in antigen processing have employed engineered model antigens in which a well-defined epitope is affixed to a protein that naturally localizes to the autophagosome. For example, autophagy-dependent presentation was demonstrated for an influenza matrix protein 1-derived epitope fused to the LC3B protein, which is involved in autophagosome formation (60).
While a contribution by macroautophagy to class II processing is intuitively appealing, several considerations complicate the picture. (i) In many cases, the aggressive proteolysis associated with autophagy may be counterproductive for antigen processing. The presentation of many epitopes is diminished or ablated with elevated endosomal proteolysis (6, 58). Likewise, it has been reported that dendritic cells with reduced endosomal proteolytic activity are more efficient antigen presenters (6, 58). (ii) Results that suggest a role for autophagy may have alternative explanations. For example, autophagy was implicated in the class II-restricted presentation of antigens derived from nuclear and cytosolic proteins (8) due to the positive effects of serum starvation, but this condition also enhances proteasomal activity (14). In addition, the gene ablation of a key autophagy component has been reported to affect not just autophagy but also the presentation of exogenous antigens (1, 34). (iii) Some autophagy substrates may be atypical. For example, EBNA-1 is unusual in containing a glycine-alanine (Gly-Ala) repeat region that appears to be involved in immune evasion (36, 37) and also renders the protein prone to aggregation (19, 40), thereby increasing its susceptibility to autophagy (52, 68). (iv) Finally, in studying the presentation of class II-restricted epitopes from the influenza neuraminidase and hemagglutinin (HA) glycoproteins, our laboratory has identified an endogenous pathway that depends upon the multicatalytic proteasome, the macromolecular complex that is responsible for most protein turnover within the cytosol and nucleus (13, 17, 20). Remarkably, enzyme-linked immunospot (ELISpot) assays revealed that 30 to 40% of the total MHC class II-restricted response to influenza is specific for proteasome-dependent epitopes (66), a fraction that, when added to the contribution of exogenous processing, may largely discount other processing pathways. Thus, elucidating the relative roles of various processing pathways will require a more global approach.
Here, we have taken such a global approach in assessing the CD4+ T cell responses to influenza virus infection. Although influenza virus infection activates functional autophagy and we were able to detect autophagy-dependent presentation with engineered processing substrates, we could discern no significant contribution of this pathway to the CD4+ T cell response.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
L929 fibroblasts stably expressing the H2d-restricted MHC class II molecule I-Ed (L-IEd) or B6 fibroblasts stably expressing I-Ed and CIITA (B6-IEd) were cultured in Dulbecco's modified essential medium (DMEM) (Mediatech Inc., Manassas, Va) supplemented with 10% fetal calf serum (FCS) (Invitrogen, Carlsbad, CA) and 1× 2-mercaptoethanol (2ME; Invitrogen) at 9% CO2. Atg5 wild-type or knockout (KO) fibroblasts were cultured in DMEM supplemented with 10% FCS and 1× nonessential amino acids and maintained at 9% CO2 (fibroblasts were a kind gift of David Leib, Dartmouth Medical School). The A20 B cell lymphoma line was maintained in Iscoves modified Dulbecco's medium (IMDM) with 10% FCS, and the Raw264.7 cell line was maintained in DMEM with 10% FCS at 9% CO2. A lacZ T cell hybridoma specific for the Site-1 epitope of influenza hemagglutinin (62, 66) was used in 4-methylumbelliferyl ß-d-glucuronide (MUG) assays and passaged in RPMI-1640 (Mediatech, Inc.) supplemented with 10% FCS and 1× 2-mercaptoethanol and maintained at 6% CO2.
For plasmid generation, the S1 epitope was inserted at the C terminus of neomycin phosphotransferase or the N terminus of LC3B by PCR using primers that encoded the S1 epitope at the indicated position. The PCR product was inserted into the pGEM-T Easy vector (Thermo Fisher Scientific) and subsequently cloned into the pMSCV vector (a generous gift of Jianke Zhang, Thomas Jefferson University). All DNA constructs were verified by sequencing.
The influenza viruses A/PR/8/34 (PR8; H1N1), A/Japan/305/57 (Jap; H2N2), and B/Lee/40 (B/Lee) were grown as previously described (66). For experiments involving UV-inactivated virus, live PR8 was diluted in PBS and subjected to UV light for 10 min with gentle shaking. The inactivation of the virus was confirmed by staining fibroblasts pulsed with UV PR8 virus for influenza virus proteins via flow cytometry. Viral titers were determined using chicken red blood cells and were expressed in hemagglutinin units (HAU). Adenovirus expressing S1LC3 (Ad-S1LC3) was generated by cloning the S1LC3 construct into the pENTR4 entry vector (Invitrogen), followed by being subcloned into pAd/CMV/V5-DEST (Invitrogen). Virus was produced in 293A cells (Invitrogen) by transfection with linearized S1LC3-pAd/CMV/V5, followed by expansion and purification with the Adeno-X purification kit according to the manufacturer's recommendations (BD Biosciences, San Jose CA).
Mice.
Female BALB/c (H-2d) and CB6F1 (H-2bxd) mice aged 6 to 8 weeks were obtained from Jackson Laboratories (Bar Harbor, ME) and maintained at Thomas Jefferson University (Philadelphia, PA). Mice were primed intraperitoneally with 100 HAU of infectious PR8 or B/Lee diluted in cold phosphate-buffered saline (PBS) or intranasally with 0.1, 0.01, or 0.0001 HAU of infectious PR8 in cold PBS. At least 10 days after being primed, mice were euthanized using CO2 and spleens were harvested. CD4+ T cells were purified using the Dynal magnetic bead negative selection kit according to the manufacturer's instructions (Invitrogen), and the purified cells were used subsequently in all ELISpot experiments. All experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee at Thomas Jefferson University.
Generation and retroviral transduction of bone marrow-derived primary dendritic cells (BMDCs).
Bone marrow was isolated from the femurs and tibias of naïve female BALB/c or CB6F1 mice as previously described (66). Bone marrow was plated in 10-cm culture dishes at 2 × 106 cells/plate in a final volume of 10 ml of RPMI supplemented with 10% FCS, 1× penicillin-streptomycin, 1× l-glutamine, 1× 2ME, and granulocyte-macrophage colony-stimulating factor (GM-CSF) (10 ng/ml). Forty-eight h later the cells were harvested, counted, and plated in 24-well plates at 1 × 106 cells per well. Short hairpin RNA (shRNA) directed against Atg7 was designed using RNAiCODEX and cloned into the MSCV/LTRmiR30-PIG (LMP) retroviral vector encoding green fluorescent protein (GFP) under an internal ribosome entry site (IRES) promoter (OpenBiosystems; Thermo Fischer Scientific). Retrovirus encoding Atg7 shRNA or an empty LMP vector was overlaid onto the primary cells, and the cells were spun at 1,000 × g for 2 h. After the transduction, fresh growth medium was added to the cells. Forty-eight to 72 h after transduction (66), the cells were harvested and sorted for GFP expression for use in ELISpot assays. As in other knockdown experiments, the reduction of Atg7 was confirmed by Western blotting.
siRNA transfections.
Short interfering RNAs (siRNAs) against Atg7, Rpt2, or a scrambled control were obtained from Ambion (Applied Biosystems, Austin, TX). L-IEd fibroblasts were transfected with the indicated siRNAs using an Amaxa nucleofection kit V (Lonza, Allendale, NJ) once (48 h before assay) or twice in a 4-day period (once at day 0, then again at day 2). B6-IEd fibroblasts were transfected with the indicated siRNAs 3 days prior to assays using RNAiMAX transfection reagent (Invitrogen). A20 and Raw264.7 cell lines were transfected with siRNA using the Amaxa nucleofection kit V (Lonza) 48 h before use (for A20) or twice within a 48-h period (for Raw264.7). For the final 24 h of culture, the Raw264.7 cells were treated with gamma interferon (IFN-γ) to upregulate class II machinery. For the siRNA knockdown of the autophagy construct S1LC3, B6-IEd fibroblasts stably expressing S1LC3 were transfected twice within a 3-day period to ensure the complete knockdown of preexisting S1pMHC complexes.
Assessment of autophagy induction and knockdown.
To examine the induction of autophagy after influenza virus infection, L-IEd or B6-IEd fibroblasts were infected with PR8 (4 to 50 HAU/106 cells) for 30 min at 37°C in PBS–0.1% bovine serum albumin (BSA). At the indicated time points or after an overnight (∼16 h) culture, the cells were trypsinized, washed, and lysed in lysis buffer (100 mM NaCl, 100 mM Tris, pH 8.0, 1% Triton-X) in the presence of HALT protease inhibitor cocktail (Thermo Fischer Scientific) for 15 min on ice. Samples were centrifuged for 15 min at 13,000 × g at 4°C to remove debris, and protein concentrations were determined by BCA assay (Thermo Fisher Scientific). Equal amounts of protein were loaded onto a 12% SDS gel, subjected to electrophoresis, and transferred to a nitrocellulose membrane (GE Healthcare, Piscataway, NJ) for blotting. Anti-LC3b, anti-Atg7, and anti-actin were obtained from Sigma Aldrich (St. Louis, MO).
In vitro antigen presentation assays.
B6-IEd fibroblasts were transiently transfected with the indicated constructs using GeneJuice (EMD Biosciences, Gibbstown, NJ). Twenty-four h after transfection, autophagy was induced by treating cells with 200 nM rapamycin (EMD Biosciences) overnight. Cells were harvested, fixed with 0.05% paraformaldehyde (PFA; Electron Microscopy Services, Hatfield, PA) in 1× Hanks balanced salt solution (HBSS) (Thermo Fisher Scientific) for 15 min at room temperature, washed extensively, and cultured overnight with S1 T hybridomas. T cell activation was assessed by measuring beta-galactosidase activity using the fluorescent substrate MUG, with the amount of fluorescence being directly proportional to T cell activation (56, 62).
For chemical inhibition, the autophagy construct S1LC3 was transfected into B6-IEd as described above. Twenty-four h later the cells were washed in PBS, acid washed to remove existing pMHC-II complexes from the surface, washed extensively, and cultured overnight in the presence of 3-methyladenine (3MA; Sigma Aldrich) or wortmannin (EMD Biosciences). Cells were harvested and cultured for 6 h with S1 T hybridomas in a MUG assay. For siRNA-mediated inhibition, the autophagy construct S1LC3 was stably transfected into B6-Ied, and siRNA was transfected as described above. Cells were harvested and cultured with S1 T hybridomas overnight in a MUG assay.
ELISpot assay.
L-IEd or B6-IEd fibroblasts, A20 B cell lymphomas, RAW264.7 macrophages, or primary BMDCs were transfected/transduced with the indicated siRNAs or shRNAs. After the optimal time periods for maximal knockdown, the cells were harvested and pulsed with infectious PR8 or UV-inactivated PR8 for 30 to 60 min at 37°C in PBS–0.1% BSA. Cells were washed and cultured overnight with purified CD4+ T cells from the spleens of PR8-primed mice in an IFN-γ ELISpot assay. The next day the assay was developed according to the manufacturer's instructions (BD Biosciences). For experiments involving AdenoS1LC3, L-IEd fibroblasts were transfected with the indicated siRNAs. Six hours after the second transfection, media were changed and the cells were infected with Ad-S1LC3 (3 × 108 IFU/0.2 × 106 cells). Thirty-six to 48 h after infection, the infected L-IEd fibroblasts were harvested and cultured overnight with purified CD4+ T cells from a PR8-primed mouse in an IFN-γ ELISpot assay. The AdS1LC3-infected L-IEd fibroblasts also were cultured overnight with S1 T hybridomas in a MUG assay. For experiments involving epoxomicin, cells were pretreated with 0.5 μM epoxomicin (Enzo Life Sciences, Plymouth Meeting, PA) for 15 min prior to infection.
Statistical analysis.
Statistics were calculated using a paired Student t test. P < 0.05 was considered significant.
RESULTS
Influenza infection induces functional macroautophagy.
Autophagy can be induced by a variety of viral and bacterial infections (26, 30, 48). Two recent publications reported that influenza virus infection can induce autophagy (15, 73); however, one of these indicated that the process is incomplete due to the inhibition of vesicular fusion (15). To explore autophagy induction and maturation in our system, L929 fibroblasts stably transfected with the I-Edα and I-Edβ genes (L-IEd) or B6 fibroblasts stably transduced with I-Edα and I-Edβ (B6-IEd) were infected with influenza virus A/PR/8/34 (PR8). As a positive control, the cell lines were treated with the known autophagy inducer rapamycin (47). Autophagy induction then was assessed by measuring the conversion of LC3-I to LC3-II via Western blotting (41, 55). In both fibroblast lines, PR8 infection induced the conversion of LC3-I to LC3-II in a time-dependent manner beginning at 8 h after infection (Fig. 1A). The treatment of infected cells with bafilomycin A1, which neutralizes endosomal vesicles, thereby preventing the turnover of converted LC3, accentuated this effect (Fig. 1B), indicating that LC3 conversion is not an artifact of increased LC3 synthesis. The inhibition of autophagy via siRNA targeting Atg7, a ubiquitin ligase-like protein essential for autophagosome expansion, resulted in decreased LC3-II accumulation compared to that of control cells in response to influenza virus infection (Fig. 1C). Atg7 siRNA had a similar impact when rapamycin was used to induce autophagy (data not shown).
Fig. 1.
Autophagy is induced in fibroblasts after PR8 infection. (A) L-IEd or B6-IEd fibroblasts were infected with PR8 (4 to 50 HAU/106 cells) or stimulated with rapamycin (200 nM) in the presence of E64d (10 μg/ml) for the indicated times. (B) Fibroblasts were infected with PR8 (4 to 50 HAU/106 cells) overnight, and 2 h before harvesting cells for lysate collection bafilomycin A1 was added. (C) L-IEd or B6-IEd fibroblasts were transfected with siRNA to Atg7 or a scrambled control. To achieve optimal (100%) knockdown, L-IEd fibroblasts were transfected twice within a 4-day period, while B6-IEd fibroblasts were transfected once. Forty-eight to 72 h after the final transfection of siRNA, cells were harvested to assess the knockdown (KD) and functional inhibition of autophagy. In all cases, autophagy induction was assessed by Western blotting, probing lysates for LC3-II levels. Results are representative of three independent experiments.
To determine whether macroautophagy progressed to maturation, we generated B6-IEd fibroblasts that stably express a GFP-LC3 fusion construct (42). Autophagy induction via PR8 infection in B6-IEd-GFP-LC3 cells resulted in a significant decrease in GFP fluorescence (Fig. 2A, left), reflecting the successful fusion of the autophagosome and destruction of the contents (61). A reduction in fluorescence was not observed in cells expressing unconjugated GFP (Fig. 2A, right). Furthermore, the chemical inhibition of autophagy via wortmannin prevented the reduction of GFP levels after influenza virus infection (Fig. 2B) as well as the accumulation of endogenous LC3-II (Fig. 2C). In addition, the PR8 infection of L-IEd fibroblasts resulted in decreased levels of p62, a protein specifically degraded by the autophagy pathway (51 and data not shown). Taken together, these results indicate that the cell lines used for subsequent experiments are capable of being induced to functional autophagy by influenza virus infection and that our methods of silencing this process are effective.
Fig. 2.
Autophagy induced by PR8 infection proceeds without a block in the pathway. (A) B6-IEd fibroblasts stably expressing GFP-LC3 (left) or GFP alone (right) were infected with PR8 (4 to 50 HAU/106 cells), and GFP fluorescence was assessed via flow cytometry. (B) GFP-LC3-expressing fibroblasts were infected with PR8 in the presence or absence of the chemical inhibitor wortmannin, and GFP fluorescence was assessed. (C) B6-IEd fibroblasts were infected with PR8 as described for panel B, and autophagy was assessed by probing lysates for LC3 conversion by Western blotting. Results are representative of three independent experiments.
Autophagy can contribute to the presentation of endogenous antigens on MHC class II molecules.
We next determined whether autophagy can deliver endogenous antigens for epitope presentation to CD4+ T cells as has been described previously (8, 23, 34, 45, 50, 53). To this end, we appended the classically presented H2-IEd-restricted Site-1 (S1) epitope of influenza hemagglutinin, which appears to have innate resistance to endocytic proteases (3, 11) of two proteins that have been well described to localize naturally to the autophagosome. S1-conjugated neomycin phosphotransferase (C-terminal attachment) or S1-conjugated LC3B (N-terminal attachment) was transiently transfected into B6-IEd fibroblasts, and S1 presentation was assessed using S1-specific T cell hybridomas. As shown in Fig. 3, the S1 epitope was presented efficiently from both constructs, unlike mock-transfected controls. T cell stimulation was enhanced when cells expressing either construct were treated with rapamycin, which is consistent with an autophagy-dependent mechanism of presentation. The more dramatic response of the S1LC3 construct likely is due to LC3B being a specific component of the autophagy pathway, whereas NeoS1 is an autophagy substrate that may be less efficiently incorporated into the autophagosome.
Fig. 3.
Site-1 presentation from NeoS1 and S1LC3 is enhanced upon metabolic induction of autophagy. B6-IEd fibroblasts were transiently transfected with NeoS1 (A) or S1LC3 (B) (S3LC3 and NeoNA served as transfection controls), and 24 h later, cells were stimulated with rapamycin (200 nM) overnight. The next day, cells were fixed in 0.05% PFA for 15 min at room temperature and cultured with S1-specific T cell hybridomas overnight. The legend at the bottom of the figure indicates the number of APCs/well. Results are representative of three independent experiments.
The overnight treatment of S1LC3-expressing cells with a chemical inhibitor of autophagy, 3MA or wortmannin, significantly reduced the presentation of the S1 epitope from B6-IEd fibroblasts (Fig. 4A). 3MA treatment inhibited S1 presentation to a greater extent than wortmannin; however, it also was observed to be more toxic to the cells in culture (data not shown). The Atg7 siRNA transfection of B6IEd cells stably expressing S1LC3 also resulted in decreased S1 presentation compared to that of control siRNA-transfected cells (Fig. 4B). Additionally, we mutated the glycine at residue 120 of LC3 to an alanine, which prevents LC3 conversion (25). This substantially reduced S1 presentation from the fusion protein (see Fig. S1 in the supplemental material).
Fig. 4.
Presentation of S1 derived from S1LC3 is dependent upon autophagy. (A) B6-IEd fibroblasts transiently transfected with S1LC3 for 24 h were acid stripped with citric acid buffer, pH 2.0, washed extensively, and cultured overnight in the presence of autophagy inhibitors. The cells then were cultured with S1-specific T cell hybridomas for 6 h to assess presentation. (B) B6-IEd fibroblasts stably expressing S1LC3 were transfected with the indicated siRNAs twice for 2 days and cocultured overnight with S1 T cell hybridomas in a MUG assay. (C) L-IEd fibroblasts were transfected with the indicated siRNAs twice during a period of 4 days. Six hours after the second siRNA transfection, the cells were infected with S1LC3 adenovirus. Two days later, cells were harvested and cocultured with CD4+ T cells isolated from the spleens of PR8-primed mice overnight in an IFN-γ ELISpot assay. (D) Transfection of fibroblasts as described for panel C, except that the L-IEd fibroblasts were cultured with S1-specific T cell hybridomas in a MUG assay. Results are representative of at least two independent experiments. *, P < 0.05; **, P < 0.005.
Establishing a system to assess the contribution of autophagy to the CD4+ T cell response to influenza.
To determine the contribution of autophagy to the in vivo influenza virus-specific CD4+ T cell responses, we elected to use an assay that we previously developed for assessing the influence of proteasome activity on the magnitude of the CD4+ T cell response (66). This involved the infection of wild-type mice and analyzing the resulting CD4+ T cell response with an interferon-γ ELISpot assay in which influenza-infected stimulator cells were left untreated or were treated with proteasome inhibitor. The difference in spot number reflected the number of T cells specific for proteasome-dependent epitopes. Even if proteasome-deficient mice had been available, we would have chosen this configuration rather than comparing responses in infected wild-type and knockout mice. Our rationale was that the limited homeostatic space allowed greater and lesser quantities of naïve T cells in the wild-type and knockout mice, respectively, to expand to the same absolute numbers. Thus, although mice have been generated with autophagy deficiencies in certain cell types (34), we opted to dissect the response that occurs in wild-type mice. Aiding this decision was the report that DCs from these knockout mice appear to be impaired in exogenous antigen processing as well (34). To ensure that the assay was sufficiently sensitive, we determined the change in the responding T cell population that can be statistically detected. BALB/c mice were primed with either PR8 or B/Lee influenza viruses, which are immunologically distinct. Ten days after being primed, CD4+ T cells were isolated from the spleens, mixed in different proportions, and cultured with L-IEd fibroblasts infected with either PR8 or B/Lee virus. Antigen-specific CD4+ T cell responses above background levels were detected when 10% of the total T cell population was specific for the restimulating virus (see Fig. S2 in the supplemental material).
Also in preparation for the analysis of the in vivo response, we constructed a positive-control adenovirus that expresses S1LC3 (Adeno-S1LC3). We immunized mice intranasally with live PR8 virus, and at least 10 days after immunization we isolated CD4+ T cells from the spleen via negative selection. Under these conditions, the S1 epitope is immunodominant (21). The CD4+ T cells isolated from influenza virus-primed mice were cultured with siRNA-transfected L-IEd fibroblasts infected with Adeno-S1LC3 virus. Because only S1 is shared between the two viruses, only S1-specific T cells respond in the assay. Atg7 knockdown in Adeno-S1LC3-infected L-IEd fibroblasts significantly reduced the number of responding CD4+ T cells in an IFN-γ ELISpot assay (Fig. 4C). A decrease in S1-specific T cell activation in an in vitro T-hybridoma assay (Fig. 4D) also was observed. Taken together, these results demonstrate that ELISpot assays are capable of detecting with considerable sensitivity a CD4+ T cell population specific for autophagy-dependent epitopes expressed in the context of a viral infection.
Proteasomal activity, but not autophagy, contributes to global MHC class II-restricted presentation in an influenza virus infection.
Having observed that influenza virus does induce functional autophagy, and having validated various aspects of our approach, we asked whether macroautophagy plays an appreciable role in the global CD4+ T cell response to this complex antigen. L-IEd or B6-IEd fibroblasts were transfected with control or Atg7-specific siRNA, infected with live PR8 virus, and cultured with purified CD4+ T cells from influenza virus-primed mice in IFN-γ ELISpot assays. Under these conditions, more than 90% of the restimulating fibroblasts were infected with PR8 virus (see Fig. S3 in the supplemental material). If macroautophagy plays a major role in the generation of MHC class II-restricted influenza virus-derived epitopes in vivo, then CD4+ T cells that recognize autophagy-dependent epitopes will have been induced, and the inhibition of this pathway should result in a decrease in the number of responding CD4+ T cells in the ELISpot assay. However, despite the validation of knockdown (Fig. 4c) and sensitivity of the assay, Atg7 siRNA had no detectable effect on the number of responding CD4+ T cells (Fig. 5A to D). Similar results were obtained utilizing intraperitoneal and intranasal routes of immunization and also when limiting doses of influenza were used both for the priming and for the infection of the antigen-presenting cells (Fig. 5A versus C and B versus D) (data not shown). When utilized as stimulators in the ELISpot assay, the A20 B cell lymphoma line, the Raw264.7 macrophage cell line (see Fig. S4 in the supplementary material), and fibroblasts deficient in the autophagy protein Atg5 (data not shown) yielded similar results.
Fig. 5.
Autophagy does not significantly contribute to MHC class II-mediated presentation during an influenza viral infection. L-IEd (A) or B6-IEd (B) fibroblasts were transfected with the indicated siRNAs harvested at optimally determined time points and infected with 100 HAU/106 cells of PR8. Infected cells were cultured with CD4+ T cells isolated from splenocytes of mice primed intraperitoneally (i.p.) with PR8. (C and D) Treatments were similar to those described for panels A and B, except the fibroblasts were infected with a lower dose of influenza virus (4 HAU/106 cells) and were cultured with CD4+ T cells isolated from splenocytes of mice primed intranasally (i.n.) with PR8. Results are representative of at least three independent experiments.
To address the possibility that experimental conditions bias the response toward exogenously processed epitopes, thereby masking contributions by endogenously presented epitopes, stimulators were pulsed with equivalent doses of infectious or UV-inactivated influenza virus. An appreciably greater number of CD4+ T cells were activated in response to live virus stimulation, indicating that endogenous epitopes are well represented in the ELISpot assays (see Fig. S5 in the supplemental material). We further investigated possible masking effects by eliminating the response to the glycoproteins (HA and NA), which are major targets of the CD4+ T cell response (see below) and which, by virtue of their membrane tethering, are not expected to be substrates for autophagy. This was accomplished by stimulating PR8 (H1N1)-primed CD4+ T cells in the ELISpot assay with JAP (H2N2)-infected L-IEd fibroblasts, thereby restricting the response to the untethered internal and nonstructural proteins that are virtually identical in all influenza A viruses. This heterologous restimulation reduces the total number of responding T cells by ∼50% compared to the level for PR8 restimulation (see Fig. S5 in the supplemental material). However, autophagy inhibition using Atg7 siRNA does not significantly reduce the number of responding T cells after JAP restimulation. Thus, a substantial portion of the CD4+ T cell response is directed against internal proteins whose epitopes are produced principally, if not completely, by processes other than autophagy.
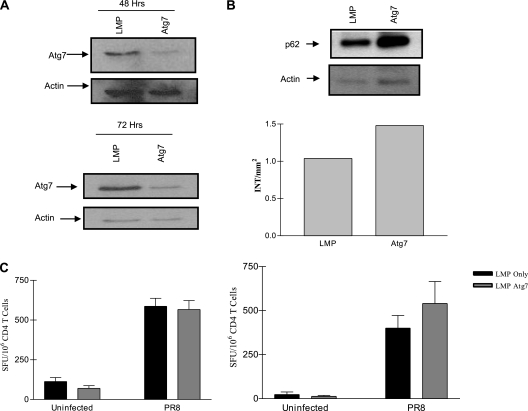
DCs play a major role in priming both class I- and class II-restricted responses in vivo, and it was possible that DCs are more active in macroautophagy than the ELISpot stimulator cells that had been tested and/or are distinct in their selection of autophagy substrates. To address this concern, BMDCs were transduced with the shRNA retroviral vector LMP or LMP encoding an shRNA for Atg7. Forty-eight to 72 h after transduction, BMDCs were sorted for high GFP expression (GFPhi), infected with PR8 virus, and cultured with CD4+ T cells purified from influenza virus-infected mice in an IFN-γ ELISpot assay. Flow cytometry confirmed the synthesis of viral proteins within DCs pulsed with infectious virus but not equivalent amounts of UV-inactivated virus, indicating the availability of endogenous antigens (see Fig. S3 in the supplemental material and data not shown). Furthermore, the stimulation of influenza virus-specific CD4+ T cells with DCs infected with live virus resulted in larger amounts of T cell activation than that when UV-inactivated virus was used (data not shown), again indicating that a substantial portion of the class II response is directed against endogenous epitopes. The knockdown of Atg7 in GFPhi cells was substantial (Fig. 6A), and this resulted in functionally impaired autophagy, as reflected by p62 accumulation (Fig. 6B). As with all other cell types tested, these primary DCs demonstrated no significant contribution of the macroautophagy pathway to in vivo CD4+ T cell activation (Fig. 6C). Importantly, in contrast to Atg5-deficient DCs (34), the presentation of exogenous antigens in our system was not affected (see Fig. S6A and B in the supplemental material).
Fig. 6.
Autophagy is not a significant pathway of MHC class II-mediated presentation during influenza viral infection in dendritic cells. BMDCs from BALB/c or CB6F1 mice were transduced with LMP alone or LMP-ATG7 shRNA retrovirus. (A) Forty-eight to 72 h after transduction cells were harvested and sorted for high GFP expression (GFPhi), and knockdown was assessed by Western blotting. (B) Functional knockdown of autophagy was determined by probing infected cell lysates for p62 accumulation via Western blotting (top, raw data; bottom, densitometry analysis). (C) GFPhi BMDCs were infected with PR8 and cultured with CD4+ T cells purified from influenza virus-primed mice overnight in an IFN-γ ELISpot assay (left, 48 h knockdown; right, 72 h knockdown). Results are representative of at least two independent experiments at each time point.
As previously reported (66), the treatment of stimulatory APCs with the proteasome inhibitor epoxomicin resulted in a significant decrease in the number of CD4+ T cells responding in the ELISpot assay (Fig. 7A and B). This was not due to nonspecific effects of epoxomicin, as the presentation of synthetic peptide and the classically presented S1 epitope were not affected. Additionally, while the inhibition of protein translation is a potential off-target effect of proteasome inhibitors, the production of influenza proteins was only marginally inhibited (see Fig. S7 in the supplemental material). To further substantiate the contribution of proteasome-dependent processing and to obtain a closer comparison with our autophagy-related results, we carried out the siRNA knockdown of Rpt2, an ATPase subunit of the 19S regulatory cap of the proteasome (54). The impact on the ELISpot assay was comparable to that of epoxomicin treatment (without an effect on protein synthesis), while the siRNA inhibition of Atg7 again had no effect (Fig. 7C and D).
Fig. 7.
Proteasome activity contributes to MHC class II-restricted presentation during influenza virus infection. L-IEd (A) or B6-IEd (B) fibroblasts were left untreated or were pretreated with 0.5 μM epoxomicin (Epox) for 15 min, infected with PR8, and cultured with CD4+ T cells isolated from splenocytes of mice primed intranasally with PR8. Results shown in A and B are pooled data from three mice per group. (C) L-IEd fibroblasts were transfected with the indicated siRNAs for 48 h, infected with PR8, and cultured with CD4+ T cells isolated from splenocytes of CB6F1 (H2bxd) mice primed intranasally. (D) Flow cytometry analysis of fibroblasts expressing NA at high levels on the cell surface after siRNA transfection. Results are representative of at least two independent experiments.
In summary, although we were able to demonstrate the autophagy-dependent presentation of individual epitopes, both after transfection and viral infection we were unable to detect a significant contribution of this pathway to the global presentation of class II-restricted epitopes derived from influenza virus infection. At the same time, we did observe a significant contribution of the proteasome-dependent pathway to the global class II-restricted response using two different approaches. These results reinforce the notion that, at least for influenza virus, the majority of the endogenous epitopes presented on MHC class II are processed via a proteasome-dependent pathway(s).
DISCUSSION
With recent work highlighting the potential for macroautophagy to orchestrate the endogenous MHC class II-restricted presentation of individual antigens (16, 35, 44, 45, 50, 60), our goal was to determine the role in the CD4+ T cell response to a complex pathogen, specifically influenza virus. We focused on this virus due to our previous elucidation of a proteasome-dependent endogenous pathway that is responsible for a substantial portion of the class II-restricted response to influenza virus infection (66). Considering the large contribution that the classical exogenous pathway also makes to the response, we wondered if there would be sufficient opportunity for another major pathway. Before addressing this central question, preliminary work in three general areas was required to validate both the relevance of studying autophagy in the context of an influenza infection and the experimental system that we had selected.
First, although influenza virus infection has been shown to induce LC3 conversion in a number of cell lines, it remains unclear whether functional autophagy, resulting in membrane fusion with active lysosomes, is stimulated (15, 33, 73). It has been reported that the influenza infection of human lung epithelial cells prevents autophagosomes from fusing with lysosomes (15), with the viral M2 protein acting as an inhibitor of Beclin-1, a key protein in the autophagy pathway. However, others have concluded that functional autophagy, as measured by the degradation of the autophagy substrate p62, does occur in the face of influenza virus infection (33). The discrepancies may be due to the cell types and/or the influenza virus strains that were utilized. As assessed by both GFP-LC3 and p62 degradation, we observed that influenza virus infection does induce functional macroautophagy in several different cell types, including primary DCs, with no detectable block in the pathway.
Second, using a standard approach in the field, we ascertained our ability to observe macroautophagy-dependent antigen processing by tracking the presentation of an individual epitope (2, 35, 45, 50). Appending the S1 epitope onto neomycin phosphotransferase or LC3B and expressing the constructs via transfection resulted in autophagy-dependent presentation that could be enhanced by inducing autophagy with rapamycin or significantly decreased by the chemical and genetic inhibition of the autophagy pathway. We also constructed an adenovirus expressing the S1LC3 fusion protein and obtained similar results.
Third, we utilized a recombinant adenovirus to validate the ELISpot assay that was used subsequently to assess the global contribution of autophagy. Results demonstrated that the dependence of a polyclonal T cell population on functionally active autophagy within the APC could be readily appreciated. Because this assay represented a best-case scenario in which all of the responding T cells were specific for the autophagy-dependent epitope, we carried out additional assays with heterologous influenza viruses that confirmed that even minor populations within the pool of primed CD4+ T cells (≤10%) could be detected.
With the general framework of the assay having been validated, analyses could have been carried out in two general ways: (i) the infection of wild-type and autophagy-deficient mice (which have autophagy defects in only certain cell types) and the subsequent determination of the differences in the CD4+ T cell responses, or (ii) the infection of wild-type mice only, with the subsequent identification of the autophagy-dependent component. We chose the latter for two reasons. First, despite possible differences in precursor frequencies, CD4+ T cells in wild-type and mutant mice might have expanded to the same absolute numbers, requiring us to analyze differences by less meaningful criteria, such as T cell receptor diversity. Furthermore, because multiple subsets of antigen-presenting cells can initiate an immune response, the selective autophagy knockout system that is currently available would complicate interpretation. In fact, mice that lack the Atg7 gene only in the hematopoietic stem cell compartment (either due to conditional knockout or the reconstitution of an irradiated wild-type mouse with Atg7-deficient bone marrow) display reduced numbers of multiple immune cell subsets and die within weeks following the elimination of this protein (43). Furthermore, the recent analysis of Atg5-deficient DCs from the knockout mice indicates a deficiency in exogenous antigen processing (34). Our results demonstrate that the short-term knockdown of autophagy, in contrast, does not have this effect.
Having carried out these preliminary steps, we asked whether macroautophagy plays a major role in the generation of epitopes during influenza virus infection. Despite the potent induction of functional autophagy by influenza virus infection and a sufficiently sensitive assay, we detected no significant contribution of this pathway to the global CD4+ T cell response to influenza virus. This was true of all antigen-presenting cell types tested, including primary DCs and also when the assay was restricted to the internal and nonstructural proteins of the virus. In contrast, as previously reported (66) a substantial portion of the CD4+ T cell response was observed to be proteasome dependent by two independent methods for modulating proteasome activity (chemical inhibition and siRNA treatment).
Several factors may limit the utility of autophagy for class II-restricted epitope generation. Although largely considered a nonselective pathway of protein degradation, recent data suggest that the efficient delivery of individual soluble proteins to the autophagosomes is dependent upon a specific recognition mechanism (27, 29, 46). In agreement with this, the S1 epitope is more robustly presented from the S1LC3 construct, which is directly targeted to the autophagy pathway via LC3B, unlike the NeoS1 construct, which likely is targeted by less selective mechanisms. It seems unlikely that individual viral proteins would self target to this compartment, as has been reported in some cases (69), unless it were advantageous. Both poliovirus and rhinovirus induce the formation of double membrane autophagosomes to facilitate replication, and autophagy inhibition via siRNA was observed to decrease the amount of poliovirus released from infected cells. Indeed, conditions necessary for processing likely are absent, since induced autophagosomes are not mobile and presumably do not fuse efficiently with the late endosome compartment (65). Likewise, hepatitis C virus induces autophagy to initiate the early stages of viral replication (9, 10, 64), but the degradation of autophagy substrates, including p62, is blocked (63).
Once captured by the autophagosome, an epitope must survive in the highly proteolytic environment of the late endosome/lysosome. This may be a losing prospect in many cases. Enhanced proteolytic activity has been reported generally to be detrimental to antigen presentation, which is consistent with the idea that proteases and empty class II molecules compete for access to unfolded proteins (7, 59). What is more, autophagy inhibition via siRNA has been reported to reduce cathepsin activity in the late endosome (34), suggesting that the fusion of the autophagosome with the endosomal compartment creates an even more aggressive environment. We have previously reported on the negative impact of heightened proteolysis. The presentation of the hemagglutinin-based S3 epitope from exogenous sources of PR8 is enhanced up to 10-fold when endosomal cathepsins are inhibited (3), while the NA79 epitopes appear to be so susceptible to endosomal proteases that presentation from exogenous virus is undetectable in most cases (11, 66). In contrast to S1, neither of these epitopes was presented when appended to neomycin phosphotransferase with or without rapamycin treatment (data not shown), suggesting that they are immediately destroyed upon delivery to the autophagosome. Thus, only those class II-restricted epitopes that are sufficiently resistant to endolysosomal proteolysis can be candidates for autophagy-mediated presentation.
We cannot rule out the possibility that there is a small autophagy-dependent fraction of the class II-restricted influenza virus response that is below the detection limit of our assays. In our system, we determined the sensitivity of the ELISpot assay to detect antigen-specific T cells to be <10%, suggesting that if autophagy does play a role in the class II response, the contribution is minimal. In addition, we cannot discount the possibility that epitopes are generated via parallel pathways such that the inhibition of autophagy would shift the substrate to other processing routes.
In preliminary experiments the global class II responses to vaccinia and adenovirus infections also are not substantially altered by the inhibition of autophagy (J. D. Comber, unpublished observations). We are, nevertheless, cautious about generalizing these findings to other viruses. All three of the viruses that have been tested mediate acute and limiting infections in mice. In contrast, herpesviruses such as Epstein-Barr virus generally are chronic, and evasion from CD8+ T cell recognition appears to be a major priority (5). The EBNA-1 protein contributes directly to this effort by blocking proteasomal degradation and subsequent MHC class I-restricted presentation via an N-terminal Gly-Ala repeat (36, 37). Other herpesviruses encode proteins with similar functions (31, 71). The inhibition of the proteasome induces aggregate formation, which may shift the balance of global protein degradation toward the autophagy pathway (28, 32, 68). Thus, similar analyses of viruses with fundamentally different replication strategies are warranted.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01AI069192 to L.C.E.
We thank Christopher Snyder, James Testa, Nancy Luckashenak, and Michael Miller for helpful comments and suggestions during the preparation of the manuscript and Michael Bevan (University of Washington) for providing reagents. We also thank the Department of Biostatistics, Thomas Jefferson University (Philadelphia, PA), for help with statistical analysis.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Blanchet F., et al. 2010. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 32:654–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brazil M., Weiss S., Stockinger B. 1997. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur. J. Immunol. 27:1506–1514 [DOI] [PubMed] [Google Scholar]
- 3. Chianese-Bullock K. A., et al. 1998. Antigen processing of two H2-IEd-restricted epitopes is differentially influenced by the structural changes in a viral glycoprotein. J. Immunol. 161:1599–1607 [PubMed] [Google Scholar]
- 4. Crotzer V., Blum J. 2008. Cytosol to lysosome transport of intracellular antigens during immune surveillance. Traffic 9:10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davis-Poynter N., Farrell H. 1996. Masters of deception: a review of herpesvirus immune evasion strategies. Immunol. Cell Biol. 74:513–522 [DOI] [PubMed] [Google Scholar]
- 6. Delamarre L., Couture R., Mellman I., Trombetta E. S. 2006. Enhancing immunogenicity by limiting susceptibility to lysosomal proteolysis. J. Exp. Med. 203:2049–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delamarre L., Pack M., Chang H., Mellman I., Trombetta E. S. 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307:1630–1634 [DOI] [PubMed] [Google Scholar]
- 8. Dengjel J., et al. 2005. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. U. S. A. 102:7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dreux M., Chisari F. 2009. Autophagy proteins promote hepatitis C virus replication. Autophagy 5:1224–1225 [DOI] [PubMed] [Google Scholar]
- 10. Dreux M., Gastaminza P., Wieland S. F., Chisari F. V. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eisenlohr L., Gerhard W., Hackett C. 1988. Individual class II-restricted antigenic determinants of the same protein exhibit distinct kinetics of appearance and persistence on antigen-presenting cells. J. Immunol. 141:2581–2584 [PubMed] [Google Scholar]
- 12. Eisenlohr L., Hackett C. 1989. Class II major histocompatibility complex-restricted T cells specific for a virion structural protein that do not recognize exogenous influenza virus. Evidence that presentation of labile T cell determinants is favored by endogenous antigen synthesis. J. Exp. Med. 169:921–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuertes G., Martin De Llano J., Villarroya A., Rivett A., Knecht E. 2003. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 375:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gannagé M., et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gannagé M., Munz C. 2009. Autophagy in MHC class II presentation of endogenous antigens. Curr. Top. Microbiol. Immunol. 335:123–140 [DOI] [PubMed] [Google Scholar]
- 17. Glickman M. H., Ciechanover A. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373–428 [DOI] [PubMed] [Google Scholar]
- 18. He C., Klionsky D. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43:67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hennessy K., Kieff E. 1983. One of two Epstein-Barr virus nuclear antigens contains a glycine-alanine copolymer domain. Proc. Natl. Acad. Sci. U. S. A. 80:5665–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 21. Hurwitz J., Herber-Katz E., Hackett C., Gerhard W. 1984. Characterization of the murine TH response to influenza virus hemagglutinin: evidence for three major specificities. J. Immunol. 133:3371–3377 [PubMed] [Google Scholar]
- 22. Jacobson S., Sekaly R. P., Jacobson C. L., McFarland H. F., Long E. O. 1989. HLA class II-restricted presentation of cytoplasmic measles virus antigens to cytotoxic T cells. J. Virol. 63:1756–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jagannath C., et al. 2009. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 15:267–276 [DOI] [PubMed] [Google Scholar]
- 24. Jaraquemada D., Marti M., Long E. 1990. An endogenous processing pathway in vaccinia virus-infected cells for presentation of cytoplasmic antigens to class II-restricted T cells. J. Exp. Med. 172:947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kabeya Y., et al. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirkegaard K. 2009. Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 335:323–333 [DOI] [PubMed] [Google Scholar]
- 27. Klionsky D. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 [DOI] [PubMed] [Google Scholar]
- 28. Kopito R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524–530 [DOI] [PubMed] [Google Scholar]
- 29. Korolchuk V., Menzies F., Rubinsztein D. 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 584:1393–1398 [DOI] [PubMed] [Google Scholar]
- 30. Kudchodkar S., Levine B. 2009. Viruses and autophagy. Rev. Med. Virol. 19:359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kwun H. J., et al. 2007. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics epstein-barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 81:8225–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lamark T., Johansen T. 2010. Autophagy: links with the proteasome. Curr. Opin. Cell Biol. 22:192–198 [DOI] [PubMed] [Google Scholar]
- 33. Law A., Lee D., Yuen K., Peiris M., Lau A. 2010. Cellular response to influenza virus infection: a potential role for autophagy in CXCL10 and interferon-alpha induction. Cell Mol. Immunol. 7:263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee H., et al. 2010. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32:227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leung C. S., Haigh T. A., Mackay L. K., Rickinson A. B., Taylor G. S. 2010. Nuclear location of an endogenously expressed antigen, EBNA1, restricts access to macroautophagy and the range of CD4 epitope display. Proc. Natl. Acad. Sci. U.S.A. 107:2165–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levitskaya J., et al. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375:685–688 [DOI] [PubMed] [Google Scholar]
- 37. Levitskaya J., Sharipo A., Leonchiks A., Ciechanover A., Masucci M. G. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. U. S. A. 94:12616–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lich J. D., Elliott J. F., Blum J. S. 2000. Cytoplasmic processing is a prerequisite for presenation of an endogenous antigen by major histocompatability complex class II proteins. J. Exp. Med. 191:1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loss G. E., et al. 1993. Major histocompatibility complex class II-restricted presentation of an internally synthesized antigen displays cell-type variability and segregates from the exogenous class II and endogenous class I presentation pathways. J. Exp. Med. 178:73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luka J., Lindahl T., Klein G. 1978. Purification of the Epstein-Barr virus-determined nuclear antigen from Epstein-Barr virus-transformed human lymphoid cell lines. J. Virol. 27:604–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mizushima N., Yoshimori T. 2007. How to interpret LC3 immunoblotting. Autophagy 3:542–545 [DOI] [PubMed] [Google Scholar]
- 42. Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15:1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mortensen M., et al. 2011. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 208:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Münz C. 2009. Enhancing immunity through autophagy. Annu. Rev. Immunol. 27:423–449 [DOI] [PubMed] [Google Scholar]
- 45. Nimmerjahn F., et al. 2003. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur. J. Immunol. 33:1250–1259 [DOI] [PubMed] [Google Scholar]
- 46. Noda N., Ohsumi Y., Inagaki F. 2010. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584:1379–1385 [DOI] [PubMed] [Google Scholar]
- 47. Noda T., Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963–3966 [DOI] [PubMed] [Google Scholar]
- 48. Ogawa M., Sasakawa C. 2006. Bacterial evasion of the autophagic defense system. Curr. Opin. Microbiol. 9:62–68 [DOI] [PubMed] [Google Scholar]
- 49. Oxenius A., et al. 1995. Presentation of endogenous viral proteins in association with major histocompatibility complex class II: on the role of intracellular compartmentalization, invariant chain and the TAP transporter system. Eur. J. Immunol. 25:3402–3411 [DOI] [PubMed] [Google Scholar]
- 50. Paludan C., et al. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307:593–596 [DOI] [PubMed] [Google Scholar]
- 51. Pankiv S., et al. 2007. p62/SQSTM1 Binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 [DOI] [PubMed] [Google Scholar]
- 52. Ravikumar B., Duden R., Rubinsztein D. C. 2002. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11:1107–1117 [DOI] [PubMed] [Google Scholar]
- 53. Riedel A., et al. 2008. Endogenous presentation of a nuclear antigen on MHC class II by autophagy in the absence of CRM1-mediated nuclear export. Eur. J. Immunol. 38:2090–2095 [DOI] [PubMed] [Google Scholar]
- 54. Rubin D., Glickman M., Larsen C., Dhruvakumar S., Finley D. 1998. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17:4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rubinsztein D., et al. 2009. In search of an “autophagomometer”. Autophagy 5:585–589 [DOI] [PubMed] [Google Scholar]
- 56. Sanderson S., Shastri N. 1994. LacZ inducible, antigen/MHC-specific T cell hybrids. Int. Immunol. 6:369–376 [DOI] [PubMed] [Google Scholar]
- 57. Sant A. 1994. Endogenous antigen presentation by MHC class II molecules. Immunol. Res. 13:253–267 [DOI] [PubMed] [Google Scholar]
- 58. Savina A., Amigorena S. 2007. Phagocytosis and antigen presentation in dendritic cells. Immunol. Rev. 219:143–156 [DOI] [PubMed] [Google Scholar]
- 59. Savina A., et al. 2006. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126:205–218 [DOI] [PubMed] [Google Scholar]
- 60. Schmid D., Pypaert M., Munz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 26:79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shvets E., Fass E., Elazar Z. 2008. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy 4:621–628 [DOI] [PubMed] [Google Scholar]
- 62. Sinnathamby G., Eisenlohr L. C. 2003. Presentation by recycling MHC class II molecules of an influenza hemagglutinin-derived epitope that is revealed in the early endosome by acidification. J. Immunol. 170:3504–3513 [DOI] [PubMed] [Google Scholar]
- 63. Sir D., et al. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tanida I., et al. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945 [DOI] [PubMed] [Google Scholar]
- 65. Taylor M. P., Burgon T. B., Kirkegaard K., Jackson W. T. 2009. Role of microtubules in extracellular release of poliovirus. J. Virol. 83:6599–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tewari M., Sinnathamby G., Rajagopal D., Eisenlohr L. 2005. A cytosolic pathway for MHC class II-restricted antigen processing that is proteasome and TAP dependent. Nat. Immunol. 6:287–294 [DOI] [PubMed] [Google Scholar]
- 67. van Luijn M., et al. 2010. Alternative II-independent antigen-processing pathway in leukemic blasts involves TAP-dependent peptide loading of HLA class II complexes. Cancer Immunol. Immunother. 59:1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. 2003. α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278:25009–25013 [DOI] [PubMed] [Google Scholar]
- 69. Wileman T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875–878 [DOI] [PubMed] [Google Scholar]
- 70. Wing S., Chiang H., Goldberg A., Dice J. 1991. Proteins containing peptide sequences related to Lys-Phe-Glu-Arg-Gln are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem. J. 275:165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zaldumbide A., Ossevoort M., Wiertz E., Hoeben R. 2007. In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma herpes virus. Mol. Immunol. 44:1352–1360 [DOI] [PubMed] [Google Scholar]
- 72. Zhou D., et al. 2005. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity 22:571–581 [DOI] [PubMed] [Google Scholar]
- 73. Zhou Z., et al. 2009. Autophagy is involved in influenza A virus replication. Autophagy 5:321–328 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.