Abstract
mRNA-processing bodies (P bodies) are cytoplasmic foci that contain translationally repressed mRNA. Since they are important for the retrotransposition of Ty elements and brome mosaic virus in yeast cells, we assessed the role of P bodies in the movement of endogenous intracisternal A particles (IAPs) in mammalian cells. In contrast to the case for these other systems, their disruption via knockdown of RCK or eukaryotic initiation factor E transporter (eIF4E-T) increased IAP retrotransposition as well as levels of IAP transcripts, Gag proteins, and reverse transcription products. This increase was not mediated by impairing the microRNA pathway. Rather, the removal of P bodies shifted IAP mRNA from nonpolysomal to polysomal fractions. Although IAP mRNA localized to P bodies, Gag was targeted to the endoplasmic reticulum (ER), from which IAP buds. Thus, by sequestering IAP mRNA away from Gag, P bodies inhibit rather than promote IAP retrotransposition.
INTRODUCTION
Intracisternal A particles (IAPs) are endogenous retroviruses. The mouse genome contains 1,000 to 2,000 IAPs, of which a hundred or so are still active for autonomous intracellular retrotransposition (RTP) (28). These elements contain 5′ and 3′ long terminal repeats (LTRs) with functional gag, pr, and pol genes (28, 34). They produce virus-like particles (VLPs) from membranes of the endoplasmic reticulum (ER) and bud into its cisternae (28). Due to the absence of a functional env gene, IAPs lack an extracellular phase and are not infectious (34). Expression of IAPs is detected in germ line and somatic tissues (12, 13, 27, 29, 39, 46). IAP insertions can lead to mutations and contribute to pathological processes. Therefore, to maintain genome stability, it is critical for host cells to maintain their RTP at low levels.
mRNA-processing bodies (P bodies) are discrete cytoplasmic foci that contain nontranslating mRNAs and mRNA degradation machineries. The nontranslating mRNAs can be degraded or stored and return for translation (5, 37, 42, 43). The translationally repressed messenger ribonucleoprotein complexes (mRNPs) in mammalian P bodies contain components of the 5′-3′ mRNA decay machinery [5′-3′ exonuclease Xrn1, decapping enzymes DCP1/DCP2, and decapping coactivators Hedls (GE1)/Lsm1/EDC3/RCK(p54)] (24), the RNA-induced silencing complex (RISC) (GW182, microRNA [miRNA], and argonaute) (16, 31, 32, 41), eukaryotic translation initiation factor 4E (eIF4E) and its transporter (eIF4E-T), and several RNA-binding proteins associated with mRNA translation and decay, which include TTP, BRF1, CPEB, and Smaug. The formation of P bodies is a dynamic process, which affects levels of transcripts available for translation (10, 43). Retrotransposons like IAPs utilize host cell factors for the expression of their genes. Thus, it is reasonable to hypothesize that P bodies could alter IAP RTP in negative or positive ways.
Indeed, P bodies play a major role during the replicative cycles of retrovirus-like element Ty3 in yeast (4). Its mRNA, proteins, and VLPs were found to accumulate within P bodies. Although P-body components are also important for Ty1 RTP, it is controversial whether Ty1 RNA and proteins colocalize to P bodies (8, 14). Similarly to yeast Ty elements, brome mosaic virus (BMV) genomic RNA2 and RNA3 accumulate in P bodies (2), which is required for efficient BMV RTP (33, 36). In contrast to the case for yeast Ty elements and BMV, P bodies inhibit HIV-1 replication, where P-body disruption increases viral production and infectivity (7, 35). P-body inhibition of HIV-1 involves RISC, where targeted HIV-1 mRNA translation is repressed (35). However, it is not clear what roles P bodies play in the replication of other retroviruses and retroelements. In this study, we report that P-body disruption increases IAP RTP.
MATERIALS AND METHODS
Cells, constructs, and siRNAs.
Human 293 cells were maintained at 37°C with 5% CO2 in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS), 100 mM l-glutamine, and 50 μg/ml of penicillin and streptomycin. The IAP reporter plasmid pFL was a kind gift from Kyoji Horie. Human RCK small interfering RNA (siRNA) (sc-72246), mouse Dicer siRNA (sc-40490), and control siRNA-A (sc-37007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Customized eIF4E-T siRNA (5′-GAACAAGAUUAUCGACCUAdTdT-3′) (18), human Dicer siRNA (5′-UGCUUGAAGCAGCUCUGGAdTdT-3′) (44), and human Drosha siRNA (5′-CGAGUAGGCUUCGUGACUUdTdT-3′) (44) were synthesized by Dharmacon (Lafayette, CO), and nontargeting siRNA 1 (D-001810-01-05) was purchased from Dharmacon.
Immunoblotting.
293 cells were harvested and washed with phosphate-buffered saline (PBS) before resuspended in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) and a mixture of protease inhibitors (Roche Diagnostics). Immunoblotting was performed as described previously (30). Goat polyclonal antibody to RCK (sc-51415 from Santa Cruz Biotechnology), goat polyclonal antibody to eIF4E-T (sc-13453), mouse monoclonal antibody to G3BP1 (sc-81940), goat polyclonal antibody to actin (sc1615), mouse monoclonal antibody to Dicer (ab14601; Abcam), rabbit polyclonal antibody to Drosha (ab12286), and rabbit anti-IAP Gag antibody (a kind gift from Bryan Cullen and described in reference 11) were used as primary antibodies.
RNA-IP.
RNA immunoprecipitation (RNA-IP) was performed as described by Peritz et al. (38). Briefly, cells were lysed in polysome buffer, and supernatants were collected and divided into two aliquots. Each aliquot was incubated with normal goat IgG or goat anti-RCK antibodies at 4°C overnight, and protein G-Sepharose beads were added afterwards. Protein G-Sepharose beads were pelleted. RNAs and proteins binding to the beads were eluted, and RNAs were isolated from the elution. Quantitative PCR was performed to detect IAP mRNA.
Immunofluorescence and RNA-FISH.
Immunofluorescence was performed as described previously with minor modification. Briefly, 293 cells were grown on coverslips in 6-well plates and fixed with 4% paraformaldehyde in PBS at room temperature for 15 min. After being washed with PBS twice, cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min and washed 3 times with 0.05% Tween 20 in PBS (PBST). Samples were blocked for 30 min in PBST containing 5% donkey serum. Primary and secondary antibodies were diluted in blocking buffer. Primary antibodies used included anti-RCK, anti-eIF4E-T, anti-IAP Gag antibodies. Secondary antibodies included donkey anti-goat and donkey anti-rabbit IgGs conjugated to Alexa Fluor dye 488 or 594 (Molecular Probes, Eugene, OR). After a final wash, samples were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) and visualized with a Zeiss LSM 510 confocal microscope, with a 63× 1.4-numerical-aperture lens (Carl Zeiss Inc.).
For RNA fluorescence in situ hybridization (RNA-FISH), 293 cells were grown on coverslips in 6-well plates. Samples were fixed in 4% paraformaldehyde in PBS for 20 min. After being washed, cells were permeabilized in 0.1% Triton X-100 in PBS for 10 min, followed by treatment with 10 U/ml DNase I (Invitrogen) for 2.5 min at room temperature. After being washed with PBS, slides were dehydrated in 70%, 80%, 95%, and 100% ethanol for 3 min each and then rehydrated in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 50% formamide. The probe preparation was done by diluting the probe to a stock solution of 1 μM in 50% deionized formamide, 2× SSC, 10% dextran sulfate, and 50 mM sodium phosphate (pH 7). One microliter of the stock solution was added to a 40-μl mixture including 10% dextran sulfate, 2 mM vanady-ribonucleoside complex, 0.02% RNase-free bovine serum albumin (BSA), 40 μg of yeast tRNA, 2× SSC, and 50% formamide. The mixture was applied on each slide, and the slides were incubated at 37°C overnight in a humidified chamber (light protected). After the hybridization, slides were washed 3 times in 2× SSC and 50% formamide at 42°C, followed by 3 washes in 2× SSC. The slides were either mounted with DAPI-containing medium or used for immunofluorescence assays (IFAs). The probe used in this study is an oligonucleotide (complementary to the gag region of IAP) labeled with Alexa Fluor 488 at the 5′ end: 5′-/5Alex488N/TTCTTGATGTCCTAACCCTTTTCCTTC-3′.
Cytoplasmic extract analysis on sucrose gradients.
At 2 days after the initial transfection of control siRNA or RCK siRNA, 293 cells were cotransfected with pFL together with control siRNA or RCK siRNA for each well in 6-well plates. At 3 days after the transfection, cells were incubated with 100 μg/ml cycloheximide at 37°C for 15 min. Cells were lysed in polysome lysis buffer (10 mM Tris-HCl, 1% Triton X-100, 15 mM MgCl2, 0.1 μg/ml cycloheximide, 0.3 M NaCl, 1 mg/ml heparin, 0.2 U/μl RNasin, and 1× EDTA-free protease inhibitor) and incubated on ice for 10 min before centrifugation at 10,000 × g for 10 min at 4°C. For preparation of sucrose gradients, 10, 20, 30, 40, and 50% sucrose solutions were dissolved in buffer containing 10 mM Tris-HCl (pH 7.4), 15 mM MgCl2, 0.1 μg/ml cycloheximide, 0.3 M NaCl, and 1 mM dithiothreitol (DTT). A 10-ml gradient was made by adding 2 ml of each solution (50% on the bottom and 10% on the top) in an ultracentrifuge tube and left at 4°C overnight to obtain a linear sucrose gradient. Cytoplasmic extracts (0.5 ml) were loaded on the top of the sucrose gradient and centrifuged for 3 h at 38,000 rpm in an SW41Ti rotor. After ultracentrifugation, 20 fractions were collected, and the optical density (OD) at 254 nm was measured in aliquots of each fraction using the SpectraMax M5 multiplate reader (Molecular Devices). The rest of each fraction was used for total RNA isolation.
RTP assay.
293 cells in 6-well plates were transfected with 100 pmol control siRNA, RCK, or eIF4E-T siRNA using Lipofectamine 2000 according to the manufacturer's protocol for reverse transfection. At 2 days after the initial siRNA transfection, 1 μg pFL was cotransfected with 100 pmol control siRNA, RCK, or eIF4E-T siRNA, and at 3 days after the cotransfection, 293 cells were collected and subjected to fluorescence-activated cell sorter (FACS) analysis of green fluorescent protein (GFP)-positive cells to determine the rate by using a BD LSR II cytometer (BD Biosciences).
qPCR.
Total RNA or DNA was extracted, and quantitative PCR (qPCR) was performed. The primers used in this study were for IAP Gag (forward, 5′-ACCCAGGAAGCAGTCAGAGA-3′; reverse, 5′-CCTTTAGGGCTTGAGCACAG-3′), GFP (forward, 5′-TGAAGAAGTCGCTGATGTCCT-3′; reverse, 5′-GAGCAAGCAGATCCTGAAGAA-3′), human β-actin (forward, 5′-AAAGACCTGTACGCCAACAC-3′; reverse, 5′-GTCATACTCCTGCTTGCTGAT-3′), and human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (forward, 5′-GAGCCACATCGCTCAGACAC-3′; reverse, 5′-CATGTAGTTGAGGTCAATGAAGG-3′).
RESULTS
Knockdowns of RCK/p54 or eIF4E-T disrupt P bodies and increase IAP RTP.
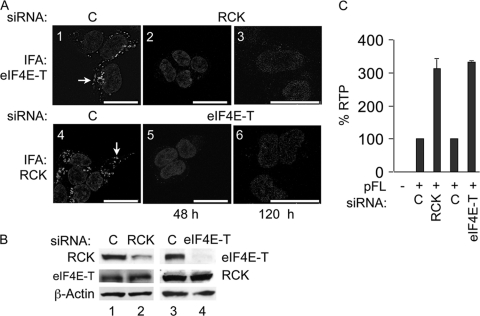
Factors that are required for P-body formation in human cells include the RNA helicase RCK/p54 and eIF4E-T (1). RCK is a decapping coactivator in P bodies. The depletion of RCK by siRNA disrupts P bodies and increases HIV-1 replication in human cells (1, 7, 9, 35). To examine whether P bodies play a role in IAP RTP in human cells, we knocked down RCK by siRNA and followed this with an IAP RTP assay. To track IAP RTP, we used a reporter IAP plasmid (pFL), which expresses GFP only after RTP (Fig. 1) (22). Thus, we detected GFP-positive cells by fluorescence-activated cell sorting (FACS). RCK or control siRNAs were transfected into 293 cells using Lipofectamine 2000. Two days after the transfection, cells were harvested for Western blotting of RCK and eIF4E-T or subjected to immunofluorescence assays (IFAs) of eIF4E-T to detect P bodies. Consistent with previous publications, depletion of RCK disrupted P bodies as demonstrated by IFAs of P-body marker eIF4E-T, whose expression remained constant (Fig. 2A1, A2, and B). Similar results were also obtained with the knockdown of eIF4E-T as demonstrated by IFAs of P-body marker RCK (Fig. 2A4 and A5). The expression of RCK was not affected by eIF4E-T knockdown (Fig. 2B). We then carried out experiments to determine effects of P-body disruption on IAP RTP. Two days after the initial transfection of siRNAs, pFL was cotransfected with the same siRNAs into 293 cells. Three days later, i.e., 120 h after the initial siRNA transfection, cells were examined by FACS. Figure 2C reveals that this disruption of P bodies increased IAP RTP 3.1- and 3.3-fold, respectively. The depletion of RCK or eIF4E-T (data not shown) and the disruption of P bodies at the time of FACS were confirmed (Fig. 2A3 and A6). Thus, P bodies inhibit IAP RTP.
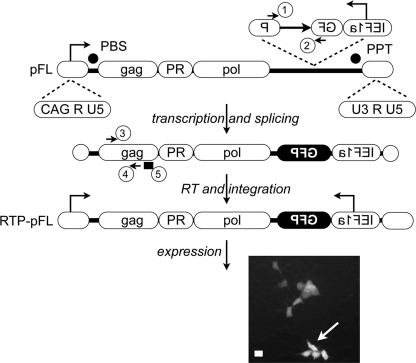
Fig. 1.
Structure of the marked IAP with the GFP-intron cassette (pFL) and a description of the RTP assay. The genomic organization of the full-length IAP (top panel) includes two LTRs (U3-R-U5) bordering three open reading frames (ORFs) homologous to the retroviral genes gag, PR (protease), and pol, which are separated from one another by a −1-nucleotide (nt) frameshift. The transcription start site is marked with a black arrow on the 5′ LTR, and the signals necessary for retroviral RT are indicated. PBS, primer-binding site; PPT, polypurine tract. In the reporter GFP-intron cassette, GFP is placed in the reverse orientation with its promoter (IEF1α) and is rendered inactive by the presence of a forward γ-globin intron. The intron is spliced out in the transcribed RNA intermediate (middle panel), resulting in an active GFP gene in the newly integrated IAP (RTP-pFL, retrotransposed pFL [bottom panel]). GFP-positive cells can be visualized by fluorescence microscopy or scored by FACS. The microscopy reveals that GFP is expressed in the nucleus and cytoplasm of cells where RTP had occurred, which is indicated by a white arrow. Arrows 1 and 2 on GFP exons and 3 and 4 on gag indicate primers for GFP and gag, respectively; the black rectangle (5) on Gag indicates the location of the probe for RNA-FISH. CAG, cytomegalovirus (CMV) early enhancer element and chicken β-actin promoter.
Fig. 2.
Depletion of P-body component RCK or eIF4E-T disrupts P bodies and increases IAP RTP. (A) Depleting RCK or eIF4E-T disrupts P bodies. Panels 1 and 2, IFA of eIF4E-T in 293 cells at 48 h after the transfection with control (panel 1) or RCK (panel 2) siRNA. Panels 4 and 5, IFA of RCK in 293 cells at 48 h after the transfection with control (panel 4) or eIF4E-T (panel 5) siRNAs. Panel 3, IFA of eIF4E-T in 293 cells. These cells were cotransfected with pFL together with the same siRNAs at 48 h after the initial transfection of control or RCK siRNAs and then incubated for additional 72 h. Panel 6, IFA of RCK in 293 cells. These cells were cotransfected with pFL together with the same siRNAs at 48 h after the initial transfection of control or eIF4E-T siRNA and then incubated for additional 72 h. Cells were counterstained with DAPI. Scale bars, 20 μm. Arrows indicate representative P bodies in panels 1 and 4. (B) Depletion of RCK (bars 1 and 2) or eIF4E-T (bars 3 and 4) by siRNA. Western blotting of RCK and eIF4E-T in 293 cells was performed at 48 h after the siRNA transfection. β-Actin was used as the loading control. (C) Depletion of RCK or eIF4E-T increases IAP RTP. RTP frequency was determined by FACS of GFP-positive cells at 120 h after the initial transfection, as described for panels A3 and A6. Data are presented as averages ± standard deviations (SD) from 3 to 5 independent experiments. The average RTP frequency in control samples was 0.15%.
Whereas IAP mRNA and RCK colocalize to P bodies, Gag displays a characteristic ER staining.
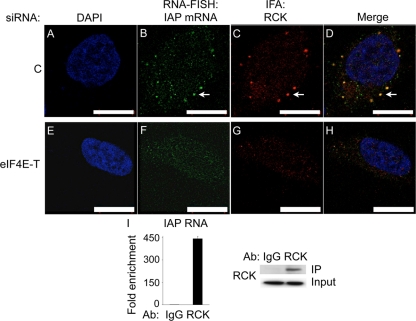
Given that the disruption of P bodies increased IAP RTP and that they play a critical role in mRNA metabolism, we examined whether IAP mRNA localizes to P bodies using RNA fluorescence in situ hybridization (RNA-FISH) followed by IFAs of P bodies. IAP mRNA was detected using an Alexa Fluor 488-labeled oligonucleotide probe complementary to the gag sequence (Fig. 1). Anti-RCK antibodies followed by Alexa Fluor 594-conjugated secondary antibodies were used to detect P bodies. Indeed, IAP mRNA colocalized with RCK in P bodies (Fig. 3A to D). After depleting eIF4E-T and disrupting P bodies, RCK and IAP mRNA were no longer found in any cytoplasmic foci (Fig. 3E to H). Moreover, RNA-IP with RCK antibodies revealed that RCK was associated with IAP mRNA (Fig. 3I). These data suggest that RCK and IAP mRNA are in the same mRNP in P bodies.
Fig. 3.
RCK and IAP mRNA colocalize to P bodies. (A to H) IAP mRNA colocalizes with RCK to P bodies. pFL was cotransfected with control (top panel) or eIF4E-T (bottom panel) siRNA into 293 cells. Two days after the cotransfection, cells were subjected to RNA-FISH to detect IAP mRNA (B and F), followed by IFAs of RCK to detect P bodies (C and G). Cells were stained with DAPI to visualize nuclei (A and E). Images were digitally merged (D and H). Arrows indicate colocalization of IAP mRNA and RCK in a representative P body. Scale bars, 10 μm. (I) IAP RNA was coimmunoprecipitated with endogenous RCK protein with anti-RCK antibodies by RNA-IP. Real-time qPCR was performed to detect IAP RNA. Data are presented as averages ± SD from 2 independent experiments. Western blotting was performed to detect RCK in cell lysates (input) and IPs.
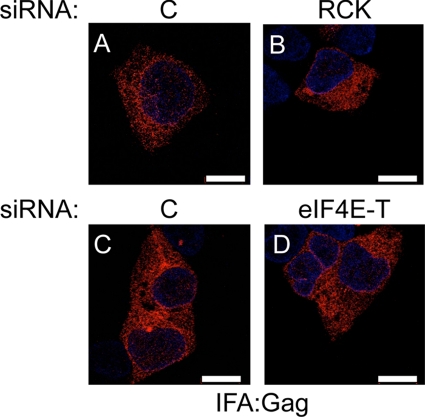
In contrast to IAP mRNA, IAP Gag protein was not concentrated in discrete cytoplasmic foci but instead exhibited a characteristic ER pattern: reticular staining of the cytoplasm and staining of the nuclear membrane (Fig. 4A and C). This observation is consistent with previously published data (17; reviewed in reference 28). Disruption of P bodies did not change the reticular and nuclear membrane staining pattern (Fig. 4B and D). Taken together, these results demonstrate that whereas IAP mRNA concentrates in P bodies, IAP Gag is found elsewhere in the cytoplasm. Moreover, the subcellular localization of IAP mRNA and Gag suggests that P bodies inhibit the assembly of IAPs by sequestering their mRNA away from structural proteins.
Fig. 4.
Subcellular localization of IAP Gag. (A and C) IAP Gag in 293 cells cotransfected with pFL and control siRNA shows the characteristic ER pattern: reticular staining in the cytoplasm and staining of the nuclear membrane. (B and D) P-body disruption by depleting RCK or eIF4E-T did not change the subcellular localization of Gag. 293 cells were cotransfected with pFL, together with RCK (B) or eIF4E-T (D) siRNA. IAP Gag was detected by IFA using rabbit anti-Gag primary and Alexa Fluor 594-conjugated secondary antibodies at 2 days after the transfection. Scale bars, 10 μm.
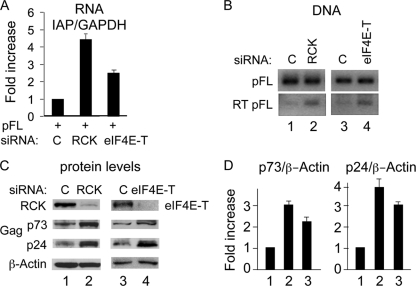
P-body disruption increases levels of steady-state IAP mRNA, Gag protein, and RT products.
To determine at which point(s) of IAP's replicative cycle P-body disruption influences its RTP, IAP transcripts, reverse transcription (RT) products, and Gag were examined. As described above, at 2 days after the initial transfection of RCK, eIF4E-T, or control siRNAs, pFL was cotransfected with the same siRNAs. Three days later, cells were harvested. Total RNA was isolated, and this was followed by RT-qPCR to determine levels of IAP mRNA. IAP mRNA was increased 4.4- and 2.5-fold upon P-body disruption by RCK or eIF4E-T siRNAs, respectively (Fig. 5A). DNA was also isolated from these samples, and conventional qPCR was performed to detect IAP RT products, which were also increased in cells depleted of RCK or eIF4E-T (Fig. 5B). Consistent with increased levels of IAP mRNA, amounts of Gag were also increased. Whereas p73 Gag was increased 3- and 2.2-fold (Fig. 5C and D), its processed p24 subunits were increased 4- and 3-fold upon the depletion of RCK and eIF4E-T, respectively (Fig. 5C and D). Taken together, these data suggest that P bodies sequester and degrade IAP mRNA and inhibit its translation and RT. Since the RISC complex localizes to P bodies, it is possible that increased levels of IAP mRNA, which could lead to increased levels of Gag proteins and RT product, are due to an impaired microRNA (miRNA) pathway.
Fig. 5.
P-body disruption increases levels of steady-state IAP mRNA, RT products, and Gag. (A) P-body disruption by depleting RCK or eIF4E-T increases levels of steady-state IAP mRNA. GAPDH was used as the internal control. Data are presented as averages ± SD from 2 independent experiments. (B) P-body disruption by depletion of RCK (lanes 1 and 2) or eIF4E-T (lanes 3 and 4) increases IAP RT products. IAP RT product was detected using genomic DNA as the template. The upper bands (pFL) represent PCR products from transfected IAP plasmids containing the GFP intron, and the lower bands (RT-pFL) represent PCR products from integrated IAP DNAs which do not contain the GFP intron. (C) P-body disruption by depletion of RCK (lanes 1 and 2) or eIF4E-T (lanes 3 and 4) increases levels of IAP Gag, particularly its processed p24 subunits. Gag proteins (p73 and p24) were detected by Western blotting using rabbit anti-Gag antibodies. β-Actin was used as the loading control. (D) Densitometric analysis of Gag (p73 and p24) Western blots upon P-body disruption is presented as averages ± SD from 3 independent experiments. Primers used for panel A were primers 3 and 4, and primers used for panel B were primers 1 and 2 (as indicated in Fig. 1).
Increased IAP RTP upon P-body depletion is not mediated by an impaired miRNA pathway.
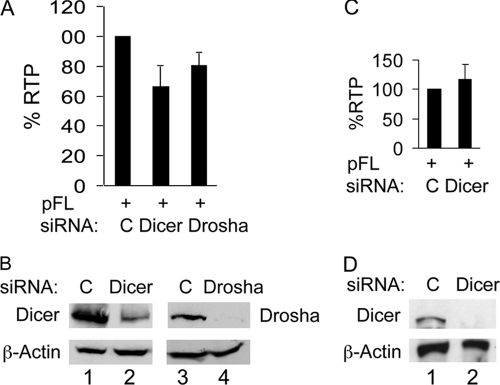
RCK is required for translational repression by miRNA-induced gene silencing (9), and this pathway can contribute to antiviral defenses (15). Furthermore, the replication of HIV-1 upon P-body disruption involves miRNAs and RISC (7, 35). Thus, it was possible that the disruption of P bodies led to impaired gene silencing by the miRNA pathway. To test this possibility, we knocked down Dicer or Drosha with specific siRNAs and examined their effects on IAP RTP. To our surprise, knocking down these essential components of the miRNA pathway did not increase IAP RTP (Fig. 6A and B). On the contrary, knocking down Dicer or Drosha slightly decreased IAP RTP (Fig. 6A and B). Since IAP-specific miRNAs are most likely present in mouse cells, we performed the same assay in transformed mouse 3T3-SV40 cells. miRNAs have a half-life that can range from hours to days (26). A mathematical model based on 6 miRNAs predicts an average half-life of 119 h (19). To ensure that Dicer knockdown was long enough to confer an effect on endogenous miRNAs, we performed two sequential transfections of mouse Dicer siRNA and analyzed IAP RTP at 8 days after the initial transfection. However, with this treatment, we did not observe any effect of Dicer knockdown on IAP RTP (Fig. 6C and D). Dicer depletion was also confirmed at day 2 after the initial Dicer siRNA transfection and day 4 after the second Dicer siRNA transfection (data not shown).These observations exclude the possibility that increased IAP RTP upon P-body disruption was mediated via an impaired miRNA pathway.
Fig. 6.
An impaired miRNA pathway does not increase IAP RTP. (A) Impairment of the miRNA pathway by depleting Dicer or Drosha slightly decreases IAP RTP in 293 cells. (B) Dicer or Drosha was depleted by siRNA, and β-actin was used as the loading control. 293 cells were cotransfected with pFL, control siRNA, Dicer siRNA, or Drosha siRNA. Three days later, cells were collected for FACS (A) or Western blotting of Dicer or Drosha (B). Data in panel A are presented as averages ± SD from 4 independent experiments. (C) Knockdown of Dicer in mouse 3T3-SV40 cells does not affect IAP RTP. Mouse 3T3-SV40 cells were transfected with mouse Dicer siRNA or control siRNA. Four days later, Dicer siRNA or control siRNA was cotransfected with pFL, and FACS analysis was performed 4 days after the cotransfection. (D) Western blotting revealed Dicer depletion at 4 days after the initial Dicer siRNA transfection.
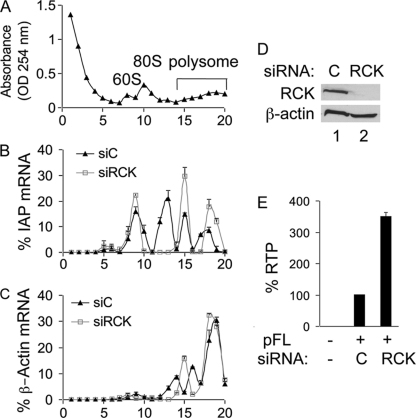
P-body disruption causes a shift of IAP mRNA from nonpolysomal to polysomal fractions.
Transcripts in P bodies can be degraded or returned to polysomes for translation. Therefore, in addition to rates of mRNA degradation, P bodies can influence levels of mRNA in polysomes (37). Indeed, we observed increased levels of IAP mRNA and Gag upon P-body disruption. To determine if it shifts from nonpolysomal to polysomal fractions, we analyzed the cytoplasmic distribution of IAP mRNA by sucrose gradient centrifugation. Twenty fractions were collected. The OD at 254 nm was measured, and total RNA was isolated from each fraction. Indeed, P-body disruption by RCK siRNA shifted IAP mRNA from nonpolysomal to polysomal fractions compared to control siRNA (Fig. 7A and B). In contrast, the distribution of endogenous β-actin mRNA was not affected by RCK depletion (Fig. 7A and C). RCK depletion and increased IAP RTP were confirmed in samples analyzed by sucrose gradient centrifugation (Fig. 7D and E). These observations suggest that P bodies prevent IAP mRNA translation by sequestering it away from polysomes.
Fig. 7.
P-body disruption shifts IAP mRNA from nonpolysomal to polysomal fractions. (A) Representative profile of the 254-nm UV absorption across sucrose gradients in cells transfected with control siRNA. Profiles obtained from cells with depletion of RCK are almost identical (data not shown). The absorption peaks corresponding to the 60S-, 80S-, and polysome-containing fractions are indicated. (B and C) P-body disruption shifted IAP mRNA from nonpolysomal to polysomal fractions and did not result in such a shift of β-actin mRNA. Total RNAs were isolated from each fraction, and IAP or β-actin mRNA was quantified by RT-qPCR. (D and E) Western blotting of RCK confirmed the depletion of RCK by siRNA (D), and FACS confirmed increased RTP upon RCK depletion in samples analyzed in panels A to C (E). Transfection was performed as described for Fig. 2C.
DISCUSSION
The functional study of retrotransposons is often challenging because of their large numbers in the genome and sequence heterogeneity of individual copies. To overcome this difficulty, we used the previously characterized IAP reporter plasmid pFL, which expresses GFP upon RTP, to investigate the role of P bodies in this process (22). Previous studies on such IAP RTP demonstrated that IAP transposes in mouse and human cell lines at similar rates (11, 21). In this study, we determined that by depleting RCK or eIF4E-T, P-body disruption increased IAP RTP and levels of IAP mRNA, Gag proteins, and RT products. This increased IAP RTP was not a consequence of decreased miRNA silencing. Further investigations demonstrated that the depletion of P bodies shifted IAP mRNA from nonpolysomal to polysomal fractions. Finally, whereas IAP mRNA localized to P bodies, Gag displayed a characteristic ER staining. We conclude that P bodies play an inhibitory role in IAP RTP.
IAPs assemble at the ER membrane and bud into its cisternae, where we found Gag. In contrast, IAP mRNA was localized to P bodies and shifted from nonpolysomal to polysomal fractions upon their disruption. This finding suggests that P bodies may function to inhibit IAP RTP by keeping their mRNA in translationally repressed RNPs and away from their structural proteins. IAP mRNA in P bodies can be degraded or returned to polysomes for translation. This notion is also supported by our findings that steady-state levels of IAP mRNA and Gag were increased upon P-body disruption. More efficient processing of Gag ensued. Of note, the overexpression of MOV10, another P-body component, decreases HIV-1 replication and the efficiency of Gag processing (6). These data suggest that the efficiency of Gag processing could depend on levels of available precursors. Of interest is that stress granules, which are related cytoplasmic foci (20, 25), were not detected in these cells, and thus they play no role in this process (data not shown).
We also excluded the possibility that increased IAP RTP upon P-body depletion was due to the impaired miRNA pathway. Indeed, depleting Dicer or Drosha did not increase IAP RTP. Thus, increased IAP RTP upon P-body disruption is not mediated via an impaired miRNA pathway. It is intriguing that whereas disabling the miRNA pathway decreases IAP RTP, it increases HIV-1 production and infectivity (7, 35, 44). The effect of the miRNA pathway on HIV-1 is in agreement with the existence of cellular miRNAs that target HIV-1 and inhibit its replication (23, 45). Thus, the miRNA pathway is not involved in IAP RTP.
P bodies play a variety of roles to influence viral replication (reviewed in reference 3). Their components are required for efficient RTP of yeast Ty elements and for the translation and subsequent recruitment to replication of BMV (2, 33, 36). These studies suggested that P bodies are subcellular foci where viral genomic RNAs (gRNAs) and proteins aggregate to facilitate their assembly and replication. In contrast, P-body disruption increases HIV-1 production and infectivity. In addition, the antiviral host factor APOBEC3G, which inhibits HIV-1 and retrotransposons, is found in P bodies. These observations suggest that P bodies can also function in host defenses against viruses and mobile genetic elements. Phylogenetic analyses based on their pol genes indicate that IAPs belong to the retrovirus family and are distinct from the other LTR retrotransposons, such as the Ty elements in yeast or copia and gypsy in Drosophila (40). This difference might explain why P bodies play a similar inhibitory role for HIV-1 and IAP. Indeed, the IAP genome shares significant homology to the type B mouse mammary tumor virus (MMTV) and type D simian retroviruses (SRV), suggesting that they are evolutionarily related.
It will be interesting to determine if P bodies play a similar role in the replicative cycles of these viruses. In addition, determining how IAP mRNA is targeted to P bodies will elucidate this host restriction of endogenous and exogenous retroviruses.
ACKNOWLEDGMENTS
We thank members of the Peterlin laboratory for helpful advice and discussions, Zeping Luo for technical help, and Kyoji Horie for the pFL plasmid.
Chunye Lu was supported in part by a fellowship from the GIVI-UCSF Center for AIDS Research. This work was funded by a grant from the NIH (P50 GMO82250).
Footnotes
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Andrei M. A., et al. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11:717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beckham C. J., et al. 2007. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 81:9759–9768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beckham C. J., Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beliakova-Bethell N., et al. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brengues M., Teixeira D., Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burdick R., et al. 2010. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J. Virol. 84:10241–10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chable-Bessia C., et al. 2009. Suppression of HIV-1 replication by microRNA effectors. Retrovirology 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Checkley M. A., Nagashima K., Lockett S. J., Nyswaner K. M., Garfinkel D. J. 2010. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 30:382–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chu C. Y., Rana T. M. 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4:e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coller J., Parker R. 2005. General translational repression by activators of mRNA decapping. Cell 122:875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dewannieux M., Dupressoir A., Harper F., Pierron G., Heidmann T. 2004. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 36:534–539 [DOI] [PubMed] [Google Scholar]
- 12. Dupressoir A., Heidmann T. 1996. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol. Cell. Biol. 16:4495–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dupressoir A., Puech A., Heidmann T. 1995. IAP retrotransposons in the mouse liver as reporters of ageing. Biochim. Biophys. Acta 1264:397–402 [DOI] [PubMed] [Google Scholar]
- 14. Dutko J. A., Kenny A. E., Gamache E. R., Curcio M. J. 2010. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 84:5052–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dykxhoorn D. M. 2007. MicroRNAs in viral replication and pathogenesis. DNA Cell Biol. 26:239–249 [DOI] [PubMed] [Google Scholar]
- 16. Eystathioy T., et al. 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13:1338–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fehrmann F., Jung M., Zimmermann R., Krausslich H. G. 2003. Transport of the intracisternal A-type particle Gag polyprotein to the endoplasmic reticulum is mediated by the signal recognition particle. J. Virol. 77:6293–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferraiuolo M. A., et al. 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 170:913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gantier M. P., et al. 29 March 2011. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goodier J. L., Zhang L., Vetter M. R., Kazazian H. H., Jr 2007. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27:6469–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heidmann O., Heidmann T. 1991. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell 64:159–170 [DOI] [PubMed] [Google Scholar]
- 22. Horie K., et al. 2007. Retrotransposons influence the mouse transcriptome: implication for the divergence of genetic traits. Genetics 176:815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang J., et al. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 24. Kedersha N., Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81 [DOI] [PubMed] [Google Scholar]
- 25. Kedersha N., Tisdale S., Hickman T., Anderson P. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521–552 [DOI] [PubMed] [Google Scholar]
- 26. Krol J., et al. 2010. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141:618–631 [DOI] [PubMed] [Google Scholar]
- 27. Kuff E. L., Fewell J. W. 1985. Intracisternal A-particle gene expression in normal mouse thymus tissue: gene products and strain-related variability. Mol. Cell. Biol. 5:474–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuff E. L., Lueders K. K. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51:183–276 [DOI] [PubMed] [Google Scholar]
- 29. Leiter E. H., Fewell J. W., Kuff E. L. 1986. Glucose induces intracisternal type A retroviral gene transcription and translation in pancreatic beta cells. J. Exp. Med. 163:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lenassi M., et al. 2010. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11:110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu J., et al. 2005. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7:1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mas A., Alves-Rodrigues I., Noueiry A., Ahlquist P., Diez J. 2006. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 80:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mietz J. A., Grossman Z., Lueders K. K., Kuff E. L. 1987. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J. Virol. 61:3020–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nathans R., et al. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 34:696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Noueiry A. O., Diez J., Falk S. P., Chen J., Ahlquist P. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parker R., Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635–646 [DOI] [PubMed] [Google Scholar]
- 38. Peritz T., et al. 2006. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 1:577–580 [DOI] [PubMed] [Google Scholar]
- 39. Puech A., Dupressoir A., Loireau M. P., Mattei M. G., Heidmann T. 1997. Characterization of two age-induced intracisternal A-particle-related transcripts in the mouse liver. Transcriptional read-through into an open reading frame with similarities to the yeast ccr4 transcription factor. J. Biol. Chem. 272:5995–6003 [DOI] [PubMed] [Google Scholar]
- 40. Ribet D., et al. 2008. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. 18:597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sen G. L., Blau H. M. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633–636 [DOI] [PubMed] [Google Scholar]
- 42. Sheth U., Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Triboulet R., et al. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 45. Wang X., et al. 2009. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 113:671–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wivel N. A., Smith G. H. 1971. Distribution of intracisternal A-particles in a variety of normal and neoplastic mouse tissues. Int. J. Cancer 7:167–175 [DOI] [PubMed] [Google Scholar]