Abstract
Autophagy is a conserved eukaryotic mechanism that mediates the removal of long-lived cytoplasmic macromolecules and damaged organelles via a lysosomal degradative pathway. Recently, a multitude of studies have reported that viral infections may have complex interconnections with the autophagic process. The findings reported here demonstrate that hepatitis B virus (HBV) can enhance the autophagic process in hepatoma cells without promoting protein degradation by the lysosome. Mutation analysis showed that HBV small surface protein (SHBs) was required for HBV to induce autophagy. The overexpression of SHBs was sufficient to induce autophagy. Furthermore, SHBs could trigger unfolded protein responses (UPR), and the blockage of UPR signaling pathways abrogated the SHB-induced lipidation of LC3-I. Meanwhile, the role of the autophagosome in HBV replication was examined. The inhibition of autophagosome formation by the autophagy inhibitor 3-methyladenine (3-MA) or small interfering RNA duplexes targeting the genes critical for autophagosome formation (Beclin1 and ATG5 genes) markedly inhibited HBV production, and the induction of autophagy by rapamycin or starvation greatly contributed to HBV production. Furthermore, evidence was provided to suggest that the autophagy machinery was required for HBV envelopment but not for the efficiency of HBV release. Finally, SHBs partially colocalized and interacted with autophagy protein LC3. Taken together, these results suggest that the host's autophagy machinery is activated during HBV infection to enhance HBV replication.
INTRODUCTION
Hepatitis B virus (HBV) is a noncytopathic, enveloped DNA virus that belongs to the family Hepadnaviridae (57, 70). It is one of the most successful human pathogens, with an estimated 2 billion people infected worldwide, of whom 400 million have chronic HBV infection (54). Chronic HBV infection is correlated with a strongly increased risk for the development of liver cirrhosis and hepatocellular carcinoma (HCC) (49, 54).
Effective preventive vaccines against HBV have been available for nearly three decades; however, their effectiveness in preventing blood-borne transmission from an infected mother to her newborn is about 90% (77), and therapeutic vaccines for the treatment of established hepatitis B infection are not available (65, 67). Currently, two types of antiviral therapies are approved: pegylated alpha interferon and nucleoside/nucleotide analogues (60). However, these antivirals cannot completely eradicate the virus, and their efficacy in preventing liver cirrhosis and HCC is limited (21, 64). Thus, the details of the host-virus relationship during HBV infection urgently need to be further clarified to facilitate the development of efficient therapeutic strategies for the control of HBV infection.
Autophagy, an evolutionarily conserved intracellular process, involves the formation of a double membrane structure, called the autophagosome, which engulfs long-lived cytoplasmic macromolecules and damaged organelles and delivers them to lysosomes for degradation and recycling (33). The dysfunction of autophagy has been implicated in multiple diseases, including neurodegenerative diseases, muscle diseases, cancer, cardiac diseases, and infectious diseases (46). Autophagy can contribute to innate and adaptive immunity against intracellular microbial pathogens (11, 43). However, this intracellular process has been exploited by some viruses to benefit their replication, such as poliovirus, coxsackievirus, dengue virus, hepatitis C virus, human immunodeficiency virus, and influenza A virus (14, 28, 42, 44, 75, 80, 95, 100).
The aim of this study was to investigate the relationship between HBV and autophagy. We show that HBV enhanced the autophagic process, which required HBV small surface protein (SHBs) and depended on the induction of endoplasmic reticulum (ER) stress. Moreover, we demonstrate that this process greatly enhanced virus envelopment. In addition, we show that SHBs partially colocalized and coimmunoprecipitated with autophagy protein LC3.
MATERIALS AND METHODS
Plasmids.
The plasmid pHBV1.3, which contains a 1.3-fold-overlength genome of HBV, was described previously (47). The plasmid pHBV3.8, which contains a 1.1-fold-overlength genome of HBV, was described previously (90). pHBV1.3-Pol− was derived from pHBV1.3 by a frameshift mutation introduced into the P gene after codon 108 and is defective in viral polymerase expression. pHBV1.3-HBx−, which is defective in the expression of HBx, was described previously (91). pHBV1.3-ENV−, a derivative of pHBV1.3, has a single point mutation that changes codon 15 of the S gene from UUA to the stop codon UGA. This mutation abrogates the expression of all HBV surface proteins but is silent in the overlapping P gene. pHBV1.3-SHBs− was derived from pHBV1.3 by a single point mutation in the S gene that converts the translation initiation codon AUG into the codon of ACG. This mutation abolishes the synthesis of SHBs but is silent in the overlapping P gene. pHBV3.8-SHBs− was derived from pHBV3.8 and constructed as described above. To construct pcDNA3.1-Flag-SHBs and pcDNA3.1-Flag-HBx, DNA sequences coding for SHBs and HBx were amplified from pHBV1.3 and cloned into pcDNA3.1-Flag (Invitrogen), respectively. A secretion-deficient SHBs construct that contains the Cys48Ala mutation was constructed in the context of pcDNA3.1-Flag-SHBs, as reported by Mangold et al. (53). To construct green fluorescent protein (GFP)-tagged or Myc-tagged LC3, human LC3 mRNA was amplified by reverse transcription-PCR (RT-PCR) and cloned into pEGFP-C2 (Invitrogen) and pCMV-Myc (Invitrogen), respectively. All of the mutations were performed by inverse PCR using a KOD-Plus mutagenesis kit (Koyobo) according to the manufacturer's instructions. All primer sequences are available upon request. All of the constructs were confirmed by DNA sequencing.
Chemicals, antibodies, and other reagents.
Rapamycin, 3-methyladenine (3-MA), and anti-β-actin antibody were obtained from Sigma-Aldrich. Anti-LC3 antibody was obtained from Cell Signaling Technology or MBL International. Anti-p62, anti-phospho-eIF2α (eIF2α is the α subunit of eukaryotic initiation factor 2), and anti-GRP94 antibodies were obtained from Cell Signaling Technology. Anti-phospho-PERK, anti-total PERK, and anti-IRE1α antibodies were obtained from Santa Cruz Biotechnology. Anti-ATF6 and anti-HBx antibodies were obtained from Millipore. Anti-Beclin1 antibody was obtained from MBL International. Anti-ATG5 antibody was obtained from Proteintech Group. Dithiothreitol (DTT) was obtained from Sangon (Shanghai, China). Anti-HBV surface antigen (HBsAg) antibody was obtained from Dako or Gene Company Limited (Hong Kong, China). Anti-HBV core antigen (HBcAg) antibody was obtained from Dako. Plasma HBsAg was described in our previous study (92) and obtained from Kehua (Shanghai, China).
Cell culture and transfection.
The hepatoma cell line Huh7 and the HBV-expressing stable cell line HepG2.2.15 were maintained as described previously (47). Transient transfection was performed with the indicated plasmids using Fugene 6 or Fugene HD (Roche) according to the manufacturer's instructions.
Western blot analysis.
Western blot analysis was performed as described previously (47). Briefly, after transfection or treatment, cells were washed with phosphate-buffered saline (PBS) and lysed with 2× SDS protein loading buffer (100 mM Tris-HCl, pH 6.8, 20% glycerol, 10% β-mercaptoethanol, 4% SDS, and 0.2% bromophenol blue). For phosphorylation detection, cells were lysed in the lysis buffer described above and supplemented with 0.1 mM sodium vanadate (Na3VO4), 60 mM β-glycerophosphate, 50 mM NaF, and 1× protease inhibitor mixture (Roche Applied Science). After being boiled for 10 min, the cell lysates were centrifuged, and equivalent amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then electrotransferred to nitrocellulose membranes (Roche Applied Science). The membranes were incubated with the indicated primary antibodies for 2 h at room temperature or overnight at 4°C after being blocked with 5% nonfat milk in 1× PBS-Tween. The membranes were washed with 1× PBS-Tween and incubated with the corresponding secondary antibodies (Sigma). Immunoreactive bands were visualized by an enhanced chemiluminescence system (ECL; PerkinElmer Life Sciences).
Confocal microscopy.
For the detection of autophagosomes, cells were transfected with GFP-LC3, and after 12 h they were treated or transfected as indicated for 48 h. The cells were fixed, and the nuclei were stained with 4′-6-diamidino-2-phenylindole (DAPI; blue stain; Invitrogen). The fluorescence of GFP-LC3 was observed under a confocal fluorescence microscope (Leica TCS SP2). Cells containing three or more GFP-LC3 dots were defined as autophagy-positive cells. The percentage of cells with characteristic punctate GFP-LC3 fluorescence relative to all GFP-positive cells was calculated as described previously (88). A minimum of 200 GFP-positive cells per sample was counted, and three independent experiments were performed.
For immunofluorescent staining, cells were grown on coverslips and transfected with pHBV1.3. After transfection for 48 h, cells were fixed with 3.5% paraformaldehyde for 15 min. The fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and blocked with 3% bovine serum albumin in PBS for 2 h. Cells then were incubated with anti-HBsAg antibody (1:100; Gene Company Limited) or anti-HBx antibody (1:200) together with anti-LC3 antibody (1:100; MBL International) at 4°C overnight, followed by being stained for 2 h with Alexa Fluor 488-conjugated anti-mouse secondary antibody (1:200; Invitrogen) and Cy3-conjugated anti-rabbit secondary antibody (1:200; Jackson). The nuclei were stained with DAPI, and the colocalization of SHBs or HBx (green stain) with LC3 (red stain) was observed with a confocal fluorescence microscope.
Transmission electron microscopy.
Electron microscopy was performed as described previously (73). Briefly, after the indicated transfection or treatment, cells were washed three times with 1× PBS, trypsinized, and collected by centrifugation at 1,000 × g for 5 min. The cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M Na-phosphate buffer (pH 7.4) at 4°C overnight. The cells were washed in the same buffer three times for 15 min each and postfixed in 1% OsO4 in 0.1 M cacodylate buffer (pH 7.4) for 2 to 3 h. After being washed in 0.1 M Na-phosphate buffer, the cells were dehydrated with a graded series of ethanol and gradually infiltrated with epoxy resin. The samples were sequentially polymerized at 37°C overnight, 45°C for 12 h, and 60°C for 24 h. Ultrathin sections (about 60 nm) were cut on an LKB microtome and mounted on copper slot grids. Sections were doubly stained with uranyl acetate and lead citrate and observed under a Philips CM-120 transmission electron microscope. About 10 cells per sample were counted, and for each cell the number of double-membrane vesicles (autophagosomes) was examined as described previously (1).
RT-PCR analysis.
The activation of IRE1α was assessed by quantitatively measuring the splicing of its substrate, the mRNA encoding the XBP1 transcription factor, using a modified version of the method described by Shang et al. (72). In brief, total RNA was isolated by the use of TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA synthesis was performed with the Superscript first-strand synthesis system (Invitrogen) by following the manufacturer's directions. To amplify XBP1 mRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, a PCR with the cDNA was conducted for 30 cycles (94°C for 50 s, 58°C for 30 s, and either 72°C for 30 s or a 10-min incubation in the final cycle) using 5′-CTGGAAAGCAAGTGGTAGA-3′ and 5′-CTGGGTCCTTCTGGGTAGAC-3′ for XBP1 and 5′-GGTATCGTGGAAGGACTCATGAC-3′ and 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′ for GAPDH with ExTaq DNA polymerase (Takara). The amplified products of the spliced (XBP1S; 398 bp) and unspliced (XBP1U; 424 bp) XBP1 were resolved on 2% agarose gels with ethidium bromide and visualized using the Gene Genius Bio-imaging system (Syngene).
RNA interference.
Huh7 cells were grown to 30 to 40% confluence and then transiently transfected with the indicated small interfering RNA (siRNA) duplexes (Invitrogen or Santa Cruz Biotechnology) at a final concentration of 40 nM using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and as described previously (47). At 24 h posttransfection, the cells were transfected with 20 nM the same siRNA duplexes and the indicated plasmids. After the second transfection for 48 h, the cells were collected and the silencing efficiency of the siRNA duplexes was detected by Western blot analysis.
Northern blot and Southern blot analyses.
Intracellular viral RNA and DNA analyses were performed as described previously (47). For viral RNA analysis, total cellular RNA was extracted with TRIzol reagents. Fifteen micrograms of total RNA was resolved in a 1% agarose gel containing 2.2 M formaldehyde and transferred onto a positively charged nylon membrane in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). For viral DNA analysis, intracellular viral nucleocapsid-associated DNA was extracted as described previously (50), resolved by electrophoresis on a 1% agarose gel, and transferred onto a positively charged nylon membrane. All membranes were detected with a 32P-radiolabeled HBV DNA probe prepared using the Random Priming labeling kit (Roche) in Ultrahyb ultrasensitive hybridization buffer (Ambion). For Northern blot analysis, the blots then were stripped and rehybridized with a 32P-radiolabeled GAPDH DNA probe for loading normalization. Hybridization signals were visualized by phosphorimaging (Fujifilm).
Coimmunoprecipitation analysis.
Coimmunoprecipitation analysis was performed as described previously, with minor modifications (63). Cells were plated in 6- or 10-cm dishes and transfected with the indicated plasmids. After transfection for 48 h, cells were lysed in immunoprecipitation buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% NP-40, 5 mM EDTA, 1 mM Na3VO4, 1 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail (Roche). The cell lysates were rotated at 4°C for 1 h, and insoluble material was pelleted at 12,000 × g at 4°C for 30 min. The supernatants were precleared with 30 μl of protein A/G plus agarose beads (Santa Cruz Biotechnology) at 4°C for 1 h and immunoprecipitated with 2 μg of normal control IgG (Santa Cruz Biotechnology) or specific primary antibodies. The mixtures were rotated overnight at 4°C, followed by incubation with 40 μl of protein A/G plus agarose beads for 1 to 2 h at 4°C. After four washes in immunoprecipitation buffer, the immunoprecipitates were subjected to Western blot analysis as described above.
Endogenous polymerase assay.
The detection of intracellular nucleocapsids and intracellular or extracellular virions by the endogenous polymerase assay (EPA) was performed as described previously, with some modifications (86, 93). For the detection of intracellular nucleocapsids, cells were lysed in immunoprecipitation buffer; for the detection of intracellular virions, cells were lysed by repetitive cycles of freezing and thawing (using liquid nitrogen to freeze and a 37°C bath to thaw) as described previously (19). The immunoprecipitation of nucleocapsids by anti-HBcAg antibody or virions by anti-HBsAg antibody was performed as described above. The immunoprecipitates were incubated with dATP, dGTP, dTTP (0.1 mM each), and 10 μCi of [α-32P]dCTP (Beijing Free, China) in an EPA reaction buffer (50 mM Tris-HCl, pH 7.0, 40 mM NH4Cl, 20 mM MgCl2, 1% NP-40, and 0.3% β-mercaptoethanol) overnight at 37°C. Nonlabeled dCTP was added to a final concentration of 0.1 mM for additional incubation (1 h). Viral nucleic acids, labeled by the incorporation of [α-32P]dCTP, were deproteinized with proteinase K treatment and phenol extraction. The labeled DNA was separated on a 1% agarose gel, which then was dried by a gel dryer (Bio-Rad) and imaged by phosphorimaging.
Statistical analysis.
Results are presented as means ± standard deviations. The significance of the differences was determined by Student's t tests. P < 0.05 was considered statistically significant.
RESULTS
HBV increases autophagosome formation.
LC3 is the mammalian homologue of an essential yeast autophagy protein, autophagy-related protein 8 (Atg8) (29, 35). Following translation, the unprocessed form of LC3 (proLC3) is cleaved by Atg4 protease, yielding the cytosolic LC3-I form (89). Upon the induction of autophagy, LC3-I is converted to LC3-II through lipidation by a ubiquitin-like system that allows for LC3 to become associated with autophagic vesicles (89). The conversion of LC3-I to the form of LC3-II that migrates at a lower level and the relocalization of LC3 from a diffuse cytoplasmic distribution to distinct puncta have been used as indicators of autophagy (3, 36, 55, 56).
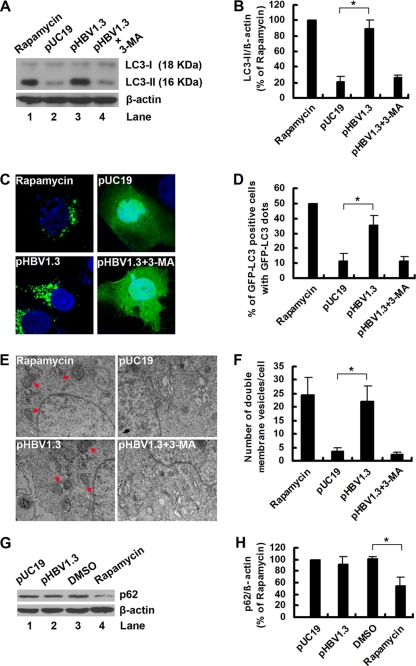
To investigate whether HBV could regulate autophagy, we transfected pHBV1.3, or its backbone vector pUC19, into Huh7 cells, and the conversion of endogenous LC3-I to LC3-II was examined by Western blot analysis. The results showed that HBV induced a clear increase in LC3-II levels (Fig. 1A, lane 3 versus lane 2) and that the LC3-II/β-actin ratio was significantly increased in pHBV1.3-transfected cells (Fig. 1B). Accordingly, the increases in LC3-II levels and the LC3-II/β-actin ratio induced by HBV could be reversed by treatment with the autophagy inhibitor 3-MA (Fig. 1A, lane 4 versus lane 3, and B), suggesting that authentic autophagy was induced. As a positive control, the autophagy inducer rapamycin dramatically induced LC3-II expression (Fig. 1A, lane 1, and B). To test whether LC3 modification correlated with its relocalization to autophagic vesicles during HBV infection, we monitored the subcellular localization of GFP-tagged LC3 (GFP-LC3) protein by confocal microscopy. As shown in Fig. 1C, GFP-LC3 redistribution into discrete dots was markedly increased by HBV. The percentage of GFP-LC3-positive cells containing green punctate dots in the pHBV1.3-transfected cells was increased approximately 3-fold compared to that of the pUC19-transfected cells (Fig. 1D). Notably, this enhanced autophagy by HBV also was inhibited by 3-MA treatment (Fig. 1C and D), thereby confirming that the GFP-LC3 punctate dots in the pHBV1.3-transfected cells represent autophagy signals.
Fig. 1.
HBV induces autophagosome formation in hepatoma cells without promoting protein degradation by the lysosome. (A) Huh7 cells were treated with rapamycin (50 nM) or transfected with pUC19 or pHBV1.3. After transfection for 46 h, pHBV1.3-transfected cells were left untreated or were treated with 3-MA (10 mM) for 2 h. After 48 h of transfection or treatment, the levels of LC3-II expression were determined by Western blot analysis. β-Actin expression was examined as a protein loading control. (B) The LC3-II/β-actin ratios (% of rapamycin) were determined by Western blot analysis, and the bands were quantified by densitometric analysis using NIH ImageJ software. Results represent the mean data from three independent experiments. *, P < 0.05. (C) Huh7 cells were transfected with GFP-LC3, and after 12 h they were treated or transfected as described for panel A. The nuclei were stained by DAPI (blue stain), and the distribution of GFP-tagged LC3 protein was visualized with a confocal fluorescence microscope. Representative confocal images are shown. (D) Quantification of the frequency of Huh7 cells displaying a punctate distribution of GFP-tagged LC3 protein was performed as described in Materials and Methods. Results represent the mean data from three independent experiments. *, P < 0.05. (E) Huh7 cells were treated or transfected as described for panel A. After 48 h of transfection or treatment, cells were subjected to transmission electron microscopy. Arrows indicate representative autophagosomes. (F) Quantification of numbers of autophagosomes was performed as described in Materials and Methods. Results represent the mean data from two independent experiments. *, P < 0.05. (G) Huh7 cells were transfected with pUC19 or pHBV1.3 or treated with DMSO or rapamycin. After 48 h of transfection or treatment, the levels of p62 protein expression were determined by Western blot analysis. β-Actin expression was examined as a protein loading control. (H) The p62/β-actin ratios (% of rapamycin) were determined as described for panel B. Results represent the mean data from three independent experiments. *, P < 0.05.
To provide further evidence of the HBV-induced autophagosome formation, we detected HBV-induced autophagy at the ultrastructural level by transmission electron microscopy. Compared to the pUC19-transfected cells, whose autophagic vacuoles were rarely observed, we noticed an accrual of membrane vacuoles in the pHBV1.3-transfected cells and that cytosolic components or organelles were sequestered in some of these vacuoles (Fig. 1E and F).
Some pathogens induce early stages of autophagy but block later stages (i.e., autophagolysosomal fusion) (28, 75, 81, 88, 95), so we investigated whether HBV could induce a complete autophagic process (i.e., autophagic flux) by measuring the level of the autophagic substrate p62, an adaptor protein that interacts with LC3 and is specifically degraded by the autophagy pathway (4, 59). The results showed that HBV could not change the expression of p62, in contrast to rapamycin (Fig. 1G and H). Taken together, these results indicate that an incomplete autophagic process is induced by HBV.
SHBs is required for HBV to induce autophagy.
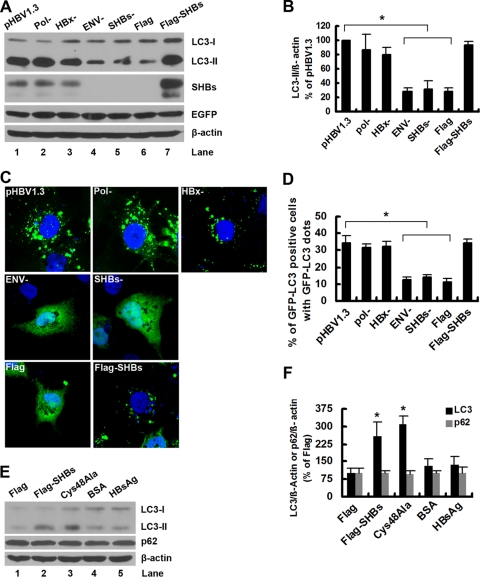
The HBV genome encodes four overlapping open reading frames (ORFs) that are translated to make the viral core protein, the envelope proteins, viral polymerase, and HBx (57, 70). There are three envelope proteins: large (LHBs), medium (MHBs), and small (SHBs) surface proteins, all encoded by the S gene ORF (57, 70). To determine which viral protein is responsible for the induction of autophagy by HBV, the expression of polymerase, envelope proteins, and HBx were separately abolished by mutation in the context of pHBV1.3. The wild-type or resultant mutants were transfected into Huh7 cells, and both the lipidation of endogenous LC3-I to yield LC3-II and the relocalization of GFP-LC3 from a diffuse to a punctate pattern were detected. The results showed that the deletion of the HBV envelope proteins, but not the deletion of viral polymerase and HBx, abrogated the HBV-induced lipidation of LC3-I (Fig. 2A, lanes 2 to 4 versus lane 1, and B) and relocalization of GFP-LC3 (Fig. 2C and D), suggesting that the HBV envelope proteins mediate the HBV-induced autophagy. Notably, these results were not due to variations in the transfection efficiency of the wild type or its mutants, as similar enhanced GFP (EGFP) expression levels were observed (Fig. 2A).
Fig. 2.
SHBs is required for HBV to induce autophagy. (A) Huh7 cells were transfected with pHBV1.3, pHBV1.3-Pol−, pHBV1.3-HBx−, pHBV1.3-ENV−, pHBV1.3-SHBs−, pcDNA3.1-Flag, or pcDNA3.1-Flag-SHBs together with pEGFP-C2 plasmid. After 48 h of transfection, the levels of LC3-II expression were determined by Western blot analysis. β-Actin and EGFP expression levels were examined to control for loading and transfection efficiency. (B) The LC3-II/β-actin ratios (% of pHBV1.3) were determined as described in the legend to Fig. 1B. Results represent the mean data from three independent experiments. *, P < 0.05. (C) Huh7 cells were transfected with GFP-LC3 plasmid, and after 12 h they were transfected with the plasmids described for panel A (excluding pEGFP-C2). The nuclei were stained by DAPI, and the distribution of GFP-tagged LC3 protein was visualized with a confocal fluorescence microscope. Representative confocal images are shown. (D) Quantification of the frequency of Huh7 cells displaying a punctate distribution of GFP-tagged LC3 protein was performed as described in Materials and Methods. Results represent the mean data from three independent experiments. *, P < 0.05. (E) Huh7 cells were transfected with pcDNA3.1-Flag, pcDNA3.1-Flag-SHBs, or pcDNA3.1-Flag-SHBs (Cys48Ala) or treated with BSA or HBsAg (200 ng/ml). After 48 h of transfection or treatment, the levels of LC3-II and p62 expression were determined by Western blot analysis. β-Actin expression was examined as a protein loading control. (F) The ratios of LC3-II/β-actin or p62/β-actin (% of pcDNA3.1-Flag) were determined as described in the legend to Fig. 1B. Results represent the mean data from two independent experiments. *, P < 0.05.
Among the HBV envelope proteins, SHBs is the most abundant, while LHBs and MHBs are expressed at levels of about 5 to 15% and 1 to 2%, respectively, of the expression of SHBs (94). To investigate whether SHBs is required for HBV to induce autophagy, an SHBs expression-deficient mutant from pHBV1.3 was transfected into Huh7 cells. The mutant lost the ability to induce the lipidation of LC3-I (Fig. 2A, lane 5 versus lane 1, and B) and the relocalization of GFP-LC3 (Fig. 2C and D). Moreover, the overexpression of SHBs alone was sufficient to induce autophagy (Fig. 2A, lane 7 versus lane 6, and B, C, and D). In addition, the ability of SHBs to mediate the HBV-induced autophagy is not limited to the pHBV1.3 isolate; a similar result was observed for another HBV isolate, pHBV3.8 (data not shown).
SHBs initially is synthesized as a transmembrane protein spanning the membrane of the ER and can be secreted as a subviral particle (57, 70). To determine the contributions of intracellular and extracellular SHBs in the induction of autophagy, a secretion-deficient SHBs-expressing plasmid was transfected into Huh7 cells. As shown in Fig. 2E (lane 3 versus lane 2) and F, the deficiency of SHBs in secretion had no effect on its ability to induce autophagy, which was demonstrated by the lipidation of LC3-I. To exclude the possibility that the induction of autophagy by this mutant is due to a trace amount of SHBs in the culture supernatant, Huh7 cells were cultured with HBsAg (200 ng/ml) at a concentration comparable to that of the HBsAg in the culture supernatant of transfected cells, and the lipidation of LC3-I was detected. The results showed that no apparent increase in the lipidation of LC3-I was observed (Fig. 2E, lane 5 versus lane 4, and F), suggesting that autophagy was not induced by extracellular SHBs. In addition, similarly to HBV, SHBs did not change the expression of p62 (Fig. 2E, lane 2 versus lane 1, and F), indicating that an incomplete autophagic process was induced by intracellular SHBs. In summary, these results show that intracellular SHBs but not extracellular SHBs is required for HBV to induce autophagy.
SHBs induces autophagy via ER stress.
To determine how SHBs induces autophagy, we investigated whether SHBs can trigger ER stress, which was reported to trigger autophagy (25, 39, 45, 48, 58, 66, 69, 97, 98). A number of cellular stress conditions can perturb protein folding in the ER, leading to a condition known as ER stress (23, 68). ER stress triggers the unfolded protein response (UPR), which involves at least three distinct signaling pathways (68). One involves activating PERK, which, when induced, phosphorylates eIF2α to attenuate mRNA translation. In the ATF6 pathway, the ATF6 precursor (90 kDa) is cleaved in response to ER stress, and the cleaved ATF6 N-terminal fragment (50 kDa) translocates to the nucleus to activate the transcription of genes encoding chaperones that refold unfolded or misfolded proteins. In the IRE1 pathway, XBP1 mRNA is spliced by activated IRE1 to generate mature XBP1 mRNA, the translation product of which leads to the transcription of genes encoding protein degradation enzymes.
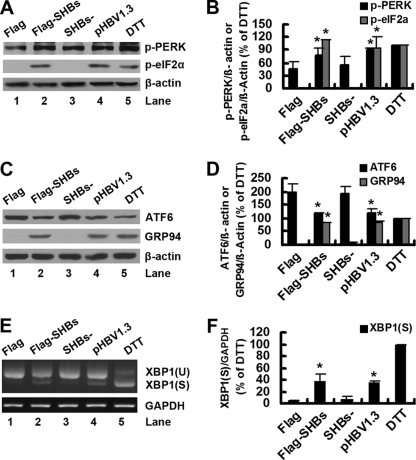
To determine whether SHBs could induce ER stress, pcDNA3.1-Flag-SHBs (or its control vector) and pHBV1.3 (or its SHBs deletion mutant) were transfected into Huh7 cells. As a control, one set of Huh7 cells was treated with DTT, an ER stress inducer. As shown in Fig. 3A (lane 2 versus lane 1) and B, the overexpression of SHBs induced the phosphorylation of PERK and its downstream effector eIF2α. Importantly, the expression of SHBs from pHBV1.3 also increased the phosphorylation of PERK and eIF2α (Fig. 3A, lane 4 versus lane 3, and B). As expected, the phosphorylation of PERK and eIF2α was induced following treatment with DTT (Fig. 3A, lane 5, and F). In parallel, we examined ATF6 cleavage by Western blot analysis and found that both exogenous SHBs from pcDNA3.1-Flag-SHBs and endogenous SHBs from pHBV1.3 induced a slight reduction in the expression levels of ATF6 precursor (Fig. 3C, lane 2 versus lane 1 and lane 4 versus lane 3, and D). Unfortunately, we could not detect the 50-kDa cleavage products in the transfected cells or even in the DTT-treated cells (Fig. 3C). To determine whether ATF6 target genes would be activated in pcDNA3.1-Flag-SHBs- or pHBV1.3-transfected cells, we assessed the expression of GRP94, an ER homologue of HSP90 protein (15), by Western blot analysis. As shown in Fig. 3C and D, exogenous or endogenous SHBs and DTT induced a comparable increase in GRP94 levels, suggesting that the ATF6 pathway was activated following the expression of exogenous or endogenous SHBs. We then examined the splicing of XBP1 by RT-PCR. In pcDNA3.1-Flag- or pHBV1.3-SHBs−-transfected control cells, only the unspliced form of XBP1 was detected (Fig. 3E, lanes 1 and 3). However, in addition to the unspliced form of XBP1, the spliced form of XBP1 also was detectable in pcDNA3.1-Flag-SHBs- or pHBV1.3-transfected cells (Fig. 3E, lanes 2 and 4, and F) and at a higher level in DTT-treated cells (Fig. 3E, lane 5, and F).
Fig. 3.
SHBs triggers ER stress. (A) Huh7 cells were transfected with pcDNA3.1-Flag, pcDNA3.1-Flag-SHBs, pHBV1.3-SHBs−, or pHBV1.3 for 48 h or were treated with DTT (2 mM) for 2 h. The phosphorylation levels of PERK and eIF2α were determined by Western blot analysis. β-Actin expression was examined as a protein loading control. (B) The ratios of p-PERK/β-actin or p-eIF2α/β-actin (% of DTT) were determined as described for Fig. 1B. Results represent the mean data from two independent experiments. *, P < 0.05. (C) Huh7 cells were transfected or treated as described for panel A. The cleavage of ATF6 was determined by Western blot analysis. β-Actin expression was examined as a protein loading control. (D) The ratios of ATF6/β-actin or GRP94/β-actin (% of DTT) were determined as described for Fig. 1B. Results represent the mean data from two independent experiments. *, P < 0.05. (E) Huh7 cells were transfected or treated as described for panel A. Total cellular RNA was analyzed for XBP1 mRNA by RT-PCR as described in Materials and Methods. XBP1 (U) and XBP1 (S) represent DNA fragments derived from unspliced and spliced XBP1 RNA, respectively. The GAPDH mRNA was analyzed to serve as an internal control. (F) The XBP1 (S)/GAPDH ratios (% of DTT) were determined as described for Fig. 1B. Results represent the mean data from two independent experiments. *, P < 0.05.
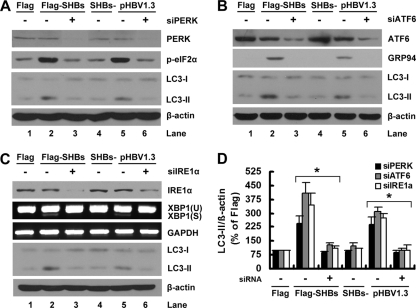
These results showed that SHBs could induce ER stress; therefore, we next asked whether SHBs induced an autophagic response via ER stress. We knocked down the expression of PERK, IRE1α, or ATF6 by siRNA duplexes in pcDNA3.1-Flag-SHBs- or pHBV1.3-transfected cells and evaluated the lipidation of LC3-I. As shown in Fig. 4A (lane 3 versus lane 2 and lane 6 versus lane 5) and D, the PERK siRNA duplex, but not the control siRNA duplex, greatly reduced the LC3-II expression levels in pcDNA3.1-Flag-SHBs- or pHBV1.3-transfected cells. The effectiveness of the siRNA duplex targeted against PERK was confirmed by Western blot analysis of the total expression of PERK and the phosphorylation of eIF2α (Fig. 4A). Similarly, the silencing of ATF6 or IRE1α inhibited the SHBs-induced lipidation of LC3-I (Fig. 4B and C, lane 3 versus lane 2, lane 6 versus lane 5, and D). The effectiveness of the siRNA duplex targeted against ATF6 was confirmed by Western blot analysis of the expression of ATF6 and GRP94 (Fig. 4B). The effectiveness of the siRNA duplex targeted against IRE1α was confirmed by Western blot analysis of the expression of IRE1α and RT-PCR analysis of XBP1 splicing (Fig. 4C). In addition, we also assessed the effects of PERK, ATF6, and IRE1α siRNA duplexes on the lipidation of LC3-I in naïve Huh7 cells. None of these siRNA duplexes had any effect on the lipidation of LC3-I (data not shown). Taken together, these results clearly show that SHBs induces autophagy via ER stress.
Fig. 4.
Blockage of UPR signaling pathways by RNA interference abrogated the SHBs-induced lipidation of LC3-I. (A) Huh7 cells were transfected with 40 nM siRNA duplexes targeting EGFP or PERK (siPERK). At 24 h posttransfection, the cells were transfected with 20 nM the same siRNA duplexes together with pcDNA3.1-Flag, pcDNA3.1-Flag-SHBs, pHBV1.3, or pHBV1.3-SHBs− for 48 h as indicated. The levels of LC3-II expression were determined by Western blot analysis. The effectiveness of siRNA duplexes was detected by Western blot analysis using the antibodies against PERK or eIF2α. β-Actin expression was examined as a protein loading control. (B) Huh7 cells were transfected as described for panel A, except that siRNA duplexes targeting ATF6 (siATF6) were used in place of the siRNA duplexes targeting PERK. The levels of LC3-II expression were determined by Western blot analysis. The effectiveness of siRNA duplexes was detected by Western blot analysis using antibodies against ATF6 or GRP94. β-Actin expression was examined as a protein loading control. (C) Huh7 cells were transfected as described for panel A, except that siRNA duplexes targeting IRE1α (siIRE1α) were used in place of the siRNA duplexes targeting PERK. The levels of LC3-II expression were determined by Western blot analysis. The effectiveness of siRNA duplexes was detected by the Western blot analysis of the expression of IRE1α or by RT-PCR analysis of XBP1 splicing. β-Actin expression was examined as a protein loading control. (D) The ratios of LC3-II/β-actin (% of pcDNA3.1-Flag) in panels A, B, and C were determined as described for Fig. 1B. Results represent the mean data from two independent experiments. *, P < 0.05.
The autophagy machinery is required for HBV production.
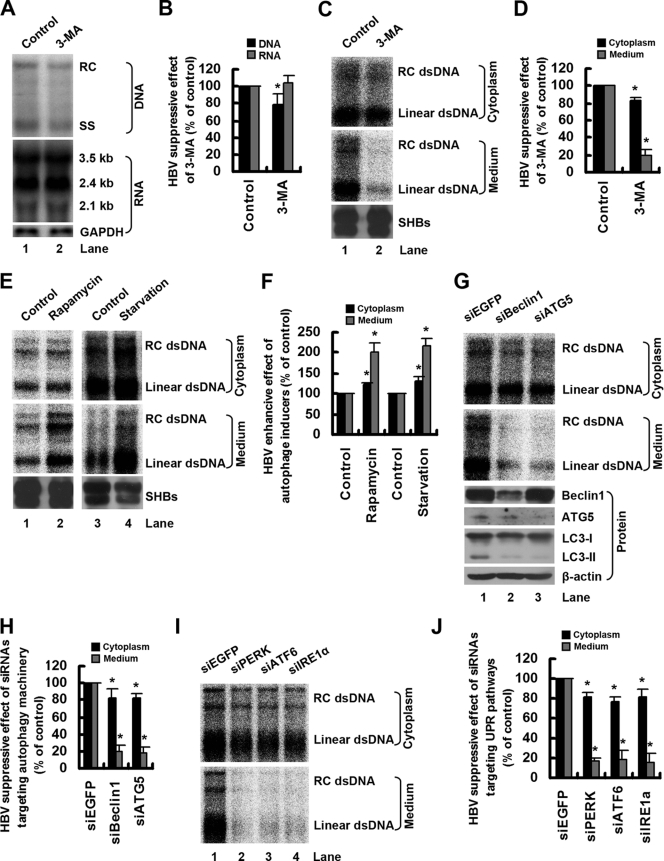
An increasing body of research has shown that autophagy functions in antiviral or proviral capacities in the life cycles of a broad range of viruses (11, 40). To determine whether the autophagy machinery performs an antiviral or proviral function during HBV infection, we tested the effect of the autophagy inhibitor 3-MA on HBV replication in Huh7 cells. The amounts of nucleocapsid-associated DNA and viral RNA were determined by Southern blot and Northern blot analyses, respectively. As shown in Fig. 5A (lane 2 versus lane 1) and B, 3-MA treatment did not change the level of viral RNA but resulted in a slight decrease in the level of nucleocapsid-associated DNA. To define the effect of 3-MA on the production of extracellular virions, enveloped HBV virions in culture supernatant were precipitated with antibody against HBsAg and assayed by EPA. Consistently with the results of the Southern blot analysis, EPA analysis also showed that 3-MA treatment had only a slight inhibitory effect on the level of nucleocapsid-associated DNA (Fig. 5C, lane 2 versus lane 1, top, and D). 3-MA treatment markedly inhibited the production of extracellular virions (Fig. 5C, lane 2 versus lane 1, middle, and D), suggesting that autophagy contributes to HBV envelopment and/or release. The inhibitory effect of 3-MA on HBV production is not specific to Huh7 cells, as similar results were obtained in HepG2.2.15 cells (data not shown). To further determine the role of autophagy in the life cycle of HBV, we investigated the effect of the autophagy inducer rapamycin or starvation on the production of intracellular nucleocapsids and extracellular virions by EPA. As shown in Fig. 5E (lane 2 versus lane 1 and lane 4 versus lane 3) and F, the induction of autophagy by rapamycin or starvation significantly increased the level of extracellular virions but only slightly upregulated the level of nucleocapsid-associated DNA, further indicating the role of autophagy in HBV envelopment and/or release. Interestingly, we found that both the autophagy inhibitor (3-MA) and inducers (rapamycin and starvation) had no effect on the production of SHBs in the culture supernatant (Fig. 5C and E, bottom), suggesting a specific effect of autophagy on HBV production.
Fig. 5.
Autophagy machinery is required for HBV production. (A) Huh7 cells were transfected with pHBV1.3 and after 36 h were left untreated or were treated with 3-MA for 12 h. The amounts of nucleocapsid-associated DNA and viral RNA were determined by Southern blot and Northern blot analyses, respectively, using a 32P-radiolabeled HBV DNA probe. After the detection of HBV RNA, the blots were stripped and rehybridized with a 32P-radiolabeled GAPDH DNA probe to control for gel loading. The positions of HBV relaxed circular (RC), single-stranded (SS) DNAs and the positions of viral pregenomic RNA (3.5 kb), pre-S1/S RNA (2.4 kb), and pre-S2/S RNA (2.1 kb) are indicated. (B) The percentage of the inhibitory effect of 3-MA (% of control) on the abundance of HBV DNA and RNA shown in panel A was calculated. The bands were quantified by densitometric analysis using NIH ImageJ software. Results represent the mean data from two independent experiments. *, P < 0.05. (C) Huh7 cells were transfected and treated as described for panel A. Enveloped HBV virions in culture supernatant and nucleocapsids in cell lysates were precipitated with antibody against HBsAg or antibody against HBcAg, respectively, and assayed by EPA as described in Materials and Methods. SHBs in the precipitate from culture supernatant was determined by Western blot analysis. dsDNA, double-stranded DNA. (D) The percentage of inhibitory effect of 3-MA (% of control) on the abundance of extracellular virions and intracellular nucleocapsids shown in panel C was calculated as described for panel B. Results represent the mean data from two independent experiments. *, P < 0.05. (E) Huh7 cells were transfected with pHBV1.3. After 12 h of transfection they were treated with rapamycin for 36 h, or after 46 h of transfection they were transferred to a starvation medium (Earle's balanced salts) for 2 h. Enveloped HBV virions in culture supernatant and nucleocapsids in cell lysates were assayed by EPA as described in Materials and Methods. SHBs in the precipitate from culture supernatant was determined by Western blot analysis. (F) The percentage of the enhancive effect of autophagy inducers (% of control) on the abundance of extracellular virions and intracellular nucleocapsids shown in panel E was calculated as described for panel B. Results represent the mean data from two independent experiments. *, P < 0.05. (G) Huh7 cells were transfected with 40 nM siRNA duplexes targeting EGFP (siEGFP), Beclin1 (siBeclin1), or ATG5 (siATG5). At 24 h posttransfection, the cells were transfected with 20 nM the same siRNA duplexes together with pHBV1.3 for 48 h. Enveloped HBV virions in culture supernatant and nucleocapsids in cell lysates were assayed by EPA as described in Materials and Methods. The effectiveness of siRNA duplexes targeting Beclin1 or ATG5 was detected by Western blot analysis of the expression of Beclin1, ATG5, or LC3-II. (H) The percentage of the inhibitory effect of siRNA duplexes (% of control) on the abundance of extracellular virions and intracellular nucleocapsids shown in panel G was calculated as described for panel B. Results represent the mean data from two independent experiments. *, P < 0.05. (I) Huh7 cells were transfected as described for panel G, except that the siRNA duplexes targeting PERK (siPERK), ATF6 (siATF6), or IRE1α (siIRE1α) were used in place of the siRNA duplexes targeting Beclin1 or ATG5. Enveloped HBV virions in culture supernatant and nucleocapsids in cell lysates were assayed by EPA as described in Materials and Methods. (J) The percentage of the inhibitory effect of siRNA duplexes (% of control) on the abundance of extracellular virions and intracellular nucleocapsids shown in panel I was calculated as described for panel B. Results represent the mean data from two independent experiments. *, P < 0.05.
Because pharmacological agents such as 3-MA and rapamycin may not affect only the autophagic pathway (34), we monitored the effect of the siRNA duplex-mediated knockdown of key autophagy proteins on viral replication in Huh7 cells. As shown in Fig. 5G (lane 2 versus lane 1, top and middle) and H, the knockdown of Beclin1 expression greatly diminished the level of extracellular virions but had only a slight inhibitory effect on the level of nucleocapsid-associated DNA. A knockdown of the expression of another essential autophagy factor, ATG5, resulted in a similar effect on HBV replication (Fig. 5G, lane 3 versus lane 1, top and middle, and H). The effectiveness of siRNA duplexes targeted against Beclin1 or ATG5 was confirmed by Western blot analysis (Fig. 5G, bottom). In addition, we confirmed that Beclin1 and ATG5 knockdowns inhibited autophagy in Huh7 cells, as determined by LC3-II levels (Fig. 5G, bottom).
Because HBV induced the accumulation of autophagosomes via the induction of ER stress (Fig. 4), we reasoned that the blockage of three pathways of the UPR should inhibit the production of HBV. To test this hypothesis, we knocked down the expression of PERK, IRE1α, and ATF6 by siRNA duplexes and detected intracellular nucleocapsids and extracellular virions by EPA. The results showed that the suppression of any of these three pathways inhibited the levels of extracellular virions to a greater extent than those of nucleocapsid-associated DNA (Fig. 5I and J).
Taken together, these results demonstrate that the autophagy machinery is required for the envelopment and/or release of HBV and thus represents a proviral factor for HBV replication.
The autophagy machinery is required for HBV envelopment.
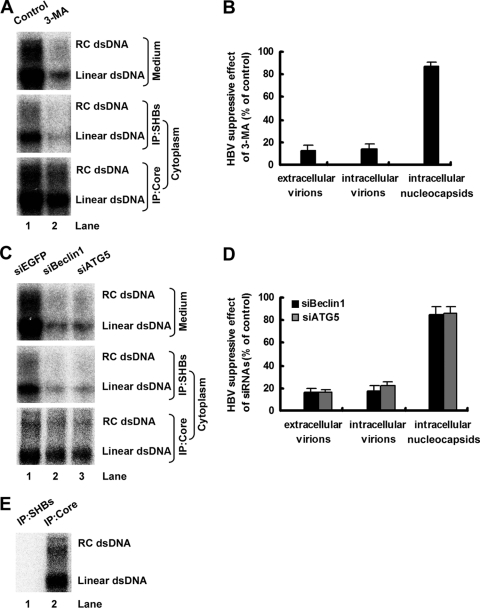
To determine whether the autophagy machinery is required only for HBV envelopment or if it also is required for viral release, cell lysates and culture supernatants from pHBV1.3-transfected Huh7 cells with or without 3-MA treatment were precipitated with antibody against HBsAg. After precipitation by antibody against HBsAg, cell lysates were recaptured with antibody against HBcAg. The abundance of intracellular or extracellular enveloped HBV virions and intracellular nucleocapsids was analyzed by EPA. Consistently with the results presented in Fig. 5C, 3-MA treatment markedly inhibited the level of extracellular enveloped HBV virions (Fig. 6A, top, and B), whereas it only slightly suppressed the level of intracellular nucleocapsids (Fig. 6A, bottom, and B). Furthermore, the abundance of intracellular enveloped HBV virions was significantly reduced by 3-MA treatment in a manner commensurate with the level of extracellular enveloped virions (Fig. 6A, middle, and B), suggesting that the autophagy machinery is required for HBV envelopment but not for the efficiency of virus release.
Fig. 6.
Autophagy machinery is required for HBV envelopment. (A) Huh7 cells were transfected and treated as described for Fig. 5A. The abundance of extracellular or intracellular enveloped virions and intracellular nucleocapsids was analyzed by EPA as described in Materials and Methods. dsDNA, double-stranded DNA; IP, immunoprecipitation. (B) The percentage of the inhibitory effect of 3-MA (% of control) on the abundance of extracellular or intracellular enveloped virions and intracellular nucleocapsids was calculated as described for Fig. 5B. Results represent the mean data from two independent experiments. (C) Huh7 cells were transfected as described for Fig. 5G. The abundance of extracellular or intracellular enveloped virions and intracellular nucleocapsids was analyzed by EPA as described in Materials and Methods. siEGFP, siBeclin1, and siATG5 indicate siRNA complexes targeting EGFP, Beclin1, and ATG5, respectively. (D) The percentage of the inhibitory effect of siRNA duplexes targeting Beclin1 or ATG5 (% of control) on the abundance of extracellular or intracellular enveloped virions and intracellular nucleocapsids was calculated as described for Fig. 5B. Results represent the mean data from two independent experiments. (E) pHBV1.3-ENV− was transfected into Huh7 cells. The cell lysates were precipitated with antibodies against HBsAg or HBcAg, respectively, and then subjected to EPA as described in Materials and Methods.
To further confirm these results, we analyzed the effect of the knockdown of Beclin1 or ATG5 on HBV envelopment in pHBV1.3-transfected Huh7 cells. As shown in Fig. 6C and D, while the accumulation of intracellular nucleocapsids was only slightly inhibited in Beclin1 or ATG5 knockdown cells, the abundance of intracellular and extracellular enveloped HBV virions was greatly inhibited and reduced to the same extent (about 80%), confirming that the autophagy machinery is required for HBV envelopment.
It is usually assumed that virion secretion is fast and there are few intracellular virions. To confirm that the signals in Fig. 6A and C (middle) were derived from intracellular virions and not from intracellular nucleocapsids, cell lysates from pHBV1.3-ENV−-transfected Huh7 cells were precipitated with antibodies against HBsAg or HBcAg, respectively, and then subjected to EPA. As shown in Fig. 6E, the immunoprecipitate from antibody against HBsAg showed no signal similar to the one from antibody against HBcAg.
SHBs is associated with autophagosome marker LC3 during HBV replication.
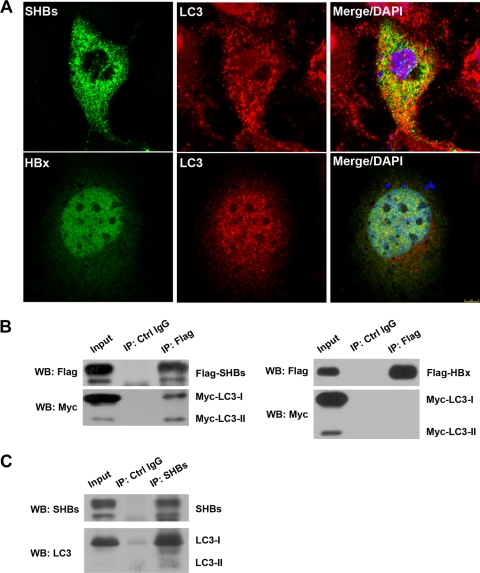
To test whether autophagy is directly involved in HBV envelopment, the relative distributions of SHBs as a key component of virion maturation and autophagosome marker LC3 were investigated by fluorescence microscopy. As shown in Fig. 7A, HBsAg showing a cytoplasmic punctate pattern of immunofluorescence displayed moderate colocalization with LC3, while HBx did not colocalized with LC3.
Fig. 7.
SHBs partially colocalized and coimmunoprecipitated with autophagy protein LC3. (A) Huh7 cells were transfected with pHBV1.3 for 48 h. Cells were fixed, blocked, and incubated with anti-SHBs or anti-HBx antibodies together with anti-LC3 antibody, followed by being stained with Alexa Fluor 488-conjugated anti-mouse secondary antibody and Cy3-conjugated anti-rabbit secondary antibody. The nuclei were stained with DAPI, and the colocalization of SHBs or HBx (green stain) with LC3 (red stain) was observed with a confocal fluorescence microscope. Representative confocal images are shown. (B) Huh7 cells were transfected with pcDNA3.1-Flag-SHBs or pCDNA3.1-Flag-HBx together with pCMV-Myc-LC3 for 48 h. Coimmunoprecipitation analysis was performed with anti-Flag antibody as described in Materials and Methods. The presence of exogenous LC3 in the immunoprecipitate (IP) was detected by Western blot analysis (WB) with anti-Myc antibody. (C) Huh7 cells were transfected with pHBV1.3 for 48 h. Coimmunoprecipitation analysis was performed with anti-SHBs antibody as described in Materials and Methods. The presence of endogenous LC3 in the immunoprecipitate was detected by Western blot analysis with anti-LC3 antibody.
We next determined whether SHBs is physically associated with LC3 by coimmunoprecipitation analysis. In Huh7 cells, exogenous Flag-tagged SHBs was cotransfected with Myc-tagged LC3. Both LC3-I and LC3-II could be precipitated by anti-Flag antibody (Fig. 7B, left). Interestingly, we found that relative to the input protein levels, a significantly larger proportion of LC3-II was captured with SHBs than that of LC3-I (Fig. 7B, left), suggesting a higher affinity of LC3-II for SHBs than that of LC3-I in Huh7 cells. A converse experiment using anti-Myc antibody also revealed the interaction of SHBs with LC3 (data not shown). However, LC3 could not coprecipitate with HBx (Fig. 7B, right). We tested whether the interaction of SHBs with endogenous LC3 could be detected during HBV replication. As shown in Fig. 7C, antibody against HBsAg precipitated both LC3-I and LC3-II, and again a higher proportion of LC3-II than LC3-I in the cell lysate from pHBV1.3-transfected Huh7 cells was immunoprecipitated. In all immunoprecipitation experiments, an IgG control showed negative results for the specific proteins analyzed (Fig. 7B and C). Our results collectively demonstrate that SHBs is associated with autophagosome marker LC3 during HBV replication.
DISCUSSION
A growing number of viruses have been shown to affect autophagy (11, 31, 40). In this study, we show that HBV induced an incomplete autophagic process in hepatoma cells without promoting protein degradation by lysosomes (Fig. 1), which is in agreement with a recent report (76). It was reported that human immunodeficiency virus (HIV) induces the formation of autophagosomes in macrophages, while HIV Nef protein blocks the maturation of early autophagic organelles into autolysosomes through interaction with the autophagy regulatory protein Beclin1, thus avoiding autophagic protein degradation and protecting HIV from degradation (42). Therefore, it is possible that HBV also developed such a strategy to perturb the maturation of autophagic vacuoles. Autophagy generally has been considered a nonselective process. However, recent studies indicate that, in addition to targeting proteins for degradation by the 26S proteasome, ubiquitin also represents a selective degradation signal and directs protein aggregates, membrane-bound organelles, and microbes to autophagosomes (30, 32, 37, 38, 87). Therefore, it also is likely that HBV cannot induce the labeling of cargo and hence does not enhance autophagic protein degradation.
A recent report showed that HBx is required for HBV-induced autophagy (76). However, our observation that the deletion of SHBs abrogated the HBV-induced autophagy while the deletion of HBx did not, suggests that the enhancement of the autophagic response by HBV was dependent on SHBs but not on HBx in our system (Fig. 2). We indeed confirmed that the overexpression of SHBs alone was sufficient to induce autophagy (Fig. 2). The reason for this discrepancy is unknown. One possibility is the use of a different virus isolate. Both point mutations and deletion mutants of the X gene have been observed in HCC and in cirrhotic livers with chronic HBV infection (8, 101). HBx variants may display different distribution patterns, expression levels, and functions (41, 51). Consistently with this point, an initial report on the effect of HBV on autophagy suggested that HBx itself is not sufficient to induce autophagy but can only enhance starvation-induced autophagy (79). In fact, we observed that the overexpression of HBx induced the lipidation of LC3-II in our system (data not shown). Notably, the ability of SHBs to mediate HBV-induced autophagy is not limited to one virus isolate; a similar result was observed for another HBV isolate, pHBV3.8 (data not shown). In addition, HIV also utilizes envelope proteins to induce autophagy (10, 16, 17). Similarly, poliovirus induces autophagosome-like vesicles via its ER membrane-associated viral proteins (78). In addition, it also is possible that in different situations the virus utilizes different strategies to induce autophagy. For example, when HBx is not enough to induce autophagy for its low expression or other reasons, HBV might trigger autophagy via SHBs. Nevertheless, our results show that SHBs is required for HBV to induce autophagy, at least in our system. The relationship between SHBs and HBx in the induction of autophagy deserves further study.
The process of autophagosome formation is tightly regulated. A multitude of nutritional and stress inputs transduced through protein and lipid kinase signaling cascades that regulate autophagy converge upon two key signaling nodes, mammalian target of rapamycin (mTOR) and the Beclin1/hVps34 complex (11, 24, 61). We found that the overexpression of SHBs did not change the phosphorylation levels of the mTOR substrates S6K1 and 4EBP1 (data not shown), suggesting that mTOR signaling is not involved in HBV-induced autophagy. While HBx was reported to enhance starvation-induced autophagy by the upregulation of Beclin1 expression (79), the overexpression of SHBs did not increase Beclin1 expression (data not shown).
Recently, an increasing body of research has shown that ER stress can induce autophagy (25, 39, 45, 48, 58, 66, 69, 97, 98). As a processing plant for the folding and posttranslational modification of proteins, the ER is an essential organelle for viral replication and maturation. In the course of virus productive infection, a large amount of viral proteins is synthesized in infected cells, where unfolded or misfolded proteins result in ER stress (23). The accumulation of viral envelope proteins in the ER has been implicated as the trigger of the UPR in several virus infections (6, 7, 12, 52, 84). For HBV, there are three envelopment proteins. Among them, SHBs is the most abundantly produced and may have a greater potential to induce ER stress. A previous study showed that SHBs can greatly increase the expression of binding immunoglobulin protein (BiP), a central regulator for ER stress (68), and was suggested to induce ER stress (26). In this study, we extended this finding and found that these three signaling pathways of PERK, ATF6, and IRE1, downstream of BiP, were activated by SHBs, although to a different extent (Fig. 3). The blockage of any of these three UPR signaling pathways almost abrogated the SHBs-induced lipidation of LC3-I (Fig. 4). Similarly, a recent report showed that hepatitis C virus (HCV) triggers autophagosome formation by inducing ER stress and that all three of these UPR signaling pathways are required for the induction of autophagy (75). These observations suggested that the target genes induced by these three different UPR signaling pathways must work in concert to induce autophagy. In addition, LHBs with pre-S mutations was reported to inhibit the syntheses of SHBs and MHBs, with the accumulation of LHBs in the ER, and to induce ER stress (85), so it is possible that in this situation LHBs also induce autophagy via ER stress.
Although the autophagy pathway is emerging as a component of host defense, certain viruses have developed strategies to counteract these antiviral mechanisms, and others appear to have coopted the autophagy machinery as proviral host factors favoring viral replication (13). A recent study showed that the inhibition of the autophagy pathway by 3-MA or by hVps34 siRNA duplex or ATG7 siRNA duplex suppressed HBV DNA replication (76). In this study, we also showed that the inhibition of autophagy by treatment with 3-MA or the knockdown of Beclin1 or ATG5 decreased HBV DNA replication, but it did so to a lesser extent (Fig. 5). This discrepancy might arise as a consequence of the different method used to block autophagic processes or the different cell lines used. Another possibility is that the autophagy-independent functions of 3-MA and autophagy-related genes, which have been shown previously (9, 18, 34, 99), could be involved in the HBV life cycle. Importantly, we demonstrated that the inhibition of autophagy had a significant inhibitory effect on viral envelopment (Fig. 6), which resulted in a marked reduction of enveloped virion levels in the culture supernatant (Fig. 5 and 6). Recently, autophagy was shown to contribute to the effective production of HCV particles with little effect on the intracellular production of HCV mRNA and HCV-related proteins (80), but whether autophagy is involved in viral assembly, release, or both still is unclear. Nevertheless, to our knowledge, our report is the first demonstration that the autophagy process is involved in viral envelopment.
HBV envelopment is believed to occur at post-ER/pre-Golgi membranes, where cytosolic nucleocapsids are packaged inside a lipid envelope integrated with viral envelope proteins (5, 27, 62, 71). The mechanism by which nucleocapsids receive their envelope has not been not resolved. Recently, a model that suggests an important role for ER, Golgi, and their adjacent specialized compartments in the formation of the autophagosomal membranes has been proposed (2, 20, 22, 74, 82, 102). In addition, it was suggested that the autophagic processes induced by ER stress are aimed at disposing of unfolded or misfolded proteins aggregated in the ER lumen for ER quality control (83, 96), although it seems that HBV has developed some strategies to block this degradation (Fig. 1G). Interestingly, we found that SHBs, a key component of virion maturation, is partially associated with the autophagosome marker LC3 during HBV replication (Fig. 7). Therefore, the autophagosomes or the possible intermediates between autophagosomes and ER or Golgi might provide a physical scaffold for HBV envelopment. Moreover, it also is possible that autophagosome membranes could be used as a source of membranes for viral envelopment, although the envelope of HBV is poorly characterized. Finally, it is tempting to speculate that the autophagy pathway enhances HBV envelopment by sequestering the restriction factor(s) of HBV envelopment.
In summary, our study shows that HBV induced an incomplete autophagic process in hepatoma cells. Furthermore, the induction of autophagy requires SHBs and depends on the induction of ER stress. Moreover, we demonstrate that autophagy machinery is required for HBV envelopment. Finally, our results show that SHBs is associated with autophagosome marker LC3. The elucidation of how HBV manipulates autophagy to enhance replication might ultimately lead to the development of new therapeutics for acute and chronic HBV infection.
ACKNOWLEDGMENTS
This work was supported by a Chinese State Basic Research Foundation grant (2009CB522504), the National Natural Science Fund for distinguished scholars (30425041), the National Megaprojects for Infectious Diseases (2008ZX10203), the Program for Outstanding Medical Academic Leader of Shanghai, and the China Postdoctoral Science Foundation (20100480031).
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Alexander D. E., Ward S. L., Mizushima N., Levine B., Leib D. A. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 81:12128–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Axe E. L., et al. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barth S., Glick D., Macleod K. F. 2010. Autophagy: assays and artifacts. J. Pathol. 221:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bjørkøy G., et al. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruss V. 2004. Envelopment of the hepatitis B virus nucleocapsid. Virus Res. 106:199–209 [DOI] [PubMed] [Google Scholar]
- 6. Chan S. W., Egan P. A. 2009. Effects of hepatitis C virus envelope glycoprotein unfolded protein response activation on translation and transcription. Arch. Virol. 154:1631–1640 [DOI] [PubMed] [Google Scholar]
- 7. Chan S. W., Egan P. A. 2005. Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J. 19:1510–1512 [DOI] [PubMed] [Google Scholar]
- 8. Chen G. G., et al. 2005. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J. Clin. Virol. 34:7–12 [DOI] [PubMed] [Google Scholar]
- 9. de Haan C. A., Molinari M., Reggiori F. 2010. Autophagy-independent LC3 function in vesicular traffic. Autophagy 6:994–996 [DOI] [PubMed] [Google Scholar]
- 10. Denizot M., et al. 2008. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy 4:998–1008 [DOI] [PubMed] [Google Scholar]
- 11. Deretic V., Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5:527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dimcheff D. E., Faasse M. A., McAtee F. J., Portis J. L. 2004. Endoplasmic reticulum (ER) stress induced by a neurovirulent mouse retrovirus is associated with prolonged BiP binding and retention of a viral protein in the ER. J. Biol. Chem. 279:33782–33790 [DOI] [PubMed] [Google Scholar]
- 13. Dreux M., Chisari F. V. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295–1307 [DOI] [PubMed] [Google Scholar]
- 14. Dreux M., Gastaminza P., Wieland S. F., Chisari F. V. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eletto D., Dersh D., Argon Y. 2010. GRP94 in ER quality control and stress responses. Semin. Cell Dev. Biol. 21:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Espert L., et al. 2006. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Investig. 116:2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Espert L., et al. 2009. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection. PLoS One 4:e5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujiki Y., Yoshimoto K., Ohsumi Y. 2007. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143:1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gastaminza P., Kapadia S. B., Chisari F. V. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geng J., Klionsky D. J. 2010. The Golgi as a potential membrane source for autophagy. Autophagy 6:950–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghany M. G., Doo E. C. 2009. Antiviral resistance and hepatitis B therapy. Hepatology 49:S174–S184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayashi-Nishino M., et al. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 [DOI] [PubMed] [Google Scholar]
- 23. He B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 13:393–403 [DOI] [PubMed] [Google Scholar]
- 24. He C., Klionsky D. J. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43:67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Høyer-Hansen M., Jaattela M. 2007. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 14:1576–1582 [DOI] [PubMed] [Google Scholar]
- 26. Huang I. C., Chien C. Y., Huang C. R., Lo S. J. 2006. Induction of hepatitis D virus large antigen translocation to the cytoplasm by hepatitis B virus surface antigens correlates with endoplasmic reticulum stress and NF-kappaB activation. J. Gen. Virol. 87:1715–1723 [DOI] [PubMed] [Google Scholar]
- 27. Huovila A. P., Eder A. M., Fuller S. D. 1992. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 118:1305–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jackson W. T., et al. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kabeya Y., et al. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim P. K., Hailey D. W., Mullen R. T., Lippincott-Schwartz J. 2008. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U. S. A. 105:20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kirkegaard K. 2009. Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 335:323–333 [DOI] [PubMed] [Google Scholar]
- 32. Kirkin V., McEwan D. G., Novak I., Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol. Cell 34:259–269 [DOI] [PubMed] [Google Scholar]
- 33. Klionsky D. J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 [DOI] [PubMed] [Google Scholar]
- 34. Klionsky D. J., et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klionsky D. J., et al. 2010. A comprehensive glossary of autophagy-related molecules and processes. Autophagy 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klionsky D. J., Cuervo A. M., Seglen P. O. 2007. Methods for monitoring autophagy from yeast to human. Autophagy 3:181–206 [DOI] [PubMed] [Google Scholar]
- 37. Komatsu M., Ichimura Y. 2010. Selective autophagy regulates various cellular functions. Genes Cells 15:923–933 [DOI] [PubMed] [Google Scholar]
- 38. Korolchuk V. I., Menzies F. M., Rubinsztein D. C. 2009. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 5:862–863 [DOI] [PubMed] [Google Scholar]
- 39. Kouroku Y., et al. 2007. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14:230–239 [DOI] [PubMed] [Google Scholar]
- 40. Kudchodkar S. B., Levine B. 2009. Viruses and autophagy. Rev. Med. Virol. 19:359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwun H. J., Jang K. L. 2004. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Res. 32:2202–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kyei G. B., et al. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186:255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee H. K., Iwasaki A. 2008. Autophagy and antiviral immunity. Curr. Opin. Immunol. 20:23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee Y. R., et al. 2008. Autophagic machinery activated by dengue virus enhances virus replication. Virology 374:240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lepine S., et al. 2011. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 18:350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li J., et al. 2010. Inhibition of hepatitis B virus replication by MyD88 involves accelerated degradation of pregenomic RNA and nuclear retention of pre-S/S RNAs. J. Virol. 84:6387–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li J., et al. 2008. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 15:1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liang T. J. 2009. Hepatitis B: the virus and disease. Hepatology 49:S13–S21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin S., et al. 2007. Inhibition of hepatitis B virus replication by MyD88 is mediated by nuclear factor-kappaB activation. Biochim. Biophys. Acta 1772:1150–1157 [DOI] [PubMed] [Google Scholar]
- 51. Lin X., et al. 2005. Biological impacts of “hot-spot” mutations of hepatitis B virus X proteins are genotype B and C differentiated. World J. Gastroenterol. 11:4703–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu N., et al. 2004. Possible involvement of both endoplasmic reticulum- and mitochondria-dependent pathways in MoMuLV-ts1-induced apoptosis in astrocytes. J. Neurovirol. 10:189–198 [DOI] [PubMed] [Google Scholar]
- 53. Mangold C. M., Streeck R. E. 1993. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J. Virol. 67:4588–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McMahon B. J. 2005. Epidemiology and natural history of hepatitis B. Semin. Liver Dis. 25(Suppl. 1):3–8 [DOI] [PubMed] [Google Scholar]
- 55. Mizushima N. 2004. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 36:2491–2502 [DOI] [PubMed] [Google Scholar]
- 56. Mizushima N., Yoshimori T., Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nguyen D. H., Ludgate L., Hu J. 2008. Hepatitis B virus-cell interactions and pathogenesis. J. Cell. Physiol. 216:289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ogata M., et al. 2006. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26:9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pankiv S., et al. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 [DOI] [PubMed] [Google Scholar]
- 60. Papatheodoridis G. V., Manolakopoulos S., Dusheiko G., Archimandritis A. J. 2008. Therapeutic strategies in the management of patients with chronic hepatitis B virus infection. Lancet Infect. Dis. 8:167–178 [DOI] [PubMed] [Google Scholar]
- 61. Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. 2008. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 90:313–323 [DOI] [PubMed] [Google Scholar]
- 62. Patzer E. J., Nakamura G. R., Simonsen C. C., Levinson A. D., Brands R. 1986. Intracellular assembly and packaging of hepatitis B surface antigen particles occur in the endoplasmic reticulum. J. Virol. 58:884–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peng L., Liang D., Tong W., Li J., Yuan Z. 2010. Hepatitis C virus NS5A activates the mammalian target of rapamycin (mTOR) pathway, contributing to cell survival by disrupting the interaction between FK506-binding protein 38 (FKBP38) and mTOR. J. Biol. Chem. 285:20870–20881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Perrillo R. 2009. Benefits and risks of interferon therapy for hepatitis B. Hepatology 49:S103–S111 [DOI] [PubMed] [Google Scholar]
- 65. Pol S., Michel M. L. 2006. Therapeutic vaccination in chronic hepatitis B virus carriers. Expert Rev. Vaccines 5:707–716 [DOI] [PubMed] [Google Scholar]
- 66. Qin L., Wang Z., Tao L., Wang Y. 2010. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6:239–247 [DOI] [PubMed] [Google Scholar]
- 67. Roggendorf M., Schulte I., Xu Y., Lu M. 2007. Therapeutic vaccination in chronic hepatitis B: preclinical studies in the woodchuck model. J. Viral Hepat. 14(Suppl. 1):51–57 [DOI] [PubMed] [Google Scholar]
- 68. Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529 [DOI] [PubMed] [Google Scholar]
- 69. Sakaki K., Wu J., Kaufman R. J. 2008. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J. Biol. Chem. 283:15370–15380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Seeger C., Mason W. S. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Seitz S., Urban S., Antoni C., Bottcher B. 2007. Cryo-electron microscopy of hepatitis B virions reveals variability in envelope capsid interactions. EMBO J. 26:4160–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shang J., Lehrman M. A. 2004. Discordance of UPR signaling by ATF6 and Ire1p-XBP1 with levels of target transcripts. Biochem. Biophys. Res. Commun. 317:390–396 [DOI] [PubMed] [Google Scholar]
- 73. Shi C. S., Kehrl J. H. 2008. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283:33175–33182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Simonsen A., Stenmark H. 2008. Self-eating from an ER-associated cup. J. Cell Biol. 182:621–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sir D., et al. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sir D., et al. 2010. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. U. S. A. 107:4383–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stevens C. E., et al. 1985. Perinatal hepatitis B virus transmission in the United States. Prevention by passive-active immunization. JAMA 253:1740–1745 [PubMed] [Google Scholar]
- 78. Suhy D. A., Giddings T. H., Jr., Kirkegaard K. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tang H., et al. 2009. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology 49:60–71 [DOI] [PubMed] [Google Scholar]
- 80. Tanida I., et al. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945 [DOI] [PubMed] [Google Scholar]
- 81. Taylor M. P., Kirkegaard K. 2007. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 81:12543–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tooze S. A., Yoshimori T. 2010. The origin of the autophagosomal membrane. Nat. Cell Biol. 12:831–835 [DOI] [PubMed] [Google Scholar]
- 83. Verfaillie T., Salazar M., Velasco G., Agostinis P. 2010. Linking ER stress to autophagy: potential implications for cancer therapy. Int. J. Cell Biol. 2010:930509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. von dem Bussche A., et al. 2010. Hepatitis C virus NS2 protein triggers endoplasmic reticulum stress and suppresses its own viral replication. J. Hepatol. 53:797–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang H. C., et al. 2003. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am. J. Pathol. 163:2441–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Watanabe T., et al. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. U. S. A. 104:10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wild P., Dikic I. 2010. Mitochondria get a Parkin' ticket. Nat. Cell Biol. 12:104–106 [DOI] [PubMed] [Google Scholar]
- 88. Wong J., et al. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Xie Z., Klionsky D. J. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 [DOI] [PubMed] [Google Scholar]
- 90. Xiong W., et al. 2004. Interferon-inducible MyD88 protein inhibits hepatitis B virus replication. Virology 319:306–314 [DOI] [PubMed] [Google Scholar]
- 91. Xu J., et al. 2010. Hepatitis B virus X protein blunts senescence-like growth arrest of human hepatocellular carcinoma by reducing Notch1 cleavage. Hepatology 52:142–154 [DOI] [PubMed] [Google Scholar]
- 92. Xu Y., et al. 2009. HBsAg inhibits TLR9-mediated activation and IFN-alpha production in plasmacytoid dendritic cells. Mol. Immunol. 46:2640–2646 [DOI] [PubMed] [Google Scholar]
- 93. Xu Z., Ou J. H. 2004. Endogenous polymerase assay for the analysis of hepatitis B virus in transgenic mice. Methods Mol. Med. 95:295–302 [DOI] [PubMed] [Google Scholar]
- 94. Yokosuka O., Arai M. 2006. Molecular biology of hepatitis B virus: effect of nucleotide substitutions on the clinical features of chronic hepatitis B. Med. Mol. Morphol. 39:113–120 [DOI] [PubMed] [Google Scholar]
- 95. Yoon S. Y., et al. 2008. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J. Virol. 82:11976–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yorimitsu T., Klionsky D. J. 2007. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 17:279–285 [DOI] [PubMed] [Google Scholar]
- 97. Yorimitsu T., Klionsky D. J. 2007. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy 3:160–162 [DOI] [PubMed] [Google Scholar]
- 98. Yorimitsu T., Nair U., Yang Z., Klionsky D. J. 2006. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281:30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhao Z., et al. 2008. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4:458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhou Z., et al. 2009. Autophagy is involved in influenza A virus replication. Autophagy 5:321–328 [DOI] [PubMed] [Google Scholar]
- 101. Zhu P., Tan D., Peng Z., Liu F., Song L. 2007. Polymorphism analyses of hepatitis B virus X gene in hepatocellular carcinoma patients from southern China. Acta Biochim. Biophys. Sin. (Shanghai) 39:265–272 [DOI] [PubMed] [Google Scholar]
- 102. Zoppino F. C., Militello R. D., Slavin I., Alvarez C., Colombo M. I. 2010. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11:1246–1261 [DOI] [PubMed] [Google Scholar]