Abstract
The induction of type I interferons (alpha/beta interferon [IFN-α/β]) in response to viral infection is a crucial step leading to the antiviral state in the host. Viruses produce double-stranded RNA (dsDNA) during their replication cycle that is sensed as nonself by host cells through different receptors. A signaling cascade then is activated to block viral replication and spread. Foot-and-mouth disease virus (FMDV) is a picornavirus that is highly sensitive to IFN, and it causes one of the world's most important animal diseases. In this study, we showed the ability of structural domains predicted to enclose stable dsRNA regions in the 5′- and 3′-noncoding regions (NCRs) of the FMDV genome to trigger an IFN-α/β response in porcine kidney cultured cells and newborn mice. These RNAs, generated by in vitro transcription, were able to stimulate IFN-β transcription and induce an antiviral state in SK-6 cells. The induction levels elicited by the different NCR RNAs were compared. Among them, the 3′NCR was identified as a potent IFN activator, and the features in this region involved in signaling have been analyzed. To address whether the FMDV NCR transcripts were able to trigger the innate immune response in vivo, Swiss suckling mice were inoculated intraperitoneally with the RNAs. All transcripts induced the innate response in transfected animals, measured as IFN-α/β protein levels, antiviral activity in sera, and reduced susceptibility to FMDV infection. Our work provides new insight into innate responses against FMDV and identifies these small noninfectious RNA molecules as potential adjuvants for vaccine improvement and antiviral strategies against picornaviruses.
INTRODUCTION
The innate immune response is a first line of defense against invading pathogens, and it depends on several sensors and signaling pathways. The detection of viral products as pathogen-associated molecular patterns (PAMPs) initiates a signaling cascade that leads to a rapid antiviral response involving the secretion of type I IFNs (IFN-α and IFN-β) that have antiviral, antiproliferative, and immunomodulatory functions. PAMPs presented during viral infections include single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA). dsRNA is generated in infected cells as genomic fragments, replicative intermediate, or by stem-loop structures and is recognized by viral sensors. Toll-like receptors (TLRs), expressed on the surface and endosomal compartments of some cell types, and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), ubiquitous cytosolic RNA helicases, are the two major systems for virus detection (5, 21, 49, 51, 52). The RLRs RIG-I and melanoma differentiation antigen 5 (MDA-5) play critical roles in triggering immune defenses against RNA virus infection (22). Recently, class A scavenger receptors (SR-As) have been shown to recognize extracellular dsRNA and mediate its entry and delivery to intracellular sensors (12).
Foot-and-mouth disease virus (FMDV) is a member of the Picornaviridae family and the causative agent of an acute vesicular disease affecting pigs, ruminants, and other cloven-hoofed livestock (23, 39). The viral genome consists of a single-stranded positive-sense RNA molecule of about 8.5 kb in length with the 5′ end covalently linked to the viral protein VPg and a poly(A) tract at the 3′ end. The viral products are translated from a single open reading frame, which is flanked by two noncoding regions (NCRs) containing specific structures involved in the control of the replication and translation of the viral genome (4). The 5′-terminal S fragment is a 360-nucleotide (nt)-long region predicted to form a stable long hairpin structure (13, 50). The S region is followed by a heteropolymeric poly(C) tract, several pseudoknots, the cis-acting replication element (cre) (25), and the internal ribosome entry site (IRES), a complex multidomain structure about 450 nt long, mediating the cap-independent translation of the viral RNA through the recruitment of cellular translation initiation factors and other trans-acting factors (3, 14). The 3′NCR is a 90-nt-long region predicted to fold into two well-defined stem-loop structures highly conserved among isolates (43). The deletion of the complete 3′NCR or replacement for its swine vesicular disease virus (SVDV) counterpart on an FMDV infectious clone was deleterious for viral replication (38). Further studies showed that while proximal SL1 was dispensable for infectivity, acting as a replication enhancer, the deletion of distal SL2 completely abrogated replication (35). The FMDV 3′NCR exerts a stimulatory effect on IRES-dependent translation (24) and is able to interact with the IRES and S regions located at the 5′ end through direct RNA-RNA interactions (43). Cellular proteins binding to the 3′NCR also have been described (24, 34). Some of these factors are able to interact with the S region and undergo proteolytic cleavage upon FMDV infection (34).
FMDV is very sensitive to alpha, beta, and gamma interferon (IFN-α/β/γ but has evolved mechanisms to evade those innate responses (18, 19). The leader protease Lpro is the first viral protein to be translated and has been identified as a virulence factor that blocks the host immune innate response. Lpro also blocks host cell translation by the cleavage of eIF-4G (11) and inhibits the induction of IFN-β mRNA and the expression of IFN-α/β-stimulated genes in swine cells (8). This inhibitory effect was further associated with Lpro translocation to the nucleus and the degradation of p65/RelA, a subunit of NF-κB (9). A recent report describes the suppression of dsRNA-induced IFN-β induction through the degradation of interferon regulatory factor 3/7 by Lpro (48).
RNA motifs in the NCRs of flaviviruses have been reported as type I IFN inducers when transfected into cultured cells and mice (37, 46).
Here, we analyzed the ability of defined structural domains in the NCRs of the viral RNA to induce an immune innate response in swine cells and suckling mice. Our studies suggest the presence of PAMP motifs in the viral genome and define the FMDV 3′NCR as a potent IFN-β activator in porcine cells. Transfection with the S or IRES region also elicited an antiviral state in transfected cells. The role of the secondary structure for 3′NCR signaling has been demonstrated for both FMDV and SVDV, another swine picornavirus. Moreover, RNA transcripts corresponding to the FMDV S, IRES, and 3′NCR regions all were able to trigger the innate immune response when inoculated into suckling mice and remarkably reduced their susceptibility to subsequent FMDV infection. Further studies based on these small noninfectious molecules might lead to the development of new antiviral strategies and vaccine improvement.
MATERIALS AND METHODS
Cells and viruses.
Porcine kidney cell lines IBRS-2 and SK-6 were obtained from the Centro de Investigación en Sanidad Animal (CISA-INIA), Valdeolmos, Spain. Murine L-929 cells were obtained from Antonio Alcamí's laboratory in Centro de Biología Molecular Severo Ochoa, Madrid, Spain. All three cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, penicillin-streptomycin (pen-strep), and l-glutamine (l-glu). Foot-and-mouth disease virus (FMDV) O1K, swine vesicular disease virus (SVDV) SPA 93, and vesicular stomatitis virus (VSV) Indiana were used for infection experiments in IBRS-2, SK-6, or L-929 cells.
Preparation of RNAs and transfections.
RNA transcripts corresponding to the S fragment, 3′NCR, and its derivatives SL1 (3′NCRΔSL2), SL2 (3′NCRΔSL1), and ΔAn of the FMDV O1K genome, as well as the 3′NCR of SVDV, were generated by in vitro transcription with T3 RNA polymerase (NEB) from previously described plasmids that were linearized with NotI (43). RNA corresponding to the IRES of FMDV C-S8 was generated from a pGEM-derived clone (31), a gift of E. Martínez-Salas, after linearization with XhoI and in vitro transcription with T7 RNA polymerase (NEB). FMDV full-length transcripts assayed were derived from cDNA clones pO1K VP3-R56 and pO1K VP3-H56 (35) after linearization with HpaI (NEB) and in vitro transcription with SP6 RNA polymerase (Promega). A derivative of the pO1K clone bearing a 3′-terminal deletion spanning the coding sequence for the C terminus of 3Dpol and the entire 3′NCR (nt 7304 to 8167; EMBL X00871), but maintaining poly(A), was constructed by the excision of the 864 bp-CelII/EcoRV fragment from the pDM construct (38). RNA corresponding to the complete 5′NCR enclosing the S fragment, IRES, and 212 nt of the Lpro coding sequence was synthesized with SP6 RNA polymerase using pO1K clone linearized with AccI as the template. All viral transcripts contain 5′ppp, and FMDV- and SVDV-3′NCR and derivative RNAs carry a 58-nt poly(A) tract unless specified otherwise.
After transcription, RNAs were treated with RQ1 DNase (1 U/μg; Promega), extracted with phenol-chloroform, and precipitated with ethanol. All transcripts were resuspended in water and quantified by spectrometry. The RNA integrity and size were analyzed in denaturing 6% acrylamide, 7 M urea gel electrophoresis (15), or agarose gels.
The removal of 5′ phosphates from FMDV 3′NCR transcripts was performed by incubation with 0.5 U/μg of calf intestinal alkaline phosphatase (CIP; NEB) for 90 min. After CIP treatment, RNAs were phenol extracted twice, and ethanol was precipitated and treated as described above. The RNA integrity of CIP-treated transcripts was tested on denaturing 6% polyacrylamide, 7 M urea gels.
Prior to transfection, RNAs were heated at 92°C for 5 min, cooled down to room temperature for 10 min, and then chilled on ice. In some experiments aimed to address the effect of RNA structure on IFN induction, transcripts were heated as described above and then treated as described in reference 31, with modifications. Briefly, RNAs were chilled on ice before a 5-fold-concentrated buffer was added to a final concentration of either (i) 10 mM Tris, pH 7.8, 1.5 mM MgCl2, 300 mM KCl to preserve the native RNA structure (transcripts then were renatured at room temperature for 15 min) or (ii) 10 mM Tris, pH 7.8, 0.5 mM EDTA to keep RNAs denatured (in this case, transcripts were kept on ice until transfection).
SK-6 cells were transfected using 20 μg/ml of each RNA with Lipofectin (Invitrogen) or 10 μg/ml of poly(I:C) (Sigma) using Lipofectamine (Invitrogen).
IFN-β expression analysis.
At different times of transfection, cells were lysed and RNA extracted and quantified as described previously (35). Reverse transcription (RT) was performed using 500 ng of total RNA and Transcriptor RT (Roche). Quantitative PCR was carried out using aliquots of the RT reactions (1/10) and LightCycler FastStart DNA master SYBR green I (Roche). All reactions were conducted in triplicate. Data were analyzed using the ΔΔCT method. In all assays, IFN-β gene expression was normalized to that of the housekeeping gene GAPDH and was expressed as the fold increase above the level of mock-transfected cells. The following primers were used to amplify porcine IFN-β: 5′-CTTTGAGGTCCCTGAGGAGATTATGCAACC-3′ and 5′-AGGCACAGCTTCTGTACTCCTTGG-3′. GAPDH amplification was performed using primers previously described (16).
IFN bioassay.
The antiviral activity of the supernatants from transfected SK-6 cells was determined by a VSV infection inhibition assay on IBRS-2 cells. Briefly, SK-6 cells were transfected for 24 h with 40 μg/ml of FMDV or SVDV NCR transcripts or with 10 μg of poly(I:C) or tRNA. IBRS-2 cells were incubated with different dilutions of transfection supernatants for 24 h, washed, and infected with ∼100 PFU of VSV for plaque assay. Plaques were counted 24 h postinfection. In some cases, the transfection supernatants were incubated previously for 1 h at 37°C with 1 μg of specific neutralizing monoclonal antibodies against swine IFN-α (K9; purchased from PBL InterferonSource) or IFN-β (26), a gift of A. Garmendia, University of Connecticut. No inhibitory effect on VSV infectivity was observed for MAb amounts of up to 10 μg. Antiviral activity was expressed as the highest supernatant dilution needed to reduce the number of plaques by 50%.
Mouse experiments.
Litters of Swiss mice (3 to 7 days old) were intraperitoneally (i.p.) injected with 100 μl containing 100 μg of 3′NCR, S, and IRES transcripts or poly(I:C) in phosphate-buffered saline (PBS). In the case of RNA transcripts, 20 μg of Lipofectine (Invitrogen) also was added. Serum samples were collected at 0, 4, 8, 24, and 48 h postinoculation, and sera from four to five mice were pooled for each RNA and kept as aliquots at −80°C until used. IFN-α and IFN-β in sera were measured using enzyme-linked immunosorbent assay (ELISA) kits from PBL InterferonSource.
To test the antiviral activity of pooled sera from the transfected mice, a cytopathic-effect inhibition assay was performed (36). Briefly, monolayers of L-929 cells in 96-well microtiter plates were incubated with serial 2-fold dilutions of the corresponding sera for 24 h. Media then were removed and replaced with 100 μl of fresh media containing 100 50% tissue culture infectious doses (TCID50) of VSV. Cytopathic effect at 72 h postinfection was monitored by microscopic examination. Antiviral activity was expressed as the reciprocal of the highest serum dilution protecting cells from cytotoxicity in 50% of the wells.
To test the protective effect of transfection with the FMDV NCRs against viral infection, litters of Swiss newborn mice (10 to 12 animals) were transfected with the FMDV transcripts or poly(I:C) as described above, and 24 h later they were inoculated i.p. with 7 × 102 or 7 × 104 PFU of FMDV O1K in a final volume of 100 μl in PBS. Dead animals were scored up to 11 days after infection. Mice showing severe signs of disease (tremors ataxia, paralysis of the hind limbs, etc.) were euthanized.
All animals in this study were handled in strict accordance with the guidelines of the European Community 86/609/CEE. The protocol was approved by the Committee on the Ethics of Animal Experiments of INIA (permit number CBS 2008/016).
RESULTS
FMDV NCRs encode PAMP motifs that trigger innate immune signaling in the host cell.
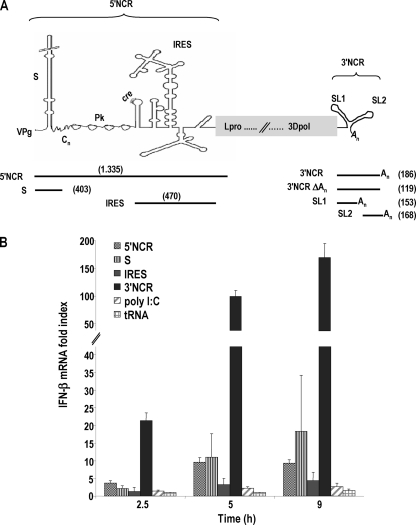
We initially analyzed the eliciting of the IFN-β expression in porcine cells of highly structured domains in the 5′- and 3′-terminal NCRs of FMDV RNA, which are known to play relevant roles in the replication and translation of the viral genome. RNA transcripts corresponding to the 5′- and 3′NCRs, as well as the S fragment and IRES independently, were generated by in vitro transcription and used to transfect SK-6 cells (Fig. 1A). Swine kidney SK-6 cells, unlike hamster BHK-21 or swine IBRS-2 cell lines, are known to have an active type I IFN system responsive to FMDV infection (6, 8). The expression of IFN-β induced by the NCR RNAs was measured by real-time RT-quantitative PCR (qPCR) at three different time points (Fig. 1B). Remarkable differences were observed among the levels of induction triggered by the different RNAs. At all time points assayed, the 3′NCR transcripts proved to be the most potent IFN-β transcriptional inducers, reaching almost 200-fold increases above the level of mock-transfected cells after 9 h. The S fragment induced a 10-fold increase in IFN-β mRNA transcription, and the entire 5′NCR triggered slightly lower levels of induction. For the RNA transcript corresponding to the FMDV IRES, a maximum of a 4.5-fold induction was detected, which was slightly higher than the level of the dsRNA analogue poly(I:C). These results suggest that the RNA structures composing the 3′NCR are recognized more efficiently than those in the 5′NCR by the cytoplasmic viral sensors expressed in porcine cells. Also, the S RNA hairpin mostly accounts for the induction exerted by the entire 5′NCR, being a stronger inducer when transfected by itself.
Fig. 1.
Structural motifs in the NCRs of FMDV genome trigger IFN-β signaling in porcine cells. (A) Schematic representation of the 5′- and 3′NCR motifs in the FMDV RNA and the corresponding in vitro transcripts. The length of the RNAs is indicated in parentheses. (B) SK-6 cells were transfected with 20 μg/ml of FMDV NCR-derived transcripts or 10 μg/ml of poly(I:C) or tRNA. The fold induction of IFN-β mRNA in RNA-transfected cells above that of mock-transfected cells at the indicated time points was determined by RT-qPCR normalized to GAPDH. Error bars show the standard deviations of the averages from three independent experiments performed in triplicate.
The low levels of induction observed with poly(I:C) are in agreement with several studies reporting it as a very poor IFN-β inducer in pig kidney cells in the range of 10 to 50 μg/ml (1, 47).
Features in the 3′NCR RNA relevant for IFN-β induction in porcine cells.
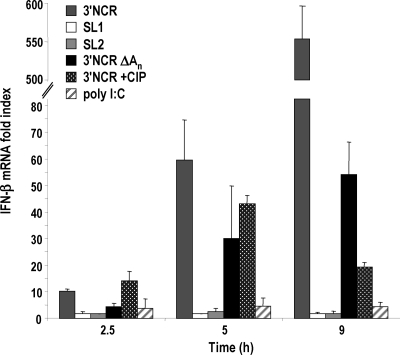
With the aim of gaining knowledge of the requirements in the 3′NCR for the induction of IFN response, the contribution of the two independent stem-loop structures composing the 3′NCR, as well as the poly(A) tail, was analyzed by the transfection of the corresponding transcripts into SK-6 cells as described above (Fig. 1A and 2). Interestingly, neither of the stem-loop structures, SL1 and SL2, was able to induce relevant levels of IFN-β when transfected individually, being minimally active and always below the induction values achieved with poly(I:C). Poly(A) motifs of >50 nt have been reported as determinants that confer efficient RIG-I binding and signaling (37). However, the 3′NCR without the poly(A) was still a potent activator (Fig. 2). The deletion of the poly(A) tail had a less drastic effect on induction than the deletion of each SL, reducing it by 2-fold at 5 h and causing a maximum decrease of 1 log at later times of transfection. The levels of induction shown in Fig. 2 did not correlate with the RNA size or molar ratio, as 3′NCRΔAn was the smallest transcript tested (Fig. 1A).
Fig. 2.
Examining the role of the FMDV 3′NCR sequence composition in IFN-β induction. SL1, SL2, 3′NCRΔAn, as well as CIP-treated or untreated 3′NCR RNAs (20 μg/ml) and poly(I:C) (10 μg/ml) were transfected into SK-6 cells. The fold induction of IFN-β mRNA in transfected cells at the indicated time points was determined as described for Fig. 1. Error bars show standard deviations between triplicates.
As all of the in vitro-transcribed RNAs assayed, showing different levels of potency as IFN-β inducers, had a 5′-triphosphate end, this feature seemed unlikely to be responsible by itself for the high IFN-β induction levels triggered by the 3′NCR. However, we tested the sensitivity of the response to a phosphatase treatment of the 3′NCR RNA (Fig. 2). The dephosphorylation of the 5′ end of the 3′NCR transcript did not completely abrogate IFN-β induction and had a minor effect at early times of transfection. However, after 9 h of transfection, a strong reduction was observed, but a 20-fold induction above the level for mock-transfected cells still was triggered by the dephosphorylated 3′NCR RNA. The integrity of CIP-treated RNA was verified in both cases. These results indicated that a potent IFN induction depended on the presence of not only the 5′ triphosphate but both SLs in the 3′NCR, suggesting a role for the RNA structure of the whole element for the cytoplasmic recognition of FMDV 3′NCR as a PAMP.
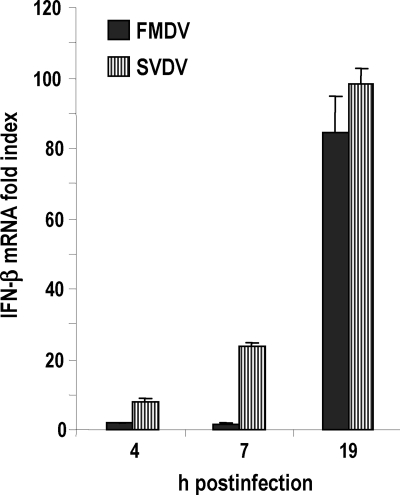
To assess whether the presence of secondary structures in the 3′NCR was relevant for IFN induction, prior to transfection the RNAs were heated and then treated either to promote the stable refolding of the structures or to keep the RNA denatured (see Materials and Methods). In these experiments, the 3′NCR of a different swine picornavirus, SVDV, was included to analyze the ability of an analogous element to induce IFN-β in porcine cells. The 100-nt-long 3′NCR of SVDV is different in sequence and secondary structure from its FMDV counterpart. It is predicted to fold into three stem-loops, with two of them interacting to form a pseudoknot (30). Consistently with that, in previous work we showed the remarkable tendency of the SVDV 3′NCR to form dimers in RNA-RNA interaction experiments, suggesting stable intermolecular interactions (43). We first compared the levels of IFN-β induced by FMDV and SVDV at different times postinfection in SK-6 cells. As shown in Fig. 3, both picornaviruses were able to efficiently trigger IFN-β transcription in porcine cells, as expected. At all of the time points assayed, SVDV infection elicited higher levels of induction, and this difference was more evident at early times postinfection.
Fig. 3.
Comparative analysis of the levels of induction of IFN-β signaling by FMDV and SVDV infections. SK-6 cells were infected with FMDV or SVDV at an MOI of 1. Cellular RNA was harvested at the indicated times postinfection, and the fold induction of IFN-β mRNA in virus-infected cells above that of mock-infected cells was determined by RT-qPCR normalized to GAPDH. Error bars show standard deviations between triplicates.
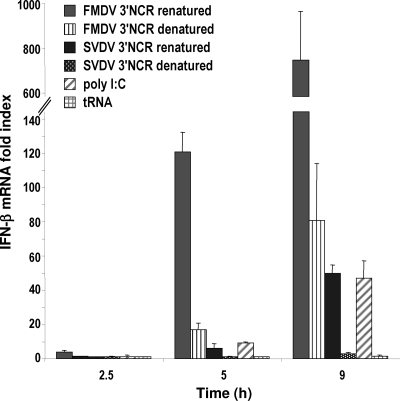
When transfected into SK-6 cells, the SVDV 3′NCR proved to be a potent IFN-β inducer as well, eliciting in all cases levels up to 20-fold lower than that of the FMDV 3′NCR (Fig. 4). The denaturing of both RNAs had a strong effect on IFN-β induction even at early times of transfection. A decrease of 7- to 9-fold was observed for FMDV 3′NCR, while a larger decrease was found for SVDV, reaching 17-fold at 9 h of transfection (Fig. 4). In this experiment, higher levels of IFN-β induction than average were observed in cells transfected with poly(I:C) or FMDV 3′NCR, while tRNA was unable to elicit detectable induction, indicating a potent induction mediated by exogenous RNA. The remarkable differences found in the ability to induce IFN-β between the folded and denatured forms of the swine picornaviral 3′NCRs strongly suggest that secondary, and likely higher-order, structures present in those RNAs enhance their recognition as PAMPs by the cytoplasmic viral sensors of porcine cells and then trigger intracellular signaling.
Fig. 4.
RNA structures in FMDV and SVDV 3′NCRs enhance IFN-β induction. SK-6 cells were transfected with 20 μg/ml of FMDV and SVDV 3′NCR transcripts (denatured or renatured as described in Materials and Methods), with 10 μg/ml of poly(I:C) or 10 μg of tRNA. The fold induction of IFN-β in RNA-transfected cells above that in mock-transfected cells was determined by RT-qPCR normalized to GAPDH. Error bars show standard deviations between triplicates.
Deletion of the 3′NCR from the viral genome decreases IFN-β induction.
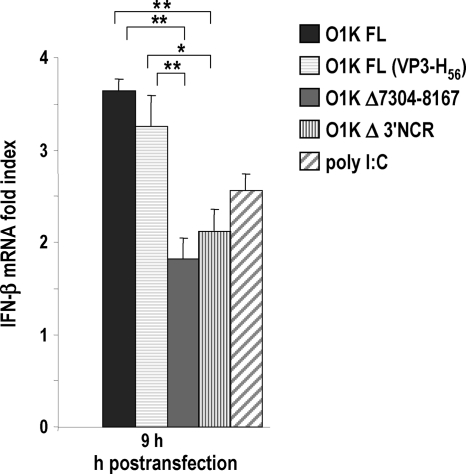
To determine whether the presence of the 3′NCR in the context of a full-length (FL) viral genome had an activator effect, IFN-β expression induced by RNAs with the 3′NCR deleted was compared to that triggered by FL-O1K FMDV transcripts by the transfection of SK-6 cells (Fig. 5). RNAs carrying a 3′NCR deletion or a larger 864-nt deletion, enclosing the whole 3′NCR and part of the 3Dpol gene, both preserving the poly(A) tail, were significantly less active than FL genomes, including a replication-defective VP3-H56 version (35). Similar results were observed when transfections were performed in the presence of the translation inhibitor cycloheximide (data not shown). These results strongly suggest that the 3′NCR is an RNA feature inducing the transcriptional activation of IFN-β within the viral genome, and that this effect is independent of viral replication. Since the 5′ end of all of the O1K transcripts analyzed, unlike viral genomes, are phosphorylated, the differences found among them must be due to alternative activator features other than the 5′-triphosphate.
Fig. 5.
Deletion of the 3′NCR in the FMDV genome decreases IFN-β induction. Comparison of FL, FL (VP3-H56), Δ3′NCR, and (Δ7304-8167)-FMDV O1K transcripts as activators of IFN-β signaling. SK-6 cells were transfected with 20 μg/ml of each RNA or 10 μg/ml of poly(I:C). After 9 h of transfection, the fold induction of IFN-β in RNA-transfected cells above that of mock-transfected cells was determined by RT-qPCR normalized to GAPDH. Error bars show standard deviations between triplicates. Statistical analysis was performed by a one-way analysis of variance with a Bonferroni's post hoc test (*, P < 0.05; **, P < 0.01).
The FMDV FL RNAs were poor activators compared to the 5′- and 3′NCR elements, for which higher induction levels were observed independently (Fig. 1B). The size and structure of the NCR RNAs could be enhancing the recognition of PAMP motifs in a more effective manner than that in the FL RNAs. The low levels of IFN-β induction observed in transfections with O1K FL RNA compared to that of FMDV infections may be due in part to the accumulation of replication intermediates eliciting the innate immune response in virus-infected cells that might be present at lower levels in transfected cells at the time point assayed.
FMDV and SVDV PAMP RNAs stimulate innate immunity and antiviral response in porcine cells.
To determine whether the observed induction of IFN-β mRNA correlated with the activation of the cellular antiviral state in host cells, we transfected SK-6 cells with S, IRES, and 3′NCR RNAs and then tested the antiviral activity in the transfection supernatants (Table 1). For this purpose, IBRS-2 cells, with an impaired type-I IFN system but responsive to exogenous IFN, were treated for 24 h with dilutions of the supernatants of transfected SK-6 cells and then infected with VSV. Poly(I:C)- and tRNA-transfected cells were used as controls. As shown in Table 1, antiviral activity was detected and measured in the supernatants of cells transfected with all of the NCR RNAs, while no activity was found in cells transfected with poly(I:C) or tRNA. The inhibition of VSV infection correlated well with the levels of IFN-β mRNA induced by each RNA (Fig. 1B and 4). Consistently with this, the maximum inhibitory effect, 27, was found in FMDV 3′NCR-transfected cells, with lower values, 9 to 13, for FMDV S, IRES, and SVDV 3′NCR. To confirm that the antiviral activity was type I IFN specific, transfection supernatants containing the highest activities were pretreated with specific neutralizing monoclonal antibodies against IFN-α, IFN-β, or a combination of both (Table 1). The antibodies against IFN-α were able to neutralize completely the antiviral activity, while the anti-IFN-β antibodies neutralized only about one-fourth of the activity. None of the antibodies showed any inhibitory effect on VSV infection of untreated cells, proving the specificity of the neutralizations observed. These results indicate that mainly IFN-α activity was present in the supernatant of transfected cells. In agreement with our results, de los Santos et al. reported the complete neutralization of antiviral activity present in the supernatants of SK-6 cells infected with a leaderless FMDV using anti-IFN-α monoclonal antibodies (8). However, we were able to detect a partial neutralization of antiviral activity in NCR RNA-transfected cells with the antibodies against IFN-β.
Table 1.
Antiviral activity in transfected porcine cellsa
| RNA | Antiviral activity of: |
|||
|---|---|---|---|---|
| Untreated supernatant | Anti-IFN-α MAb | Anti-IFN-β MAb | Anti-IFN-α+β MAbs | |
| FMDV 3′NCR | 27 | <2 | 6 | <2 |
| FMDV S | 13 | − | 3.8 | − |
| FMDV IRES | 9 | − | − | − |
| SVDV 3′NCR | 11 | − | − | − |
| Poly(I:C)/tRNA | <2 | NA | NA | NA |
SK-6 cells were transfected for 24 h with 40 μg/ml of FMDV or SVDV NCR transcripts or 10 μg/ml of poly(I:C) or tRNA. Antiviral activity is expressed as the reciprocal of the highest dilution of supernatants from SK6-transfected cells needed to reduce the number of VSV plaques on IBRS-2 cells by 50%. Data are averages of duplicates from three independent transfection experiments. In some cases, supernatants were incubated previously for 1 h at 37°C with 1 μg/ml of neutralizing MAbs against INF-α, INF-β, or both. −, not determined; NA, not applicable.
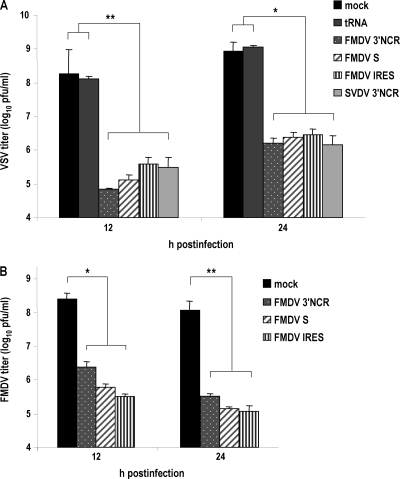
The antiviral state induced in transfected cells by the NCR RNAs also was studied. SK-6 cells transfected with the different RNAs were infected with VSV at an MOI of 1, and the infection supernatants were collected at 12 and 24 h. The viral titers in supernatants were determined by plaque assay of IBRS-2 cells (Fig. 6A). The treatment with all NCR RNAs reduced VSV yield by about 3 log compared to that of tRNA- or mock-transfected cells at both times postinfection. Nonsignificant differences of up to 5.5-fold in VSV titers were found between the FMDV 3′NCR-treated cells and those treated with the other NCR RNAs at 12 h postinfection, while viral titers at 24 h after infection were more similar. An equivalent effect was observed when transfected cells were infected with FMDV (Fig. 6B), showing a reduction of viral production of up to 3 log between mock- and NCR-transfected cells and nonsignificant differences among the different transcripts, which were slightly higher at 12 h than at 24 h postinfection. These results suggest that although they induce IFN-β transcription at different levels, all of the NCR RNAs after 24 h of transfection were able to reach the induction threshold required to trigger an effective antiviral response in transfected cells. Moreover, viral mechanisms counteracting the host cell innate response are likely to interfere with and mask the antiviral activity induced by the different NCR RNAs at 12 and 24 h postinfection. This is in agreement with antiviral activities detected in supernatants of transfected cells (Table 1), and taken together these findings prove the autocrine and paracrine antiviral effect of FMDV and SVDV NCR RNAs on porcine host cells.
Fig. 6.
Induction of an antiviral state in cells transfected with NCR RNAs. After 24 h of transfection with 40 μg/ml of FMDV or SVDV NCR transcripts or 10 μg/ml of tRNA, SK-6 cells were infected with VSV (A) or FMDV (B) at an MOI of 1. Supernatants from infected cells were collected at 12 and 24 h postinfection, and viral titers were determined in IBRS-2 cells. The bars show average values from two independent experiments performed in triplicate plus standard deviations. Statistical analysis was performed by a one-way analysis of variance with a Bonferroni's post hoc test (*, P < 0.05; **, P < 0.01).
Inoculation of FMDV genome PAMPs triggers antiviral response in suckling mice.
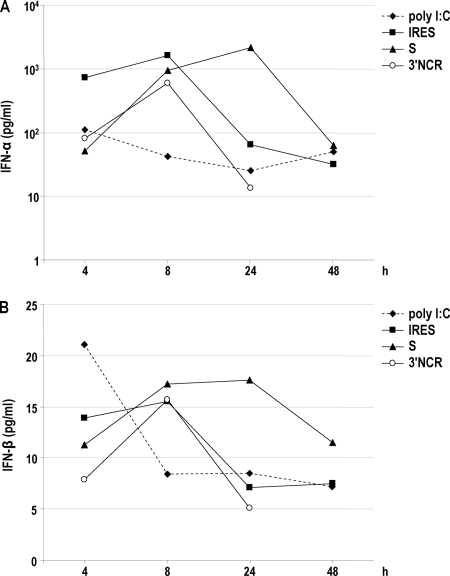
To determine whether the FMDV NCR RNAs were able to trigger innate immune responses in vivo, we inoculated the S, IRES, and 3′NCR transcripts i.p. in litters of Swiss newborn mice. Poly(I:C) also was inoculated as a control. Early reports described the poly(I:C)-induced resistance to FMDV in 9-day-old (suckling) and 23-day-old (weanling) mice when inoculated i.p. at microgram quantities 18 h before challenge as well as the enhancement on the early antibody production of swine to FMDV vaccine (32, 33). The levels of IFN-α and IFN-β in pools of sera collected at 4, 8, 24, and 48 h postinoculation were determined for each RNA by ELISA (Fig. 7A and B). All of the FMDV NCRs acted as potent IFN-α/β inducers in suckling mice. Interestingly, even NCR RNAs giving detectable but lower levels of IFN-β mRNA induction in cultured porcine cells were able to trigger a potent and long-lasting IFN response in suckling mice. The 3′NCR and the IRES induced a peak of IFN-α in serum at 8 h after injection. In the case of the S RNA, the time of IFN-α/β detection was longer, with a maximum at 24 h postinoculation (Fig. 7A). At 8 h after inoculation, the levels of IFN-α in sera were about 15- to 40-fold higher for the NCR RNAs than the poly(I:C)-induced levels. At 24 h postinoculation, the S RNA induced levels 90-fold higher than that of poly(I:C). The same kinetics was observed for the levels of IFN-β in sera of mice transfected with each RNA, with a peak at 8 h postinoculation, maintained up to 24 h in the case of the S RNA (Fig. 7B).
Fig. 7.
FMDV NCRs trigger innate immunity in suckling mice. Groups of four to five Swiss suckling mice were inoculated i.p. with 100 μg of 3′NCR, S, and IRES transcripts or poly(I:C). The levels of IFN-α (A) and IFN-β (B) in pools of sera collected at different times postinoculation for each RNA were determined by ELISA. The average values of IFN-α and IFN-β for pools of sera collected at 0 h postinoculation were 25 and 6 pg/ml, respectively.
To confirm the biological relevance of the increased levels of IFN-α/β detected in sera from suckling mice transfected with the NCR RNAs, the antiviral activity in those serum samples was analyzed by the inhibition of VSV infection in L-929 murine cells (Table 2). The levels of antiviral activity detected were in good correlation with the levels of IFN-α/β measured by ELISA (Fig. 7A and B). At 8 h postinoculation, antiviral activity was detected for all of the NCR RNAs as well as poly(I:C), with the IRES and S RNAs being the most potent inducers. At 24 h postinoculation, the antiviral activity was still detected at high levels in sera from S-transfected mice, while lower levels were found for animals transfected with the IRES.
Table 2.
Antiviral activity in sera from transfected suckling micea
| RNA | Activity at time point (h postinoculation): |
|||
|---|---|---|---|---|
| 4 | 8 | 24 | 48 | |
| S | 2 | 128 | 162 | <2 |
| IRES | 64 | 128 | 8 | <2 |
| 3′NCR | <2 | 32 | <2 | ND |
| Poly(I:C) | 8 | 8 | <2 | <2 |
Groups of four to five mice were inoculated with 100 μg of FMDV transcripts or poly(I:C). Sera were collected and pooled at the indicated times postinoculation for each RNA. Antiviral activity is expressed as the reciprocal of the highest dilution of sera able to suppress VSV-induced cytopathic effect on L-929 cells in 50% of the wells assayed. No antiviral activity was detected in the pools of sera collected at 0 h postinoculation (<2). ND, not done.
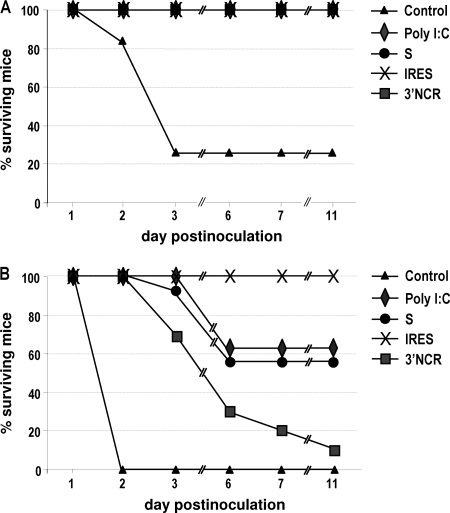
To address whether the innate immune response induced in vivo by inoculation with the NCR RNAs correlated with protection against FMDV challenge, suckling mice inoculated with the corresponding RNAs were infected 24 h later with two different FMDV infectious doses. As shown in Fig. 8A, all mice inoculated with the NCR transcripts, as well as poly(I:C), survived FMDV challenge with 102 PFU, whereas 28% of the group of animals inoculated with PBS survived. When the viral dose was raised to 104 PFU, none of the mice in the control group survived at day 2 postinfection, while survival was 100% for the groups of RNA-inoculated mice. However, relevant differences in susceptibility to FMDV were observed among the different groups from 3 to 11 days after infection (Fig. 8B). Mice inoculated with the 3′NCR showed the lowest rate of survival after 11 days of infection (10%). The susceptibility of poly(I:C)- and S-inoculated animals was similar, with maximum survival rates of about 60%. In the case of IRES-inoculated mice, all animals were protected against viral challenge, as no signs of disease or death were observed after infection. The susceptibility to FMDV infection of the different groups of mice was consistent with the different levels of antiviral activity detected in sera from the corresponding RNA-inoculated mice (Table 2).
Fig. 8.
Inoculation of FMDV NCRs in suckling mice has a protective effect against viral challenge. Newborn Swiss mice were inoculated i.p. with 100 μg of 3′NCR, S, and IRES transcripts or poly(I:C) (n = 10 to 12) and infected 24 h later with 7 × 102 (A) or 7 × 104 (B) PFU of FMDV. Survival percentages at different days postinfection are shown.
Taken together, these results indicate that synthetic RNA transcripts corresponding to structural elements present in the FMDV 5′ and 3′NCRs, playing essential roles in viral genome expression, can trigger the innate immune response and induce an antiviral state in vivo.
DISCUSSION
Type I IFN induction as an early consequence of viral infection in host cells is crucial for the establishment of an antiviral state aimed at blocking virus replication and spread. Viruses have evolved to circumvent this host defense system, and the balance between innate response and viral antagonism may determine the outcome of disease and pathogenesis. Moreover, the occurrence of in silico-predicted genome-scale ordered RNA structures (GORS) might correlate with the ability of some positive-stranded RNA viruses, including aphthoviruses, to persist in their natural hosts (44). In the case of FMDV, a small RNA virus that is highly sensitive to IFN, early studies suggested that the lack of virulence in cattle was correlated with increased IFN production (42). Indeed, the ability of the virus to form plaques in cell culture is associated with the suppression of IFN-α/β (7), which are equally effective at inhibiting FMDV replication. The leader protein Lpro, a papain-like proteinase, has been identified as a virulence factor that blocks the host immune innate response. Lpro translocates to the nucleus of infected cells in correlation with a decrease in the amount of nuclear p65/RelA, a subunit of NF-κB (10), then inhibiting NF-κB-dependent gene expression (53). The suppression of dsRNA-induced IFN-β induction through the degradation of interferon regulatory factor 3/7 by Lpro has been described recently (48).
In this work, we have analyzed transcripts corresponding to functional domains in the 5′- and 3′NCRs of the FMDV genome, which are predicted to fold into stable secondary structures, for their ability to trigger immune innate responses in porcine cells and suckling mice. Detectable levels of IFN-β induction were observed for all of the NCR transcripts assayed. Among them, the entire 3′NCR, including both stem-loops and the poly(A) tail, was the best inducer in transfected SK-6 cells, followed by the S fragment and the IRES. Relevant differences were observed between the IFN-β induction levels elicited by the different transcripts, all of them having a 5′-triphosphate, a potential substrate for RIG-I detection (20, 28), suggesting that this is not the major feature mediating induction by the NCR RNAs. However, small differences in base pairing at the 5′-triphosphate end of short dsRNAs have been shown to strongly affect IFN induction (40, 41). The immunostimulatory activity of aberrant nontemplated RNA hairpins generated during in vitro transcription with T7 RNA polymerase has been reported under some reaction conditions (41, 45). The NCR transcripts used in this study all migrated as unique bands of the corresponding expected sizes as analyzed by denaturing PAGE. The phosphatase treatment of the 3′NCR transcript strongly decreased but did not completely abolish induction.
The removal of the poly(A) tail, known to elicit the IFN response through RIG-I signaling (37), had a detrimental effect on IFN-β induction, but it was milder than CIP treatment at later times posttransfection. However, the deletion of any of the 3′NCR stem-loops rendered RNAs minimally active for IFN-β signaling. The size of the transcripts was not in correlation with the IFN-β induction levels detected, and small differences in the molar ratio of the transfected transcripts, up to 2.5-fold, are unlikely to account for the large differences in the levels of induction detected.
To address whether the IFN-β induction potential of FMDV 3′NCR could be extended to other picornaviruses, we compared it to the 3′NCR of SVDV, a swine picornavirus closely related to the human pathogen coxsackievirus B5. The SVDV 3′NCR, divergent in sequence and predicted secondary structure, proved to be an IFN-β inducer at levels up to 20 fold lower than that of the FMDV counterpart. We next showed the relevance of the secondary structure in the 3′NCR RNAs for induction, as denatured transcripts greatly reduced their signaling capacity. Interestingly, denaturing affected SVDV RNA more dramatically. This correlates with the dimerization ability of this transcript (43), likely due to tertiary interactions (30).
A nice correlation was found between the levels of IFN-β induction and antiviral activity present in the supernatant of transfected SK-6 cells with the different NCR RNAs. This activity was specifically blocked by anti-IFN-α/β MAbs, and an effective antiviral state was induced in transfected cells, reducing more than 100-fold their susceptibility to VSV or FMDV infection.
The 100-nt polyuridine motif [poly(U/UC)] of the hepatitis C virus (HCV) genome 3′NCR has been identified as a potent PAMP, a substrate of RIG-I recognition and immune triggering in human and murine cells (37). In contrast, the structured 3′-terminal X region failed to trigger signaling. The HCV 3′NCR seems to have a higher level of activity than those of the other flaviviruses. The 5′- and 3′NCRs of dengue virus (DEN) elicited low but measurable stimulation of innate immune signaling, while the smaller, highly structurally conserved 3′-terminal stem-loop RNAs of DEN, West Nile, and yellow fever viruses were minimally active (46).
Our results suggest the presence of PAMPs in the FMDV RNA and define the 3′NCR as a potent IFN-β activator in porcine cultured cells. The stimulatory effect of the FMDV 3′NCR in the context of the full viral genome was further proven. A significant decrease was found for FMDV RNAs carrying the 3′NCR deletion. This effect was not dependent on replication, as similar results were observed with replication-defective transcripts. An equivalent decrease in IFN-β induction was reported for HCV transcripts bearing a 3′NCR deletion (37). However, FMDV FL transcripts induced much lower levels of IFN-β than the NCR RNAs alone in porcine cells. The size and structure of the NCR transcripts could be promoting the exposure of PAMP motifs in a more effective manner than that in the FL RNAs, where they might be partly masked. Additionally, the small size and base-pairing extent of the 5′-triphosphate of the NCR RNAs may have an enhancing effect on RLR recognition and signaling (40, 41). It still is unclear what specific features of picornavirus are required for the activation of the RLR pathway. MDA-5 has been shown to be responsible for the recognition of infection with picornaviruses, whose RNA 5′ ends are linked to the VPg viral protein lacking a 5′-triphosphate (17, 22). Interestingly, the cleavage of RIG-I during picornavirus infections has been reported and associated with viral protease 3Cpro (2, 27), suggesting that RIG-I signaling plays a role in the innate immune response against picornaviruses. The stimulatory activity of MDA-5 appears to reside in higher-order structures containing single strands and double strands in a web-like structure generated during virus infection (29).
Encouraged by the results of IFN-β induction in swine-cultured cells transfected with the NCR transcripts, we addressed the potential of such molecules as type I IFN inducers in vivo. For this purpose, the FMDV NCRs were inoculated i.p. into Swiss suckling mice and the levels of IFN-α/β proteins and antiviral activity in sera were measured. Newborn mice are a suitable model for innate immune responses, as their adaptive immunity still is immature. All of the FMDV NCRs were able to induce a peak of IFN-α/β in sera of the inoculated animals at 8 h after injection, and the peak was remarkably higher than those observed for poly(I:C)-transfected mice. This peak was maintained for up to 24 h in the case of the S RNA. The presence of antiviral activity in sera from NCR-transfected mice also was detected and measured, and a good correlation with IFN-α/β levels tested by ELISA was found. Interestingly, even those transcripts showing a lower capacity for IFN-β induction in porcine cultured cells were able to induce an innate immune response in mice. On one hand, this suggests that the effect of low-level inductions of type I IFN observed in cultured cells can be magnified in vivo. On the other hand, the action of other viral sensors in vivo, mainly TLRs, may account for the enhancing effect observed. Thus, the specific immunostimulatory activity of each NCR RNA may be different depending on the host cell context assayed. This was the case for the IRES: despite its complex structure, it was a poor inducer in SK-6 cells, as it was less effective than the entire 5′NCR RNA. The high-affinity binding of cellular factors to the IRES at early times of transfection might prevent the recognition of domains or features potentially detectable by porcine cytoplasmic viral sensors. However, the IRES acted as a strong IFN inducer in suckling mice. We further showed that the innate immune responses triggered by the NCRs in suckling mice resulted in a reduced susceptibility to FMDV infection in all cases, in particular the IRES, which induced protection in all animals inoculated against 104 PFU of FMDV.
Although additional work to explore the efficiency of the NCR RNAs on IFN induction in natural host species needs to be done, our findings suggest that the use of these small noninfectious RNAs is useful to induce a rapid antiviral state, in combination with effective FMD vaccines, to overcome the problem of susceptibility until protective levels of antibodies are produced by vaccinated animals. Additionally, a potential role in pathogenesis for the innate immune responses triggered by the recognition of the FMDV NCR RNAs cannot be ruled out. While the specific mechanisms of signaling pathways remain to be determined, our results provide new insight into broad-spectrum antiviral development strategies.
ACKNOWLEDGMENTS
This work was supported by grants CSD2006-2007 and BIO2008-04487-C03-01 and by the ICTS program from the Spanish Ministry of Science and Innovation.
We thank Antonio Garmendia (University of Connecticut) for the kind provision of reagents and Encarnación Martínez-Salas for helpful discussion, reagents, and critical reading of the manuscript. We also thank Francisco Mateos and Nuria de la Losa for their assistance in mouse experiments at the CISA-INIA animal facility.
Footnotes
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Amadori M., Bugnetti M., Tironi D. 1987. Induction and characterization of swine beta interferon. Comp. Immunol. Microbiol. Infect. Dis. 10:71–78 [DOI] [PubMed] [Google Scholar]
- 2. Barral P. M., Sarkar D., Fisher P. B., Racaniello V. R. 2009. RIG-I is cleaved during picornavirus infection. Virology 391:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belsham G. J. 2009. Divergent picornavirus IRES elements. Virus Res. 139:183–192 [DOI] [PubMed] [Google Scholar]
- 4. Belsham G. J., Martinez-Salas E. 2004. Genome organisation, translation and replication of FMDV RNA, p. 19–52 In Sobrino F., Domingo E. (ed.), Foot-and-mouth disease: current perspectives. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 5. Creagh E. M., O'Neill L. A. 2006. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 27:352–357 [DOI] [PubMed] [Google Scholar]
- 6. Chinsangaram J., Koster M., Grubman M. J. 2001. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J. Virol. 75:5498–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chinsangaram J., Piccone M. E., Grubman M. J. 1999. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J. Virol. 73:9891–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de los Santos T., de Avila Botton S., Weiblen R., Grubman M. J. 2006. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 80:1906–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de los Santos T., Diaz-San Segundo F., Grubman M. J. 2007. Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J. Virol. 81:12803–12815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de los Santos T., et al. 2009. A conserved domain in the leader proteinase of foot-and-mouth disease virus is required for proper subcellular localization and function. J. Virol. 83:1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Devaney M. A., Vakharia V. N., Lloyd R. E., Ehrenfeld E., Grubman M. J. 1988. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 62:4407–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeWitte-Orr S. J., Collins S. E., Bauer C. M., Bowdish D. M., Mossman K. L. 2010. An accessory to the ‘Trinity’: SR-As are essential pathogen sensors of extracellular dsRNA, mediating entry and leading to subsequent type I IFN responses. PLoS Pathog. 6:e1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Escarmís C., Toja M., Medina M., Domingo E. 1992. Modifications of the 5′ untranslated region of foot-and-mouth disease virus after prolonged persistence in cell culture. Virus Res. 26:113–125 [DOI] [PubMed] [Google Scholar]
- 14. Fernández-Miragall O., Lopez de Quinto S., Martinez-Salas E. 2009. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 139:172–182 [DOI] [PubMed] [Google Scholar]
- 15. Fernández-Miragall O., Martinez-Salas E. 2003. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 9:1333–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. García-Briones M. M., et al. 2004. Immunogenicity and T cell recognition in swine of foot-and-mouth disease virus polymerase 3D. Virology 322:264–275 [DOI] [PubMed] [Google Scholar]
- 17. Gitlin L., et al. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Golde W. T., Nfon C. K., Toka F. N. 2008. Immune evasion during foot-and-mouth disease virus infection of swine. Immunol. Rev. 225:85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grubman M. J., Moraes M. P., Diaz-San Segundo F., Pena L., de los Santos T. 2008. Evading the host immune response: how foot-and-mouth disease virus has become an effective pathogen. FEMS Immunol. Med. Microbiol. 53:8–17 [DOI] [PubMed] [Google Scholar]
- 20. Hornung V., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997 [DOI] [PubMed] [Google Scholar]
- 21. Johnsen I. B., et al. 2006. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 25:3335–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 23. Kitching R. P. 2005. Global epidemiology and prospects for control of foot-and-mouth disease. Curr. Top. Microbiol. Immunol. 288:133–148 [DOI] [PubMed] [Google Scholar]
- 24. López de Quinto S., Saiz M., de la Morena D., Sobrino F., Martinez-Salas E. 2002. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 30:4398–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mason P. W., Bezborodova S. V., Henry T. M. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Overend C., et al. 2007. Recombinant swine beta interferon protects swine alveolar macrophages and MARC-145 cells from infection with porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 88:925–931 [DOI] [PubMed] [Google Scholar]
- 27. Papon L., et al. 2009. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393:311–318 [DOI] [PubMed] [Google Scholar]
- 28. Pichlmair A., et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001 [DOI] [PubMed] [Google Scholar]
- 29. Pichlmair A., et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pöyry T., et al. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699–1717 [DOI] [PubMed] [Google Scholar]
- 31. Ramos R., Martinez-Salas E. 1999. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES). RNA 5:1374–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richmond J. Y. 1971. Mouse resistance against foot-and-mouth disease virus induced by injections of pyran. Infect. Immun. 3:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richmond J. Y., Hamilton L. D. 1969. Foot-and-mouth disease virus inhibition induced in mice by synthetic double-stranded RNA (polyriboinosinic and polyribocytidylic acids). Proc. Natl. Acad. Sci. U. S. A. 64:81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodríguez Pulido M., Serrano P., Saiz M., Martinez-Salas E. 2007. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology 364:466–474 [DOI] [PubMed] [Google Scholar]
- 35. Rodríguez Pulido M., Sobrino F., Borrego B., Saiz M. 2009. Attenuated foot-and-mouth disease virus RNA carrying a deletion in the 3′ noncoding region can elicit immunity in swine. J. Virol. 83:3475–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rubinstein S., Familletti P. C., Pestka S. 1981. Convenient assay for interferons. J. Virol. 37:755–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sáiz M., Gomez S., Martinez-Salas E., Sobrino F. 2001. Deletion or substitution of the aphthovirus 3′ NCR abrogates infectivity and virus replication. J. Gen. Virol. 82:93–101 [DOI] [PubMed] [Google Scholar]
- 39. Sáiz M., Nunez J. I., Jimenez-Clavero M. A., Baranowski E., Sobrino F. 2002. Foot-and-mouth disease virus: biology and prospects for disease control. Microbes Infect. 4:1183–1192 [DOI] [PubMed] [Google Scholar]
- 40. Schlee M., et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmidt A., et al. 2009. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 106:12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sellers R. F. 1963. Multiplication, interferon production and sensitivity of virulent and attenuated strains of the virus of foot-and-mouth disease. Nature 198:1228–1229 [DOI] [PubMed] [Google Scholar]
- 43. Serrano P., Pulido M. R., Saiz M., Martinez-Salas E. 2006. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 87:3013–3022 [DOI] [PubMed] [Google Scholar]
- 44. Simmonds P., Tuplin A., Evans D. J. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: implications for virus evolution and host persistence. RNA 10:1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Triana-Alonso F. J., Dabrowski M., Wadzack J., Nierhaus K. H. 1995. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 270:6298–6307 [DOI] [PubMed] [Google Scholar]
- 46. Uzri D., Gehrke L. 2009. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 83:4174–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vengris V. E., Mare C. J. 1972. Swine interferon. II. Induction in pigs with viral and synthetic inducers. Can. J. Comp. Med. 36:288–293 [PMC free article] [PubMed] [Google Scholar]
- 48. Wang D., et al. 2010. Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced type I interferon transcription by decreasing interferon regulatory factor 3/7 in protein levels. Biochem. Biophys. Res. Commun. 399:72–78 [DOI] [PubMed] [Google Scholar]
- 49. Wilkins C., Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Witwer C., Rauscher S., Hofacker I. L., Stadler P. F. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29:5079–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yoneyama M., Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20:4–22 [DOI] [PubMed] [Google Scholar]
- 52. Yoneyama M., Onomoto K., Fujita T. 2008. Cytoplasmic recognition of RNA. Adv. Drug Deliv. Rev. 60:841–846 [DOI] [PubMed] [Google Scholar]
- 53. Zhu J., Weiss M., Grubman M. J., de los Santos T. 2010. Differential gene expression in bovine cells infected with wild type and leaderless foot-and-mouth disease virus. Virology 404:32–40 [DOI] [PubMed] [Google Scholar]