Abstract
The influenza virus RNA-dependent RNA polymerase is capable of initiating replication but mainly catalyzes abortive RNA synthesis in the absence of viral and host regulatory factors. Previously, we reported that IREF-1/minichromosome maintenance (MCM) complex stimulates a de novo initiated replication reaction by stabilizing an initiated replication complex through scaffolding between the viral polymerase and nascent cRNA to which MCM binds. In addition, several lines of genetic and biochemical evidence suggest that viral nucleoprotein (NP) is involved in successful replication. Here, using cell-free systems, we have shown the precise stimulatory mechanism of virus genome replication by NP. Stepwise cell-free replication reactions revealed that exogenously added NP free of RNA activates the viral polymerase during promoter escape while it is incapable of encapsidating the nascent cRNA. However, we found that a previously identified cellular protein, RAF-2p48/NPI-5/UAP56, facilitates replication reaction-coupled encapsidation as an NP molecular chaperone. These findings demonstrate that replication of the virus genome is followed by its encapsidation by NP in collaboration with its chaperone.
INTRODUCTION
The genome of influenza type A viruses consists of eight-segmented and single-stranded RNAs of negative polarity. Transcription from the viral RNA (vRNA) genome is initiated using the oligonucleotide containing the cap-1 structure from cellular pre-mRNAs as a primer, whereas genome replication is primer independent and generates full-length vRNA through cRNA (full-sized complementary copy of vRNA) (reviewed in reference 17). Generally, each viral DNA or RNA genome is not present as a naked form but as a complex with viral basic proteins. The influenza virus genome exists as a ribonucleoprotein (termed vRNP) complex with nucleoprotein (NP), one of the basic viral proteins, and viral RNA-dependent RNA polymerases consisting of three subunits (PB1, PB2, and PA). NP binds single-stranded RNA without sequence specificity and is required for maintaining the RNA template in an ordered conformation suitable for viral RNA synthesis and packaging into virions (6, 23, 34). In the case of Mononegavirales, nonsegmented and negative-stranded RNA viruses, it is proposed that the nucleocapsid (N) protein forms a trimeric complex with the viral RNA polymerase large (L) protein and phosphoprotein (P) to form a replicase complex to produce the progeny viral genome with concomitant encapsidation of nascent RNA by N protein and that encapsidation is mediated by the chaperone activity of P protein (2, 7, 14, 24). In the case of influenza virus, it is also postulated that NP might regulate the viral polymerase function and encapsidate the virus genome through its interaction with PB1 and/or PB2 (1, 23). Genetic analyses suggest that NP participates in the replication process (15). Recently, it was also shown that NP that is saturated with single-stranded DNA (ssDNA), resulting in the lack of RNA binding activity, stimulates virus genome replication from a model template without primer (18). It is possible that NP stimulates virus genome replication through interaction with the viral polymerase in an RNA binding activity-independent manner. Moreover, the in vitro cRNA synthesis using infected cell extracts as an enzyme source depends on a supply of NP free of RNA (27). This finding has been interpreted as indicating that NP prevents the premature termination of RNA synthesis, possibly by binding to nascent RNA chains, that is, encapsidating them. Based on these observations, it could be hypothesized that NP facilitates virus genome replication by both RNA binding- and viral polymerase binding-dependent mechanisms. It is proposed that encapsidation is initiated by successive targeting of exogenous NP monomer to a replicating RNA through the interaction between NP and the viral polymerase, which is distinct from the replicative enzyme bound to the 5′ end of nascent RNA (1, 8, 11, 22), and then additional NP molecules are subsequently recruited by the NP-NP oligomerization (3, 23). It is also reported that nascent cRNA is degraded by host cellular nucleases unless it is stabilized by newly synthesized viral RNA polymerases and NP (33). However, the precise molecular mechanisms involved in virus genome replication and encapsidation by NP are yet unclear.
The cRNA synthesis occurs from incoming vRNA in infected cells, but vRNP complexes isolated from virions by themselves hardly synthesize cRNA (9). Thus, it was reasonable to examine whether a host factor(s) and/or a viral factor(s) is required for the replication process. We reconstituted a cell-free virus genome replication system with virion-associated vRNP and nuclear extracts prepared from uninfected HeLa cells (9). Using biochemical fractionation and complementation assays, we identified influenza virus replication factor 1 (IREF-1) that enabled the viral polymerase to synthesize full-sized cRNA. Otherwise, the viral RNA polymerase produces mainly abortive short RNA chains in the absence of IREF-1. IREF-1 was found to be identical with a minichromosome maintenance (MCM) heterohexamer complex. IREF-1/MCM stabilizes replicating polymerase complexes by promoting the interaction between the nascent cRNA and the PA subunit.
Here, we examined the molecular function of NP in influenza virus genome replication using a previously established cell-free virus genome replication system and virion-associated vRNP. Exogenously added NP free of RNA stimulated virus genome replication with MCM in an additive manner. Further, we found that NP activates the viral polymerase during its transition from initiation to elongation to synthesize the unprimed full-length cRNA, but NP by itself is incapable of encapsidating the nascent cRNA. However, we found that RAF-2p48/NPI-5/UAP56/BAT1, which was identified as a host factor for activation of viral RNA synthesis (16), is required for the encapsidation of nascent cRNA with exogenously added NP free of RNA and for the stimulation of the elongation process of virus genome replication. We observed that the level of the virus genome replication was decreased in infected cells when the expression of the RAF-2p48/UAP56 gene was knocked down by small interfering RNA (siRNA)-mediated gene silencing. Based on these observations, we propose an NP- and host factor-dependent mechanism of virus genome encapsidation in concert with its replication.
MATERIALS AND METHODS
Biological materials.
vRNP was prepared from purified influenza A/Puerto Rico/8/34 virus as previously described (28). For the expression of His-tagged NP (His-NP), we cloned the open reading frame (ORF) corresponding to the NP gene into pET14b. Rabbit polyclonal antibody against NP was generated by immunization of a 2-month-old female rabbit with His-NP protein. HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum.
Preparation of recombinant proteins.
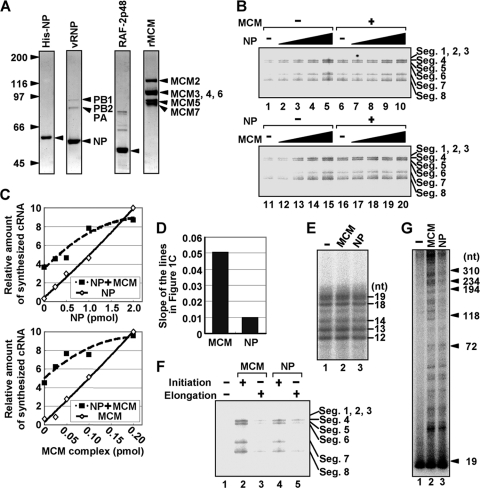
His-tagged recombinant proteins were prepared and purified according to the manufacturer's protocol. In addition, to remove the bacterial RNA possibly bound to NP, we treated recombinant proteins with RNase A before purification and washed them with a buffer containing 1 M NaCl. Recombinant RAF-2p48/UAP56 was prepared from glutathione S-transferase (GST)-tagged RAF-2p48/UAP56 by PreScission protease (GE Health Care) digestion. Purified proteins were stored in a buffer containing 50 mM HEPES-NaOH (pH 7.9), 300 mM KCl, 20% glycerol, and 1 mM dithiothreitol (DTT) at −80°C until use. Recombinant MCM complex was prepared as previously described (9). These purified recombinant proteins were separated by SDS-PAGE and visualized by staining with Coomassie brilliant blue in Fig. 1A.
Fig. 1.
NP and MCM additively stimulate virus genome replication. (A) Purified recombinant proteins and vRNP. Purified His-NP, vRNP, RAF-2p48/UAP56, and MCM complexes were separated by 7.5% SDS-PAGE and visualized by staining with Coomassie brilliant blue. (B) Stimulatory activity of NP and MCM in cell-free virus genome replication. RNA synthesis was carried out in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of recombinant MCM complex (0.05 pmol of MCM complex) with 0 (lanes 1 and 6), 0.25 (lanes 2 and 7), 0.5 (lanes 3 and 8), 1.0 (lanes 4 and 9), and 2.0 pmol (lanes 5 and 10) of recombinant NP (upper panel). For the experiments shown in the lower panel, we performed the RNA synthesis assay in the absence (lanes 11 to 15) or presence (lanes 16 to 20) of recombinant NP (0.50 pmol) with 0 (lanes 11 and 16), 0.025 (lanes 12 and 17), 0.05 (lanes 13 and 18), 0.10 (lanes 14 and 19), and 0.20 pmol (lanes 15 and 20) of MCM complex (lower panel). (C) Quantitative summary of panel A. The amounts of newly synthesized cRNA corresponding to segment 7 were determined by the ImageJ software. (D) Stimulatory activity per molecule of MCM and NP. The slopes of the lines in the presence of NP or MCM in panel C were determined. (E) Limited elongation assays. Unprimed limited elongation assays were carried out in the absence (lane 1) or presence (lane 2; 0.5 pmol) of MCM or NP (lane 3; 3.0 pmol). (F) NP functions during transition from initiation to elongation reaction. Unprimed limited elongation reactions were performed without (lanes 1, 3, and 5) or with (lanes 2 and 4) either MCM (lane 2; 0.5 pmol) or NP (lane 4; 3.0 pmol). After incubation for 1 h, elongation reactions were restarted by the addition of UTP. For lanes 3 and 5, MCM (0.5 pmol) and NP (3.0 pmol) were added at the restart of elongation reaction, respectively. (G) MCM stimulates the elongation process more effectively than NP. RNA synthesis was carried out in the absence (lane 1) or presence of either MCM (lane 2; 0.5 pmol) or NP (lane 3; 3.0 pmol) with 0.3 μM UTP, 250 μM each ATP, CTP, and GTP, and 10 μCi of [α-32P]UTP (3,000 Ci/mmol). The purified products were separated through 4 to 15% linear gradient PAGE containing 8 M urea and visualized by autoradiography.
Cell-free virus genome replication system.
Cell-free virus genome replication was carried out at 30°C for 90 min in a final volume of 25 μl containing 50 mM HEPES-NaOH (pH 7.9), 5 mM MgCl2, 50 mM KCl, 1.5 mM dithiothreitol, 500 μM each ATP, CTP, and UTP, 25 μM GTP, 5 μCi of [α-32P]GTP (3,000 Ci/mmol), 8 U of RNase inhibitor, and vRNP (10 ng of NP equivalents) in the presence or absence of purified proteins. RNA products were purified, subjected to 4% PAGE in the presence of 8 M urea, and visualized by autoradiography. For limited elongation assays, RNA synthesis was performed with vRNP (150 ng of NP equivalents) in the absence of UTP, and RNA products were separated by 15% PAGE containing 8 M urea. To address the encapsidation of nascent cRNA with NP, RNA synthesis was carried out by following the standard protocol described above except that 0.3 μM UTP, 250 μM each ATP, CTP, and GTP, and 10 μCi of [α-32P]UTP (3,000 Ci/mmol) were used in a final volume of 200 μl. The coprecipitated RNA products with NP or MCM were separated through 10% PAGE containing 8 M urea.
Gene silencing mediated by siRNA.
An siRNA against the RAF-2p48/UAP56 gene corresponding to its open reading frame (5′-AGUACUACGUGAAACUGAAGGACAA-3′) and control double-stranded RNA (dsRNA) targeting none of the cellular mRNAs were designed and synthesized by iGENE Therapeutics Inc. HeLa cells (1 × 105 cells) were transfected with 40 pmol of siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. At 48 h posttransfection, the cells were infected with influenza A/PR/8/34 at a multiplicity of infection (MOI) of 10 in the absence or presence of 100 μg/ml of cycloheximide (CHX). The RAF-2p48/UAP56 knockdown cells were also transfected with viral protein expression plasmids encoding PB1, PB2, PA, and NP and pHH21-vNS-Luc reporter plasmid to reconstitute a model viral replicon (19, 30). This reporter plasmid carries the luciferase (Luc) gene in reverse orientation sandwiched between 23-nucleotide (nt)-long 5′-terminal and 26-nucleotide-long 3′-terminal promoter sequences of the influenza virus segment 8, which is placed under the control of the human polymerase I (Pol I) promoter.
Indirect immunofluorescence assay.
HeLa cells on coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS). The cells were permeabilized in 0.5% Triton X-100 and incubated in PBS containing 1% bovine serum albumin (BSA). The coverslips were incubated with anti-RAF-2p48/UAP56 rabbit polyclonal antibody (16) for 1 h. After a washing step with 0.1% Tween 20 in PBS, coverslips were incubated with Alexa Fluor 568-conjugated anti-rabbit IgG (Invitrogen) for 1 h. Images were acquired under the same exposure time by a fluorescence microscope system (Axiovision; Carl Zeiss).
Primer extension assay.
Total RNAs isolated from control and RAF-2p48/UAP56 knockdown cells at 0, 3, 6, and 9 h postinfection (hpi) were subjected to reverse transcription at 42°C for 1 h with primers specific for segment 5 vRNA (5′-GGGAATACAGAGGGGAGAA-3′) corresponding to the NP cDNA between nucleotide sequence positions 1336 and 1354, segment 5 m/cRNA (5′-GATTTCAGTGGCATTCTGGC-3′) complementary to the NP cDNA between nucleotide sequence positions 101 and 120, and 5S rRNA (5′-GGGGTACCTTCGAAAGCCTACAGCACCCGGTA-3′), which were labeled at their 5′ ends with [γ-32P]ATP and T4 polynucleotide kinase (Toyobo). The products purified with phenol-chloroform extraction and ethanol precipitation were separated through 6% polyacrylamide gel containing 7 M urea and visualized by autoradiography.
Real-time quantitative PCR.
Total RNAs isolated from control and RAF-2p48/UAP56 knockdown cells at 12 h posttransfection for construction of the model viral replicon were subjected to reverse transcription with primers to determine the level of vRNA (5′-TCCATCACGGTTTTGGAATGTTTACTACAC-3′, which corresponds to the luciferase coding region between nucleotide sequence positions 728 and 757), cRNA (5′-AGTAGAAACAAGGGTGTTTTTTAGTA-3′, which is complementary to the 3′ portion of the segment 8 cRNA), and viral mRNA [oligo(dT)20 for poly(A) tail] synthesized from the reconstituted model viral replicon. The synthesized single-stranded cDNAs were subjected to real-time quantitative PCR analysis (Thermal Cycler Dice Real Time System TP800; TaKaRa) with two specific primers, 5′-TCCATCACGGTTTTGGAATGTTTACTACAC-3′, which corresponds to the luciferase coding region between nucleotide sequence positions 728 and 757, and 5′-GTGCGCCCCCAGAAGCAATTTC-3′, complementary to the luciferase coding region between nucleotide sequence positions 931 and 952. The amount of NP mRNA transcribed from the expression plasmid, which is transcribed by cellular RNA polymerase II, was detected as an internal control.
RESULTS AND DISCUSSION
Stimulation of de novo cRNA synthesis by NP.
Exogenously added recombinant NP free of RNA (here, designated exogenous NP) stimulated de novo virus genome replication in the absence of MCM and any kind of primer (Fig. 1B, lanes 1 to 5). We confirmed by RNase H digestion analyses with primers corresponding to each segment that RNA products corresponded to those synthesized from each segment (data not shown). Then, we examined whether exogenous NP and MCM coordinately stimulate the virus genome replication reaction. MCM stimulated virus genome replication additively with recombinant NP, suggesting that NP and MCM function through distinct mechanisms (Fig. 1B, lanes 6 to 10 and 16 to 20). The stimulatory activity per molecule of MCM was five times higher than that of NP, as judged by the slopes of the lines in Fig. 1C (Fig. 1D). We observed that authentic NP free of RNA purified from virions by CsCl glycerol density gradient centrifugation (5, 34) stimulates activity equally as well as recombinant NP (data not shown). We used as the enzyme source the vRNP containing authentic NP that is bound to the template RNA. Thus, it is quite likely that RNA-free NP but not template-bound NP is required for de novo virus genome replication. The RNA synthesis level varied among segments, as previously described (9). For instance, segments 1, 2, and 3 were hardly replicated compared with replication of other segments. The reason for this variation in cRNA synthesis is presently unknown.
NP facilitates the promoter escape of the viral RNA polymerase.
Previously, we demonstrated that MCM does not enhance the frequency of replication initiation, but rather makes a nonproductive viral polymerase override the step for abortive synthesis. To examine whether NP is involved in the initiation reaction of virus genome synthesis, we carried out a limited elongation assay, in which UTP is omitted from the reaction mixture and the RNA polymerase pauses at the first adenine residue on the template. The expected lengths of limited elongation products are 12 nt for segments 1, 3, and 7, 13 nt for segments 5 and 8, 14 nt for segment 6, 18 nt for segment 4, and 19 nt for segment 2. Since we detected comparable amounts of each RNA product in the absence or presence of exogenous NP (Fig. 1E), it is concluded that NP, like MCM, does not stimulate the initiation reaction (9). Thus, NP may be required for a step(s) after the initiation and the early elongation steps, in which short cRNAs are synthesized.
To examine whether NP stimulates the transition of the viral polymerase from initiation to elongation, that is, the promoter escape of the viral polymerase, unprimed limited elongation assays were first performed in the absence of UTP, and elongation reactions were restarted by the addition of UTP (Fig. 1F). MCM (0.5 pmol) or exogenous NP (3 pmol) was also added either before or after the limited elongation. The full-length cRNA was synthesized by restarting the limited elongation reaction performed in the presence of MCM (lane 2) or exogenous NP (lane 4) during the limited elongation reaction. Thus, it is quite likely that, to avoid abortive RNA synthesis by the viral polymerase, MCM and NP are required for the viral polymerase prior to its movement along a 12- to 19-nt-long vRNA template from the 3′ terminus of vRNA, where the hairpin loop and double-stranded promoter region are located. Since the initiation reaction was not stimulated by NP (Fig. 1E) and since the viral polymerase could not transit from initiation to elongation in the absence of NP (Fig. 1F), it is possible that NP stimulates elongation complexes during the promoter escape of the viral polymerase, as does MCM (9). A cell-free virus genome replication reaction was also carried out (Fig. 1G) in the presence of MCM (lane 2; 0.5 pmol) or NP (lane 3; 3 pmol) with a low concentration of UTP to slow down the reaction and synthesize a ladder of nascent cRNA chains in order to examine the length of elongated nascent cRNA chains. We found that comparable amounts of cRNA with a shorter length (∼100 nt) are synthesized in the presence of either MCM or NP. In contrast, the amount of longer cRNAs (>100 nt) stimulated by MCM was greater than that stimulated by NP (Fig. 1G, compare lane 2 with lane 3). Therefore, it is quite likely that MCM promotes the elongation process more effectively than NP, possibly due to the weak interaction of exogenously added NP with long nascent cRNA, as described later. Taking these results together, it is strongly suggested that NP, like MCM, stimulates the promoter escape of the viral polymerase. Previous reports showed that the target of MCM is PA (9), whereas that of NP is PB1 and PB2 (1). Therefore, it is possible that the replication stimulation mechanisms of NP and MCM are distinct from each other.
Encapsidation of newly synthesized virus genome by NP.
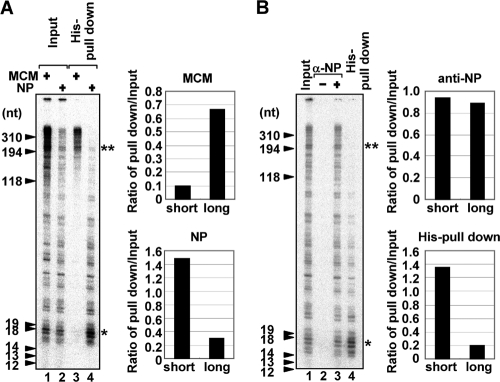
Previously, we proposed that MCM stimulates virus genome replication by acting as a scaffold between nascent cRNA chains and the viral polymerase during the promoter escape of the polymerase (9). Since NP has both RNA and viral polymerase binding activities, it should be speculated that NP, like MCM, also functions as a scaffold between newly synthesized RNA and the viral polymerase. To address this, we tried to pull down the replicated cRNA chains associated with His-tagged MCM or NP using Ni-nitrilotriacetic acid (NTA) resin (Fig. 2A). The cell-free virus genome replication reaction was carried out in the presence of an equal molar amount of MCM (lanes 1 and 3) or NP (lanes 2 and 4) with a low concentration of UTP in order to examine the length of copurified RNA as shown in Fig. 1G. As shown in input lanes, MCM stimulated the elongation process more effectively than NP (Fig. 1E and 2A, lanes 1 and 2). Further, longer nascent cRNA chains were preferentially copurified with MCM (Fig. 2A, lane 3), suggesting that MCM stabilizes the elongation complex and thereby makes the viral polymerase escape the promoter successfully. It also seems likely that MCM has a role in the elongation process, but its precise mechanism is still unknown. In contrast, rather shorter cRNA chains were associated with exogenous NP (lane 4). After or along with virus genome replication, the newly synthesized virus genome has to be encapsidated by exogenous NP to form RNP complexes as templates for the next phase of virus genome replication and to protect the virus genome from degradation by cellular nucleases (33). It is hypothesized that encapsidation proceeds by targeting exogenous NP to the nascent RNA through the interaction between NP and the viral polymerase bound to the 5′ end of the nascent RNA to allow NP to interact with the viral RNA preferentially with respect to other cellular RNA species (1, 8, 11, 22), and then subsequently NP is recruited through NP-NP oligomerization (3, 23). In our cell-free system, we found that exogenous NP interacts with shorter cRNA (Fig. 2A, lane 4) without the addition of soluble viral polymerases, which could bind to the 5′ end of the nascent RNA and be a target of NP. It might be explained that the primary targeting of NP to the nascent RNA easily occurs since there is no RNA target other than the nascent RNA in our system. However, it is worth noting that encapsidation of longer nascent cRNA by NP was not achieved when NP was simply added to the system (lane 4). This raises a question of how the newly synthesized virus genome is encapsidated with NP free of RNA.
Fig. 2.
Encapsidation of nascent cRNA with NP. (A) De novo RNA synthesis was carried out in the presence of His-MCM (lanes 1 and 3; 20 pmol) or His-NP (lanes 2 and 4; 20 pmol) with 0.3 μM UTP and 250 μM each ATP, CTP, and GTP and 10 μCi of [α-32P]UTP (3,000 Ci/mmol) in a final volume of 200 μl. The products were purified with His-MCM (lane 3) or His-NP (lane 4) by using Ni-NTA resin. Lanes 1 and 2 represent 20% of the input amounts. The band intensities of short (*) and long (**) nascent cRNA products were quantitatively measured with ImageJ software, and the relative intensity of newly synthesized cRNA coprecipitated with MCM or NP against the input fraction is indicated. (B) De novo RNA synthesis was carried out with the authentic vRNP in the presence of His-NP as described for panel A. The newly synthesized RNA products were coimmunoprecipitated without (lane 2) or with (lane 3) anti-NP antibody. Lane 1 shows 20% of the input amount. The product purified by Ni-NTA resin is also represented in lane 4. The band intensities of short (*) and long (**) nascent cRNA products were quantitatively measured with ImageJ software, and the relative intensity of newly synthesized cRNA precipitated by using anti-NP antibody or Ni-NTA resin against input fraction is indicated. α, anti.
NP recognizes the phosphodiester backbone of ssRNA in a specific sequence-independent manner. We used, as the enzyme source, the vRNP containing authentic NP, which is bound to the template RNA. Thus, it is reasonably hypothesized that newly synthesized cRNA chains remain associated with the template RNP, possibly by partial hybridization of the nascent cRNA with template vRNA and/or the interaction of nascent cRNA with template-bound authentic NP instead of exogenous NP. To address this, we immunopurified the template-bound authentic NP of vRNP in the presence of exogenous His-NP using anti-NP antibody (Fig. 2B). The length of RNA products associated with authentic NP or both authentic NP and exogenous His-NP (lane 3) was clearly distinct from that of RNA products interacting with only exogenous His-NP (lane 4). From these results, it is assumed that the nascent cRNA product is hardly encapsidated with exogenous NP since the nascent cRNA tends to interact more with template-bound NP than exogenous NP and might partially hybridize with the template.
Encapsidation with NP mediated by RAF-2p48/UAP56.
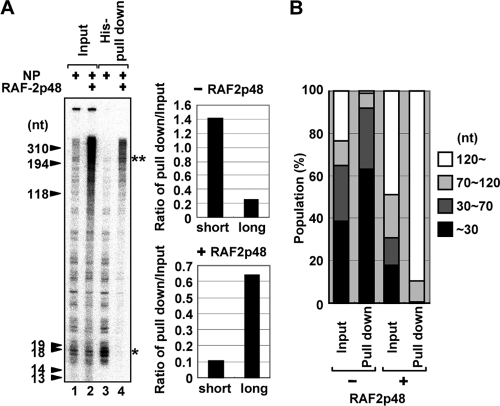
As shown in Fig. 2, it is assumed that some factor(s) may be missing in the encapsidation of nascent cRNA products with exogenous NP. Previously, RAF-2p48/UAP56/BAT1 (here, designated RAF-2p48/UAP56) belonging to the DExD-box family of ATP-dependent RNA helicase (13), also reported as NPI-5 (20), was identified as a host factor that binds to NP and stimulates influenza virus RNA synthesis from exogenously added model vRNA templates (16) and that is involved in splicing of cellular pre-mRNAs and messenger RNP maturation of cellular and viral transcripts (4, 25, 29). RAF-2p48/UAP56 binds to NP free of RNA but not to an NP-RNA complex and facilitates NP-RNA complex formation as a molecular chaperone for NP. Therefore, it was proposed that RAF-2p48/UAP56 is involved in the arrangement of NP on the template. However, its precise roles, including the requirement for the encapsidation process, have not yet been uncovered. Thus, we tried to examine whether RAF-2p48/UAP56 facilitates the encapsidation of newly synthesized RNA with exogenous NP (Fig. 3A). We found that long nascent cRNA was encapsidated with exogenous NP by the addition of RAF-2p48/UAP56 (Fig. 3A, compare lane 4, in which RAF-2p48/UAP56 is present, with lane 3, in which RAF-2p48/UAP56 is absent). The ATP-dependent RNA unwinding activity of RAF-2p48/UAP56 was not required for the encapsidation of nascent chains since the encapsidation occurred in the presence of ATPγS, which is a nonhydrolyzable analog of ATP (data not shown). Therefore, we propose a model whereby RAF-2p48/UAP56 facilitates the formation of RNP complexes by coreplicationally transferring exogenous NP to the nascent cRNA chain. It is unlikely that RAF-2p48/UAP56 remodels secondary structures of template and newly synthesized cRNA by its potential RNA helicase activity (Fig. 4). Furthermore, RAF-2p48/UAP56 stimulated the elongation activity of the viral polymerase, possibly by facilitating the encapsidation of nascent cRNA (Fig. 3, lane 2). It is speculated that the coreplicational encapsidation of nascent cRNA by NP may prevent the premature termination of RNA synthesis by avoiding a secondary structure of nascent RNA, which is hypothesized to be one of the causative factors in the termination process of other RNA polymerases (10, 27). Therefore, it is possible that the encapsidation of the nascent RNA with exogenous NP mediated by RAF-2p48/UAP56 increases the processivity of the viral polymerase by avoiding inappropriate secondary structures of nascent cRNA.
Fig. 3.
The stimulatory activity of RAF-2p48/UAP56 in encapsidation of nascent cRNA. (A) RNA synthesis was performed in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of recombinant RAF-2p48/UAP56 with His-NP as described in the legend of Fig. 2. The products were purified with His-NP by using Ni-NTA resin (lanes 3 and 4). Twenty percent of the input amounts is shown in lanes 1 and 2. The band intensities of short (*) and long (**) nascent cRNA products were quantitatively measured with ImageJ software, and the relative intensity of cRNA coprecipitated with NP in the absence or presence of RAF-2p48 against input fraction is indicated. (B) The band intensities of the regions corresponding to RNAs of less than 30 nt, 30 to 70 nt, 70 to 120 nt, and more than 120 nt in each lane in panel A were quantitatively measured with ImageJ software, and the results are indicated as a percentage of the total intensity of each lane.
Fig. 4.
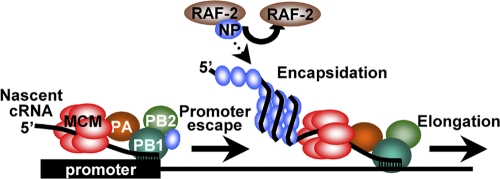
Proposed model. NP facilitates the promoter escaping from the viral polymerase through the interaction between NP and the viral polymerase in an RNA binding activity-independent manner. During elongation step, RAF-2p48/UAP56 stimulates the coreplicational encapsidation of newly synthesized cRNA with exogenous NP, thereby increasing the processivity of the viral polymerase.
Involvement of RAF-2p48/UAP56 in influenza virus genome replication in infected cells.
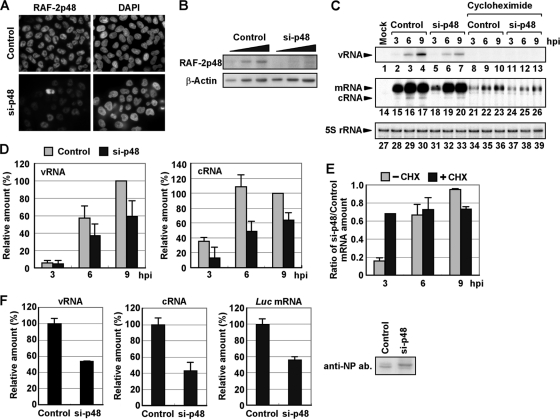
Finally, we tried to examine whether RAF-2p48/UAP56 functions in influenza virus genome replication in cultured cells using siRNA-mediated gene silencing. At 48 h posttransfection of siRNA corresponding to the RAF-2p48/UAP56 ORF, the expression level of RAF-2p48/UAP56 in knockdown cells decreased to approximately 30% of that of cells transfected with the nontargeting siRNA used as a negative control (Fig. 5A and B). We carried out quantitative primer extension assays with appropriate primers specific for each vRNA and mRNA/cRNA of segment 5 (Fig. 5C and D). We confirmed that the product corresponding to cRNA was not found from a fraction bound with oligo(dT) cellulose (data not shown). This result showed that the accumulation of vRNA and cRNA was reduced and delayed in RAF-2p48/UAP56 knockdown cells compared with levels in control cells (Fig. 5C, lanes 1 to 7 and 14 to 20, and D). The same results were obtained for other segments (data not shown). It is proposed that nascent cRNA is degraded unless it is encapsidated with viral RNA polymerase and NP (33). In addition, the results shown in Fig. 3 and a previous report (16) demonstrated that RAF-2p48/UAP56 stimulates the viral polymerase activity. Thus, RAF-2p48/UAP56 might be involved in virus genome replication and encapsidation in infected cells. We also found that the level of NP mRNA in RAF-2p48/UAP56 knockdown cells decreased to 15% in control cells at 3 hpi (Fig. 5C, lanes 15 and 18, and E). In contrast, comparable amounts of NP mRNA were found in both control and RAF-2p48/UAP56 knockdown cells at 6 and 9 hpi (Fig. 5C, lanes 16, 17, 19, and 20) since the amount of vRNA template sufficient for viral mRNA synthesis might be accumulated at 6 and 9 hpi, but the replication activity was reduced and delayed in RAF-2p48/UAP56 knockdown cells. To confirm the effect of RAF-2p48/UAP56 on viral transcription, we utilized cycloheximide (CHX), a potent protein synthesis inhibitor (Fig. 5C, lanes 8 to 13 and 21 to 26, and E). A previous report showed that CHX suppresses viral protein synthesis and thereby leads to degradation of replicated viral RNA but not mRNA since new vRNP formation was repressed (33). Therefore, we could examine the amount of viral mRNA synthesized from incoming vRNP independent of the level of vRNA accumulation in the presence of CHX (Fig. 5C, lanes 8 to 13, and E). The level of NP mRNA in RAF-2p48/UAP56 knockdown cells was reduced to 70% in control cells in the presence of CHX at 3 hpi (Fig. 5C, lanes 21 and 24, and E). Therefore, it is likely that the reduction of viral mRNA synthesis in RAF-2p48/UAP56 knockdown cells is mainly due to the decrease of vRNP accumulation in the absence of CHX although RAF-2p48/UAP56 has a stimulatory role in viral transcription, possibly by arrangement of NP on template and/or the nuclear export-competent messenger RNP formation (25). To rule out the possibility that the reduction of vRNA and cRNA synthesis was caused by the reduction of viral protein synthesis, we carried out a viral model replicon assay (19, 30) in which active vRNP complexes were reconstituted with PB1, PB2, PA, and NP and the model vRNA encoding the luciferase gene, as described in Materials and Methods (Fig. 5F). With this system, we could examine the viral polymerase activity independent of the expression level of viral proteins since viral proteins were expressed from plasmids under the control of cellular RNA polymerase II promoter in this assay. Results shown in Fig. 5F indicate that vRNA, cRNA, and viral mRNA synthesis was decreased in RAF-2p48/UAP56 knockdown cells compared with that in control cells even in the presence of comparable amounts of NP in both cells. We found that NP synthesized in RAF-2p48/UAP56 knockdown cells migrates differently from that in control cells (Fig. 5F). Previous reports showed that NP is modified by phosphorylation (23) and that its N-terminal region is digested by caspase (35), but the involvement of RAF-2p48/UAP56 in these is not known at present.
Fig. 5.
Involvement of RAF-2p48/UAP56 in influenza virus genome replication in infected cells. (A) At 48 h posttransfection, cells transfected with either a control or siRNA against the RAF-2p48/UAP56 ORF (si-p48) were subjected to indirect immunofluorescence assay with anti-RAF-2p48/UAP56 antibody. Nuclear DNA stained with 4′,6-diamidino-2-phenylindole (DAPI) is also shown. Images were acquired under the same exposure time by a fluorescence microscope system (Axiovision; Carl Zeiss). (B) Expression level of RAF-2p48/UAP56. The lysates prepared from control and RAF-2p48/UAP56 knockdown cells (5 × 103, 1 × 104, and 2 × 104 cells) were separated by SDS-PAGE and then visualized by Western blotting assays with anti-NP and β-actin antibodies. (C, D, and E) Level of viral RNAs in infected RAF-2p48/UAP56 knockdown cells. Control and RAF-2p48/UAP56 knockdown cells were infected with influenza virus in the absence (lanes 1 to 7 and 14 to 20) or presence of cycloheximide (lanes 7 to 13 and 21 to 26) for 0, 3, 6, and 9 h. Primer extension assays were carried out with primers specific for segment 5 vRNA or m/cRNA as described in Materials and Methods. As a loading control, 5S rRNA was also detected (lanes 27 to 39). The band intensities were quantitatively measured by ImageJ software, and the results of three independent experiments are summarized in panel D and are indicated in panel E as the ratio of the mRNA amount in RAF-2p48/UAP56 knockdown cells to that in control cells with or without CHX. (F) The level of viral RNAs synthesized from a reconstituted model replicon in RAF-2p48/UAP56 knockdown cells. Control and RAF-2p48/UAP56 knockdown cells were transfected with plasmids expressing PB1, PB2, PA, and NP and model vRNA encoding the luciferase gene as described in Materials and Methods. At 12 h posttransfection, total RNAs were purified and then subjected to reverse transcription, followed by quantitative real-time PCR with primer sets specific for vRNA, cRNA, and luciferase mRNA. The expression level of NP protein in control and RAF-2p48/UAP56 knockdown cells was also detected by a Western blotting assay with anti-NP antibody (ab).
It is well known that NP is one of proteins responsible for virus genome replication (15, 18, 27, 33). Recently, it is reported that ubiquitination of NP regulates virus genome replication (12). It is proposed that the soluble viral polymerase might act as a replicative enzyme in trans, but transcription occurs from template-bound viral polymerase in cis (8). In this study and recent reports (9, 31–33), de novo cRNA synthesis is found from template-bound viral polymerase; thus, it could be explained that the soluble viral polymerase might have stimulatory activity but is not completely essential for the synthesis of nascent cRNA. The viral nuclear export protein (NEP/NS2) is also involved in the accumulation level of vRNA and cRNA (26). Further, it is reported that small noncoding RNAs derived from the influenza virus genome might regulate viral transcription and replication through their interaction with viral polymerase complexes (21). To further understand the mechanism of influenza viral genome replication, precise analyses of a functional replicative enzyme including viral and cellular factors are required.
ACKNOWLEDGMENTS
We thank Y. Ishimi (Ibaraki University) for the generous gifts of baculoviruses expressing MCM proteins. This research was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K.N. and F.M.) and Research Fellowships of the Japanese Society for the Promotion of Science (to A.K.).
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Biswas S. K., Boutz P. L., Nayak D. P. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blumberg B. M., Leppert M., Kolakofsky D. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837–845 [DOI] [PubMed] [Google Scholar]
- 3. Chan W. H., et al. 2010. Functional analysis of the influenza virus H5N1 nucleoprotein tail loop reveals amino acids that are crucial for oligomerization and ribonucleoprotein activities. J. Virol. 84:7337–7345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fleckner J., Zhang M., Valcarcel J., Green M. R. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864–1872 [DOI] [PubMed] [Google Scholar]
- 5. Honda A., et al. 1990. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J. Biochem. 107:624–628 [DOI] [PubMed] [Google Scholar]
- 6. Honda A., Ueda K., Nagata K., Ishihama A. 1988. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J. Biochem. 104:1021–1026 [DOI] [PubMed] [Google Scholar]
- 7. Horikami S. M., Curran J., Kolakofsky D., Moyer S. A. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jorba N., Coloma R., Ortin J. 2009. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5:e1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawaguchi A., Nagata K. 2007. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 26:4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Komissarova N., Becker J., Solter S., Kireeva M., Kashlev M. 2002. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell 10:1151–1162 [DOI] [PubMed] [Google Scholar]
- 11. Labadie K., Dos Santos Afonso E., Rameix-Welti M. A., van der Werf S., Naffakh N. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271–282 [DOI] [PubMed] [Google Scholar]
- 12. Liao T. L., Wu C. Y., Su W. C., Jeng K. S., Lai M. M. 2010. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 29:3879–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Linder P., Stutz F. 2001. mRNA export: travelling with DEAD box proteins. Curr. Biol. 11:R961–R963 [DOI] [PubMed] [Google Scholar]
- 14. Masters P. S., Banerjee A. K. 1988. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 62:2658–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Medcalf L., Poole E., Elton D., Digard P. 1999. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J. Virol. 73:7349–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Momose F., et al. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagata K., Kawaguchi A., Naito T. 2008. Host factors for replication and transcription of the influenza virus genome. Rev. Med. Virol. 18:247–260 [DOI] [PubMed] [Google Scholar]
- 18. Newcomb L. L., et al. 2009. Interaction of the influenza a virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J. Virol. 83:29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Obayashi E., et al. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131 [DOI] [PubMed] [Google Scholar]
- 20. Palese P., Wang P., Wolff T., O'Neill R. E. 1997. Host-viral protein-protein interactions in influenza virus replication, p. 327–340 In McCrae M. A. (ed.), Molecular aspects of host-pathogen interaction. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 21. Perez J. T., et al. 2010. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U. S. A. 107:11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poole E., Elton D., Medcalf L., Digard P. 2004. Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology 321:120–133 [DOI] [PubMed] [Google Scholar]
- 23. Portela A., Digard P. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83:723–734 [DOI] [PubMed] [Google Scholar]
- 24. Qanungo K. R., Shaji D., Mathur M., Banerjee A. K. 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. U. S. A. 101:5952–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Read E. K., Digard P. 2010. Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J. Gen. Virol. 91:1290–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robb N. C., Smith M., Vreede F. T., Fodor E. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90:1398–1407 [DOI] [PubMed] [Google Scholar]
- 27. Shapiro G. I., Krug R. M. 1988. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J. Virol. 62:2285–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shimizu K., Handa H., Nakada S., Nagata K. 1994. Regulation of influenza virus RNA polymerase activity by cellular and viral factors. Nucleic Acids Res. 22:5047–5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strasser K., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304–308 [DOI] [PubMed] [Google Scholar]
- 30. Sugiyama K., et al. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vreede F. T., Brownlee G. G. 2007. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J. Virol. 81:2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vreede F. T., Gifford H., Brownlee G. G. 2008. Role of initiating nucleoside triphosphate concentrations in the regulation of influenza virus replication and transcription. J. Virol. 82:6902–6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vreede F. T., Jung T. E., Brownlee G. G. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78:9568–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamanaka K., Ishihama A., Nagata K. 1990. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J. Biol. Chem. 265:11151–11155 [PubMed] [Google Scholar]
- 35. Zhirnov O. P., Konakova T. E., Garten W., Klenk H. 1999. Caspase-dependent N-terminal cleavage of influenza virus nucleocapsid protein in infected cells. J. Virol. 73:10158–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]